Abstract
Stem rust, caused by Puccinia graminis (Pgt), is a damaging disease of wheat that can be controlled by utilizing effective stem rust resistance genes. ‘Thatcher’ wheat carries complex resistance to stem rust that is enhanced in the presence of the resistance gene Lr34. The purpose of this study was to examine APR in ‘Thatcher’ and look for genetic interactions with Lr34. A RIL population was tested for stem rust resistance in field nurseries in Canada, USA, and Kenya. BSA was used to find SNP markers associated with reduced stem rust severity. A major QTL was identified on chromosome 3BL near the centromere in all environments. Seedling testing showed that Sr12 mapped to the same region as the QTL for APR. The SNP markers were physically mapped and the region carrying the resistance was searched for sequences with homology to members of the NB-LRR resistance gene family. SNP marker from one NB-LRR-like sequence, NB-LRR3 co-segregated with Sr12. Two additional populations, including one that lacked Lr34, were tested in field nurseries. NB-LRR3 mapped near the maximum LOD for reduction in stem rust severity in both populations. Lines from a population that segregated for Sr12 and Lr34 were tested for seedling Pgt biomass and infection type, as well as APR to field stem rust which showed an interaction between the genes. We concluded that Sr12, or a gene closely linked to Sr12, was responsible for ‘Thatcher’-derived APR in several environments and this resistance was enhanced in the presence of Lr34.
Introduction
Stem rust of wheat, caused by the fungus Puccinia graminis Pers.:Pers. f. sp. tritici Eriks. & E. Henn. (Pgt), is capable of causing devastating grain yield losses. For example, during the last North American stem rust epidemic between 1953–55 up to 40% of the spring wheat crop was lost because of the virulent Pgt race 15B [1]. Resistant cultivars have effectively controlled stem rust for several decades. More recently, new races of Pgt were detected in Africa, specifically Ug99 and its variants that are broadly virulent on a majority of the wheat varieties grown globally [2–6]. This resulted in increased interest and investment in understanding the genetics of stem rust resistance in wheat.
The hard red spring wheat cultivar ‘Thatcher’ [7] was the dominant cultivar in the Canadian Prairies from 1939 to the mid-1960s mainly due to its resistance to stem rust and excellent end-use quality [8]. Stem rust resistance in ‘Thatcher’ has been the subject of several genetic studies. Four seedling stem rust resistance (Sr) genes, Sr5, Sr9g, Sr12, and Sr16 have been reported in ‘Thatcher’ [9,10]. However, the seedling resistance characterized in ‘Thatcher’ does not explain the adult-plant resistance (APR) observed in the field to North American races of Pgt [11]. Furthermore, ‘Thatcher’ shows a moderately resistant to moderately susceptible response in Kenyan field tests in the presence of Ug99 and its variants while showing a susceptible infection type at the seedling stage to the same field races demonstrating some level of APR to Ug99 [12].
Lr34 is an important disease resistance gene as it confers confers resistance to leaf rust, stem rust, stripe rust, powdery mildew and barley yellow dwarf virus [13–18]. It has also been characterized as a ‘non-suppressor’ of stem rust resistance genes in ‘Thatcher’ and its derivatives [13–19]. A locus on chromosome arm 7DL suppresses some seedling stem rust resistance in ‘Thatcher’-derived wheats that are expressed when the suppressor is removed by chromosome deficiencies or by mutagenesis [20, 21]. The presence of Lr34 appears to negate the effect of the suppressor [19], though a direct link between the actions of these loci has not been established. Regardless, Lr34 enhances field resistance to Pgt in the ‘Thatcher’ background and in the cultivar ‘Roblin’ and ‘Pasqua’, and has been shown to interact with other Sr genes [22–24]. In our preliminary experiments and in other studies, a ‘Thatcher’ near-isogenic line (NIL) carrying Lr34 (RL6058 = Thatcher*6/PI58548; Tc-Lr34) showed enhanced stem rust resistance compared to ‘Thatcher’ in North America, Australia and Kenya [25].
Other studies have also examined the interactions between Lr34 and Sr genes found in ‘Thatcher’. One study suggested that Sr16 and Sr12 may have contributed to resistance of adult plants in their ‘Thatcher’-derived population but focused on resistance associated with markers on chromosome arm 2BL [26]. In a follow-up study, a QTL was identified in ‘Thatcher’ on chromosome arm 2BL that was enhanced by Lr34 [25]. The peak of the QTL on 2BL was near the map location of Sr9 [27, 28]. In a similar study, a QTL was found on chromosome 3B which corresponded to the map position of Sr12 as determined by testing their population at the seedling stage with a race of Pgt that was avirulent to Sr12 [12]. The above studies used populations that segregated for Lr34 and suggested that Sr12 plays an important role in the APR of ‘Thatcher’ [12, 26]. Sr12 was assigned to the short arm of chromosome 3B by telocentric mapping; however the authors state that it is possible that Sr12 could be near the centromere on the long arm [29]. None of the QTL mapping studies above addressed which arm of chromosome 3B is involved in Thatcher’s APR.
In this study, our objectives were to map stem rust resistance that interacts with Lr34 in ‘Thatcher’ at the adult plant stage and to develop DNA markers that facilitate further characterization of the resistance and are useful for marker-assisted breeding.
Materials and Methods
Populations
An F6:7 recombinant inbred line (RIL) population consisting of 94 lines was developed from the cross of RL90/RL6058, which will be referred to as the RL population hereafter. RL6058 is a ‘Thatcher’ near-isogenic line (NIL) that carries Lr34 and has a high level of resistance to stem rust. RL90 is a line selected from a RIL population developed from the cross of RL6071 (‘Prelude’/8*‘Marquis’*2/3/‘Prelude’//‘Prelude’/8*‘Marquis’) and RL6058 [25]. RL90 carries Lr34 but shows a moderately susceptible to susceptible reaction to stem rust. An F4 family derived from a single F3 plant from the RL population (RL90-B17) that was heterozygous for markers flanking resistance on chromosome 3B was grown to maturity. The seed from this population was treated as an “F2-equivalent” population (RL-F2 population) and 110 lines were used to improve the map resolution of markers mapped in the RL population.
A doubled haploid (DH) population consisting of 137 lines was generated from the cross ‘Chinese Spring’/RL6058 using the maize pollination method [30] and will be referred to as the CS population hereafter. ‘Chinese Spring’ carries Lr34 [14], but it is susceptible to the Pgt races used in this experiment while RL6058 is resistant.
Another RIL population of 95 lines was developed from the cross of ‘Kenyon’/861SMN-2137 and will be referred to as the KN population hereafter. ‘Kenyon’ (Neepawa*5/Buck Manantial) is a Canadian hard red spring wheat cultivar registered in the 1985 [31] and is resistant to stem rust in field tests in Canada and Kenya. Line 861SMN-2137 is resistant to stem rust in field tests in Canada but is susceptible to stem rust in Kenya. Neither parent carries Lr34.
To study interactions with Lr34, RL90 (carries Lr34 and is susceptible to stem rust) was crossed to RL6077 (‘Thatcher’*6/PI250413; a near-isogenic line of ‘Thatcher’ carrying Lr67) to generate F2 seed which was screened as half seeds for markers flanking the chromosome 3B resistance locus (IWA6086 and IWA4613) and the gene-based marker csLr34SNP for Lr34 [32]. Three to four F2 plants were identified for each genotypic category (null, Lr34, Sr12 region, and Lr34+Sr12 region). F3 families were scored for stem rust resistance at the seedling stage using Australian Pgt race 98–1,2,(3),(5),6 and screened in the field nurseries in St. Paul in 2014 and Morden in 2015 (see below).
A final RIL population of 160 lines from the cross ‘McNeal’/‘Thatcher’ was used in this study. The population was previously reported [28] and will be referred to as the MN population hereafter. ‘McNeal’ (RS 6880/Glenman) is susceptible to the Pgt races used in this study and carries Lr34. This population was used previously to map Sr12.
In summary, the RL, CS and KN populations were used to map major resistance to Pgt in the field. The RL-F2 population was generated to improve map resolution for the chromosome 3B region and the RL90/RL6071 population was used to study interactions with Lr34. Markers developed in this study were also tested in the MN population to confirm tight linkage to the Sr12 gene.
Testing with Pgt
The RL, CS, and KN populations and F3 families from the RL90/RL6077 population were tested for stem rust resistance in replicated and randomized field trials. The RL population was tested in field nurseries in Morden, MB, Canada in 2014 and 2015 (80 RILs), Njoro, Kenya in 2013 (89 RILs), and St. Paul, MN, USA in 2013 (87 RILs). The CS population (137 RILs) was tested in the Morden nursery in 2014. The KN population of 95 lines was tested in Njoro, Kenya in 2013 and 2014. No specific permissions were required to conduct field trials in Canada, USA and Kenya.
In Morden, entries were planted in 1 m rows with ranges of five rows flanked by susceptible spreader rows. The spreader rows were inoculated with a mixture of Pgt races (TPMKR, TMRTK, RKQSR, RHTSJ, QTHJT, RTHJT and MCCFR with AAFC-MRDC isolate numbers 1373, 1311, 1312, 1562, 1347, 1561, and 1541 respectively; from the AAFC-MRDC Pgt collection of isolates found in Canadian fields) at the jointing stage by spraying urediniospores suspended in a light mineral oil on a day preceding anticipated overnight dew. Spores from heavily infected spreader rows caused infection of the experimental plots. The populations were rated for stem rust severity and infection response once the susceptible checks showed heavy disease (severities near 80%) which was approximately at anthesis. The modified Cobb scale was used to assess the populations [33]. In St. Paul a similar procedure was used except 2 m rows were planted, spreader rows were planted perpendicular to the experimental plots, and the race mixture included Pgt races QFCSC, QTHJC, MCCFC, RCRSC, RKQQC, and TPMKC (isolates 06ND76C, 75ND717C, 59KS19, 77ND82A, 99KS76A, and 74MN1409, respectively; from the USDA-CDL Pgt collection of isolates found in US fields). Of the above races, only MCCFR (in Canada) and QFCSC and MCCFC (in the USA) were avirulent to Sr12.
In Kenya, Pgt inoculum for the field was increased on lines carrying Sr31 to select Ug99-type races. Experimental lines were planted in two 1 m rows and hills of plants containing a mixture of cultivars susceptible to Ug99 were grown next to the experimental plots. Spreader hills were inoculated at jointing. Plots were rated for stem rust severity at anthesis.
The RL, RL-F2 and MN populations were tested with Pgt at the seedling stage using races that are avirulent to Sr12. The RL population was inoculated with Pgt race MCCFR (isolate 1541, as above) by spraying a mixture of urediniospores in light mineral oil onto the seedlings after the first leaf was fully emerged. Seedlings were incubated in dew chambers for 16 hr and then treated with light for 3 hr in the dew chambers. Seedlings were grown in a greenhouse at approximately 20°C with 16 hr of light daily. Two weeks after inoculation seedlings were rated for their infection types (IT) following the commonly used ‘0’ to ‘4’ IT scale [34]. The MN population was inoculated with race SCCSC (isolate 09ID73-2) as previously described [12]. The same IT scale was used to assess the MN population. The RL and RL-F2 populations were inoculated with Pgt race 98–1,2,(3),(5),6 (an isolate from the University of Sydney, Plant Breeding Institute collection) by applying urediniospores mixed in talc onto seedlings and incubating in dew chamber for 24 hrs at approximately 18–20°C before transferring them to greenhouse set at approximately 18–20°C. Families were rated for their infection types 14 days post-inoculation (DPI) and were classified resistant, segregating, or susceptible.
The selected genotypically homozygous RL90/RL6077 F3 families were inoculated at the seedling stage using Pgt race 98–1,2,(3),(5),6 by mixing urediniospores and talc as described above. Ten days after inoculation tissue from the first leaf was harvested and chitin content was measured to determine fungal biomass as previously described [35]. Fourteen days after inoculation the ITs of plants not used for the chitin assay were recorded for the lines in each of the four genotypic classes (“null”, Sr12, Lr34, and Sr12 + Lr34).
Genetic mapping and DNA markers
DNA was extracted from young leaves of RILs/DH lines in the RL, CS, and KN populations using a modified ammonium acetate extraction [36]. DNA was isolated from MN population using a rapid extraction protocol [12, 37]. RILs from RL population that were resistant or susceptible to Pgt in the field were used to construct DNA bulks to perform bulked segregant analysis (BSA) [38]. RL90, RL6058 and the resistant and susceptible bulks were tested with SNP markers using a 9K iSelect custom wheat SNP array [39]. SNP markers were analyzed using GenomeStudio software (Illumina Inc., San Diego, USA). Markers that were polymorphic between the parental lines and between the bulks were assessed for their association with stem rust severity in the entire RL population. QTL analysis was performed with QTL IciMapping software [40] using the markers identified by BSA to determine if they were associated with field stem rust resistance. SNP markers from the iSelect array associated with stem rust resistance were converted to KASP assays [41] for convenient mapping in populations (S1 Table).
DNA sequences associated with SNP markers were positioned on the 3B pseudomolecule [42]. The genomic sequence defined by SNP markers that flanked the Sr gene was searched for nucleotide-binding leucine repeat like-sequences (NB-LRR) as members of this class commonly encode disease resistance genes. SNP markers were developed from the NB-LRR-like sequences (S1 Table) and mapped in the RL-F2 recombinants along with the SNP markers from the 9K iSelect array. A marker from one NB-LRR-like sequence (NB-LRR3) that co-segregated with stem rust resistance was tested on the CS, KN, and MN populations.
The CS population was genotyped with SSR markers on chromosome 3B [43–46] and the KASP markers (above) converted from the 9K iSelect custom SNP array [39] that were mapped in the RL population. The NB-LRR3 SNP marker was added to the CS genetic map and the association with stem rust resistance was determined.
A whole-genome map was constructed in the KN population using Diversity Array Technologies (DArT) markers [47], SSR markers [43–46] and SNP markers from the 9K iSelect custom SNP array [39] to map several qualitative and quantitative traits, including field resistance to Ug99 (McCartney, unpublished). In this study, the NB-LRR3 marker was added to the KN genetic map to determine the association with stem rust resistance.
Linkage maps of the chromosome 3B region were constructed in the RL, CS and KN populations using MapDisto [48]. The map position of the maximum LOD for reduction in stem rust severity, percent variation explained (PVE), and additive effect was calculated for the above populations in each environment using QTL IciMapping [40]. The NB-LRR3 was added to a previously published genetic map of Sr12 in the MN population [28]. NB-LRR3 was also tested on a panel of North American and Australian germplasm.
Results
Stem rust phenotyping of populations in the field
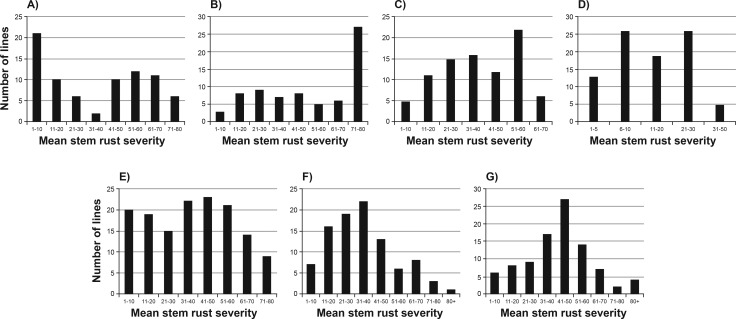
The RL population was evaluated for response to stem rust in Morden, Canada in 2014 and 2015, in Njoro, Kenya in 2013 and in St Paul, USA in 2013. The KN population was scored in Njoro, Kenya in 2013 and 2014 and the CS population in Morden, Canada in 2014. The field trials for the RL, CS, and KN populations were uniform in maturity and showed a full range (resistant to susceptible) of stem rust severities regardless of the location of the trial (Fig 1). Thus, adequate disease pressure resulted in phenotypic separation of lines within each population. With the exception of the RL population in Morden, field tests showed a continuous distribution for stem rust severity. The RL population resembled a bi-modal distribution in Morden in 2014. The rust severity across all plots in the RL90 test in Morden increased from 36.3 in 2014 to 52.5 in 2015 which was reflected in the different frequency distributions. However, the ranking of RL lines according to their response to stem rust was largely consistent across years resulting in heritability of 0.76. The overall heritability of response to stem rust for the RL population across four environments was 0.79 and 0.87 for the KN population across two environments. The CS population was tested in one environment and showed a heritability of 0.86 between replicates.
Fig 1.
Distributions of stem rust severities in the A) RL population tested in Morden, Canada in 2014, B) RL population tested in Morden, Canada in 2015, C) RL population tested in St. Paul, USA in 2013, D) RL population tested in Njoro, Kenya in 2013, E) CS population tested in Morden, Canada in 2014, F) KN population tested in Njoro, Kenya in 2013, and G) KN population tested in Njoro, Kenya in 2014.
DNA markers and genetic mapping of stem rust resistance in RL population
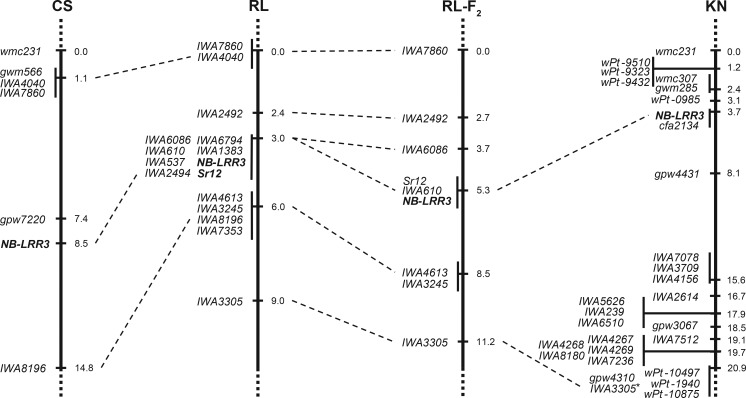
In the RL population, field scores from USA and Kenya were used to select the most resistant and susceptible lines for BSA. SNP markers from the 9K iSelect array that differentiated the parents and the bulks were located at chromosomes 3B and 2B. These results suggested that these chromosomes carried Sr gene(s) responsible for the field resistance in RL6058 (S2 Table). Some of these SNP markers were converted to KASP assays and mapped on the RL population. While there was a weak association between markers on chromosome 2B and stem rust severity, the association was much stronger with the markers on chromosome 3B. A partial linkage map of the centromeric region of chromosome 3B was constructed and the marker/trait association was calculated for each marker. The association between genotype and stem rust severity was strongest with SNP marker IWA610 in all four environments (PVE = 50.6% to 83.9%) (Table 1). A subset of the RL population was tested at the seedling stage using Australian Pgt race 98–1,2,(3),(5),6. The parental lines RL90 and RL6058 showed infection types (ITs) “3” and “12-”, respectively, and similar ITs were produced by the susceptible (“3” to “33+”) and resistant RILs (“12” to “2”). SNP markers IWA6086, IWA610 and IWA4613 that were previously associated with field resistance were also associated with seedling resistance. Markers IWA6086 and IWA3245 were predicted to flank the locus. Because the race-specific stem rust resistance gene Sr12 was previously mapped to the centromeric region of chromosome 3B [12, 29], it is possible that seedling resistance detected by race 98–1,2,(3),(5),6 corresponded to Sr12. The hypothesis was supported by testing the RL population at the seedling stage with the Canadian Pgt race MCCFR which is avirulent on Sr12. The parental lines RL90 and RL6058 showed infection types (IT) of “33+” and “;” respectively, and the susceptible and resistant progeny lines showed ITs of “33-” to “33+” and “;” to “12-” respectively. The segregation of stem rust response (45 resistant: 49 susceptible) fitted the ratio for a single gene (χ21:1 = 0.17, p = 0.68) and co-segregated with IWA610 on chromosome 3B (Fig 2). The results indicate that the genetic interval carrying stem rust resistance which was observed under field conditions in USA, Kenya and Canada overlapped with the map position of Sr12 as determined by seedling tests.
Table 1. Statistics for QTL analysis (interval mapping) of stem rust severity in the RL, CS, and KN populations.
| Population | Environment | Position (cM)a | LODb | PVE(%)c | Addd |
|---|---|---|---|---|---|
| RL | Canada 2014 | 3.0 | 28.98 | 83.93 | -24.31 |
| RL | Canada 2015 | 2.4 | 19.17 | 70.16 | -21.11 |
| RL | Kenya 2013 | 3.6 | 14.14 | 56.26 | -9.12 |
| RL | USA 2013 | 3.5 | 20.29 | 69.94 | -14.40 |
| CS | Canada 2014 | 7.0 | 12.09 | 33.68 | -13.19 |
| KN | Kenya 2013 | 3.1 | 4.13 | 18.16 | -7.88 |
| KN | Kenya 2014 | 3.1 | 5.16 | 22.12 | -9.30 |
a The “position” refers to the position on the genetic maps of chromosome arm 3BL for each population shown in Fig 2.
b The significant LOD thresholds for the RL and CS populations were set at 3.0 as permutation analysis is inappropriate in the absence of a full genome map. However, if thresholds from a permutation test were used, the significance threshold would be less stringent. Permutation analysis (1000 permutations) was performed for the KN population and the significant LOD threshold was 3.4 for 2013 and 3.5 for 2014.
c Percent variation explained for stem rust severity by the QTL on chromosome arm 3BL.
d The additive effect that the resistant allele has on stem rust severity.
Fig 2. Genetic maps of the proximal region of chromosome 3BL in the Chinese Spring/RL6058 (CS) DH, RL90/RL6059 (RL) RIL, RL90/RL6058 F2-equivalent (RL-F2), and Kenyon/8615MN-2137 (KN) RIL populations.
Map positions are in centi-Morgans. *There were 11 additional SNP markers that co-segregated with IWA3305 in the KN population–IWA3306, IWA4427, IWA4429, IWA4847, IWA5775, IWA5874, IWA6221, IWA6492, IWA6493, IWA7132, and IWA7388.
Physical mapping of stem rust resistance
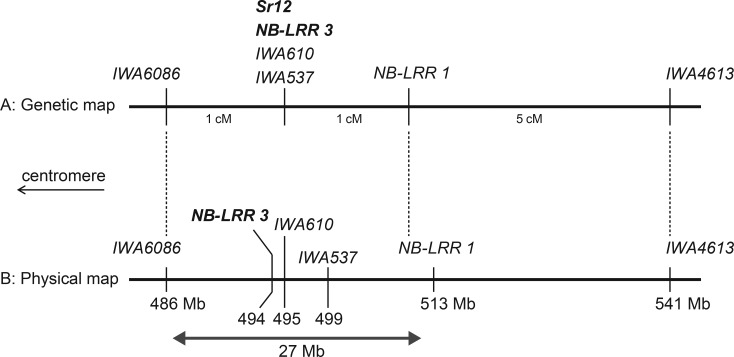
SNP markers IWA6086 and IWA3245/IWA4613 defined the genetic interval carrying the resistance on chromosome 3B. The RL-F2 consisted of 110 F2-equivalent plants and was screened with flanking markers to identify individuals that were recombinant in the region. Fifteen recombinant F2 plants were identified which were progeny-tested in F3 with Pgt race 98–1,2,(3),(5),6 at the seedling stage (see above). Stem rust resistance was mapped within the genetic interval flanked by IWA6086 and IWA4613 and confirmed to co-segregate with IWA610 (Fig 2). Flanking SNP markers were placed on the genome sequence of the chromosome 3B pseudo-molecule of ‘Chinese Spring’. The DNA sequence of approximately 55 Mb between IWA6086 and IWA4613 was searched for sequences with homology to members of the nucleotide binding-leucine rich repeat (NB-LRR) resistance gene family. Two NB-LRR sequences were identified at position 513 Mb and 494 Mb on chromosome 3B (Fig 3). The NB-LRR sequence at position 513 MB was separated from the seedling resistance by 2 recombinants, while a KASP marker developed from the partial NB-LRR sequence at position 494 Mb (NB-LRR3) co-segregated with seedling resistance. Two SNP markers, IWA610 and IWA537, co-segregated with NB-LRR3; however, physical mapping determined their order on the chromosome 3B pseudo-molecule (Fig 3). The physical interval in ‘Chinese Spring’ that corresponded to the resistance gene interval in RL6058 was approximately 27 Mb and mapped to the centromeric region on the long arm of chromosome 3B as determined by the analysis of ditelosomic aneuploid lines of ‘Chinese Spring’. It is likely that the seedling resistance mapped here corresponds to Sr12 in RL6058. ‘Chinese Spring’ contained a partial NB-LRR gene in this region which may be homologous to a candidate gene for Sr12 in RL6058.
Fig 3.
Alignment of the RL-F2 genetic map (A) with the physical map derived from the chromosome 3B pseudo-molecule (B).
NB-LRR3 associated with stem rust resistance in other populations
Markers from chromosome 3B were also tested for association with stem rust severity in the CS population. The NB-LRR3 marker was added to the genetic map for the CS population in which it co-segregated with markers that showed the best association with field resistance (Table 1; Fig 2). The KN RIL population was tested in Kenya for resistance to Ug99-type stem rust in the field. A single QTL was identified on chromosome 3B. The NB-LRR3 marker was added to the genetic map and identified the same chromosome region that was responsible for field stem rust resistance in the KN population as identified in the RL and CS populations (Table 1; Fig 2). The peak of the QTL corresponded to the map location of NB-LRR3 which is consistent with the RL and CS populations.
Furthermore, NB-LRR3 was added to a genetic map of chromosome 3B in the MN population that was previously used to map Sr12 [12]. In this population NB-LRR3 also co-located with a major QTL for stem rust resistance that was observed under field conditions on chromosome 3B and co-segregated with race SCCSC seedling resistance, which was presumably Sr12. Between the RL and MN populations there were a total 248 RILs tested which is equivalent to 496 opportunities for crossing-over, yet no recombinants were observed between Sr12 and NB-LRR3.
Lr34 effect on stem rust resistance
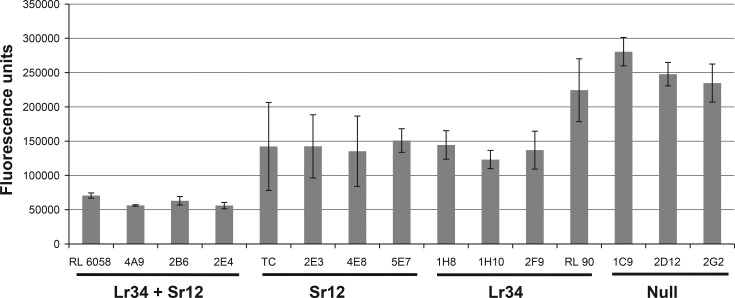
Previous reports showed that the combination of Lr34 with additional genes from ‘Thatcher’ can contribute to enhanced stem rust resistance under field conditions [12, 26]. Here, we have quantified possible interactions by examining the effect of Lr34 on Sr12 at the seedling stage. Homozygous sister lines from the RL90/RL6077 population that represented four genotypic categories; null (neither Lr34 nor Sr12 were present), Lr34, Sr12 and Lr34+Sr12 were evaluated for stem rust infection using the Australian Pgt race 98–1,2,(3),(5),6. Lr67, present in RL6077, was absent in all of the lines evaluated. The level of stem rust infection was determined by measuring chitin levels in leaf tissue at 10 DPI [35]. Single gene lines carrying either Lr34 or Sr12 showed a reduction in stem rust infection compared to null lines but lines with both genes reduced infection levels even further (Fig 4) indicating that the effect of Lr34 and Sr12 on stem rust resistance in response to an avirulent race is additive. Lr34 by itself reduced fungal biomass and resulted in lower IT scores compared to null lines. Macroscopic symptoms at 14 DPI reflected differences in fungal biomass, where “null” lines showed IT = “3”, lines with either Sr12 of Lr34 showed IT = “2”, and lines with both genes showed IT = “11-”. The same lines were also tested in North America at the seedling stage and under field conditions. Lines carrying both genes were also more resistant than single gene lines when tested in the field in Minnesota in 2014 and in Canada in 2015 (Table 2). However, using US races Lr34 by itself did not contribute to stem rust resistance at the seedling stage or in the field, indicating that stem rust resistance involving Lr34 and Sr12 is epistatic. Lines with Sr12 did exhibit significantly reduced stem rust severities in response to the bulk inoculum that included some Pgt races that were avirulent to Sr12.
Fig 4. Amount of Sr12-avirulent Pgt race 98–1,2,(3),(5),6 infection at the seedling stage after 10 DPI in four genotypic classes of homozygous lines from the RL90/RL6077 population as determined by an assay measuring chitin abundance in the leaf tissue.
TC represents ‘Thatcher’.
Table 2. Seedling and field stem rust responses of lines from the RL90/RL6077 population grouped by genotypic class.
| Seedling IT | Mean stem rust severity | ||||||
|---|---|---|---|---|---|---|---|
| Linea | Lr34 | Sr12 | SCCSC | US 2014 | SE | Canada 2015 | SE |
| RL90/RL6077 lines | - | - | 3+ | 55.0 | 1.9 | 72.2 | 2.22 |
| RL90/RL6077 lines | - | + | 0; | 25.0 | 3.3 | 65.6 | 2.22 |
| RL90/RL6077 lines | + | - | 2 to 3+ | 50.0 | 5.0 | 70.0 | 3.33 |
| RL90/RL6077 lines | + | + | 0 | 16.0 | 1.9 | 30.6 | 5.47 |
| Thatcher | - | + | 0 | 5.0 | 15.0 | 2.89 | |
| RL6058 | + | + | 0 | 1.0 | 5.0 | 1.15 | |
| RL6077 | - | + | 0 | 1.0 | 5.2 | 2.09 | |
| RL90 | + | - | 3 | 40.0 | 60.0 | 10.00 | |
a RL6077 carries Lr67 which likely contributed to a low stem rust severity, however none of the RL90/RL6077 lines carried Lr67.
Allele survey of linked SNP markers
SNP markers were identified that are tightly linked to stem rust resistance on chromosome 3B, including the marker NB-LRR3. To evaluate the usefulness of the NB-LRR3 marker for marker-assisted selection, the allele frequency was determined in ‘Thatcher’ derivatives and historic North American germplasm with known Sr12 status. In these lines, the marker genotype corresponded with the presence/absence of Sr12 including for ‘Iumillo’ durum, the donor line for Sr12 (Table 3). We also tested the allele frequency of NB-LRR3 and other linked SNP markers in 92 Australian cultivars of unknown Sr12 status. The ‘Thatcher’ allele of NB-LRR3 and IWA610 was present in approximately 50% of lines tested (S3 Table). The ‘Thatcher’ haplotype at five linked SNP marker loci was present in more than one third of cultivars, suggesting that this haplotype is relatively common in Australian germplasm. The frequency of Sr12 in these lines is unknown.
Table 3. Allele survey of NB-LRR3 marker and seedling IT with Pgt race 98-1-2,(3),(5),6 on selected ‘Thatcher’ derivatives and historic North American germplasm.
| Line/Cultivar | Seedling IT | Sr12 | NB-LRR3 KASP |
|---|---|---|---|
| Thatcher | 1+ | + | + |
| Iumillo durum | 1- | + | + |
| Marquis | 2,2+ | - | - |
| RL6077 | ; 1- | + | + |
| RL90 | 3 | - | - |
| RL6071 | 2+ | - | - |
| RL6058 | ;1 | + | + |
| Canthatch | 1- | + | + |
| Columbus | 1+ | + | + |
| Neepawa | 1- | + | + |
| Napayo | 1 C | + | + |
| Red Fife | 3 | - | - |
| NewThatch (Sr2) | 1+ | + | + |
| Glenlea | ; | - | - |
| Prelude | ; | - | - |
The NB-LRR3 marker was tested on 97 wheat cultivars and lines developed in or introduced into Canada. In total, 50 lines carried the ‘Thatcher’ allele of NB-LRR3 while 47 did not (S3 Table). The influence of ‘Thatcher’ in the genealogy of Canadian Western Red Spring (CWRS) wheat is apparent in the results. Most of these cultivars that carry the Thatcher allele of NB-LRR3 have Thatcher in their pedigrees (S3 Table) [8]. The cultivars from the Canadian Prairie Spring (CPS) class of wheat commonly carried the ‘Thatcher’ allele of NB-LRR3 despite being more genetically distinct from the CWRS cultivars. The ‘Thatcher’ allele was also present in some of the soft white spring and hard red winter cultivars. As in the Australian lines above, the distribution of Sr12 in this set of cultivars is largely unknown.
Discussion
Relationship of Sr12 with field stem rust resistance
In this study, stem rust resistance in the field co-located with the race-specific Sr12 gene on chromosome arm 3BL in the RL, RL-F2 and MN populations. Previously, it was uncertain which arm of chromosome carried Sr12 [29], however this has been resolved by the genetic and physical maps presented here. The NB-LRR3 marker, which explained a large proportion of phenotypic variation for stem rust resistance in the field, co-segregated with Sr12 in these populations. However, it is unknown if Sr12 is responsible, in whole or in part, for the APR observed on chromosome arm 3BL. Sr12 is ineffective at the seedling stage to field races in Kenya [3] and is largely ineffective to North American races of Pgt. In North America the mixture of isolates used for inoculating field trials included some isolates that were avirulent to Sr12. In Canada one out of seven races (14%) was avirulent to Sr12 and in the USA two of the six races (33%) were avirulent to Sr12. This could explain why the effect of the Sr12 region was stronger in North America than in Kenya for the RL population (Table 1), however this does not explain the effect of the Sr12 region on APR when races virulent to Sr12 are present. There are two possible explanations: APR and Sr12 seedling resistance are controlled by independent genes on 3BL, and/or under field conditions Sr12 can provide APR against races that are virulent in the seedling stage. There are reports that describe APR to stem rust in ‘Thatcher’ including the association of the centromeric 3B region, thereby lending support to the independent gene hypothesis [49]. Alternatively, Sr12 may contribute APR by itself or through interactions with other genes. Stem rust resistance in ‘Thatcher’ has been studied extensively but the resistance was found to be complex and difficult to dissect [10].
Recently, a major QTL on chromosome arm 3BL in ‘Thatcher’, which was associated with Sr12, required other unlinked genes to confer effective stem rust resistance in the field [12]. It is possible that gene interactions are relatively common but remain poorly understood. For example, the Canadian cultivar ‘Pasqua’ shows no visual symptoms to leaf rust (Puccinia triticina Eriks.) in the field in most seasons and has always been very resistant since its release [22, 50]. ‘Pasqua’ carries Lr11, Lr13, Lr14b, Lr30, and Lr34 for leaf rust resistance (Dyck 1993). Analysis of these genes in near-isogenic lines in the greenhouse and in the field has shown that none of the genes confers resistance singly to a level near the resistance observed in ‘Pasqua’ (B. McCallum, personal communication). The genes Lr11, Lr13, Lr14b, and Lr30 confer race-specific resistance while Lr34 confers race-nonspecific resistance that is of a quantitative nature. However, reconstruction of the Lr gene combination found in ‘Pasqua’ and in various two, three, and four gene combinations has shown that genes that contribute little to resistance in isolation can still contribute to APR in the field when combined with other genes, particularly in combination with Lr34. For example, lines with Lr11 singly showed an average leaf rust severity of 52% over four season and lines with Lr34 singly showed an average severity of 26%, however lines with Lr11 and Lr34 combined had an average severity of 2% (B. McCallum, personal communication). Major genes are typically assessed for effectiveness by seedling tests; however, relatively little emphasis is placed on their utility in “field resistance”. It appears that some defeated or partially defeated rust resistance genes may play a role in APR when combined with other genes.
Lr34 effect on stem rust resistance
A goal of this study was to identify and genetically map stem rust resistance in ‘Thatcher’ that is expressed or enhanced in the presence of Lr34. In the RL and CS populations, which were fixed for Lr34, a region of chromosome arm 3BL was identified that contributed to resistance in the field. To test for interactions, sister lines were developed from a population that was segregating for Lr34 and Sr12 in the RL90/RL6077 population. Using these lines we showed that Lr34 can reduce stem rust infection at the seedling stage, although Lr34 is not known to confer stem rust resistance in seedling assays. Using a chitin assay to determine the fungal biomass before macroscopic symptoms became visible provided a more sensitive assay to study the early infection process and the effect of Lr34. Field tests of these lines showed that stem rust severity in each genotypic group agreed with both the seedling test for IT and fungal biomass (Fig 4; Table 2). The field resistance on chromosome 3BL did not depend on the presence of Lr34, as demonstrated by the results obtained from the KN population which lacked Lr34 (Table 1).
Previously, one study identified resistance from ‘Thatcher’ located on chromosome arm 2BL that interacted with Lr34 but did not report resistance on chromosome 3B [26]. In a follow-up study, further mapping of the resistance on chromosome arm 2BL found that the peak of the QTL for resistance was marked by the SSR wmc175 [25]. This marker is closely associated with the Sr9 locus [27, 28]. In another study analysing the stem rust resistance in ‘Thatcher’ four QTLs were identified, which were located on chromosomes 3B, 1A, 7DS, and 2BS but no QTL was detected on chromosome arm 2BL [12]. Of the QTL reported in that study, only the QTL on chromosome 3B was significant in all three field environments and it corresponded to the mapped location of Sr12 [12]. The QTL on 7DS corresponded to Lr34, which interacted with the 3B resistance by reducing rust severity in the field. The association between the Sr12 locus and APR in the field is consistent with our findings. Here, we also identified markers on 2B that were associated with resistance, but because of the relatively weak effect from 2B which was probably masked by the strong 3B effect, it was not possible to accurately map the 2B resistance and evaluate if this region interacted with Lr34.
In the RL population, association of the NB-LRR3 marker (and other co-segregating markers) with stem rust resistance was high in all three environments, particularly in Canada. While one of the races (MCCFR) used in the Pgt field inoculum in Canada is avirulent to Sr12, approximately 85% of the field inoculum was composed of six races that are virulent to Sr12. Similar results were observed for the CS and KN populations as in the RL population, although the PVE values were lower (Table 1). If Lr34 does enhance the resistance on 3BL as previously reported [12], it is possible that the effect of the 3BL resistance was lower in the KN population due to the absence of Lr34 in the population. However, the additive effect of the Sr12 region was similar for the KN population and the RL population in Kenya. Like the RL population, the CS population was fixed for Lr34; however, the NB-LRR3 marker explained much less of the phenotypic variation in the CS population compared to the RL population. In both populations the resistant parent was RL6058 (a ‘Thatcher’ near-isogenic line carrying Lr34) thus the genetic basis of resistance should be the same. However, ‘Chinese Spring’, the susceptible parent in the CS population matures late compared to Thatcher-derived lines and as a result the DH lines varied in their maturity dates. It is most likely that the variation in maturity affected the stem rust severity scores in the CS population.
Candidate gene for Sr12
Physical mapping localized a partial NB-LRR-like sequence within the interval in the ‘Chinese Spring’ reference sequence that corresponded to the region in RL6058 carrying Sr12. The NB-LRR (NB-LRR3) sequence in ‘Chinese Spring’ was predicted to encode only part of the 3’ half of a full length NB-LRR gene. Although the partial NB-LRR gene is expressed in seedling leaves of RL6058, we have not yet been able to isolate a full length transcript. It is uncertain if the partial NB-LRR sequence is closely related to a full length NB-LRR gene and candidate gene for Sr12. Given the large physical interval in ‘Chinese Spring’, it is possible that RL6058 carries additional NB-LRR genes in the corresponding interval which are absent in ‘Chinese Spring’. Our results reinforce the notion that sequence information from resistant donor germplasm is often required to identify candidate genes for disease resistance.
Conclusion
The goal was to improve our understanding why Thatcher-Lr34 (RL6058) is significantly more resistant to stem rust than ‘Thatcher’. Our study identified a proximal region on chromosome 3BL as the most significant factor for stem rust resistance in populations which were fixed for Lr34. Unexpectedly, this region was also responsible for the resistance to race Ug99 in Kenya identified in the KN population which lacked Lr34. Further analysis of the RL, RL-F2, and MN populations showed that Sr12 is either responsible or closely linked to the field resistance observed in Kenya, the USA, and Canada. Seedling tests demonstrated that stem rust resistance on 3BL is enhanced in the presence of Lr34. Given these results, selection of Sr12 region could provide a component of durable stem rust resistance in breeding programs and the NB-LRR3 marker is a useful tool to assist in the selection process. Although the ‘Thatcher’ allele of this marker occurs in approximately 50% of Australian and Canadian cultivars, the diagnostic value cannot be assessed until the Sr12 status of these lines is determined. In the meantime, work continues to identify the Sr12 resistance gene through mutagenesis and the sequencing of NB-LRR gene sequences from mutants. The work is expected to generate a gene-based marker for Sr12 and shed light on the relationship of Sr12 and the associated APR.
Supporting Information
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors thank Mira Popovic, Ghassan Mardli, Tobi Malasiuk, Elaine Martineau, Taye Zegeye, Alison Morgan, Jessica Hyles, and Shideh Mojerlou for excellent technical assistance. The authors thank USDA-ARS Appropriated Project 5062-21220-021-00, USDA-ARS National Plant Disease Recovery System, the Durable Rust Resistance in Wheat project (MNR), Saskatchewan Agriculture Development Fund project #20080009 (CAM), Agriculture and Agri-Food Canada Growing Forward (CWH, TF, CAM), and as part of CTAG, a Genome Prairie project funded by Genome Canada, Saskatchewan Ministry of Agriculture, and Western Grain Research Foundation (CAM) for financial support. This project was also supported through a grant CIM/2007/084 provided by Australian Centre of International Agricultural Research. We also thank Dr Sridhar Bhavani for providing access to stem rust nursery at Njoro, Kenya.
Abbreviations
- APR
adult plant resistance
- BSA
bulked-segregant analysis
- SNP
single nucleotide polymorphism
- QTL
quantitative trait locus
- NB-LRR
nucleotide binding-leucine rich repeat
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors thank USDA-ARS Appropriated Project 5062-21220-021-00, USDA-ARS National Plant Disease Recovery System, the Durable Rust Resistance in Wheat project (MNR), Saskatchewan Agriculture Development Fund project #20080009 (CAM), Agriculture and Agri-Food Canada Growing Forward (CWH, TF, CAM), and as part of CTAG, a Genome Prairie project funded by Genome Canada, Saskatchewan Ministry of Agriculture, and Western Grain Research Foundation (CAM) for financial support. This project was also supported through a grant CIM/2007/084 provided by Australian Centre of International Agricultural Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Peturson B. Wheat rust epidemics in western Canada in 1953, 1954, and 1955. Can J Plant Sci. 1958; 38: 16–28. [Google Scholar]
- 2.Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 2000; 84: 203. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, Singh RP, Ward RW, Wanyera R, Kinyua M, Njau P, et al. Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 2007; 91: 1096–1099. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, Szabo L, Pretorius Z, Singh R, Ward R, Fetch T. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 2008; 92: 923–926. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, Szabo L, Rouse M, Fetch Jr. T, Pretorius ZA, Wanyera R, Njau P. Detection of virulence to resistance gene Sr36 within race TTKS lineage of Puccinia graminis f. sp. tritici. Plant Dis. 2009; 93: 367–370. [DOI] [PubMed] [Google Scholar]
- 6.Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Njau P, Wanyera R, et al. Will stem rust destroy the world’s wheat crop? Adv Agron. 2008; 98: 271–309. [Google Scholar]
- 7.Hayes HK, Ausemus ER, Stakman EC, Bailey CH, Wilson HK, Bamberg RH, et al. Thatcher wheat. Minn Agric Exp Stn Tech Bull. 1936; 325. [Google Scholar]
- 8.McCallum BD, DePauw RM. A review of wheat cultivars grown in the Canadian prairies. Can J Plant Sci. 2008; 88: 649–677. [Google Scholar]
- 9.Luig NH. A survey of virulence genes in wheat stem rust, Puccinia graminis f. sp. tritici. Advances in plant breeding, Supplement 11 to Journal of Plant Breeding, Paul Parey, Berlin; 1983.
- 10.Knott DR. The inheritance of stem rust resistance in Thatcher wheat. Can J Plant Sci. 2000; 80: 53–63. [Google Scholar]
- 11.Green GJ, Dyck PL. The reaction of Thatcher wheat to Canadian races of stem rust. Can Plant Dis Surv. 1975; 55: 85–86. [Google Scholar]
- 12.Rouse MN, Talbert LE, Singh D, Sherman JD. Complementary epistasis involving Sr12 explains adult plant resistance to stem rust in Thatcher wheat (Triticum aestivum L.). Theor Appl Genet. 2014; 127: 1549–1559. 10.1007/s00122-014-2319-6 [DOI] [PubMed] [Google Scholar]
- 13.Dyck PL. The association of a gene for leaf rust resistance with the chromosome 7D suppressor of stem rust resistance in common wheat. Genome. 1987; 29: 467–469. [Google Scholar]
- 14.Dyck PL. Genetics of adult plant leaf rust resistance in ‘Chinese Spring’ and ‘Sturdy’ wheats. Crop Sci. 1991; 31: 309–311. [Google Scholar]
- 15.Singh RP. Genetic association of leaf rust resistance gene Lr34 with adult-plant resistance to stripe rust in bread wheat. Phytopathology. 1992; 82: 835–838. [Google Scholar]
- 16.Singh RP. Genetic association between gene Lr34 for leaf rust resistance and
- 17.Singh RP. Genetic association of gene Bdv1 for tolerance to barley yellow dwarf virus with genes Lr34 and Yr18 for adult plant resistance to rusts in bread wheat. Plant Dis. 1993; 77: 1103–1106. [Google Scholar]
- 18.Spielmeyer W, McIntosh RA, Kolmer J, Lagudah ES. Powdery mildew resistance and the Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor Appl Genet. 2005; 111: 731–735. [DOI] [PubMed] [Google Scholar]
- 19.Kerber ER, Aung T. Leaf rust resistance gene Lr34 associated with nonsuppression of stem rust resistance in the wheat cultivar Canthatch. Phytopathology. 1999; 89: 518–521. 10.1094/PHYTO.1999.89.6.518 [DOI] [PubMed] [Google Scholar]
- 20.Kerber ER, Green GJ. Suppression of stem rust resistance in the hexaploid wheat cv. Canthatch by chromosome 7DL. Can J Bot. 1980; 58: 1347–1350. [Google Scholar]
- 21.Kerber ER. Stem-rust resistance in “Canthatch” hexaploid wheat induced by a nonsuppressor mutation on chromosome 7DL. Genome. 1991; 34: 935–939. [Google Scholar]
- 22.Dyck PL. Inheritance of leaf rust and stem rust resistance in ‘Roblin’ wheat. Genome. 1991; 36: 289–293. [DOI] [PubMed] [Google Scholar]
- 23.Liu JQ, Kolmer JA. Genetics of stem rust resistance in wheat cvs. Pasqua and AC Taber. Phytopathology. 1988; 88: 171–176. [DOI] [PubMed] [Google Scholar]
- 24.Hiebert CW, Fetch TG, Zegeye T, Thomas JB, Somers DJ, Humphreys DG, et al. Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ‘Peace’ and ‘AC Cadillac’. Theor Appl Genet. 2011; 122: 143–149. 10.1007/s00122-010-1430-6 [DOI] [PubMed] [Google Scholar]
- 25.Kolmer JA, Garvin DF, Yin J. Expression of a Thatcher wheat adult plant stem rust resistance QTL on chromosome arm 2BL is enhanced by Lr34. Crop Sci. 2011; 51: 526–533. [Google Scholar]
- 26.Gavin Vanegas CD, Garvin DF, Kolmer JA. Genetics of stem rust resistance in the spring wheat cultivar Thatcher and the enhancement of stem rust resistance by Lr34. Euphytica. 2008; 159: 391–401. [Google Scholar]
- 27.Tsilo TJ, Jin Y, Anderson JA. Microsatellite markers linked to stem rust resistance allele Sr9a in wheat. Crop Sci. 2007; 47: 2013–2020. [Google Scholar]
- 28.Rouse MN, Nirmala J, Jin Y, Chao S, Fetch TG, Pretorius ZA, Hiebert CW. Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor Appl Genet. 2014; 127: 1681–1688. 10.1007/s00122-014-2330-y [DOI] [PubMed] [Google Scholar]
- 29.McIntosh RA, Pertridge M, Hare RA. Telocentic mapping of Sr12 in wheat chromosme 3B. Cereal Res Comm. 1980; 8: 321–324. [Google Scholar]
- 30.Thomas J, Chen Q, Howes N. Chromosome doubling of haploids of common wheat with caffeine. Genome. 1997; 40: 552–558. [DOI] [PubMed] [Google Scholar]
- 31.Hughes GR, Hucl P. Kenyon hard red spring wheat. Can J Plant Sci. 1991; 71: 1165–1168. [Google Scholar]
- 32.Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, et al. Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor Appl Genet. 2009; 119: 889–898. 10.1007/s00122-009-1097-z [DOI] [PubMed] [Google Scholar]
- 33.Peterson RF, Campbell AB, Hannah AE. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can J Res. 1948; 26: 496–500. [Google Scholar]
- 34.Stakman EC, Stewart DM, Loegering WQ. Identification of physiologic races of Puccinia graminis var. tritici. USDA Agricultural Research Service E 617; 1962. [Google Scholar]
- 35.Ayliffe M, Periyannan SK, Feechan A, Dry I, Schumann U, Wang M-B, Pryor A, Lagudah E. A simple method for comparing fungal biomass in infected plant tissues. Mol Plant Microbe Interact. 2013; 26: 658–667. 10.1094/MPMI-12-12-0291-R [DOI] [PubMed] [Google Scholar]
- 36.Pallotta MA, Warner P, Fox RL, Kuchel H, Jefferies SJ, Langridge P. Marker assisted wheat breeding in the southern region of Australia. In: Pogna NE, Romano M, Pogna EA, Galerio G, editors. Proc 10th Int Wheat Genet Symp, Instituto Sperimentale per la Cerealcoltura, Rome; 2003. pp. 789–791.
- 37.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of genomic plant DNA for PCR analysis. Nucleic Acids Res. 1991; 19: 1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to diseaese-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci. 1991; 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci. 2013; 110: 8057–8062. 10.1073/pnas.1217133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015; 3: 269–283. [Google Scholar]
- 41.Allen AM, Barker GLA, Berry ST, Coghill JA, Gwilliam R, Kirby S, et al. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J. 2011; 9: 1086–1099. 10.1111/j.1467-7652.2011.00628.x [DOI] [PubMed] [Google Scholar]
- 42.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014; 345: 287. [DOI] [PubMed] [Google Scholar]
- 43.Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998; 149: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somers DJ, Isaac P, Edwards K. A high density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet. 2004; 109: 1105–1114. [DOI] [PubMed] [Google Scholar]
- 45.Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, et al. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics. 2004; 4: 12–25. [DOI] [PubMed] [Google Scholar]
- 46.Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, et al. Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet. 2005; 110: 550–560. [DOI] [PubMed] [Google Scholar]
- 47.Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, et al. Diversity arrays technology (DArT) for high-throughput prowling of the hexaploid wheat genome. Theor Appl Genet. 2006; 113: 1409–1420. [DOI] [PubMed] [Google Scholar]
- 48.Lorieux M. MapDisto: fast and efficient computation of genetic linkage maps. Mol Breed. 2012; 30: 1231–1235. [Google Scholar]
- 49.Nazareno NR, Roelfs AP. Adult plant resistance of Thatcher wheat to stem rust. Phytopathology. 1981; 71: 181–185. [Google Scholar]
- 50.Townley-Smith TF, Czarnecki EM, Campbell AB, Dyck PL, Samborski DJ. Pasqua hard red spring wheat. Can J Plant Sci. 1993; 73: 1095–1098. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.