Abstract
Molecular dynamics simulations employing all-atom force fields have become a reliable way to study binding interactions quantitatively for a wide range of systems. In this work, we employ two recently developed methods for the calculation of dissociation constants KD between gold nanoparticles (AuNPs) of different sizes in a near-physiological environment through the potential of mean force (PMF) formalism: the method of geometrical restraints developed by Woo et al. and formalized by Gumbart et al. and the method of hybrid Steered Molecular Dynamics (hSMD). Obtaining identical results (within the margin of error) from both approaches on the negatively charged Au18(SR)14 NP, functionalized by the negatively charged 4-mercapto-benzoate (pMBA) ligand, we draw parallels between their energetic and entropic interactions. By applying the hSMD method on Au102(SR)44 and Au144(SR)60, both of them near-spherical in shape and functionalized by pMBA, we study the effects of size and shape on the binding interactions. Au18 binds weakly with KD = 13mM as a result of two opposing effects: its large surface curvature hindering the formation of salt bridges, and its large ligand density on preferential orientations favoring their formation. On the other hand, Au102 binds more strongly with KD = 30μM and Au144 binds the strongest with KD = 3.2nM.
Keywords: Molecular Dynamics, Gold Nanoparticles, Aggregation
1. Introduction
The interactions in aqueous solutions of NPs with sizes less than 3 nm have recently become quantifiable through molecular dynamics (MD) simulations. [1, 2, 3, 4] Experimental studies performed on large NPs (100 nm) have shown that aspherical shapes require a smaller salt concentration to aggregate, due to the larger contact areas between them;[5, 6] while smaller sizes require larger salt concentrations to aggregate, due to the weaker van der Waals forces of attraction.[7, 8] Such small-sized NPs have presented novel challenges to traditional colloid science theories such as Derjaguin-Landau-Verwey-Overbeak (DLVO). In particular, since the radius of sodium rNa = 1.36 Å is more than 10% of the NP’s radius, the Poisson-Boltzmann theory for electrostatic interactions is not reasonable because it considers the ions as point charges.[9] Additionally, the DLVO assumption that the surfaces of interaction are flat cannot be applied to NPs of such large curvatures as these.[10, 11] The presence of charged surface coatings also introduces steric and bridging effects (mediated by sodium) and it thus becomes necessary to replace simple analytic expressions by a PMF obtained from atomistic numerical computations.
In recent years, the computational methods for obtaining binding affinities through molecular dynamics simulations employing all-atom force fields have matured to a point that experimental measurements can be reproduced. In this paper, we illustrate the use of two such methods, both based on the potentials of mean force (PMF) formalism.[12, 13, 14] The first of these methods, which was developed in 2005 by Woo and Roux,[15] and whose procedures were formalized by Gumbart, Roux, and Chipot in 2012, “invokes a series of geometrical restraints acting on collective variables designed to alleviate sampling limitations inherent to classical molecular dynamics simulations.”[16] The other method, which is called hybrid Steered Molecular Dynamics (hSMD)[17, 18], is a brute force approach developed in 2015 and involves mainly “simultaneously steering n centers of mass of n selected segments of the ligand using n springs of infinite stiffness”.
In this work, we employ both methods on the Au18(SR)14 NP[19, 20, 21, 22] in a near-physiological environment (see Fig. 1(a)). Gold NPs have shown such a promising potential in medical procedures that the result has been called “a golden age” of biomedical nanotechnology.[23, 24] For example, they have been found suitable as drug delivery vehicles for selective targeting of cells,[25] as passive contrast agents for labelling, visualizing and tracking specific receptors,[26] as active sensors whose optical properties change in the presence of analyte molecules,[27] or as heat sources for the treatment of cancer through absorption of light.[28] With regard to biomedical applications, a desirable property of gold nanoparticles is their ability to aggregate reversibly into nanoclusters of controlled size when placed in a special solution, and to dissociate back into individual nanoparticles when introduced into the body.[29, 30, 31, 32] Finally, we compare the binding affinity of Au18 with those of Au102[33] and Au144[34, 35, 36, 37, 38, 39, 40, 32], functionalized by the same ligand under the same near-physiological conditions.
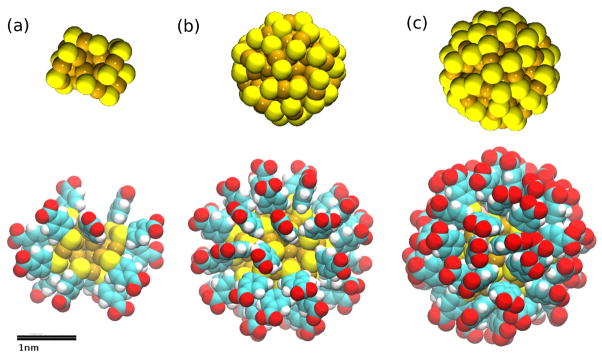
Figure 1.
Systems under study: (a) the Au18(SR)14 (diameter ~ 1nm), (b) Au102(SR)44 (diameter ~ 1.5nm) and (c) Au144(SR)60 (diameter ~ 2nm) NPs represented as spheres (with van der Waals radius). The coloring scheme throughout this paper is the following: sulfur, yellow; oxygen, red; carbon, cyan; hydrogen, white; gold, golden; nitrogen, blue. All graphics were rendered with VMD.[41]
2. Methods
Simulation Parameters
All interactions were represented with the CHARMM36 force field.[42] Water was represented with the TIP3P[43] model. The cut-off distance applied to the vdW interactions was 1.2 nm, with a switching distance of 1.0 nm and a pair list distance of 1.35 nm. Langevin dynamics were implemented with a time-step of 1.0 fs for short range interactions and 2.0 fs for long range interactions, and with a Langevin damping of 1.0 ps−1. The temperature was kept at 298 K and the pressure at 1.0 bar using the Nose-Hoover Langevin piston control.[44, 45] Periodic boundary conditions were applied in all dimensions. Full electrostatic interactions were computed through the Particle-Mesh Ewald (PME) method. All simulations were performed with NAMD.[46]
Building procedure for the AuNP
The coordinates of the Au18S14 and Au102S44 cores were obtained in Refs. [19] and [33] respectively. We chose the negatively charged 4-mercapto-benzoate ligand because of its experimental relevance. [47, 48] We bound a copy of this ligand to each sulfur and minimized the resulting structure in vacuum while fixing the core atoms (see Video 1 in the Supplementary data). The distances between each gold atom were restrained to their initial values in order to make the core rigid, while still allowing it to move freely. A copy of the NP was then placed 4 nm apart. Since Au18S14 is non-spherical, we determined the orientation of strongest attraction by placing 16 sodium ions in the plane at the middle and allowing the NPs to rotate freely in vacuum (see Fig. 2(b)). After removing the ions at the middle, the NPs were placed in a 70x70x140 Å3 water box with a 150 mM concentration of NaCl. The system comprised a total of 64 964 atoms.
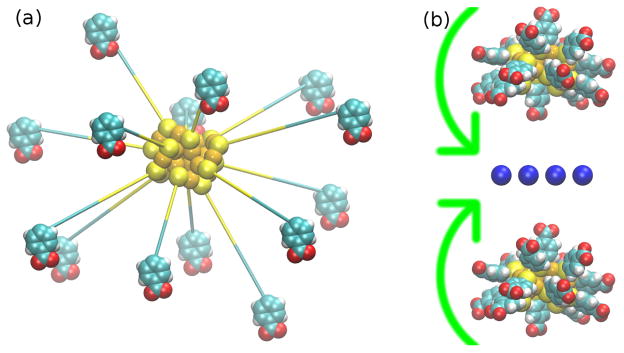
Figure 2.
Building procedure: (a) The Au18 NP structure undergoing energy minimization after attaching 14 ligands to its surface. (b) The Au18 NP pair undergoing free rotation in the presence of a layer of Na+ located at the middle, in order to find the orientation of minimum energy.
Computing binding affinities from potentials of mean force
Following the standard literature,[15] the dissociation constant KD for the process of binding between two rigid NPs can be computed through the potential of mean force 𝒲(r) formalism as:
| (1) |
where the sampling in the numerator of Eq. (1) is performed on the ensemble of microstates belonging to the bound state, while the sampling in the denominator is performed on the ensemble of microstates from the unbound state. r∞ is defined to be any microstate belonging to the unbound state, β= 1/kBT, c0 is the concentration at standard conditions, equal to 1M on the left-hand side of the equation, and on the right-hand side. Further details are presented in the supplementary information (SI), section 1.
The hSMD method
If we define r0 to be one of the states from the ensemble of bound states, r∞ to be one of the states from the ensemble of unbound states, and the free-energy difference between these two states to be Δ𝒲0,∞ ≡ 𝒲(r0) − 𝒲(r∞), then, as in Ref. [17], we have
| (2) |
where Z0 ≡ ∫site dre−β[𝒲(r)−𝒲(r0)] and Z∞ ≡ ∫bulk dre−β[𝒲(r)−𝒲(r∞)] correspond to sampling over the ensembles of bound states and unbound states, respectively, and constitute the entropic contribution to the binding affinity. The scheme of simulations employed to drag the system along the dissociation path from r0 to r∞ is presented in the SI, Fig. S1.
The hSMD method applied to the Au18 NP
The energetic interactions were studied by selecting arbitrarily three atoms (r1, r2, r3) at the center of the NP whose core was not fixed and pulling them individually along straight lines relative to the NP whose core was fixed until the NPs became aggregated. In this way, the one-dimensional PMF as function of center-to-center distance was obtained, and the NPs were not allowed to rotate at this stage. In order to study the entropic interactions, we performed two runs of 20 ns each under equilibrium conditions: one in which the NPs were bound, and another in which they were unbound. During the bound-state simulation, the core of one NP was fixed while the other NP moved freely (see Video 2 in the Supplementary data) and Z0 was approximated as a Gaussian integration:
| (3) |
where Σ is the matrix containing the variances of the coordinates in time:
| (4) |
This approximation is valid when the binding is tight. Since the Au18 NPs are non-spherical, their binding is tight only when their orientations correspond to the maximum contact area between them, that is, to a maximum formation of salt bridges. Because of this, we would not expect the NPs to roll on top of each other, but rather to remain in the initial binding orientation (the Euler angles of the free NP are plotted in Fig. 6(b)).
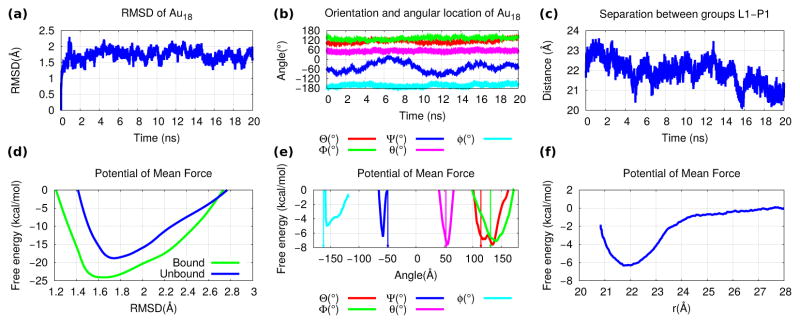
Figure 6.
Geometrical restraint results for Au18: The (a) conformational (i.e. RMSD), (b) angular (i.e. Euler angles for orientation and spherical angles for location) and (c) radial collective variables as functions of time for the 20 ns of simulation under equilibrium conditions. The PMF profiles as functions of the (d) conformational, (e) angular and (f) radial collective variables. The values employed as minima for each of the constraint potentials are indicated by arrows.
During the unbound-state simulation, only one NP was employed. Z∞ was obtained by sampling over the coordinates of groups 2 and 3 while keeping group 1 fixed:
| (5) |
The first term of equation 5 is obtained by sampling over the distance r23 between groups 2 and 3, as well as over the angle θ formed by the three groups. This corresponds to placing group 2 at the origin and group 1 at the z axis. r23, θ, ϕ are then the spherical coordinates of group 3, where ϕ is integrated out. In the second term of equation 5 we can then integrate out θ and ϕ, and sample over the distance r12 between groups 1 and 2. These integrations are also made through the Gaussian approximation:
| (6) |
where r12∞, r23∞ and θ∞ are the average values of the simulation, and (more details are presented in the SI, section 2).
The hSMD method applied to the Au102 and Au144 NPs
Since these NPs are near-spherical, the PMF is independent of the Euler angles defining their orientation, as well as of the angular spherical coordinates defining their angular position. The PMF is a one-dimensional function of the radial separation between the NPs: [49]
| (7) |
where r* is a reference position far away in the bulk (i.e. at the dissociated state, where 𝒲(r) → 0 by definition). In the case of the Au102 NP, we pulled the center of mass of the 7 gold atoms located at the center, considered as a single atomic group. In this way, we allowed the particle to rotate freely during the dissociation. As for the Au144 NP, we chose the 12 gold atoms at the center which form an icosahedron, and the same procedure was followed (see the SI, section 3 for additional details).
The method of geometrical restraints
Following Ref. [16], three atomic groups are defined on each of the binding partners. Six collective variables are then defined in terms of these groups: three Euler angles (Θ, Φ, Ψ) specifying the orientation of the body, and three spherical coordinates specifying its position (θ, ϕ, r), where r is the separation between the partners. A seventh collective variable is the body’s conformation, expressed by its root-mean-square deviation (RMSD) with respect to its time-averaged structure at the bound state. These variables each represent a component of the dissociation path. The free energy of binding is given by:
| (8) |
where I* ≡ ∫site dre−β[𝒲(r)−𝒲(r∞)] and correspond to the positional contributions to the free energy as the object is dragged away from the binding site; and correspond to the contributions of the RMSD degree of freedom at the site and the bulk regions, respectively; and correspond to the orientational degrees of freedom, and corresponds to the two spherical angles at the bound state (the term is implicit in the S*I* product). Further specifics of this methodology are presented in the SI, section 5.
The method of geometrical restraints applied to the Au18 NP
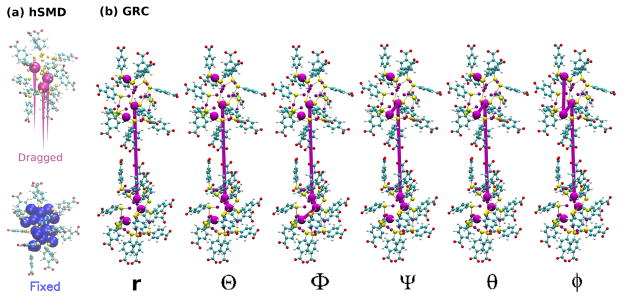
Since the shape of the Au18 NP is non-spherical, sampling over its orientational degrees of freedom becomes necessary. We defined three groups of atoms on the NP whose core was fixed, and three on the other NP. In Fig. 3(a), as well as in Video 3 in the Supplementary data, we present the collective variables defining both the orientation and the location of the free Au18(SR)14 NP relative to the fixed one.
Figure 3.
Methodology for the PMF calculations of Au18 (a) The hSMD method: The displacements of the 3 pulling gold atoms are shown as magenta cones. (b) The method of geometrical restraints: The six groups employed to define the six collective variables are represented as purple spheres. The groups involved in the definition of each collective variable are connected by purple lines.
3. Results
From the hSMD method
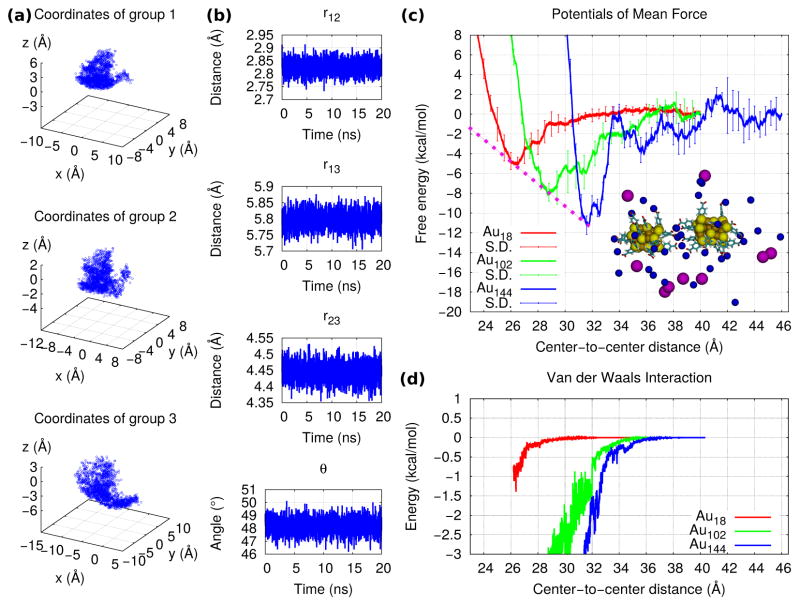
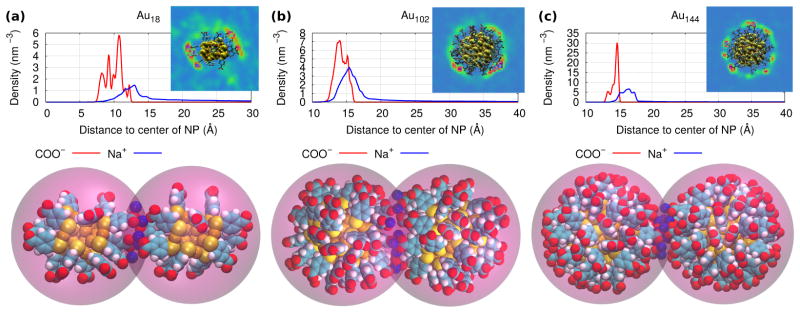
In Fig. 4(a) we plot the location over time of the three pulling atoms during a bound-state equilibration. Fig. 4(b) shows the separations among the atoms and the angle made by them during an unbound-state equilibration. From this sampling we obtained the entropic contributions to the KD of Au18, shown in Table 1. In Fig. 4(c) we show the PMF for the dissociation and compare it to those of Au102 and Au144(the results for Au144 were taken from Ref. [40]). Through these PMFs we obtained the energetic contributions to KD. These simulations are presented in Videos 4, 5 and 6 in the Supplementary data. The PMFs were obtained from the work profiles shown in the SI, Fig. S4. As shown in Fig.4(d), the smaller the NP was, the weaker the van der Waals attraction became. The non-spherical shape of Au18 originated the non-isotropic distribution of ligands and ions shown in Fig. 5(a). During the bound-state simulation, five sodium were observed to lie on average at the region in-between the two Au18 and forming salt-bridges between them. As for Au102 and Au144, we found on average eight and eleven salt-bridges in their bound states, respectively (see Figs. 5(b),(c)). The average number of salt bridges was estimated by counting the amount of ions lying within 0.35nm of both nanoparticles simultaneously (this value was taken from ref. [35] as the cutoff distance of contact between an ion and a terminal group).
Figure 4.
hSMD results : (a) Histogram of the coordinates of the three Au18 pulling atoms during the bound-state equilibration employed to compute Z0. (b) The distances among the three Au18 pulling atoms and the angle formed by them vs. time during the unbound-state equilibration employed to compute Z∞. (c) The PMFs of Au18, Au102 and Au144 as functions of distance between the cores’ centers of mass. The minima are connected by a dashed pink line. The snapshot shows the bound state of Au18 and all ions within 20 Å of the NPs colored by name: sodium, blue; chloride, magenta. (d) The energetic contributions to the PMF from the van der Waals interactions.
Table 1.
The free energy calculations for the Au18, Au102 and Au144 NPs.
| hSMD
| |||||
|---|---|---|---|---|---|
| Au18 | Au102 | Au144[40] | |||
|
| |||||
| Z0 | 1.5 × 106 | ||||
|
|
−5.0 | −8.2 | −11 | ||
| Z∞ | 5.6 × 104 | ||||
|
|
−2.6 | −6.2 | −12 | ||
| KD | 13 mM | 30 μM | 3.2 nM | ||
|
| |||||
| Geometrical Restraints (Au18) | |||||
|
| |||||
|
|
−13 |
|
−2.3 | ||
|
|
−1.4 |
|
−3.0 | ||
|
|
−1.0 |
|
6.7 | ||
|
|
−3.5 |
|
15.0 | ||
|
|
−0.33 |
|
−2.8 | ||
| KD | 9.0 mM | ||||
Figure 5.
The radial density distributions of the carboxyl groups and the sodium counterions around (a) Au18(SR)60, (b) Au102(SR)44 and (c) Au144(SR)60. The snapshots on the top show the density maps of sodium around the NP (the coloring scheme in order of increasing density is: blue, green, red). The snapshots at the bottom show the sodium lying within 3.5 Å of both NPs simultaneously at the bound state.
From the method of geometrical restraints
In Figs. 6(a),(b),(c) we plot the collective variables as functions of time during a run under equilibrium conditions as it was bound to another Au18 whose core’s location was fixed. Figs. 6(d),(e),(f) show the PMFs as functions of the collective variables. In Table 1 we present the contributions to the free energy from each collective variable, which resulted in a dissociation constant of ≈ 9.0 mM.
Spontaneity of the aggregation
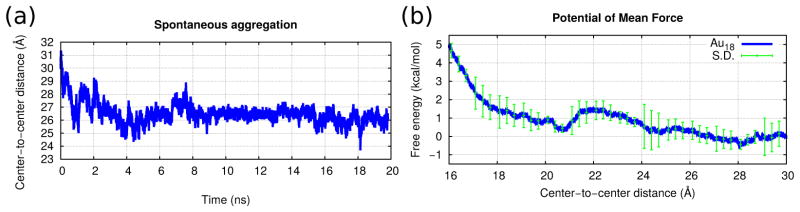
We performed a simulation under equilibrium conditions of a pair of free Au18 NPs in order to verify the spontaneity of their aggregation, in the absence of external forces or constraints. The initial center-to-center distance was set at 3 nm, since at this point the PMF of Fig. 4(c) starts becoming attractive. The center-to-center distance is plotted vs. time in Fig. 7(a) and the simulation is shown in Video 7 of the supplementary data. Notice how the separation between the NPs decreases to the value corresponding to the minimum in the PMF within the first 5 ns, and it remains stable for the next 15 ns.
Figure 7.
(a) The center-to-center separation between two free Au18 NPs vs. time. Starting with an initial value of 3 nm, their aggregation is spontaneous. (b) The PMF of Au18 as a function of the center-to-center separation, employing the opposite orientation as in Fig. 4. The net interaction becomes repulsive.
Specificity of the binding site
In the case of the Au144 and Au102 NPs, due to their spherical shape the binding site could lie anywhere at the surface. Thanks to this their dissociation constant can be obtained from eq. 7 without the need to sample over their bound and unbound states. A one-dimensional PMF as function of separation between the NPs connecting one of the bound states to one of the dissociated states contains implicitly the sampling over the full surface area of the sphere.[49] However, in the case of the Au18, their non-spherical shape causes the binding site to become specific to those orientations which allow for the formation of a maximum amount of salt bridges. In order to verify this, we measured an additional PMF using the opposite orientation from that employed in Fig. 4. The PMF profile of Fig. 7(b) became repulsive and thus no binding was observed.
4. Discussion
The aggregation process of Au18 NPs
In the SI, Fig. S2(a), we see that as the NPs approach, the direct electrostatic repulsion between them appears much sooner than the van der Waals attraction, which is weak when compared with the PMF. As the NPs approach, the electrostatic energy between them and the sodium ions decreases (see the SI, Fig. S2(b)), since the sodium is able to form salt bridges between the two NPs. However, as the sodium ions transition from being bound with one ligand to being bound with two ligands, they have to be partially dehydrated. This repulsive interaction opposes the decrease in the ion-NP electrostatic energy. Such hydration repulsion is a general property of hydrophilic surfaces.[50, 51, 52] An additional repulsive interaction of the binding process is the decrease in entropy of both the sodium ions and the ligands as their motion becomes more restricted during the formation of the salt bridges. We may observe from the method of geometrical restraints that the entropic contributions to the free energy were: for the conformational component, for the rotational component and for the translational component. Both the conformational and rotational components favor the unbound states, where the entropic freedom is larger. However, the translational component (which favors the bound state) is larger than the other two combined.
Size and Shape effects of AuNPs
Because of the small size of the Au18 NP (≈ 1 nm for the core diameter) the resulting large curvature decreases the contact area between two such NPs, thus hindering the formation of salt bridges between them. The small size also decreases the strength of the van der Waals attraction. On the other hand, the non-spherical shape of this NP increases the contact area on those sides whose curvatures are smaller, thus facilitating the formation of salt bridges at preferential orientations. Yet by limiting the orientations which are available for binding, an entropic repulsion is also originated, since there is a decrease in the amount of bound states. In Fig. 7 we saw that for some orientations the interaction actually becomes repulsive. Because of these reasons, as shown in Fig. 4(h), when we compare the PMF of Au18 with those of Au102 and Au144 (functionalized by the same ligand and in the same near-physiological conditions), the attraction between the Au18 NPs was observed to be the weakest, while that of Au144 was the strongest. This result agrees with recent experimental data from ultrasmall glutathione-coated gold nanoparticles.[53] The minima in the PMF profiles of the three NPs studied in this work was found to decrease linearly with the size:
| (9) |
where D is the diameter of the NP in units of nm.
Application of the methods on the Ligand-Protein problem
In the supplementary data we show the results from both methods on a protein-ligand binding process with known experimental value of dissociation constant: the acetazolamide (AZM) drug bound to the aquaporin 4 (AQP4) water-channel protein (see Video 8 of the Supplementary data). AZM has been found to inhibit water permeation through AQP4 with an IC50 of ≈ 3 mM.[54] Identical results (within the margin of error) were obtained from both approaches, in agreement with experimental measurements.
5. Conclusion
In this work, we compared the interactions of Au18, Au102 and Au144, all three functionalized by the same SPh(COO−) ligand under the same near-physiological saline conditions. On the one hand, the smaller size of the Au18 hinders the formation of salt bridges between two NPs because of its larger curvature and thus smaller contact area. At the same time, its smaller size decreases the strength of the van der Waals attraction. On the other hand, the non-isotropic shape of Au18 facilitates the formation of salt bridges by increasing the contact area on preferential orientations; yet it also increases the entropy of the bound state by limiting the orientations available for binding. This resulted in dissociation constants of 13mM, 30μM and 3.2nM for Au18, Au102 and Au144 respectively. The results for Au18 from two different methods agreed with each other. We therefore expect our results to be confirmed in future experiments.
We employed two computational methods for the calculation of absolute binding free energies from the PMF. The method of geometrical restraints is able to separate the contributions to the free energy into nine different geometrical components, in this way providing a great amount of detail regarding the origins of the result. The hSMD method is able to compute the free energy in a brute-force way without losing accuracy, in this way saving much effort during its implementation. Depending on the system under consideration, one may be interested in maximizing the output of information through the use of a series of biasing and constraining potentials, or in computing the result in a straightforward way through the use of steering/pulling simulations. Both methods provide a complete atomic-level picture of the interactions which lie at the origin of the result.
Supplementary Material
Acknowledgments
The authors acknowledge support from the NIH (GM 084834) and the computing resources provided by the Texas Advanced Computing Center at University of Texas at Austin. ODV acknowledges a graduate scholarship from the Mexican National Council of Science and Technology (CONACYT Grant 533262). LY acknowledges the Jiangsu Overseas Research & Training Program for University Prominent Young & Middle-aged Teachers and Presidents. Finally, the authors would like to express their gratitude to Dr. Liao Y. Chen for his continued guidance and support in their research endeavors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jabes BS, Yadav HOS, Kumar SK, Chakravarty C. Fluctuation-driven anisotropy in effective pair interactions between nanoparticles: Thiolated gold nanoparticles in ethane. J Chem Phys. 2014;141(15):154904. doi: 10.1063/1.4897541. doi:doi: http://dx.doi.org/10.1063/1.4897541. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, Yang X, Wu B, Tang L. Molecular simulation of interaction between passivated gold nanoparticles in supercritical co2. J Chem Phys. 2011;135(20):204703. doi: 10.1063/1.3661982. doi:doi: http://dx.doi.org/10.1063/1.3661982. [DOI] [PubMed] [Google Scholar]
- 3.Patel N, Egorov SA. Interactions between sterically stabilized nanoparticles in supercritical fluids: A simulation study. J Chem Phys. 2007;126(5):054706. doi: 10.1063/1.2434155. doi:doi: http://dx.doi.org/10.1063/1.2434155. [DOI] [PubMed] [Google Scholar]
- 4.Heikkil E, Martinez-Seara H, Gurtovenko AA, Javanainen M, Hkkinen H, Vattulainen I, Akola J. Cationic au nanoparticle binding with plasma membrane-like lipid bilayers: Potential mechanism for spontaneous permeation to cells revealed by atomistic simulations. The Journal of Physical Chemistry C. 2014;118(20):11131–11141. doi: 10.1021/jp5024026. arXiv: http://dx.doi.org/10.1021/jp5024026. [DOI] [Google Scholar]
- 5.Guo Z, Fan X, Xu L, Lu X, Gu C, Bian Z, Gu N, Zhang J, Yang D. Shape separation of colloidal gold nanoparticles through salt-triggered selective precipitation. Chem Commun. 2011;47(14):4180–4182. doi: 10.1039/C0CC04612D. [DOI] [PubMed] [Google Scholar]
- 6.Hao M, Liu C, Peng L, Li J, Lu W, Jiang L. Shape separation of gold nanoparticles using a ph-responsive amphiphilic dendrimer according to their shape anisotropy distinction. J Colloid Interface Sci. 2015;437:311–315. doi: 10.1016/j.jcis.2014.09.052. doi: http://dx.doi.org/10.1016/j.jcis.2014.09.052. [DOI] [PubMed] [Google Scholar]
- 7.Zhao W, Lin L, Hsing IM. Nucleotide-mediated size fractionation of gold nanoparticles in aqueous solutions. Langmuir. 2010;26(10):7405–7409. doi: 10.1021/la9044489. [DOI] [PubMed] [Google Scholar]
- 8.Yang H, Heng X, Wang W, Hu J, Xu W. Salt-induced size-selective separation, concentration, and preservation of zwitterion-modified gold nanoparticles. RSC Advances. 2012;2(7):2671–2674. doi: 10.1039/C2RA00828A. [DOI] [Google Scholar]
- 9.Silvera Batista CA, Larson RG, Kotov NA. Nonadditivity of nanoparticle interactions. Science. 350(6257) doi: 10.1126/science.1242477. arXiv: http://www.sciencemag.org/content/350/6257/1242477.full.pdf. [DOI] [PubMed] [Google Scholar]
- 10.Hotze EM, Phenrat T, Lowry GV. Nanoparticle aggregation: challenges to understanding transport and reactivity in the environment. J Environ Qual. 2010;39(6):1909–24. doi: 10.2134/jeq2009.0462. [DOI] [PubMed] [Google Scholar]
- 11.Delgado AV, Gonzlez-Caballero F, Hunter RJ, Koopal LK, Lyklema J IU of Pure, P Applied Chemistry, BCDIT Report. Measurement and interpretation of electrokinetic phenomena. J Colloid Interface Sci. 2007;309(2):194–224. doi: 10.1016/j.jcis.2006.12.075. [DOI] [PubMed] [Google Scholar]
- 12.Kirkwood JG. Statistical mechanics of fluid mixtures. J Chem Phys. 1935;3(5):300–313. doi: 10.1063/1.1749657. [DOI] [Google Scholar]
- 13.Allen TW, Andersen OS, Roux B. Molecular dynamics - potential of mean force calculations as a tool for understanding ion permeation and selectivity in narrow channels. Biophys Chem. 2006;124(3):251–67. doi: 10.1016/j.bpc.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Roux B. The calculation of the potential of mean force using computer-simulations. Comput Phys Commun. 1995;91(1–3):275–282. doi: 10.1016/0010-4655(95)00053-I. [DOI] [Google Scholar]
- 15.Woo HJ, Roux B. Calculation of absolute protein-ligand binding free energy from computer simulations. Proc Natl Acad Sci U S A. 2005;102(19):6825–30. doi: 10.1073/pnas.0409005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumbart JC, Roux B, Chipot C. Standard binding free energies from computer simulations: What is the best strategy? J Chem Theory Comput. 2012;9(1):794–802. doi: 10.1021/ct3008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen LY. Hybrid steered molecular dynamics approach to computing absolute binding free energy of ligand-protein complexes: A brute force approach that is fast and accurate. J Chem Theory Comput. 2015;11(4):1928–1938. doi: 10.1021/ct501162f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez RA, Yu LL, Chen LY. Computing protein-protein association affinity with hybrid steered molecular dynamics. J Chem Theory Comput. 2015;11(9):4427–4438. doi: 10.1021/acs.jctc.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tlahuice A, Garzón IL. On the structure of the au18(sr)14 cluster. Phys Chem Chem Phys. 2012;14(11):3737. doi: 10.1039/c2cp24016e. [DOI] [PubMed] [Google Scholar]
- 20.Das A, Liu C, Byun HY, Nobusada K, Zhao S, Rosi N, Jin R. Structure determination of [au18(sr)14] Angewandte Chemie. 2015;127(10):3183–3187. doi: 10.1002/ange.201410161. [DOI] [PubMed] [Google Scholar]
- 21.Reilly SM, Krick T, Dass A. Surfactant-free synthesis of ultrasmall gold nanoclusters. J Chem Phys C. 2010;114(2):741–745. doi: 10.1021/jp9067944. [DOI] [Google Scholar]
- 22.Jiang D-e, Chen W, Whetten RL, Chen Z. What protects the core when the thiolated au cluster is extremely small? J Chem Phys C. 2009;113(39):16983–16987. doi: 10.1021/jp906823d. [DOI] [Google Scholar]
- 23.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. The golden age: gold nanoparticles for biomedicine. Chem Soc Rev. 2012;41(7):2740–2779. doi: 10.1039/C1CS15237H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sperling RA, Rivera Gil P, Zhang F, Zanella M, Parak WJ. Biological applications of gold nanoparticles. Chem Soc Rev. 2008;37(9):1896–1908. doi: 10.1039/B712170A. [DOI] [PubMed] [Google Scholar]
- 25.Grace AN, Pandian K. Antibacterial efficacy of aminoglycosidic antibiotics protected gold nanoparticlesa brief study. Colloids Surf A: Physicochem Eng Aspects. 2007;297(13):63–70. doi: http://dx.doi.org/10.1016/j.colsurfa.2006.10.024. [Google Scholar]
- 26.Tang F, Wang C, Wang X, Li L. Preparation of biocompatible fluorescent coreshell nanoparticles for cellular imaging. Colloids Surf A: Physicochem Eng Aspects. 2015;485:49–54. doi: http://dx.doi.org/10.1016/j.colsurfa.2015.09.010. [Google Scholar]
- 27.Wang X, Wei Y, Wang S, Chen L. Red-to-blue colorimetric detection of chromium via cr (iii)-citrate chelating based on tween 20-stabilized gold nanoparticles. Colloids Surf A: Physicochem Eng Aspects. 2015;472:57–62. doi: http://dx.doi.org/10.1016/j.colsurfa.2015.02.033. [Google Scholar]
- 28.Oni Y, Hao K, Dozie-Nwachukwu S, Obayemi JD, Odusanya OS, Anuku N, Soboyejo WO. Gold nanoparticles for cancer detection and treatment: The role of adhesion. J Appl Phys. 2014;115(8):084305. doi:doi: http://dx.doi.org/10.1063/1.4863541. [Google Scholar]
- 29.Murthy AK, Stover RJ, Borwankar AU, Nie GD, Gourisankar S, Truskett TM, Sokolov KV, Johnston KP. Equilibrium gold nanoclusters quenched with biodegradable polymers. ACS Nano. 2013;7(1):239–251. doi: 10.1021/nn303937k. arXiv: http://dx.doi.org/10.1021/nn303937k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murthy AK, Stover RJ, Hardin WG, Schramm R, Nie GD, Gourisankar S, Truskett TM, Sokolov KV, Johnston KP. Charged gold nanoparticles with essentially zero serum protein adsorption in undiluted fetal bovine serum. J Am Chem Soc. 2013;135(21):7799–7802. doi: 10.1021/ja400701c. arXiv: http://dx.doi.org/10.1021/ja400701c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover RJ, Murthy AK, Nie GD, Gourisankar S, Dear BJ, Truskett TM, Sokolov KV, Johnston KP. Quenched assembly of nir-active gold nanoclusters capped with strongly bound ligands by tuning particle charge via ph and salinity. J Chem Phys C. 2014;118(26):14291–14298. doi: 10.1021/jp408715p. arXiv: http://dx.doi.org/10.1021/jp408715p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villarreal O, Chen L, Whetten R, Demeler B. Aspheric solute ions modulate gold nanoparticle interactions in an aqueous solution: An optimal way to reversibly concentrate functionalized nanoparticles. J Phys Chem B. 2015;119(50):15502–15508. doi: 10.1021/acs.jpcb.5b09864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jadzinsky PD, Calero G, Ackerson CJ, Bushnell DA, Kornberg RD. Structure of a thiol monolayer-protected gold nanoparticle at 1.1 å resolution. Science. 2007;318(5849):430–433. doi: 10.1126/science.1148624. arXiv: http://www.sciencemag.org/content/318/5849/430.full.pdf. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Acevedo O, Akola J, Whetten RL, Grönbeck H, Häkkinen H. Structure and bonding in the ubiquitous icosahedral metallic gold cluster au144(sr)60. J Chem Phys C. 2009;113(13):5035–5038. doi: 10.1021/jp8115098. arXiv: http://dx.doi.org/10.1021/jp8115098. [DOI] [Google Scholar]
- 35.Heikkilä E, Gurtovenko AA, Martinez-Seara H, Häkkinen H, Vattulainen I, Akola J. Atomistic simulations of functional au144(sr)60 gold nanoparticles in aqueous environment. J Chem Phys C. 2012;116(17):9805–9815. doi: 10.1021/jp301094m. arXiv: http://dx.doi.org/10.1021/jp301094m. [DOI] [Google Scholar]
- 36.Tlahuice-Flores A, Black DM, Bach SBH, Jose-Yacamán M, Whetten RL. Structure & bonding of the gold-subhalide cluster I-Au144Cl60[z] Phys Chem Chem Phys. 2013;15:19191. doi: 10.1039/c3cp53902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahena D, Bhattarai N, Santiago U, Tlahuice A, Ponce A, Bach SBH, Yoon B, Whetten RL, Landman U, Jose-Yacaman M. Stem electron diffraction and high-resolution images used in the determination of the crystal structure of the au144(sr)60 cluster. J Phys Chem Lett. 2013;4(6):975–981. doi: 10.1021/jz400111d. arXiv: http://dx.doi.org/10.1021/jz400111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heikkilä E, Martinez-Seara H, Gurtovenko AA, Vattulainen I, Akola J. Atomistic simulations of anionic au144(sr)60 nanoparticles interacting with asymmetric model lipid membranes. Biochim Biophys Acta. 2014;1838(11):2852. doi: 10.1016/j.bbamem.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 39.Alsharif SA, Chen LY, Tlahuice-Flores A, Whetten RL, Yacaman MJ. Interaction between functionalized gold nanoparticles in physiological saline. Phys Chem Chem Phys. 2014;16(9):3909–3913. doi: 10.1039/C3CP54503B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villarreal OD, Chen LY, Whetten RL, Yacaman MJ. Ligand-modulated interactions between charged monolayer-protected au144(sr)60 gold nanoparticles in physiological saline. Phys Chem Chem Phys. 2015;17(5):3680–8. doi: 10.1039/c4cp05137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphrey W, Dalke A, Schulten K. Vmd: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 42.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD. Charmm general force field: A force field for drug-like molecules compatible with the charmm all-atom additive biological force fields. J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. doi:doi: http://dx.doi.org/10.1063/1.445869. [Google Scholar]
- 44.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 45.Feller SE, Brown CA, Nizza DT, Gawrisch K. Nuclear over-hauser enhancement spectroscopy cross-relaxation rates and ethanol distribution across membranes. Biophys J. 2002;82(3):1396–1404. doi: 10.1016/S0006-3495(02)75494-5. doi: http://dx.doi.org/10.1016/S0006-3495(02)75494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kal L, Schulten K. Scalable molecular dynamics with namd. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong OA, Heinecke CL, Simone AR, Whetten RL, Ackerson CJ. Ligand symmetry-equivalence on thiolate protected gold nanoclusters determined by nmr spectroscopy. Nanoscale. 2012;4:4099–4102. doi: 10.1039/C2NR30259D. [DOI] [PubMed] [Google Scholar]
- 48.Heinecke CL, Ackerson CJ. Preparation of gold nanocluster bioconjugates for electron microscopy. Methods in molecular biology (Clifton, NJ) 2013;950:293. doi: 10.1007/978-1-62703-137-0_17. [DOI] [PubMed] [Google Scholar]
- 49.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009;113(8):2234–46. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gambinossi F, Chanana M, Mylon SE, Ferri JK. Stimulus-responsive au@(meo2max-co-oegmay) nanoparticles stabilized by non-dlvo interactions: Implications of ionic strength and copolymer (x:y) fraction on aggregation kinetics. Langmuir. 2014;30(7):1748–1757. doi: 10.1021/la403963c. [DOI] [PubMed] [Google Scholar]
- 51.Grasso D, Subramaniam K, Butkus M, Strevett K, Bergendahl J. A review of non-dlvo interactions in environmental colloidal systems. Rev Environ Sci Biotechnol. 2002;1(1):17–38. doi: 10.1023/A:1015146710500. [DOI] [Google Scholar]
- 52.Chang Y-I, Chang P-K. The role of hydration force on the stability of the suspension of saccharomyces cerevisiaeapplication of the extended dlvo theory. Colloids Surf, A. 2002;211(1):67–77. doi: http://dx.doi.org/10.1016/S0927-7757(02)00238-8. [Google Scholar]
- 53.Sousa AA, Hassan SA, Knittel LL, Balbo A, Aronova MA, Brown PH, Schuck P, Leapman RD. Biointeractions of ultrasmall glutathione-coated gold nanoparticles: effect of small size variations. Nanoscale. 2016;8:6577–6588. doi: 10.1039/C5NR07642K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WEC, Robbins RA, Miercke LJW, Stroud RM. Crystal structure of human aquaporin 4 at 1.8 angstrom and its mechanism of conductance. Proc Natl Acad Sci U S A. 2009;106(18):7437–7442. doi: 10.1073/pnas.0902725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.