Abstract
The shadow enhancer of the short gastrulation (sog) gene directs its sequential expression in the neurogenic ectoderm and the ventral midline of the developing Drosophila embryo. Here, we characterize three unusual features of the shadow enhancer midline activity. First, the minimal regions for the two different enhancer activities exhibit high overlap within the shadow enhancer, meaning that one developmental enhancer possesses dual enhancer activities. Second, the midline enhancer activity relies on five Single-minded (Sim)-binding sites, two of which have not been found in any Sim target enhancers. Finally, two linked Dorsal (Dl)- and Zelda (Zld)-binding sites, critical for the neurogenic ectoderm enhancer activity, are also required for the midline enhancer activity. These results suggest that early activation by Dl and Zld may facilitate late activation via the noncanonical sites occupied by Sim. We discuss a model for Zld as a pioneer factor and speculate its role in midline enhancer activity. [BMB Reports 2015; 48(10): 589-594]
Keywords: Drosophila, Embryo, Pioneer factor, Shadow enhancer, Short gastrulation, Zelda
INTRODUCTION
Most transcriptional enhancers tested to date only regulate the expression of their target genes at a particular time and in a defined space (1). Thus, many developmental genes that are used repeatedly at different times and at diverse locations during development need various enhancers by which their differential expression is controlled both temporally and spatially (2). For instance, rhomboid (rho), a dorsal-ventral (DV) patterning gene, is expressed in the presumptive neurogenic ectoderm (NE) and the ventral midline (VM) of the developing Drosophila embryo (3, 4). The sequential expression in these two different spaces is controlled by two different enhancers (5). Although these enhancers are adjacent to one another, they are functionally autonomous and geographically separable.
In contrast with previous observations, however, recent transgenic studies have shown that the short gastrulation (sog) locus contains two separate enhancers by which its expression in the NE is directed (6). The expression patterns directed by these enhancers are very similar to that of endogenous sog expression. These two enhancers are referred to as the “primary” enhancer and the “shadow” enhancer, respectively, based on the chronological order of their identification rather than any functional differences. These results suggested that expression of a gene at a particular time and in a defined space can be controlled by two redundant enhancers.
More recently, the shadow enhancer was also shown to direct sog expression in the VM (7). Like rho, sog is initially expressed in the NEs in broad lateral strips. However, by germband extension sog expression is restricted to the VM, which is composed of specialized glial cells that secrete molecules critical for nerve cord patterning (8). The early broad stripe of sog expression in the NE relies on three DV determinants, Dorsal (Dl), Zelda (Zld), and Snail (Sna) (9-11). The early broad pattern of sog expression is believed to be dictated by both synergistic activation between Dl and Zld across at least half of the syncytial blastoderm and also Sna-mediated repression in the presumptive mesoderm. Like other Single-minded (Sim) target genes, the later pattern in the VM seems to be directed by Sim (12). The sim gene encodes a transcription factor containing a basic helix-loop-helix-PAS (bHLH-PAS) domain that functions as a master regulator of VM differentiation in the Drosophila central nervous system (CNS) (13). Although previous gainand loss-of-function studies have shown that sog expression in the VM depends on Sim (7, 12), direct evidence that the midline enhancer activity of the shadow enhancer requires typical Sim-binding sites has not yet been obtained. These findings indicate that one of two redundant enhancers responsible for early sog expression can also direct late expression of sog at a different location via an unknown mechanism.
Here we present evidence that the shadow enhancer midline activity has three extraordinary features in the context of cis-regulatory DNA. First, the minimal regions for the dual enhancer activities in the shadow enhancer cannot be uncoupled. Second, the midline enhancer activity requires five Sim-binding sites, two of which are noncanonical sites. Finally, loss of either the Dl- or the Zld-binding site severely disrupts the midline enhancer activity. These results reveal that early enhancer activity in the NE may facilitate late enhancer activity in the VM, which presumably helps cope with the low quality of the cis-regulatory elements that regulate late expression.
RESULTS
The midline enhancer activity of the sog shadow enhancer cannot be uncoupled from its neurogenic ectoderm enhancer activity
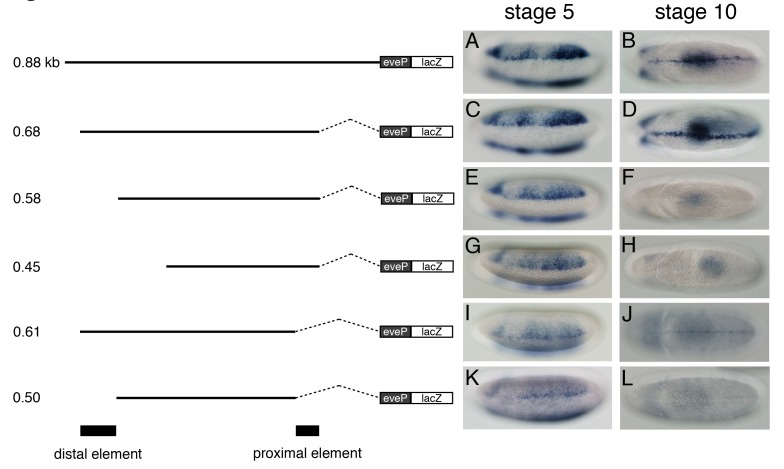
The previous finding that the two rho enhancers are functionally autonomous and geographically separable (5) raised the question of whether the ventral midline enhancer (VME) activity of the sog shadow enhancer can be geographically uncoupled from its neurogenic ectoderm enhancer (NEE) activity. To answer this question, transgenic embryos containing lacZ fusion genes directed by truncated versions of the shadow enhancers were generated, and lacZ expression in the NE and VM was tested by whole-mount in situ hybridization with an antisense lacZ RNA probe (Fig. 1). Compared with the 0.88-kb full-length construct (Fig. 1A and B), the 0.68-kb region was sufficient for each of the enhancer activities (Fig. 1C and D). However, a5’ ∼100-bp deletion of the 0.68-kb construct (0.58 kb) severely interfered with VM lacZ expression (Fig. 1E and F), while a 5’ 230-bp truncation (0.45 kb) failed to drive sog expression in the NE and VM (Fig. 1G and H). These results suggest that the 0.58-kb region is inadequate for VME activity and that the 5’ limit of the minimal NEE must lie between the 5’ ends of the 0.58-kb and 0.45-kb enhancers. The loss of ∼70 bp at the 3’ end of the 0.68-kb construct ablated sog expression in both the NE and VM (Fig. 1I and J). Moreover, simultaneous loss of the distal (5’ ∼100 bp) and proximal (3’ ∼70 bp) elements, which were previously shown to be critical for the midline enhancer activity (7), resulted in the severe reduction of sog expression in the NE and VM (Fig. 1K and L). These results indicate that both the distal and proximal elements are required to drive sog expression in the NE as well as the VM and thus that the minimal regions for the two enhancer activities appear to overlap.
Fig. 1. The midline enhancer activity of the sog shadow enhancer cannot be physically uncoupled from its neurogenic ectoderm enhancer (NEE) activity. Schematic diagrams of the six lacZ constructs used for P element-mediated germline transformation are shown. Various regions of the 0.88-kb enhancer were placed upstream of the even-skipped (eve) minimal promoter, followed by the lacZ-coding region. Wild-type (yw) embryos were injected with each construct and lacZ expression was analyzed by in situ hybridization with an antisense lacZ RNA probe. Early (stage 5) and late (stage 10) embryos are oriented to show lateral (A, C, E, G, I and K) and ventral (B, D, F, H, J and L) views, respectively, with the anterior view on the left.
The sog shadow enhancer contains three canonical Sim-binding sites, one of which is statistically functional
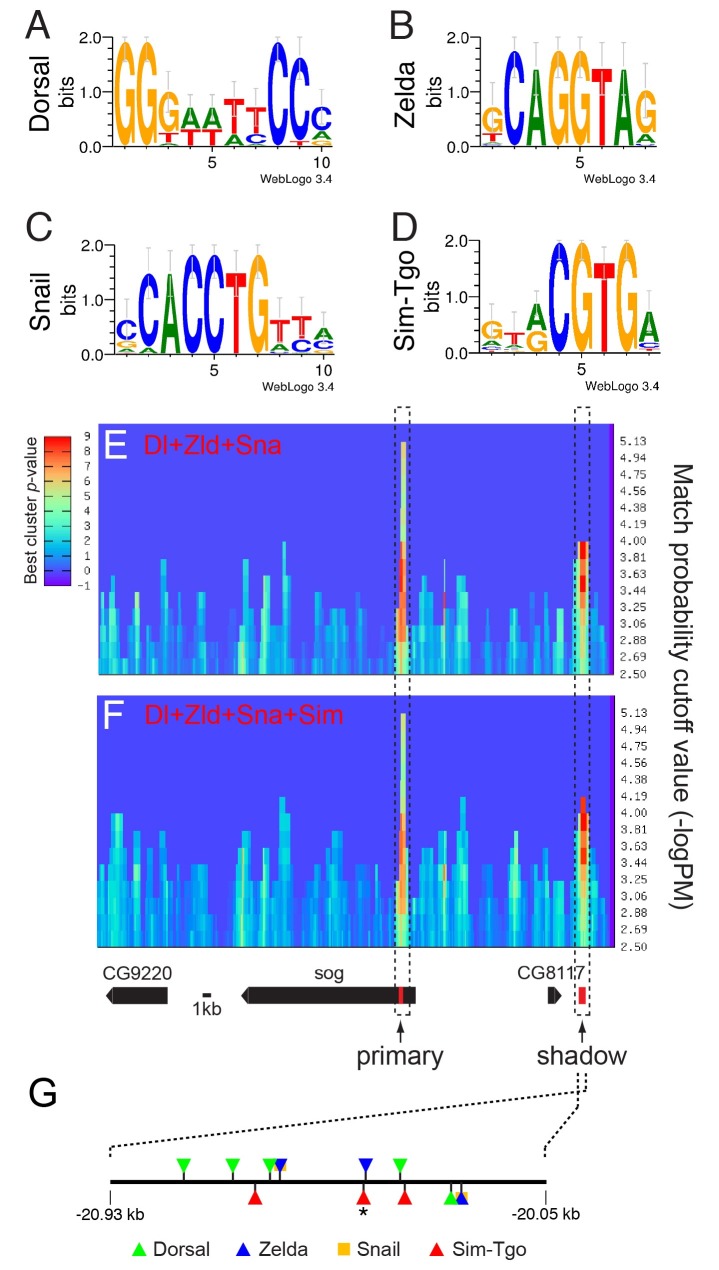
Although the proximal and distal elements, which are critical for VME activity, do not contain any Sim-binding sites, sog expression in the VM has been shown to require Sim (7, 12). This apparent contradiction led us to determine the number of Sim-binding motifs in the sog locus, how good their qualities are, and whether they can form a statistically significant cluster with the Dl, Zld, and Sna. Since no transcription factor has yet been identified that interacts cooperatively with Sim, it is extremely hard to find functional clusters of Sim-binding motifs. Thus, although it is highly unlikely that Sim, Dl, Zld, and Sna cooperate via simultaneous site occupancy during embryogenesis, assessing the statistical significance of the clusters formed by the four transcription factors may aid the identification of functional Sim-binding motif clusters. To this end, ClusterDraw analyses were performed with position frequency matrices (PFMs) (Fig. S1) of the DNA-binding motifs (Fig. 2A-D) for Dl, Zld, Sna, and Sim. The ClusterDraw algorithm is an r-scan-based program that has been used to identify the binding motifs and binding clusters of specific combinations of transcription factors (14, 15). We hypothesized that Sim-binding sites in the sog locus that yield cumulative match probability P values (y-axis) higher than the cut-off values and that are located nearby or within the Dl, Zld, and Sna cluster will generate statistically significant cluster P values (z-axis) with the Dl, Zld, and Sna PFMs. A ClusterDraw analysis of the Dl, Zld, and Sna PFMs over a ∼62-kb genomic region encompassing the sog locus (Fig. 2E) identified two clusters, which correspond precisely to the previously identified primary enhancer and shadow enhancer (Fig. 2, dotted boxes and Table S1). The similar analysis was performed with the Dl, Zld, Sna, and Sim PFMs and produced a pattern comparable with that shown in Fig. 2E, except that the match probability value corresponding to the shadow enhancer was higher than that of panel E (Fig. 2F, S2, S3, and Table S1). This finding indicates that the involvement of Sim in the preformed cluster containing Dl, Zld, and Sna increased the statistical significance of its cluster P value. This result prompted us to map the Sim-binding motifs in the shadow enhancer. Combined GenePalette (16) and ClusterDraw analyses showed that the shadow enhancer contains three Sim-binding sites within the Dl, Zld, and Sna cluster (Fig. 1G), two of which yielded match probability P values lower than the cut-off. Thus, the ClusterDraw analysis excluded these two sites upon formation of the cluster (Fig. S3). Intriguingly, the shadow enhancer does not contain the 5’-NNACGTG-3’ sequence (in which “N” represents any nucleotides), which is found at least once in each sim target enhancer experimentally tested so far (4, 17). These findings suggest that the sog shadow enhancer midline activity might be derived from these three Sim-binding sites.
Fig. 2. ClusterDraw analyses of a ∼62-kb genomic region encompassing the sog locus. DNA binding sequences for Dorsal (Dl) (A), Zelda (Zld) (B), Snail (Sna) (C), and Single-minded (Sim) (D) are graphically represented as sequence logos (21). The WebLogo 3.3 algorithm was used to build the sequence logos with the position frequency matrices (PFMs) (Fig. S1) of the Dl-, Zld-, Sna-, and Simbinding DNA sequences. ClusterDraw analysis with the Dl, Zld, and Sna PFMs (E) yielded two statistically significant match probability values, the highest and the second highest of which coincide with the primary and shadow enhancers, respectively (dotted boxes, Table S1). The best P values of these two clusters are highly similar to each other. The same analysis was performed with the Dl, Zld, Sna, and Sim PFMs (F). This analysis produced a pattern comparable to that shown in panel E, except that the match probability value corresponding to the shadow enhancer was higher than that shown in panel E (Fig. S2, S3, and Table S1). This finding suggests that the addition of the Sim PFM increased the statistical significance of the preformed cluster of Dl, Zld, and Sna in the shadow enhancer. Gene models (G) over the analyzed genomic region are depicted below panel F. Binding sites for Dl, Zld, Sna, and Sim identified by GenePalette and ClusterDraw are depicted in the sog shadow enhancer. GenePalette analysis matched three Sim sites to the consensus sequence (D), whereas ClusterDraw selected only one Sim site (marked with an asterisk) (Fig. S3 and Table S1). This discrepancy may be at least partially due to the statistically low qualities of the other two sites.
Midline enhancer activity depends on five Sim-binding sites, two of which are noncanonical sequences
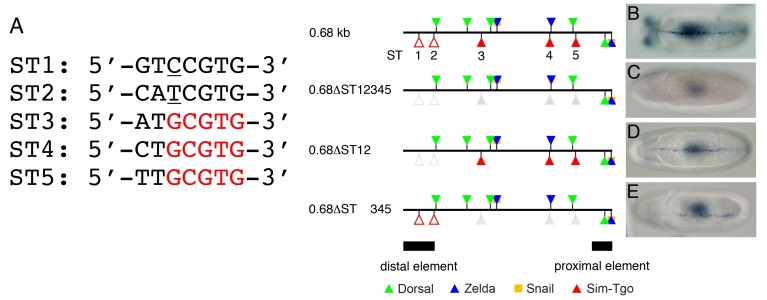
We next investigated whether the shadow enhancer contains sequences that are similar to the Sim-binding motif, even though these sequences do not perfectly match with the Sim consensus sequence. GenePalette searches permitting a one-nucleotide mismatch identified two noncanonical Sim-binding sites in the distal element (Fig. 3A, ST1, and ST2). Each Sim- binding site contained one mismatch at the third position from the 5’ end of the site, implying that the binding affinities of these sites to Sim are much lower than those of the canonical Sim-binding sites.
Fig. 3. Midline enhancer activity depends on five Sim-binding sites, two of which are noncanonical sequences. (A) The 0.68-kb sog shadow enhancer contains five Sim-binding sites. The first two sites (ST1 and ST2) appear to be noncanonical sequences because each site contains one nucleotide mismatch relative to the core consensus sequence (the Sim-binding site consensus sequence is shown in Fig. 2D). The mismatched nucleotides are underlined. The remaining three sites (ST3, ST4, and ST5) contain the core GCGTG consensus sequence. (B-E) Wild type and mutant versions of the 0.68-kb enhancer construct were used to generate lacZ transformation constructs. Cloning of the transformation constructs, germline transformation, and in situ hybridization were performed as in Fig. 1. Two noncanonical Sim-binding sites (ST1 and ST2, red open triangles) were identified by the GenePalette motif search with one nucleotide mismatch allowed. ST3, ST4, and ST5 are depicted as red closed triangles. Deletions of each Sim-binding site were introduced by PCR-mediated site-directed mutagenesis.
Although the previous ClusterDraw analyses suggested that two of the three Sim-binding sites were statistically less significant, the functionality of these sites had not been investigated in vivo. Thus, we used transgenic embryos to determine whether expression of the lacZ transgene containing the 0.68-kb enhancer in the VM requires the five Sim-binding motifs. Compared with the 0.68-kb minimal VME construct (Fig. 3B), mutation of the five Sim sites in the 0.68-kb construct (0.68ΔST12345) abrogated lacZ expression in the VM (Fig. 3C), suggesting that at least some of the five Sim sites are necessary for the expression of the reporter gene. We next investigated whether the complete repression of lacZ expression was due to the loss of the two noncanonical sites in the distal element (ST1 and ST2) or the remaining three sites (ST3-ST5). Selective mutations in the ST1 and ST2 sites (0.68ΔST12) also yielded significantly reduced expression of the lacZ transgene in the VM (Fig. 3D). However, the lacZ transcript was barely detectable in transgenic embryos containing a 0.68-kb construct harboring mutations in the ST3, ST4, and ST5 sites (0.68ΔST 345) (Fig. 3E). These results suggest that while the VME activity of the sog shadow enhancer relies on all five Sim-binding sites, the three canonical binding sites may be more crucial than the two noncanonical binding sites for driving sog expression in the VM, at least in the context of the lacZ reporter gene.
The linked Dl- and Zld-binding sites are also required for midline enhancer activity
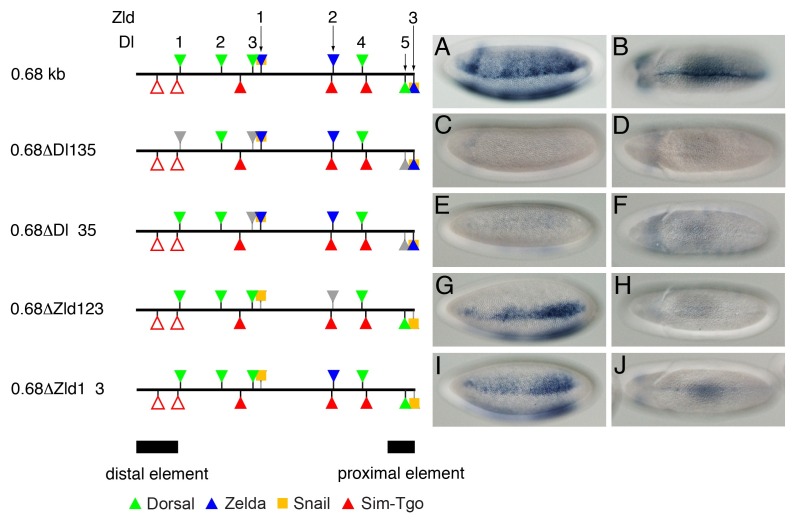
The proximal element, which corresponds to the last approximately 70 bp at the 3’ end of the 0.68-kb construct, is functionally required for the midline enhancer activity (7) but does not contain any Sim-binding sites. Instead, it has a cluster of closely linked binding sites for Dl, Zld, and Sna (Fig. 4, Tables S1 and S2). The Dl-binding sites (Dl3 and 5) are located adjacent to the Zld-binding sites (Zld1 and 3), which overlaps with the Sna-binding site. The finding that deletion of the proximal element in the 0.68-kb construct significantly repressed lacZ expression in the VM (Fig. 1I and J) raised the possibility that early determinants such as Dl and Zld may play a role in sog expression in the midline. To test this possibility, transgenic embryos containing various mutant versions of the 0.68-kb construct were generated and the levels of lacZ expression in the resultant embryos were determined by wholemount in situ hybridization using an antisense lacZ RNA probe. The 0.68-kb construct contains five Dl-binding sites and three Zld-binding sites. Of these sites, the Dl3-Zld1 and Dl5-Zld3 sites are tightly linked to one another (Fig. 4, schematic representation left of panel A). To determine whether the Dl-binding sites are necessary for the midline enhancer activity, the three Dl sites (Dl1, Dl3, and Dl5) were mutated (0.68ΔDl135). Compared with the parental 0.68-kb construct (Fig. 4A and B), the 0.68ΔDl135 construct did not drive lacZ expression in any region of the embryo (Fig. 4C and D), suggesting that the three Dl sites play critical roles for sog expression in both the NE and the VM. The loss of lacZ expression in the VM in the absence of the three Dl sites is particularly intriguing, since Dl activity declines as embryogenesis proceeds and D1 finally becomes functionally inactive before the formation of the VM (4). Moreover, the loss of two Dl sites, Dl3 and Dl5 (which are linked with the Zld sites) (0.68ΔDl 35), is sufficient to block the activities of the 0.68-kb enhancer in the NE and the VM (Fig. 4E and F).
Fig. 4. The linked Dl- and Zld-binding sites in the 0.68-kb enhancer are required for its VM enhancer activity. Wild type and mutant versions of the 0.68-kb enhancer were used to generate lacZ transformation constructs. Cloning of the transformation constructs, germline transformation, and in situ hybridization were performed as in Fig. 1. Lateral (A, C, E, G, and I, stage 5) and ventral (B, D, F, H, and J, stage 10) views of the embryos are shown; the anterior side is on the left. The 0.68-kb minimal enhancer contains five Dl-binding and three Zld-binding sites. Of these, the Dl3-Zld1 and Dl5-Zld3 sites are closely linked. Intriguingly, Zld1 and Zld3 coincide with two Sna-binding sites to form two modules, each of which is composed of a combination of Dl-, Zld-, and Sna-binding sites.
We next asked if the Zld-binding sites linked to the Dl-binding sites also contribute to the midline enhancer activity, since Zld has been shown to facilitate the occupancy of Dl to its cognate binding motif (11). Mutagenesis of all the Zld sites (0.68ΔZld123) or of the two linked sites (0.68ΔZld1 3) led to severe reduction of lacZ expression in the VM (Fig. 4H and J), consistent with the results of the 0.68ΔDl135 and 0.68ΔDl 35 constructs. The only difference in the lacZ expression patterns obtained with the 0.68ΔZld123 and 0.68ΔZld1 3 constructs is the width of the lacZ expression stripe in the NE (Fig. 4G and I). The lacZ expression pattern driven by 0.68ΔZld123 is narrower than that driven by 0.68ΔZld1 3. These different thresholds in the Dl concentration gradient are presumably due to a synergistic interaction between Zld2 and Dl4 in the 0.68ΔZld1 3 construct. These results suggest that the Dl and Zld binding motifs associated with the two Dl, Zld, and Sna clusters are indispensable for sog expression in the VM.
DISCUSSION
Shadow enhancers were initially identified as one of two separate enhancers that drive the same or similar expression patterns of many Dl target genes (6). A recent study showed that the shadow enhancer directs sequential sog expression patterns in both the NE and the VM (7). Here, we characterized for the first time three unusual DNA features of the sog shadow enhancer.
The most intriguing finding in the current study is that the Zld-binding sites in the 0.68-kb construct are required for its VME activity. The zld gene encodes a maternal transcription activator that is ubiquitously expressed in blastoderm embryos until the end of the nuclear cleavage cycle 14 (18) and is believed to drive the early broad stripe of sog expression in the NE via cooperative site occupancy with Dl (10). Why did the sog shadow enhancer lose its VME activity upon the loss of the Zld-binding sites? The simplest explanation for this finding is that Zld may function as a pioneer factor to potentiate the VME activity of the shadow enhancer. A pioneer factor is a transcription factor that confers transcriptional competency upon inactive target genes by binding condensed chromatin prior to the binding of other transcription factors (19). It is plausible that Zld establishes an open chromatin environment for NEE activity before early sog activation in the NE and enhances binding of Dl via synergistic interaction during sog activation in the NE. The established chromatin environment may even be maintained for further site occupancy by subsequent transcription factors such as Sim. The recent finding that Zld binding to regulatory DNA is strongly correlated with increased chromatin accessibility (11) supports this explanation. Alternatively, direct synergistic interaction between Zld and Sim in the VM may explain the requirement for the Zld-binding sites for the VME activity, because a zygotic Zld transcript was found in the VM during late embryogenesis (20). However, the Zld protein encoded by this zygotic transcript contains only one out of the 4 C2H2 zinc finger motifs that are known to bind typical Zldbinding DNA sequences. Thus, it is presently unclear whether the late version of Zld expressed in the VM is actually able to directly bind Zld-binding sites.
The finding that the midline enhancer activity of the shadow enhancer depends on the two noncanonical Sim-binding sites and the three canonical Sim-binding sites can also be interpreted in this context. The shadow enhancer does not contain any ‘ACGTG’ Sim-binding motifs, at least one of which is found in all known Sim target enhancers. Instead, its midline enhancer activity depends on five Sim sites that appear to have relatively low affinities for Sim. The number of Sim-binding sites in the VME has been shown to define the onset timing of its target gene expression in the VM (4). Sim target genes fall into approximately two categories, early and late, and early enhancers contain more Sim-binding sites than late enhancers. Like rho, sog appears to be an early Sim target gene because the onset of its expression is similar to that of Sim itself in the VM (7). Accordingly, the sog VME is hypothesized to have at least four high-affinity Sim-binding sites; instead, however, it has five poor quality Sim sites. Thus, the shadow enhancer may need alternative ways of coping with the inefficient capability to recruit the master regulatory protein Sim. It is conceivable that the euchromatic environment pre-established by Zld may bypass the requirement of Sim-mediated chromatin decondensation required by most Sim target enhancers. This pre-established euchromatic environment facilitates the initial recruitment of Sim via low-affinity sites and thus enhances instant communication between the shadow enhancer and its target promoter.
MATERIALS AND METHODS
Detailed information is described in online Supplementary Material.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (MSIP and MOE) (NRF-2010-0002792 and NRF-2012R1A1 A2038502).
References
- 1.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. (2014);157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. (2010);20:R754–63. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bier E, Jan LY, Jan YN. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. (1990);4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- 4.Hong JW, Park KW, Levine MS. Temporal regulation of single-minded target genes in the ventral midline of the Drosophila central nervous system. Dev Biol. (2013);380:335–343. doi: 10.1016/j.ydbio.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. (1992);6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 6.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. (2008);321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin DH, Hong JW. The shadow enhancer of short gastrulation also directs its expression in the ventral midline of the Drosophila embryo. Genes Genom. (2015);37:743–750. doi: 10.1007/s13258-015-0302-5. [DOI] [Google Scholar]
- 8.Menne TV, Luer K, Technau GM, Klambt C. CNS midline cells in Drosophila induce the differentiation of lateral neural cells. Development. (1997);124:4949–4958. doi: 10.1242/dev.124.24.4949. [DOI] [PubMed] [Google Scholar]
- 9.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. (2002);99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liberman LM, Stathopoulos A. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev Biol. (2009);327:578–589. doi: 10.1016/j.ydbio.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foo SM, Sun Y, Lim B, et al. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol. (2014);24:1341–1346. doi: 10.1016/j.cub.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinzen RP, Cande J, Ronshaugen M, Papatsenko D, Levine M. Evolution of the ventral midline in insect embryos. Dev Cell. (2006);11:895–902. doi: 10.1016/j.devcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Nambu JR, Lewis JO, Wharton KA Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loophelix protein that acts as a master regulator of CNS midline development. Cell. (1991);67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 14.Papatsenko D. ClusterDraw web server: a tool to identify and visualize clusters of binding motifs for transcription factors. Bioinformatics. (2007);23:1032–1034. doi: 10.1093/bioinformatics/btm047. [DOI] [PubMed] [Google Scholar]
- 15.Shin DH, Hong JW. Capicua is involved in Dorsal-mediated repression of zerknüllt expression in Drosophila embryo. BMB Rep. (2014);47:518–523. doi: 10.5483/BMBRep.2014.47.9.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebeiz M, Posakony JW. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev Biol. (2004);271:431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Wharton KA Jr, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. (1994);120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- 18.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. (2008);456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. (2011);25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson JC, Crews ST. Enhancer diversity and the control of a simple pattern of Drosophila CNS midline cell expression. Dev Biol. (2014);392:466–482. doi: 10.1016/j.ydbio.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. (2004);14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]