Abstract
Impairments of mitochondrial functions have been associated with failure of cellular functions in different tissues leading to various pathologies. We report here a mitochondria–targeted nanodelivery system for coenzyme Q10 (CoQ10) which can reach mitochondria, and deliver CoQ10 in adequate quantities. Multifunctional nanocarriers based on ABC miktoarm polymers (A= PEG, B = polycaprolactone (PCL) and C = triphenylphosphonium bromide (TPPBr)) were synthesized using a combination of click chemistry with ring opening polymerization, self-assembled into nano-sized micelles, and were employed for CoQ10-loading. Drug loading capacity (60 weight%), micelle size (25–60 nm) and stability were determined using a variety of techniques. The micelles had a small critical association concentration, and were colloidally stable in solution for more than 3 months. The extraordinarily high CoQ10 loading capacity in the micelles is attributed to good compatibility between CoQ10 and PCL, as indicated by low Flory-Huggins interaction parameter. Confocal microscopy studies of fluorescently labeled polymer analog together with the mitochondria-specific vital dye label, indicated that the carrier did indeed reach mitochondria. The high CoQ10 loading efficiency allowed testing of micelles within a broad concentration range, and provided evidence for CoQ10 effectiveness in two different experimental paradigms: oxidative stress and inflammation. Combined results from chemical, analytical and biological experiments suggest that the new miktoarm-based carrier provides a suitable means of CoQ10 delivery to mitochondria without loss of drug effectiveness. The versatility of the click chemistry used to prepare this new mitochondria-targeting nanocarrier offers a widely applicable, simple and easily reproducible procedure to deliver drugs to mitochondria or other intracellular organelles.
Keywords: Coenzyme Q10, mitochondria, drug targeting, miktoarm polymers, click chemistry, polymeric micelles
INTRODUCTION
Mitochondria, the power house of the cell, play a pivotal role in the homeostasis of vital physiological functions, including electron transfer, apoptosis, and calcium storage.1, 2 Mitochondrial dysfunction is associated with a variety of human disorders, such as neurodegenerative and neuromuscular diseases, obesity and diabetes, ischemia–reperfusion injury, cancer and inherited mitochondrial diseases.2 In many of these diseases a major cause of damage is mitochondrial overproduction of reactive oxygen species (ROS).3, 4 Therapeutics, such as creatine, coenzyme Q10 (CoQ10), N-acetyl cysteine (NAC) and α-lipoic acid have shown promising neuroprotective effects in in vitro and in vivo models of several neurodegenerative diseases.5–8
CoQ10, also known as ubiquinone, is a naturally occurring lipid-soluble, vitamin-like substance that is found in the inner mitochondrial and cellular membranes and in blood; both in high- and in low-density lipoproteins.9 CoQ10 is a benzoquinone derivative with 10 mono-unsaturated trans-isoprenoid units in the side chain (Figure 1).10 It functions primarily as a cofactor for the mitochondrial enzymes (complexes I, II and III) for the oxidative phosphorylation production of adenosine triphosphate (ATP), and is involved in several aspects of cellular metabolism.11 In its reduced form (CoQ10H2, ubiquinol), it acts as a potent antioxidant and free radical scavenger, protecting membranes and lipoproteins from protein oxidation and lipid peroxidation.12 In vitro studies have shown that CoQ10 pre-treatment prevented a decrease in mitochondrial transmembrane potential and reduced mitochondrial ROS generation.13
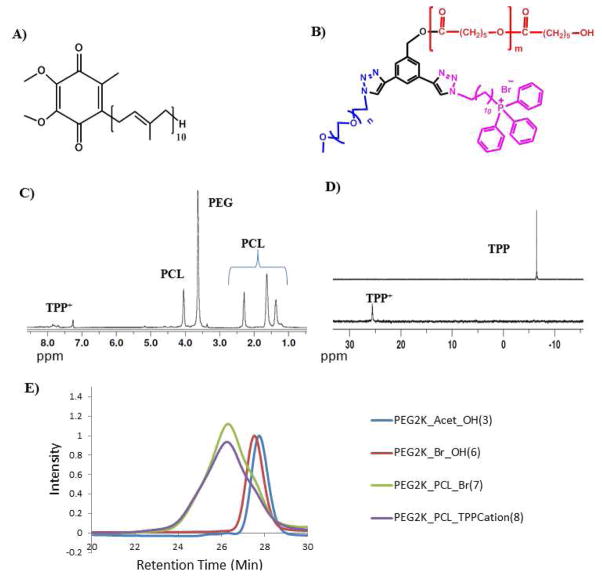
Figure 1.
A) Chemical structure of CoQ10. B) Structure of ABC miktoarm polymer having
 ,
,
 and
and
 . C) 1H NMR of polymer with PEG, PCL and TPP+. D) 31P NMR of polymer with PEG, PCL and TPP+ and of free TPP alone showing a complete shift after attachment to polymer. E) GPC chromatogram showing a shift with increase in molecular weights upon addition of each arm.
. C) 1H NMR of polymer with PEG, PCL and TPP+. D) 31P NMR of polymer with PEG, PCL and TPP+ and of free TPP alone showing a complete shift after attachment to polymer. E) GPC chromatogram showing a shift with increase in molecular weights upon addition of each arm.
Several nanocarriers are currently being investigated for targeting drugs to specific sites with improved efficacy and reduced toxicity.14–17 Polymeric micelles consist of a core-shell architecture: the core with the inner hydrophobic part of amphiphilic copolymer, which can encapsulate poorly water soluble drugs and control their release, and the outer shell or corona is generally hydrophilic which provides aqueous solubility, and prevents the recognition of micelles by reticuloendothelial system (RES). In this regard, biocompatible and biodegradable polymers have been of specific interest in designing micelles for drug delivery. Although polymeric micelles have been extensively studied for biomedical applications, most of the research has been focused on utilizing linear block copolymers.18 Amphiphilic miktoarm star-copolymers have gained considerable interest recently due to their unique aggregated morphologies in bulk, and self-assembly behavior in solution.19 Miktoarm polymers are branched macromolecules with linear polymeric chains emanating from a common central core, and these polymeric arms can vary in chemical identity and/or molecular weight.20 The composition of both the core as well as arms can be fine-tuned based on the desired application. The presence of multiple arms in miktoarm stars become advantageous for biological applications, as one could introduce multifunctionality, and covalently link targeting moieties and/or imaging molecules.21
We report here the design and construction of a mitochondria-targeting nanodelivery system for CoQ10 using ABC miktoarm star polymers which were constructed using “click” chemistry22–24 in combination with ring opening polymerization. The synthesis was achieved by designing a molecular building block with three orthogonal functionalities which facilitated the performance of sequential “click” and ring opening polymerization reactions. These star polymers self-assemble into micelles in an aqueous medium, in which the hydrophilic poly(ethylene glycol) (PEG) arm forms a corona, and the hydrophobic polycaprolactone (PCL) arm the core. Due to the ease and versatility of the CoQ10 loading and release from the miktoarm polymer micelles, as well as its extraordinary loading capacity, this carrier system can be exploited for other drugs with primary site of action within mitochondria.
EXPERIMENTAL SECTION
Materials and Methods
Water was deionized using a Millipore MilliQ system. Coenzyme Q10 was obtained from Medisca Pharmaceutical Inc., Montreal, Quebec, Canada. Lipopolysaccharides, ε-caprolactone (99%) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), Copper (II) sulfate pentahydrate (CuSO4.5H2O) (>98.0%), sodium ascorbate (NaAsc, crystalline, 98%), tetrabutylammonium fluoride (Bu4NF) (1M in THF), 11-bromo-1-undecanol (98%), tetrabromomethane (CBr4) (99%), triphenylphosphine (TPP), 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimidemethiodide(EDC), 4-(dimethylamino)pyridine (DMAP) (99%), and sodium azide (NaN3) (>99.5%) were purchased from Sigma Aldrich, St. Louis, MO, and used as received. All reactions were performed under dry conditions in an inert environment using dry and distilled solvents. Flash chromatography was performed using 60 Å (230–400 mesh) silica gel from EMD Chemicals Inc. Dialysis membranes (Spectra/por, MWCO: 6–8 kDa, unless otherwise indicated) were purchased from Fisher Scientific (Rancho Dominguez, CA). Penicillin, streptomycin and Griess Reagent (1% sulphanilamide, 0.1% N-(1-naphthyl)-ethylenediaminedihydrochloride, 5% phosphoric acid) and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA). Murine microglia (N9) cell lines were obtained from ATCC. ε-Caprolactone was dried over calcium hydride for 24 h and distilled under reduced pressure prior to use. Fluorescein isothiocyanate (FITC) (95%) was purchased from Alfa Aesar.
Synthesis of ABC miktoarm polymers
The compounds: 3-(triisopropylsilylethynyl)-5-ethynylbenzyl alcohol (1) 25, 11-Azido-undecan-1-ol (4) 26 and PEG 2K-Azide 27 were synthesized by adaptation of the previously published procedures.
Compound 2: 3-(triisopropylsilylethynyl)-5-ethynylbenzyl alcohol (1) (0.50g, 1.60mmol) and PEG 2K-azide (2.57g, 1.29mmol) were dissolved in 3 ml of tetrahydrofuran (THF), followed by the addition of sodium ascorbate (0.031g, 0.16mmol). An aqueous solution (1 ml) of CuSO4.5H2O (0.020g, 0.16mmol) was added drop wise to the solution. The reaction mixture was left to stir overnight at room temperature. THF was then evaporated, and the remaining solution was extracted with dichloromethane (DCM) (3×20 ml), and the organic layer was washed with brine (20 ml). It was dried over NaSO4, and the solvent was evaporated. The product was flushed through a silica gel column with 5% methanol in DCM. The solvent was evaporated to yield the product as a white solid (2.8g, 75%). 1H NMR (400 MHz, CDCl3): δ (ppm) 1.13 (br s, 21H, −Si(C3H7)), 3.37 (s, 3H, −OCH3), 3.54–3.87 (m, (−OCH2CH2-)n), 3.95 (t, 2H, −CH2OCH3), 4.64 (t, 2H, −OCH2CH2triazole), 4.80 (d, 2H, −CH2OH), 7.47–7.49 (m, 3H, ArH), and 7.94 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 11.3, 12.1, 18.8, 59.0, 63.0, 64.5, 69.3, 69.6, 70.5, 71.9, 90.9, 106.6, 124.3, 128.0, 130.0, 142.3 and 160.9.
Compound 3: To a solution of compound 2 (2.13g, 0.92mmol) in THF (3 ml) which was placed in a dry ice/acetone bath, a solution of Bu4NF-1M solution in THF (1.8ml, 1.85mmol) was added in a drop wise fashion. The reaction mixture was allowed to warm to room temperature and left to stir overnight. The solvent was removed under vacuum, followed by the addition of water (10 ml), and the mixture was extracted with DCM (3×20ml). The extract was dried over NaSO4, filtered and then the solvent was evaporated. Silica-gel column chromatography was performed with 7% methanol in DCM. The solvent was evaporated to yield the product as a white solid (1.9g, 95%). 1H NMR (400 MHz, CDCl3): δ (ppm) 3.11 (s, AcetH), 3.37 (s, 3H, −OCH3), 3.54–3.87 (m, (−OCH2CH2-)n), 3.89 (t, 2H, −CH2OCH3), 4.59 (t, 2H, −OCH2CH2triazole), 4.70 (d, 2H, −CH2OH), 7.44 (s, 1H, ArH), 7.86 (s, 1H, ArH), 7.91 (s, 1H, ArH), and 7.94 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 50.6, 59.0, 64.2, 69.3, 69.6, 70.5, 70.6, 71.5, 71.9, 83.2, 122.0, 122.8, 124.5, 128.2, 129.9, 130.9, and 142.5.
Compound 5: To a solution of 11-Azido-undecan-1-ol (4) (2.00g, 9.39mmol) in DCM (2ml), carbon tetrabromide (CBr4) (4.04g, 12.2mmol) was added, followed by the addition of triphenylphosphine (TPP) (3.2g, 12.2mmol). The reaction mixture was left to stir at room temperature overnight. It was subsequently diluted with DCM (30mL), and the organic phase was washed with brine (3×30 ml). It was then dried over NaSO4, and upon filtration solvent was removed under vacuum. The crude was then passed through a silica column using 10% ethylacetate in hexanes to yield a transparent liquid (2.0g, 77%). 1H NMR (200 MHz, CDCl3): δ (ppm) 1.22–1.50 (m, 10H, −CH2-), 1.52–1.65 (m, 4H, −CH2-), 1.79–1.90 (m, 4H, −CH2-), 3.26 (t, 2H, −CH2N3) and 3.41 (t, 2H, −CH2Br). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 26.7, 28.1, 28.7, 28.8, 29.1, 29.4, 32.8 34.0, and 51.5.
Compound 6: Compounds 3 (0.90g, 0.42mmol) and 5 (0.14g, 0.50mmol) were dissolved in 3 ml of tetrahydrofuran (THF), followed by the addition of sodium ascorbate (0.008g, 0.04mmol). An aqueous solution (1ml) of CuSO4.5H2O (0.005g, 0.04mmol) was added dropwise to the solution. The reaction mixture was left to stir overnight at room temperature. THF was then evaporated, and the remaining solution was extracted with dichloromethane (DCM) (3×20 ml). The organic layer was washed with brine (20 ml) dried over NaSO4, and upon filtration the solvent was evaporated. The product was flushed through a silica gel column with 5% methanol in DCM. The solvent was evaporated to yield the product as a white solid (0.90g, 89%). 1H NMR (400 MHz, CDCl3): δ (ppm) 1.22–1.50 (m, 12H, −CH2-), 1.52–1.95 (m, 6H, −CH2-), 3.37 (s, 3H, −OCH3), 3.53–3.70 (m, (−OCH2CH2-)n) & (m, 2H, −CH2Br), 3.83 (t, 2H, −CH2OCH3), 4.41 (t, 2H, −OCH2CH2triazole), 4.62 (t, 2H, −OCH2CH2triazole), 4.79 (br s, 2H, −CH2OH), 7.83 (s, 1H, ArH), 7.89 (s, 1H, ArH), 7.93 (s, 1H, ArH), 8.19 (s, 1H, triazoleH) and 8.27 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 13.7, 19.8, 24.1, 26.4, 27.8, 28.1, 28.7, 28.9, 29.3, 29.6, 30.2, 30.9, 32.6, 32.8, 34.0, 50.5, 59.0, 61.6, 64.6, 68.5, 69.0, 69.2, 69.4, 69.6, 70.4, 70.5, 70.8, 71.5, 71.9, 72.6, 120.1. 121.7, 123.5, 130.8, 131.3, 142.8, 147.2, and 206.9.
Compound 7: General procedure for ring opening polymerization (PEG2-PCL3.5-Br): A solution of compound 6 (50mg, 0.02mmol) in dry toluene (2 ml) was placed in a flame-dried two neck round bottom flask fitted with a condenser. The solution was degassed by evacuation, and distilled ε-caprolactone (0.07mL, 0.67mmol) was added with a syringe through rubber septa. A nitrogen purged solution of Sn(II) 2-ethylhexanoate (catalytic) in toluene (1 ml) was added to the reaction flask and the solution was refluxed for 24 h. The reaction mixture was then cooled to room temperature, and the solvent was removed under vacuum. The product was dissolved in dichloromethane and precipitated in cold methanol. The precipitated polymer was filtered and washed with diethylether to yield a white powder (102mg, 83% yield). 1H NMR (400 MHz, CDCl3): δ (ppm) 1.22–1.50 (m, 12H, −CH2-) & (m, −CH2PCL), 1.52–1.95 (m, 6H, −CH2-) & (m,- CH2PCL), 2.30 (t, - CH2PCL), 3.37 (s, 3H, −OCH3), 3.54 (t, 2H, −CH2Br), 3.55–3.72 (m, (−OCH2CH2-)n) & (m, 2H, −CH2Br), 3.81 (t, 2H, −CH2OCH3), 3.93 (t, 2H, −CH2), 4.05 (t, −CH2PCL), 4.41 (t, 2H, −OCH2CH2triazole), 4.61 (t, 2H, −OCH2CH2triazole), 5.19 (s, 2H, −CH2PCL), 7.77 (s, 1H, ArH), 7.85 (s, 1H, ArH), 7.90 (s, 1H, ArH), 8.10 (s, 1H, triazoleH) and 8.23 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 11.8, 13.9, 22.6, 22.9, 24.4, 24.5, 24.7, 25.2, 25.3, 25.5, 25.6, 26.4, 28.1, 28.2, 28.3, 28.5, 28.7, 29.0, 29.3, 29.5, 30.3, 31.6, 32.3, 32.8, 33.4, 34.1, 34.2, 34.3, 34.5, 46.8, 50.5, 59.0, 62.6, 63.4, 63.7, 64.0, 64.1, 64.2, 65.8, 69.3, 69.4, 70.5, 71.9, 120.1, 121.6, 122.7, 125.0, 125.1, 131.6, 137.4, 146.9, 147.0, 173.3, 173.5, 173.8, 173.7, 174.1, 175.9, and 179.4.
Compound 8 - General procedure for PEG2-PCL3.5-TPP+Br−: A solution of compound 7 (PEG2-PCL3.5-Br, 100mg, 0.02 mmol) and triphenylphosphine (TPP) (8.8mg, 0.03mmol) in acetonitrile (ACN) was refluxed for 48 h. The solvent was then evaporated and the residue was washed several times with hexanes and diethylether to remove excess of TPP. The white solid was then dried under vacuum to remove the solvents (88mg, 85% yield). 1H NMR (400 MHz, CDCl3): δ (ppm) 1.20–1.50 (m, 12H, −CH2-) & (m, −CH2PCL), 1.52–1.95 (m, 6H, −CH2-) & (m,- CH2PCL), 2.29 (t, −CH2PCL), 3.36 (s, 3H, −OCH3), 3.55–3.80 (m, (−OCH2CH2-)n), (m, 2H, −CH2Br) & (t, 2H, −CH2Br), 3.80 (t, 2H, −CH2OCH3), 3.92 (t, 2H, −CH2), 4.05 (t, −CH2PCL), 4.41 (t, 2H, −OCH2CH2triazole), 4.59 (t, 2H, −OCH2CH2triazole), 5.17 (s, 2H, −CH2PCL), 7.68–7.85 (m, 15 H, TPP+& 2H, ArH), 8.01 (s, 1H, ArH), 8.16 (s, 1H, triazoleH) and 8.22 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 22.6, 1124.5, 25.4, 25.5, 26.3, 28.3, 28.9, 30.2, 32.3, 34.0, 34.1, 50.4, 59.0, 62.6, 64.0, 64.1, 65.9, 69.5, 70.5, 70.6, 71.9, 118.8, 121.6, 122.7, 122.8, 130.4, 130.5, 133.8, 135.0, 146.9, 173.4, 204.2, and 205.9. 31P NMR (200 MHz, CDCl3): δ (ppm) 25.5.
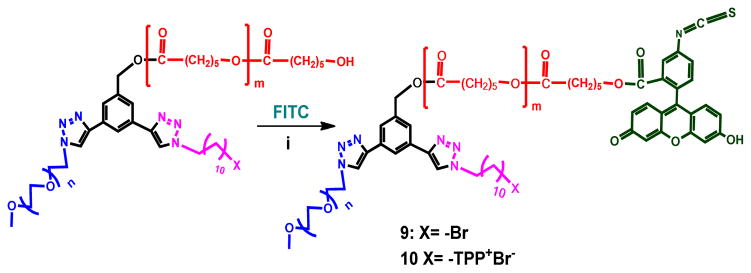
Compound 9 (PEG2-PCL4.4-FITC-Br): A solution of FITC (8.5mg, 0.01mmoles) in DMSO (1ml) was stirred with 1-[3-(Dimethylamino)propyl]-3-ethylcarbodiimidemethiodide (EDC) (4.02mg, 0.02mmoles) and 4-dimethylaminopyridine (DMAP) (1.71mg, 0.01mmoles) for 20 minutes at room temperature, followed by the addition of PEG2-PCL4.4-Br (100mg, 0.01mmoles), and stirred at room temperature overnight. The reaction mixture was then dialyzed against DMSO for 48 h by changing DMSO every 8 h to remove excess FITC. It was then dialyzed against water for 12 h. The water was then removed under vacuum and the residue was washed several times with methanol to afford orange colored solid (42mg, 43%). 1H NMR (400 MHz, CDCl3): δ (ppm) 1.22–1.50 (m, 12H, −CH2-) & (m, −CH2PCL), 1.52–1.95 (m, 6H, −CH2-) & (m,- CH2PCL), 2.30 (t, - CH2PCL), 3.37 (s, 3H, −OCH3), 3.54 (t, 2H, −CH2Br), 3.55–3.72 (m, (−OCH2CH2-)n) & (m, 2H, −CH2Br), 3.81 (t, 2H, −CH2OCH3), 3.93 (t, 2H, −CH2), 4.05 (t, −CH2PCL), 4.41 (t, 2H, −OCH2CH2triazole), 4.61 (t, 2H, −OCH2CH2triazole), 5.19 (s, 2H, −CH2PCL), 6.57–6.85 (m, 6H, FITC ArH), 7.02–7.18 (m, 3H, FITC ArH), 7.77 (s, 1H, ArH), 7.85 (s, 1H, ArH), 7.90 (s, 1H, ArH), 8.10 (s, 1H, triazoleH) and 8.23 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm)13.6, 18.1, 24.4, 24.6, 24.7, 25.3, 25.4, 25.5, 25.6, 26.5, 26.8, 28.2, 28.3, 28.5, 28.8, 29.0, 29.3, 29.4, 29.7, 30.4, 30.9, 32.3, 32.6, 33.9, 34.1, 34.2, 36.3, 45.2, 46.8, 50.6, 52.6, 55.9, 59.1, 62.6, 64.1, 65.3, 65.9, 69.6, 70.6, 71.9, 85.6, 86.2, 118.8, 124.9, 125.0, 131.6, 137.4, 146.9, 157.1, 159.3, 171.1, 171.8, 173.3, 173.5, 173.6, 173.8, 196.4, 201.8, 207.1, 208.1, 213.1, 218.5, and 219.7.
Compound 10 (PEG2-PCL4.4-FITC-TPPBr): A solution of FITC (8.5mg, 0.01mmoles) in DMSO (1ml) was stirred with 1-[3-(Dimethylamino)propyl]-3-ethylcarbodiimidemethiodide (EDC) (4.02mg, 0.02mmoles) and 4-dimethylaminopyridine (DMAP) (1.71mg, 0.01mmoles) for 20 minutes at room temperature, followed by the addition of PEG2-PCL4.4-TPPBr (100mg, 0.01mmoles) and was stirred at room temperature overnight. The reaction mixture was then dialyzed against DMSO for 48 h by changing DMSO every 8 h to remove excess FITC. It was then dialyzed against water for 12 h. The water was then removed under vacuum and the residue was washed several times with methanol to afford an orange colored solid (38mg, 36%). 1H NMR (400 MHz, CDCl3): δ (ppm) 1.20–1.50 (m, 12H, −CH2-) & (m, −CH2PCL), 1.52–1.95 (m, 6H, −CH2-) & (m,- CH2PCL), 2.29 (t, - CH2PCL), 3.36 (s, 3H, −OCH3), 3.55–3.80 (m, (−OCH2CH2-)n), (m, 2H, −CH2Br) & (t, 2H, −CH2Br), 3.80 (t, 2H, −CH2OCH3), 3.92 (t, 2H, −CH2), 4.05 (t, −CH2PCL), 4.41 (t, 2H, −OCH2CH2triazole), 4.59 (t, 2H, −OCH2CH2triazole), 5.17 (s, 2H, −CH2PCL), 6.57–6.85 (m, 6H, FITC ArH), 7.02–7.18 (m, 3H, FITC ArH), 7.68–7.85 (m, 15 H, TPP+& 2H, ArH), 8.01 (s, 1H, ArH), 8.16 (s, 1H, triazoleH) and 8.22 (s, 1H, triazoleH). 13C{1H} NMR (125 MHz, CDCl3): δ (ppm) 12.5, 13.1, 18.6, 24.4, 24.6, 24.7, 25.3, 25.5, 27.5, 28.3, 29.0, 29.2, 32.3, 33.9, 34.1, 34.2, 37.0, 37.6, 45.7, 47.1, 59.0, 59.6, 62.2, 62.6, 64.1, 67.7, 70.5, 71.9, 73.3, 76.0, 79.3, 81.3, 84.3, 86.6, 97.8, 124.0, 145.9, 155.1, 164.4, 170.1, 171.9, 173.6, 173.8, 184.4, 185.2, 187.1, 195.4, 201.1, 214.7, 217.2, 218.5, and 219.6.
Detailed synthesis and characterization of a series of targeted and non-targeted polymers are provided in the Supplementary section.
Preparation of blank and CoQ10-loaded miktoarm micelles
Blank and CoQ10-loaded miktoarm micelles were prepared by the co-solvent evaporation method.17 Specific weights of the polymer and drug (drug/polymer ratio of 5–200 wt. %) were dissolved in 1.5 ml acetone. The solution was added drop-wise (1 drop/ 10 sec) to 3 ml of magnetically stirred deionized water. The mixture was stirred in the dark for 24 h to remove acetone and trigger micelle formation. The mixture was filtered through a 0.45 μm PVDF filter to remove the free (un-encapsulated) drug. Aliquots of the micellar solutions were tested by dynamic light scattering (DLS) to determine the hydrodynamic diameter (DH) and polydispersity index (PDI) of the micelles. Aliquots of the solution were freeze dried and used to determine drug content of the micelles by an HPLC assay.
Characterization
NMR spectra were recorded on a 200, 400 or 500MHz (as specified) Varian spectrometers at ambient temperatures. The chemical shifts in ppm are reported relative to tetramethylsilane as an internal standard for 1H and 13C {1H} NMR spectra. Molecular weight and polydispersity index (PDI) were obtained from GPC (Waters Breeze) using THF as the mobile phase. The GPC was equipped with three Waters Styragel HR columns (molecular weight measurement ranges: HR1: 102 – 5 × 103 g mol−1, HR2: 5 × 102 – 2 × 104 g mol−1, HR3: 5 × 103 – 6 × 105g mol−1) and a guard column. The columns were operated at 40 °C and a mobile phase flow rate of 0.3 ml min−1 during analysis. The GPC was also equipped with both ultraviolet (UV 2487) and differential refractive index (RI 2410) detectors. The molecular weight measurements were calibrated relative to poly(styrene) narrow molecular weight standards in THF at 40 °C.
FT-IR measurements were carried out on a Perkin Elmer Instrument equipped with ATR. Transmission electron microscopy (TEM) was used to capture images of the micelles using a Phillips CM200 electron microscope equipped with an AMT 2k × 2k CCD camera at an acceleration voltage of 80 kV. TEM samples were prepared by adding 10 YL of the aqueous micelle solutions onto a formvar-coated 400 mesh grid stabilized with evaporated carbon film. The micelles were negatively stained by adding 10 YL of 1% aqueous uranyl acetate solution. The samples were allowed to dry overnight at room temperature. Dynamic light scattering (DLS) measurements were carried out at a wavelength of 532 nm on a Brookhaven photon correlation spectrometer equipped with a BI9000 AT digital correlator and a compass 315M-150 laser (Coherent Technologies). Measurements were made at ~ 25 °C and at a 90o scattering angle. Mean hydrodynamic diameter measurements were obtained from a Gaussian fit of the CONTIN analysis mode from three averaged measurements of aqueous micellar solutions. Samples were filtered through a 0.45 Ym Millex Millipore PVDF membrane prior to measurements. Steady-state fluorescence spectra were recorded using a Fluoromax-2 spectrometer. Zeta potential measurements were performed using a Brookhaven ZetaPlus Zeta Potential Analyzer. Twenty zeta potential measurements were carried out at 25 °C on aqueous solution of polymeric micelles having 1.0 mM NaCl.
HPLC analysis of CoQ10 was performed on a Waters chromatography system equipped with Waters 1525Y binary HPLC pump, Waters 717plus autosampler, Waters Symmetry® C18 5 Ym 4.6×150 mm column, Waters 2487 dual δ absorbance detector, and an IBM computer equipped with the Breeze® software. The assay was carried out at 25 oC using a 6:4 v/v mixture of ethanol-methanol at a flow rate of 1.0 ml/min. The injection volume was 20 Yl and the run time was 12 min. CoQ10, monitored by its absorbance at 275 nm, had a retention time ~ 8.9 min. A calibration curve (r2 ≥0.999) of CoQ10 was prepared using standard solutions ranging in concentration from 0.1 to 0.7mg/ml prepared immediately prior to the assay. To assay CoQ10 content of different miktoarm micelles, a given volume of aqueous micellar solution was diluted 10 times by the mobile phase. The solution was filtered through 0.2 μm Millex Millipore nylon filters and assayed by HPLC. Given volume of blank polymeric micelles was treated similarly and used as a control. CoQ10 encapsulation efficiency and loading capacity were calculated from the following equations:
| (1) |
| (2) |
Calculation of Flory Huggins interaction parameters (χsm) between CoQ10 and PCL
The Flory Huggins interaction parameter (χsm) between the micelle core and CoQ10 was calculated using equation
δCoQ10 and δPCL are solubility parameters for CoQ10 and PCL, respectively; VCoQ10 is the molar volume of drug, R is the universal gas constant, and T is the Kelvin temperature. The solubility parameters were calculated by Hansen’s approach28, which uses partial solubility parameters to calculate the total solubility parameters according to equation29, 30 :
where δd, δp, and δh refer to the partial solubility parameters accounting for van der Waals dispersion forces between atoms, dipole-dipole interactions between molecules, and proclivity of hydrogen bonding between molecules, respectively. The partial solubility parameters for CoQ10 and for PCL (Table 1) were estimated by the Hansen theory of solubility group contribution method (GCM) using Molecular Modeling Pro software (Chem SW) (Fairfield, CA).31
Table 1.
GPC analysis of miktoarm polymers
| Polymer | Mna(g/mole) | Mwa(g/mole) | PDI |
|---|---|---|---|
| PEG2-PCL3.5-Br | 5992 | 7339 | 1.22 |
| PEG2-PCL3.5-TPPBr | 6022 | 8126 | 1.34 |
| PEG2-PCL3.8-Br | 6252 | 8561 | 1.36 |
| PEG2-PCL3.8-TPPBr | 6284 | 8992 | 1.43 |
| PEG2-PCL4.4-Br | 6911 | 9150 | 1.32 |
| PEG2-PCL4.4-TPPBr | 7353 | 9863 | 1.34 |
| PEG2-PCL5.5-Br | 7932 | 11272 | 1.42 |
| PEG2-PCL5.5-TPPBr | 8374 | 11939 | 1.42 |
| PEG2-PCL7.0-Br | 9574 | 12999 | 1.35 |
| PEG2-PCL7.0-TPPBr | 11140 | 18693 | 1.67 |
| PEG2-PCL7.8-Br | 10247 | 12819 | 1.25 |
| PEG2-PCL7.8-TPPBr | 12072 | 20356 | 1.68 |
Determined from GPC measurements
Critical association concentration (CAC) of ABC miktoarm micelles
Given volumes of pyrene stock solution in acetone (180 μM) were added to a series of 4 ml vials and the acetone was allowed to evaporate overnight in the dark. Blank miktoarm micelles were prepared following the general procedure described above. Specified volumes of the micellar solutions were added to the vials having pyrene so that polymer concentration varied from 0.025 to 200μg/ml while pyrene concentration was kept constant at 6 μM. The pyrene/micellar solutions were equilibrated overnight in the dark. Excitation spectra were recorded from 260–360 nm at λem=390 nm (excitation and emission bandpass: 2 and 4 nm; respectively). The ratios of the pyrene fluorescence intensities at λ=338 and 333 nm (I338/I333) were calculated and plotted versus polymer concentration. The critical association concentration (CAC) values were determined from the graphs as the intersections of two straight lines (the horizontal line with an almost constant value of the ratio I338/I333 and the vertical line with a steady increase in the ratio value).
Stability Studies
Micelles colloidal stability
CoQ10-loaded micelles were prepared by the co-solvent evaporation method in de-ionized water and stored at 4 °C for 3 months. The particle size of micelles was measured on the freshly prepared sample and on weekly intervals after storage. The micelles were also periodically examined for signs of aggregation/precipitation.
Photostability of CoQ10
CoQ10 photostability was studied in a 35 × 35× 30 cm photoreactor equipped with 8 RPR-2537 A UV lamps operating at δmax of 253 nm (Southern New England Ultraviolet Co., Branford, CT). The studied samples were CoQ10 solution (0.5 mg/ml in 1:1 v/v ethanol acetone mixture) and CoQ10/PEG2-PCL3.8-TPPBr micelles (CoQ10 concentration: 0.5 mg/ml, polymer concentration 1 mg/ml, % CoQ10 loading: 33.34 wt. %). The samples were placed at the center of the UV chamber and 4 UV lamps were turned on. Samples were withdrawn at predetermined time intervals and analyzed by HPLC for CoQ10 content as described above.
Differential scanning calorimetry (DSC) studies
Thermal analysis was carried out with a TA Q2000 differential scanning calorimeter (DSC) (TA Instruments, Newcastle, DE). The instrument was calibrated against an indium standard. The samples (2–5 mg) were accurately weighed into DSC aluminum pans. Empty pans were used as reference. The samples were heated at rate of 10 oC/min from 20 oC to 80 oC under nitrogen flushing (flow rate of 50 ml/min).
Cell culture and media
Murine microglia (N9) cells obtained from ATCC were seeded in Iscove’s Modified Dulbecoo’s Medium (IMDM) (Gibco) containing 5% of fetal bovine serum (Gibco) and 1% penicillin–streptomycin (Gibco). For all experiments, unless otherwise stated, cells were seeded in 96-well plate (Costar) at a density of 2.5×104 cells/well maintained at 37 °C, 5% CO2 in a humidified atmosphere and were grown in serum containing media for 24 hours before cell treatments to attain confluency. Cells were used between 10 and 30 passages.
For confocal microscopy experiments, primary hippocampal neurons and glia from 3-day-old mouse pups were isolated, mechanically and enzymatically (0.25% Trypsin, Gibco) dissociated. Cells were seeded at a density of 1×105 cells/well onto coated glass coverslips. Coverslips were coated initially with laminin (0.587 ug/ml, Invitrogen) overnight. Cells were grown at 37 °C and 5% CO2 in a 24-well coated cell culture plate (Corning), initially in phenol-free Dulbecco’s Modified Eagle’s Medium (Invitrogen) with 1 mm l-Glutamine, sodium pyruvate and 1% (v/v) PSN (Invitrogen). On the second day-in-vitro (DiV), cells are cultured in Neurobasal A medium without phenol red (Invitrogen) supplemented with 2% (v/v) B-27 supplement (Invitrogen), 1% (v/v) PSN (Invitrogen), and 1 mm l-Glutamine (Sigma). Half of the culture media was changed every 5 days (note that L-Glutamine was not added to the medium after DiV 6). Cultures were maintained for 22 days in vitro prior to treatment and imaging.
Cells Viability
Mitochondrial metabolic activity of cells was measured using MTT assay. Media was aspirated and cells were treated in serum-free media with CoQ10/PEG2-PCL3.8-TPPBr (CoQ10-Targeted micelle), CoQ10, PEG2-PCL3.8-TPPBr (empty micelle) or CoQ10/PEG-PCL3.8-Br (CoQ10-Non targeted micelle) (0–50 μM, calculated in equimolar concentrations with respect to CoQ10 for 24 h).
Following treatment media was removed and replaced with fresh serum-free media (200 μl/well). MTT solution (0.5 mg/ml, Sigma) was added to each well and incubated for 30 min at 37° C. Formazan crystals was formed then dissolved by adding dimethylsulphoxide (DMSO, Sigma) and quantified by measuring the absorbance of the solution at 595 nm using Benchmark microplate reader (Bio-Rad, Canada). The extent of formazan conversion is expressed in percentages relative to the untreated control. Results are expressed as mean ± SEM obtained from at least three independent experiments performed in triplicate.
Superoxide anion and reactive oxygen species detection
Superoxide anion and reactive oxygen species were detected using dihydroethidium (DHE, Molecular probes) and 2′,7′dichlorodihydrofluorescein diacetate (DCFH-DA, Molecular probes) respectively. Briefly, Media was aspirated and cells were treated in fresh serum-free media with methyl viologen dichloride (Paraquat, 10 μM, Sigma), or hydrogen superoxide (H2O2, 2 mM, EMD) followed by CoQ10/PEG2-PCL3.6-TPPBr (CoQ10-Targeted micelle), CoQ10 or CoQ10/PEG2-PCL3.8-Br (CoQ10-Non targeted micelle) (5 μM with respect to CoQ10 concentration for 24 hr). Following treatment, media was replaced with fresh media containing DHE (20 μM) or DCFH-DA (20 μM) and incubated for 30 min at 37°C. After which, cells were washed once with media (200 μl/well) and fresh serum free media was added. Fluorescence of ethidium and 2′,7′-dichlorodihydrofluorescein (DCF) was determined with Fluostar Optima spectrofluorometer (BMG, LabTech) using excitation/emission wavelengths= 544/590 and 485/520 nm respectively. Results are expressed as mean ± SEM obtained from at least three independent experiments performed in triplicate. Representative fluorescent images of cells were acquired at 40× with a Leica DFC350FX monochrome digital camera connected to a Leica DMI4000B inverted fluorescence microscope.
Confocal microscopy
For mitochondrial membrane potential measurement experiments, primary cultures were treated with CoQ10/PEG2-PCL3.8-TPPBr (CoQ10-Targeted micelle), CoQ10 or CoQ10/PEG2-PCL3.8-Br (CoQ10-Non targeted micelle) 1 μM with respect to CoQ10 concentration for 24 h. Cells were then exposed to H2O2 (1 mM, 40 min). Media was aspirated and cells were incubated with media containing TMRE (tetramethylrhodamine ethyl ester, 100 nM, 30 min). To maintain the equilibrium of TMRE during live cells imaging, cells were incubated with fresh media containing 20 nM TMRE during imaging period. Images were acquired with a Zeiss LSM 710 confocal microscope using a W Plan Achromat 63X/1.0 M27 objective. Images were acquired at a resolution of 940 × 940 using HeNe (543 nm) and pinhole=92 μm. For each Z-stack, a total of 6–9 Z-slices were acquired using a scaling of 0.14 × 0.14 X × 0 μm (x, y, z).
For FITC-mitochondria-targeted experiments, primary cultures were treated with FITC-PEG2-PCL3.8-TPPBr (FITC-Targeted micelles) or FITC-PEG2-PCL3.8-Br (FITC- Non targeted micelles) (1 μM, 3 h). Cells were then treated with Mitotracker 633 (Molecular probes, 100 nM, 3 min). Images were acquired with a Zeiss LSM 710 confocal microscope using a W Plan Achromat 63X/1.0 M27 objective. Images were acquired at a resolution of 940 × 940 using HeNe (633 nm) and Argon (488 nm) lasers for detection of deep red and green respectively. Pinhole= 90 μm. For each Z-stack, a total of 6–8 Z-slices were acquired using a scaling of 0.14 × 0.14 × 0.30 μm (x, y, z).
Fluorescent microscopy for mitochondrial membrane potential measurement
N9 cells were co-treated with CoQ10/PEG2-PCL3.8-TPPBr (CoQ10-Targeted micelle), CoQ10 or CoQ10/PEG2-PCL3.8-Br (CoQ10-Non targeted micelle) 5 μM with respect to CoQ10 concentration together with antimycin A (A.A, 1 μM) for 24 h. Following treatment, media was aspirated and cells were incubated with media containing TMRE (tetramethylrhodamine ethyl ester, 200 nM, 30 min). To maintain the equilibrium of TMRE during live cells imaging, cells were incubated with fresh media containing 50 nM TMRE during imaging period. Fluorescent images of cells were acquired at 40× with a Leica DFC350FX monochrome digital camera connected to a Leica DMI4000B inverted fluorescence microscope. Image analysis (mean fluorescence intensity measurements) was performed using Image J software. Results are expressed as mean ± SEM obtained from at least three independent experiments performed in triplicate.
RESULTS AND DISCUSSION
Synthesis and Characterization of ABC miktoarm polymers
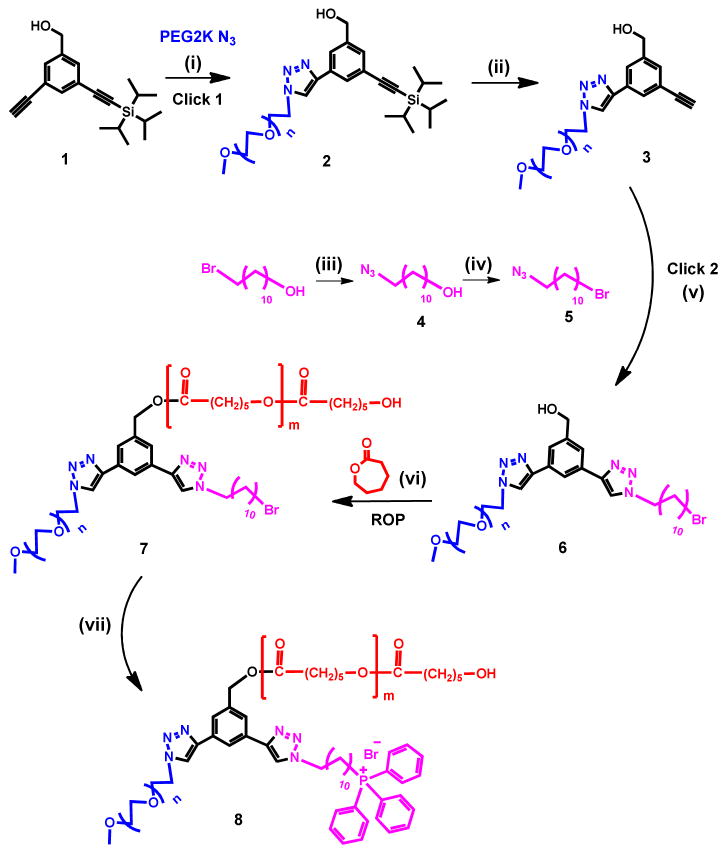
ABC amphiphilic miktoarm stars were constructed on a three arm core (1) by performing “click” and ring opening polymerization reactions, in sequence. One arm of the core building block was used to attach a hydrophilic polymer (PEG) whose molecular weight (2000) was kept constant. Another arm was conjugated with a mitochondria targeting moiety, triphenylphosphonium (TPP+), and the third arm was used to carry out ring opening polymerization of caprolactone (Scheme 1). This led to a series of miktoarm stars with variable molecular weights of polycaprocatone arm. The key building block (1) with free and protected acetylenes was synthesized using a procedure developed earlier in our group.25, 32 The free acetylene arm of 1 was utilized to covalently bind a PEG 2K chain using “click” reaction with PEG2K-azide in the presence of copper sulfate pentahydrate and sodium ascorbate. This click reaction was clean and simple, and gave a good yield of compound 2. The 1H NMR spectrum showed the disappearance of the alkyne proton, and appearance of PEG and triazole protons. It was then followed by the deprotection of triisopropylsilyl-acetylene group which made the second acetylene available in compound 3 for the next click reaction. For the attachment of the targeting moiety TPP+, we clicked a linear azide (5) containing a bromo terminal point. The latter was synthesized from 11-Azido-undecan-1-ol (4), by converting its hydroxyl group to bromo, using carbon tetrabromide and triphenylphosphine. The 1H NMR spectrum confirmed the formation of product 6 with the disappearance of acetylene proton and appearance of a triazole proton, along with protons in aliphatic region. The compound 6 with a PEG 2K chain, a bromo functional group and a third arm with free hydroxyl group, was then utilized to perform ring opening polymerization reactions using variable amounts of caprolactone monomer to get a range of molecular weights of polycaprolactone. Once again, the reaction was monitored by 1H NMR which showed the appearance of PCL protons. These polymers containing PEG, PCL and –Br units (general structure, 6) were used as non-targeting polymers without triphenylphosphonium cation. Finally, the targeting moiety was attached to 6 by reacting with excess of triphenylphosphine. 1H, 13C{1H} and 31P{1H} NMR spectra confirmed the attachment of triphenylphosphonium cation. A shift in 31P NMR was observed for TPP+ as compared to free TPP, and it also clearly indicated the absence of any excess (free) TPP (Figure 1D). GPC chromatogram showed clear shift with the addition of each arm on the core (Figure 1E). In addition, we coupled targeted and non-targeted polymers to an imaging dye, Fluorescein isothiocyanate (FITC) to trace these polymers in the cells (Scheme 2).
Scheme 1.
Synthesis of ABC miktoarm star polymers containing
 ,
,
 and
and
 .
.
i) CuSO4·5H2O/Sodium ascorbate, H2O/THF, RT, over night (O/N); ii) tetrabutylammoniumfluoride (TBAF)/THF, O/N; iii) NaN3/DMF, RT, O/N; iv) CBr4, Triphenylphosphine (TPP), DCM, O/N; v) CuSO4·5H2O/Sodium ascorbate, H2O/THF/DMF, RT, O/N; vi) Toluene, Sn(II) 2-ethylhexanoate, Reflux, 24 h; vii) TPP, ACN, Reflux, 48 h.
Scheme 2.
Synthesis of Fluorescein isothiocyanate (FITC) conjugated ABC miktoarm star polymer 9 (non-targeted) and 10 (with targeting moiety TPP+ cation)
i) 1-[3-(Dimethylamino)propyl]-3-ethylcarbodiimidemethiodide (EDC), 4-dimethylaminopyridine (DMAP), DMSO, RT, 12 h.
Preparation and characterization of miktoarm polymer micelles
We selected PEG as the hydrophilic arm of the ABC miktoarm polymers on the basis of its biocompatibility, hydrophilicity and ability to stabilize nanoparticles in aqueous solutions, whereas PCL was used as the hydrophobic arm due to its biocompatibility and ability to solubilize hydrophobic drugs in the micelle core, sustain their release and protect them against degradation.33, 34 The lipophilic cation triphenylphosphonium (TPP+) was employed as the mitochondriotropic moiety, since its conjugates with antioxidant drugs CoQ10 (MitoQ) and α-tocopherol (MitoVitE)) have been shown to enhance drug accumulation in mitochondria.35 Empty and CoQ10-loaded miktoarm micelles were prepared in deionized water by the co-solvent evaporation method which is simple and easy to scale up.17 To optimize the size of the micelles, colloidal stability and CoQ10 loading capacity, a series of ABC miktoarm polymers were prepared in which the chain length of PEG arm was kept constant at 2 kDa, whereas that of the PCL arm was varied from 3.5 to 7.8 kDa (Table 1).
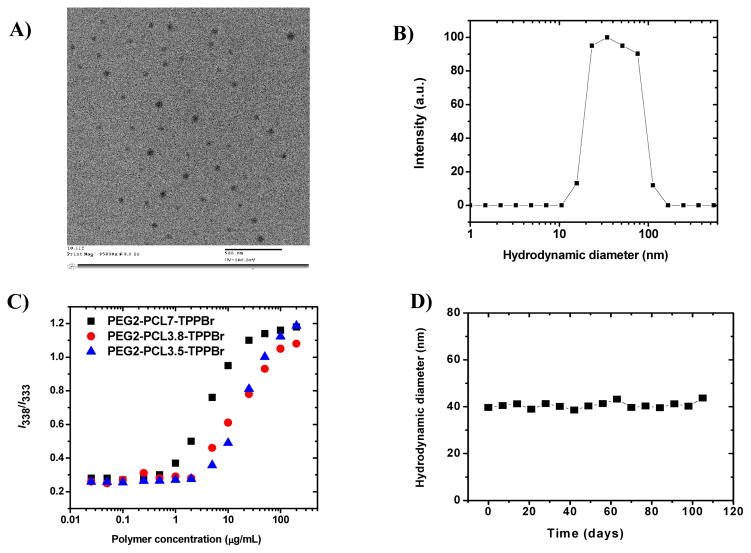
Polymers without triphenylphosphonium bromide (non-targeted polymers) were also prepared and used as control. Direct evidence of micelle formation was obtained from TEM measurements of PEG2-PCL7.0-TPPBr, which showed the presence of spherical, well-dispersed particles with an average diameter of 51.74±7.6 nm (Figure 2A). DLS measurements further confirmed the formation of monodispersed micelles as demonstrated in Figure 2B for PEG2-PCL3.8-TPPBr micelles. We subsequently studied the effect of polymer composition on micelle size (Table 2) by DLS. Increasing the chain length of PCL from 3.5 to 5.5 kDa did not result in a significant change in the hydrodynamic diameter of the micelles which remained around 30 nm (Table 2). However, the hydrodynamic diameter increased from 30.9±2.5 to 65.5±1.9 nm when the PCL chain length was increased from 3.5 to 7.8 kDa. Longer PCL chains usually lead to the formation of bigger micelles due to their incorporation in the core of the micelles.17, 36
Figure 2.
(A) TEM image of PEG2-PCL7-TPPBr micelles (polymer concentration = 1 g/L). (B) Distribution of the hydrodynamic diameter (DH) of PEG2-PCL3.8-TPPBr micelles (Deionized water; polymer concentration: 1 g/L; θ: 90°). (C) Plots of intensity ratio (I338/I333) of pyrene excitation spectra (δem = 390 nm) vs concentration of different PEG2-PCL-TPPBr miktoarm copolymers in water. (D) Hydrodynamic diameter of CoQ10-loaded/PEG2-PCL3.8-TPPBr micelles as a function of storage time at 4 °C (Deionized water; polymer concentration : 1 g/L ;CoQ10 loading :10 wt. %).
Table 2.
Properties of blank and CoQ10-loaded miktoarm polymers based micelles.
| Polymer | Micelles diametera | %DLb | %LEc | CoQ10 Std (μg/ml) | |
|---|---|---|---|---|---|
|
| |||||
| Blank micelles | CoQ10 micelles | ||||
| PEG2-PCL3.5-Br | 30.9 ± 2.5 | 40.2 ± 2.2 | 9.3 | 93 | 93 |
| PEG2-PCL3.5-TPPBr | 43.0 ± 1.9 | 57.7 ± 2.3 | 8.3 | 83 | 83 |
| PEG2-PCL3.8-Br | 31.8 ± 3.2 | 40.26 ± 1.9 | 9.2 | 92 | 92 |
| PEG2-PCL3.8-TPPBr | 36.5 ± 3.4 | 44.2 ± 2.7 | 8.5 | 85 | 85 |
| PEG2-PCL5.5-Br | 30.5 ± 3.2 | 40.26 ± 1.9 | 9.2 | 92 | 92 |
| PEG2-PCL5.5-TPPBr | 26.7 ± 3.4 | 39.7 ± 2.7 | 8.5 | 85 | 85 |
Hydrodynamic diameter (nm), mean of three measurements ±SD;
Percent drug loading = weight of CoQ10 in micelles ×100/weight of micelles tested;
Percent loading efficiency = weight of CoQ10 in micelles ×100/weight of CoQ10 used in micelles preparation.
CoQ10 solubility in water.
CoQ10 incorporation into PEG2-PCL-TPPBr miktoarm polymer micelles
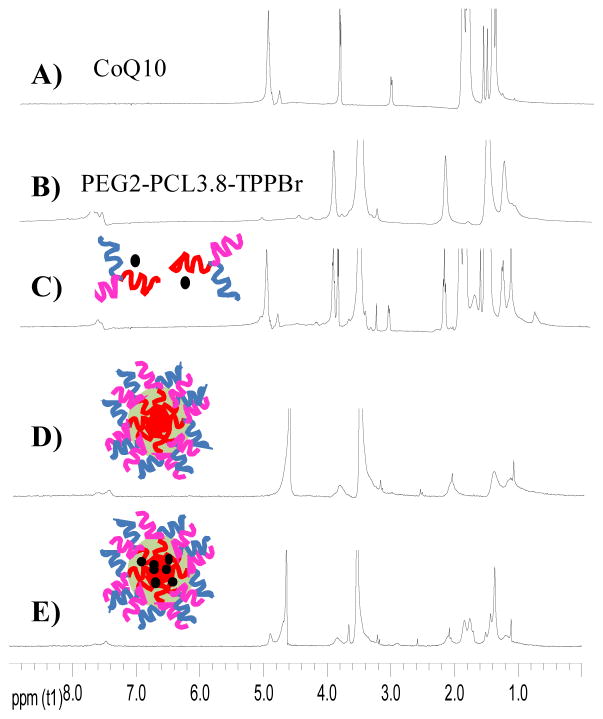
CoQ10-loaded PEG2-PCL-TPPBr miktoarm polymer micelles were prepared by a co-solvent evaporation method. Confirmation of CoQ10 incorporation in the micelles core was obtained by 1H NMR spectroscopy. 1H NMR spectra of CoQ10, PEG2-PCL3.8-TPPBr and their mixture in CDCl3, together with the blank and CoQ10-loaded micelles in D2O are shown in Figure 3. Characteristic signals of CoQ10 and PEG2-PCL3.8-TPPBr were observed when they were dissolved in CDCl3 (Figure 3A and B). A similar spectrum was observed for the CoQ10/PEG2-PCL3.8-TPPBr mixture in CDCl3 (Figure 3C) since it is a good solvent for both of them. In contrast, the spectrum of blank PEG2-PCL3.8-TPPBr micelles in D2O showed signals characteristic of PEG protons (δ 3.26 and 3.57 ppm) confirming that they are well hydrated and reserved their mobility (Figure 3D). In this spectrum the characteristic signals of the PCL arm protons appear weak and broad due to incorporation into the micelles core and severe loss of movement (Figure 3D). Similarly, the spectrum of CoQ10/PEG2-PCL3.8-TPPBr showed weak and broad CoQ10 and PCL signals and strong well-resolved PEG signals. Taken together, these results confirmed the formation of nanoparticles in D2O with core-corona structure. The core of the micelles is made up of PCL chains incorporating CoQ10, whereas PEG chains form the corona. Similar behavior has been reported earlier for other drug-loaded polymeric micelles.37 It was also important to verify the location of the TPP cation since it should be at the micelle surface to direct them to the mitochondria. Signals at δ 7.4–7.8 ppm in Figure 3D and E confirmed that the TPP cation is located in the corona of the micelles. This observation was further confirmed by zeta potential measurements of PEG2-PCL3.8-Br and PEG2-PCL3.8-TPPBr micelles in aqueous solution. The zeta potentials (-19.58 mV for PEG2-PCL3.8-Br micelles and +12.1 mV for PEG2-PCL3.8-TPPBr micelles) clearly suggested that the TPP cation is located on the surface of PEG2-PCL3.8-TPPBr micelles.38
Figure 3.
1H NMR spectra of CoQ10 in CDCl3 (A), PEG2-PCL3.8-TPPBr miktoarm in CDCl3 (B), CoQ10/PEG2-PCL3.8-TPPBr mixture in CDCl3 (C), blank PEG2-PCL3.8-TPPBr miktoarm micelles in D2O (D) and CoQ10-loaded PEG2-PCL3.8-TPPBr micelles in D2O (E).
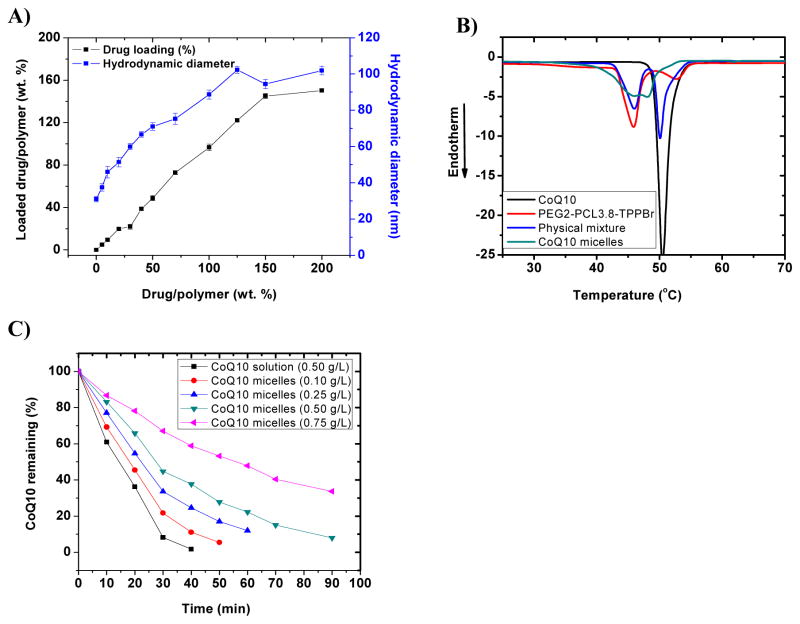
The incorporation of CoQ10 into the core of PEG2-PCL-Br and PEG2-PCL-TPPBr micelles increased the micelle hydrodynamic diameter (Table 2). For instance, PEG2-PCL3.8-TPPBr had hydrodynamic diameters of 36.5±3.4 and 44.2±2.7 nm for empty and CoQ10-loaded micelle, respectively. Other mitochondria-targeted and non-targeted miktoarm polymers with PCL chain length of 3.5–5.5 kDa showed similar results (Table 2), as well as CoQ10 loading efficiency of ≥ 83 % (Table 2). However, aggregates with a hydrodynamic diameter > 600 nm were detected for CoQ10-loaded micelles with a PCL chain length ≥ 6 kDa. The chain length of the hydrophilic PEG arm in these polymers (i.e., 2 kDa) might not be enough to stabilize the nanoparticles with a much longer hydrophobic PCL arm (≥ 6kDa) and bulky triphenylphosphonium group. This hypothesis is supported by the observation that identical non-targeted polymers formed more stable nanoparticles. For instance, no aggregates were detected for PEG2-PCL7.0-Br and PEG2-PCL7.8-Br and the hydrodynamic diameters of their micelles were 46.8±3.2 and 60.2±1.5 nm, respectively. Based on these results, the miktoarm polymer PEG2-PCL3.8-TPPBr was selected for further studies.
The effect of CoQ10/PEG2-PCL3.8-TPPBr weight feed ratio on the actual drug loading and micelle size was studied by preparing CoQ10-loaded micelles at CoQ10/polymer feed weight ratio varying from 0 to 200%. Micelle size increased linearly (R2 = 0.977) with the CoQ10/polymer feed weight ratio (Figure 4A). For instance, the micelles hydrodynamic diameter increased from 30.97±1.5 to 102.3±1.9 nm when the CoQ10/polymer weight ratio increased from 0 to 125%. The increase in micelle size with the CoQ10/polymer ratio is possibly due to drug incorporation in the micelle core which results in core expansion to accommodate the loaded drug molecules.39, 40 The micelle size leveled off at CoQ10/polymer weight ratio of 125% and it remained unchanged thereafter and up to a weight ratio of 200%. Similar trend was observed for the effect of CoQ10/polymer feed weight ratio on the actual percent CoQ10 loading. Thus, the relationship between CoQ10/polymer feed weight ratio and actual CoQ10 loading was linear (R2 = 0.997) for CoQ10/polymer weight ratio of 5–150%. The actual drug loading remained unchanged at CoQ10/polymer weight ratio ≥ 150%, indicating that maximum drug loading has been achieved at CoQ10/polymer weight ratio of 150%. The CoQ10 encapsulation efficiency was in the range of 72–99% for CoQ10/polymer feed weight ratio of 5–150%. No CoQ10 precipitation was observed for micellar solutions with CoQ10/polymer feed weight ratio of 5–150% after storage at room temperature for several weeks. This indicates that this high CoQ10 loading is not due to temporary supersaturation of the micelle. The CoQ10 content of the micelle at drug/polymer feed weight ratio of 150% is 60 weight%. This corresponds to CoQ10 concentration in aqueous solution of 1.5 mg/ml at polymer concentration of 1 mg/ml. CoQ10 is practically insoluble in water and this represents remarkable improvement in its aqueous solubility.41
Figure 4.
(A) Effect of drug/polymer weight feed ratio on drug loading capacity and micelles hydrodynamic diameter of CoQ10/PEG2-PCL3.8-TPPBr micelles prepared in deionized water at polymer concentration of 1 mg/mL. (B) DSC thermograms of CoQ10 alone, PEG2-PCL3.8-TPPBr alone, their physical mixture and micelles prepared at 35 wt% CoQ10 loading. (C) Percentage remaining of different CoQ10 samples as a function of storage time under UV light (λ = 254 nm).
The CoQ10 content in these miktoarm polymer micelles is significantly higher than that obtained for other nanoparticulate formulations. For example, poly(lactide-co-gylcolide) (PLGA) nanoparticles had a maximum CoQ10 content of 19 and 38 weight%; poly(methyl methacrylate) (PMMA) nanoparticles had a CoQ10 content of 37 weight%, whereas nanoparticles based on chitosan and N-carboxymethylchitosan cross-linked with tripolyphosphate had a CoQ10 loading capacity of 20 weight%.42–44 Moreover, we have recently found that the maximum loading capacity of nimodipine, a hydrophobic calcium channel blocker into PCL-based linear and miktoarm polymers was ca. 3–5 weight% and was not affected by polymer architecture.17 The extent of drug loading into a given polymeric micelle formulation is dependent on many factors; the most important of which is the compatibility between the drug and the polymer. Several recent studies have shown that polymer-drug compatibility is a key factor in determining the performance of polymeric micelle as drug delivery systems where high compatibility results in higher drug solubility, loading capacity and controlled drug release.30, 31, 45, 46
To explain the exceptionally high CoQ10 loading into PEG2-PCL3.8-TPPBr micelles, we used the Flory Huggins interaction parameter (χsm) to estimate the CoQ10-PCL compatibility, since it has been shown to be a good indicator of polymer–drug compatibility.31, 47 The Hansen partial solubility parameters for CoQ10, nimodipine and PCL were obtained based on the group contribution method using the Molecular Modeling Pro software (Table 1, supporting information). The Flory Huggins interaction parameter was calculated using equation 3, as described above. Maximum drug-polymer compatibility is achieved when the total solubility parameters for the drug and polymer are equal, resulting in χsm= 0. Smaller χsm values therefore indicate good compatibility between a given drug and polymer. The χsm was found to be 4.57 and 2.54 for nimodipine-PCL and CoQ10-PCL, respectively (Table 1, supporting information) confirming that CoQ10 is more compatible with PCL compared to nimodipine. This in turn explains, at least in part the superior CoQ10 loading into PEG2-PCL3.8-TPPBr micelles. The Flory Huggins theory does not take into consideration any specific interactions between the polymer and drug, and assumes that the orientation of molecules within the polymer–drug mixture is completely random. Although this may not be true for either CoQ10-PCL or nimodipine-PCL systems, the calculated χsm values could serve as a guide in evaluating the relative compatibility between a given polymer and different drugs.
Thermal analysis
To get insight into the status of CoQ10 when it is loaded into PEG2-PCL3.8-TPPBr micelle, differential scanning calorimetry (DSC) thermograms were recorded for CoQ10 and polymer alone, their physical mixture at CoQ10 content of 34 weight%, and the micelle with CoQ10 loading of 34 weight%. The crystallinity of the incorporated drug affects several aspects of the performance of the drug delivery system, such as drug loading capacity, micelles stability and drug release profile.30, 48 The thermogram of CoQ10 alone shows a sharp endothermic peak at 50.48°C ascribed to the melting of CoQ10 (Figure 4B).49, 50
The thermogram of PEG2-PCL3.8-TPPBr alone shows two distinct endothermic peaks at 45.85 and 52.79 °C, attributed respectively to the melting of PEG and PCL.51 These two melting events suggest the existence of two distinct crystalline domains in the miktoarm polymer.30 CoQ10/PEG2-PCL3.8-TPPBr physical mixture shows the same characteristic melting peaks for both CoQ10 and PEG2-PCL3.8-TPPBr copolymer (Figure 4B). In contrast, the DSC thermogram of the lyophilized CoQ10/PEG2-PCL3.8-TPPBr micelle shows a bimodal endothermic peak at 45.40 and 48.31 oC corresponding, respectively to the melting of PEG and CoQ10 (Figure 4B).
Crystallization of CoQ10 in the micelles is presumably due to the high drug content of the micelle (i.e. 34 weight %). It has been reported that lidocaine and clonazepam also showed crystallization in the micelle core at drug content of 30 weight%.52, 53 A broad endothermic shoulder is centered at 51 oC and could be attributed to the melting of PCL. The shift in the melting point of CoQ10 along with the weak melting endotherm of PCL in the thermogram of the lyophilized micelle suggest unique molecular interactions between CoQ10 and PCL arm of the miktoarm polymers. No interaction was detected between CoQ10 and PEG arm of the polymers as indicated by the unchanged melting point of PEG in the thermogram of lyophilized micelle (Figure 4B). Similar results were reported for quercetin/PEG-PCL micelles where the decrease in the PCL melting point in the drug-loaded micelle was attributed to quercetin-PCL hydrophobic interactions.54
Stability studies
Critical association concentration (CAC)
Polymeric micelles are subjected to extensive dilution in the blood stream following intravenous administration resulting in pre-mature drug release if the micelle concentration falls below their CAC. Low CAC values therefore ensure that micelles remain stable while in circulation till they reach their target and deliver their payload. CACs of PEG2-PCL-TPPBr were measured using pyrene as a fluorescent probe. Excitation spectra of aqueous polymer solutions containing 6 μM pyrene and increasing polymer concentrations were recorded from 260–360 nm at λem=390 nm. Pyrene photophysical properties depend on its microenvironment; its excitation spectrum undergoes a red shift from 333 to 338 nm when it passes from hydrophilic to hydrophobic media.55 Semi-logarithmic plots of the I338/I333 ratios versus the concentration of PEG2-PCL-TPPBr miktoarm polymers having different PCL chain lengths are shown in Figure 2C. The I338/I333 ratio remained almost constant at low polymer concentration and increased sharply when the polymer concentration reached its CAC. The CAC values determined from the graphs were 3.23, 1.91 and 0.27Yg/ml for PEG2-PCL3.5-TPPBr, PEG2-PCL3.8-TPPBr and PEG2-PCL7.0-TPPBr miktoarm polymers, respectively. The decrease in the CAC values with increasing PCL molecular weight at a given PEG molecular weight is consistent with other reports, and suggests increased micellar thermodynamic stability against dilution.55, 56
Physical stability of the CoQ10-containing micelles
CoQ10/PEG2-PCL3.8-TPPBr micelles with a CoQ10 content of 10 weight% were kept at 4 oC and their hydrodynamic diameter was determined by DLS as a function of storage time. The micelles were also visually inspected for signs of precipitation/aggregation. As shown in Figure 2D, no noticeable change in micelle size was observed and micelle diameter of about 40 nm was maintained for more than 3 months. Moreover, no signs of drug precipitation/micelle aggregation were observed during this period. This confirms micelle stability upon storage at 4 °C.
CoQ10 photostability
CoQ10 is known to be susceptible to degradation when exposed to UV irradiation, heat or oxygen. 43, 57 CoQ10 incorporated in PEG2-PCL3.8-TPPBr micelles ([CoQ10] = 0.1–0.7 mg/ml), as well as CoQ10 solution in a mixture of ethanol-acetone 1:1 v/v ([CoQ10] = 0.5 mg/ml) were exposed to UV irradiation (δ = 254 nm) at ambient temperature, and the residual CoQ10 content was determined by HPLC. As illustrated in Figure 4C, CoQ10 concentration in ethanol-acetone mixture rapidly decreased upon exposure to UV irradiation and less than 2% CoQ10 remained after 40 min. In contrast, CoQ10 incorporated in the micelle at different concentrations was much more resistant to UV degradation. For instance, after 40 min UV irradiation of the micelle with a CoQ10 concentration of 0.5 mg/ml, around 25% CoQ10 remained intact. To determine the reaction rate constant (K) and half-life (T0.5) of CoQ10 degradation, the data was fitted to zero and first order kinetics. CoQ10 degradation was found to follow first order kinetics as indicated by its higher correlation coefficient (R2) compared to that of zero order kinetics (Table 2, supplementary information). For the same CoQ10 concentration of 0.5 mg/ml, the degradation rate constant was more than three-fold higher for CoQ10 solution compared to micelle-incorporated CoQ10 (Table 2, supplementary information). This was also reflected by more than 3-fold increase in the T0.5 for the micelle-entrapped CoQ10 (Table 2, supplementary information). The enhanced stability of micelle-incorporated CoQ10 could be attributed to the entrapment of the drug in the hydrophobic micelle core which isolates CoQ10 from the aqueous environment. Moreover, the polymeric nanoparticles could act as a barrier preventing the UV rays from reaching the incorporated drug. Stability of micelle-entrapped CoQ10 was also dependent on the CoQ10 concentration in solution: higher CoQ10 concentrations had lower degradation rates (Figure 4C, Table 2, supplementary information). UV lamps used in this study has constant intensity of UV radiations which degrade certain amount of CoQ10 during a given time period. This results in lower degradation rates for higher CoQ10 concentrations. Other nanoparticulate formulations have also shown higher CoQ10 stability against degradation by UV irradiation compared to CoQ10 solution in capryliccapric triglycerides oil or dispersion in sodium dodecyl sulfate. 43, 57
Localization of miktoarm nanocarrier in mitochondria and biological activity of CoQ10 in stressed microglia
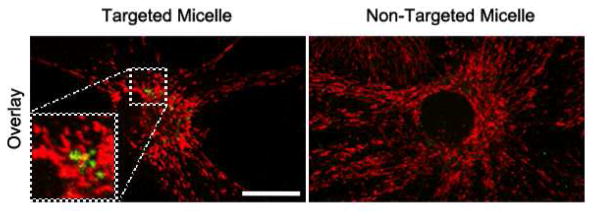
The first aim in biological experiments was to show that miktoarm polymer-targeted to mitochondria indeed reaches this intracellular site. To this end, we synthesized, characterized and tested in living cells micelles made of FITC-labeled miktoarm analog. Neural primary cultures were exposed to fluorescent micelles (1 μM, 3 h) and live cell imaging using confocal microscope was performed (Figure 5 and supplemental Figure 1). Analyses of z-stacks collected from confocal microscopy with cells co-labeled with Mitotracker 633, indicated partial co-localization (yellow fluorescence in Figure 5, see zoomed inset) of the carrier (green) with mitochondria (deep red). Micelles with non-covalently bound FITC were used as controls. Measurements of the overlap coefficients suggest consistently an increased co-localization (10–30%) of the micelles with mitochondria for the targeted micelles.
Figure 5.
Confocal micrographs of primary hippocampal neurons and glia cells treated with FITC-PEG2-PCL3.8-TPPBr (1 μM, 3 h, green fluorescence) or FITC-PEG2-PCL3.8-Br (1 μM, 3 h, green fluorescence) and Mitotracker 633 (100 nM, 3 min, deep red fluorescence). More details provided in the Supplementary section.
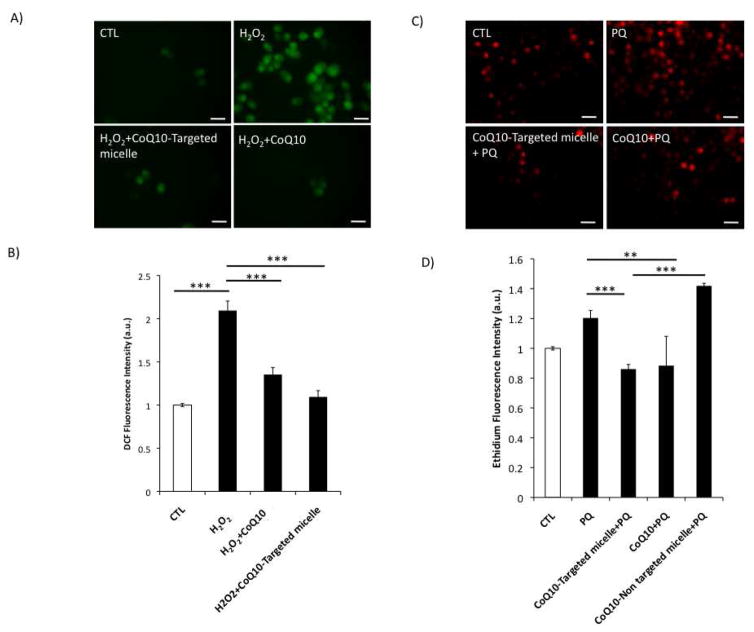
We subsequently investigated the effects of nanocarriers on mitochondrial metabolic activity using MTT assay (Supplemental Figure 2) and generation of reactive oxygen species (Figures 6A–D). Results from the MTT assay showed that targeted micelles containing CoQ10 as well as CoQ10 alone in low micromolar concentrations (< 5 YM) enhance mitochondrial metabolic activity within 24 h. This increase is significant when compared with untreated cells or those treated with the carrier without the drug. In contrast, high concentrations of polymeric micelles and CoQ10 impaired mitochondrial activity (Supplemental Figures 3–4). It is well known that there is an optimal therapeutic window for each drug which differs in different cell types. We have attempted to find this therapeutic window in neural cells of interest, and it turned out to be in relatively low concentration ranges (less than 5 uM), a desirable feature of the described nanosystem containing CoQ10. An excess of CoQ10 (as well as higher carrier concentrations) is deleterious to microglia because an excessive matrix uptake of CoQ10 can depolarize mitochondria.
Figure 6.
Production of reactive oxygen species (ROS), (A) Fluorescent micrographs showing ROS generation following H2O2 exposure. (B) Spectrofluorometric detection and quantification of ROS. (C) Fluorescent micrographs showing superoxide anion generation. (D) Spectrofluorometric detection and quantification of ethidium fluorescence intensity. More details are provided in the Supplementary section.
We next examined if CoQ10, liberated from the nanodelivery system, can reduce the impairment of mitochondria caused by H2O2 in primary hippocampal cultures. Confocal micrographs of neural cells (neurons and glia) treated with CoQ10 prior to the exposure to H2O2 (1 mM, 40 min) showed that CoQ10 treatment was effective, as measured by the fluorescence intensity of TMRE. TMRE fluorescence was consistently stronger in the presence of CoQ10 as compared to H2O2 insult alone suggesting the effectiveness of CoQ10 to maintain normal mitochondrial potential (Figure 7).
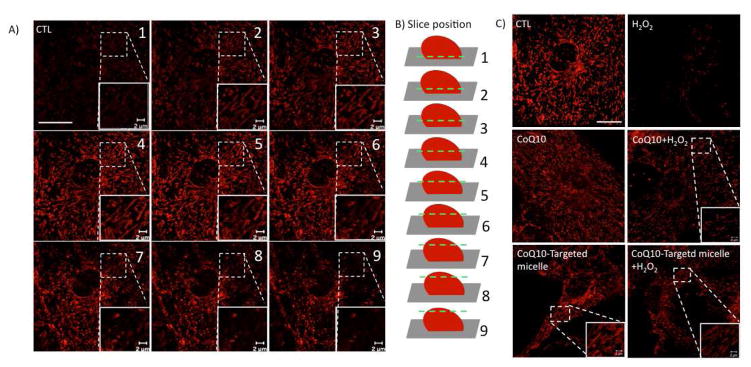
Figure 7.
Mitochondrial membrane potential measured by TMRE in H2O2 stressed cells is partially retained upon treatment with CoQ10 alone or released from the nanodelivery system. Primary hippocampal neurons and glia were treated as described in the Materials and methods section. A) Z-Stack confocal micrographs of 9 confocal sections for control cells (CTL) treated only with TMRE (100 nM, 30 min) taken at an interval of 0.3 μm. Insets represent the enlarged area (zoomed) for each corresponding section. Scale bar (20 μm) in control is representative for all panels. Scale bar (2 μm) is representative for all zoomed areas. B) Schematic illustrating the position along the Z-axis with respect to sections shown in A (numbered 1–9). C) Confocal micrograph of live cells showing loss in mitochondrial membrane potential following H2O2 exposure (1 mM, 40 min) using TMRE (100 nM, 30 min) that was partially retained in cells pretreated with CoQ10 (1 μM, 24 hr) and CoQ10-Targeted micelle (1 μM, 24 hr) compared to untreated control (CTL). Each image represents a Z-stack consisting of 6–9 confocal sections taken at an interval of 0.3 μm. Scale bar (20 μm) in control is representative for all panels. Scale bar (2 μm) is representative for all zoomed areas. Images were acquired using HeNe (543 nm) excitation laser.
To prove that micelles prepared from miktoarm-carrying CoQ10 retained the drug biological activity within the low micromolar concentrations, we used three different stress paradigms: (1) H2O2 exposure (50 YM, 24 h), (2) paraquat treatment (10 μM, 24 h) and (3) exposure to antimycin A (A.A, 1 μM, 24 h) (Figures 6B–E and supplemental Figure 2). Results from these studies clearly show that the treatment with CoQ10 with or without carrier in two stress models i.e. H2O2 treatment and exposure to paraquat, significantly reduced their impact of these insults in microglia cells, supporting previous findings that CoQ10 is an effective antioxidant agent. However, a strict control over the intracellular concentration range is required to achieve optimal mitochondrial protection and retention of physiological functions 13, 58. Exposure of microglia cells to antimycin A (A.A, 1 μM, 24 h) led to a significant reduction in mitochondrial potential as measured by TMRE fluorescence (200 nM, 30 min). CoQ10 treatment alone and CoQ10 from the targeted micelle, corrected this mitochondrial impairment, whereas CoQ10 in non-targeted micelles was ineffective.
CONCLUSIONS
We have developed an easy and efficient way of constructing multifunctional miktoarm polymer-based nanocarriers for the delivery of CoQ10, using a combination of click chemistry with ring-opening polymerisation. Results from our studies demonstrate that miktoarm polymers form micelles in an aqueous medium and are able to incorporate extraordinarily large quantities of CoQ10, not commonly seen with other carrier systems. CoQ10 micelles are suitable means of drug delivery to mitochondria, exerting beneficial antioxidant effects in insulted neural cells. The described miktoarm-based polymers provide versatile and widely applicable nanocarriers to overcome several CoQ10 limitations. In addition, they can be exploited for other novel site-directed nanodelivery systems and accommodate different pharmacological agents, individually or in combination. The latter is of particular interest for further pharmacological interventions in pathologies where multiple factors contribute to the mitochondrial impairment.
Supplementary Material
Acknowledgments
AK and DM thank Natural Sciences and Engineering Research Council (NSERC) of Canada and Canadian Institutes for Health Research (CIHR) for financial assistance. The authors also thank Mr. Sebastien Boridy for carrying confocal microscopy analyses.
Footnotes
Details of the synthesis of targeted and non-targeted miktoarm polymers, solubility and Flory-Huggins interaction parameters, zero and first order kinetics for the degradation, figures for the localization in mitochondria, mitochondrial metabolic activity, fluorescent micrographs showing mitochondrial membrane potential, confocal micrographs for non covalently bound FITC, and a detailed caption of Figure 6 in the manuscript, are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Moreira PI, Zhu X, Wang X, Lee H-g, Nunomura A, Petersen RB, Perry G, Smith MA. Biochim Biophys Acta, Mol Basis Dis. 2010;1802:212–220. doi: 10.1016/j.bbadis.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamada Y, Harashima H. Adv Drug Delivery Rev. 2008;60:1439–1462. doi: 10.1016/j.addr.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Moreira PI, Santos MS, Oliveira CR. Antioxid Redox Signaling. 2007;9:1621–1630. doi: 10.1089/ars.2007.1703. [DOI] [PubMed] [Google Scholar]
- 4.Finkel T. Nat Rev Mol Cell Biol. 2005;6:971–976. doi: 10.1038/nrm1763. [DOI] [PubMed] [Google Scholar]
- 5.Brewer GJ, Wallimann TW. J Neurochem. 2000;74:1968–1978. doi: 10.1046/j.1471-4159.2000.0741968.x. [DOI] [PubMed] [Google Scholar]
- 6.Moreira PI, Harris PLR, Zhu XW, Santos MS, Oliveira CR, Smith MA, Perry G. J Alzheimers Dis. 2007;12:195–206. doi: 10.3233/jad-2007-12210. [DOI] [PubMed] [Google Scholar]
- 7.Yang XF, Yang Y, Li G, Wang JZ, Yang ES. J Mol Neurosci. 2008;34:165–171. doi: 10.1007/s12031-007-9033-7. [DOI] [PubMed] [Google Scholar]
- 8.Weissig V. Pharm Res. 2011;28:2633–2638. doi: 10.1007/s11095-011-0588-1. [DOI] [PubMed] [Google Scholar]
- 9.Villalba JM, Parrado C, Santos-Gonzalez M, Alcain FJ. Expert Opin Invest Drugs. 2010;19:535–554. doi: 10.1517/13543781003727495. [DOI] [PubMed] [Google Scholar]
- 10.Wolf DE, Hoffman CH, Trenner NR, Arison BH, Shunk CH, Linn BO, McPherson JF, Folkers K. J Am Chem Soc. 1958;80:4752–4752. [Google Scholar]
- 11.Littarru G, Tiano L. Mol Biotechnol. 2007;37:31–37. doi: 10.1007/s12033-007-0052-y. [DOI] [PubMed] [Google Scholar]
- 12.Aberg F, Appelkvist E-L, Dallner G, Ernster L. Arch Biochem Biophys. 1992;295:230–234. doi: 10.1016/0003-9861(92)90511-t. [DOI] [PubMed] [Google Scholar]
- 13.Somayajulu M, McCarthy S, Hung M, Sikorska M, Borowy-Borowski H, Pandey S. Neurobiol Dis. 2005;18:618–627. doi: 10.1016/j.nbd.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Weissig V. Pharm Res. 2011;28:2657–2668. doi: 10.1007/s11095-011-0556-9. [DOI] [PubMed] [Google Scholar]
- 15.Boddapati SV, D’Souza GGM, Erdogan S, Torchilin VP, Weissig V. Nano Lett. 2008;8:2559–2563. doi: 10.1021/nl801908y. [DOI] [PubMed] [Google Scholar]
- 16.D’Souza GGM, Wagle MA, Saxena V, Shah A. Biochim Biophys Acta, Bioenerg. 2011;1807:689–696. doi: 10.1016/j.bbabio.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Soliman GM, Sharma R, Choi AO, Varshney SK, Winnik FM, Kakkar AK, Maysinger D. Biomaterials. 2010;31:8382–8392. doi: 10.1016/j.biomaterials.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 18.Gaucher G, Dufresne M-H, Sant VP, Kang N, Maysinger D, Leroux J-C. J Controlled Release. 2005;109:169–188. doi: 10.1016/j.jconrel.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 19.Babin J, Taton D, Brinkmann M, Lecommandoux S. Macromolecules. 2008;41:1384–1392. [Google Scholar]
- 20.Khanna K, Varshney S, Kakkar A. Polym Chem. 2010;1:1171–1185. [Google Scholar]
- 21.Soliman GM, Sharma A, Maysinger D, Kakkar A. Chem Commun. 2011;47:9572–9587. doi: 10.1039/c1cc11981h. [DOI] [PubMed] [Google Scholar]
- 22.Kolb HC, Finn MG, Sharpless KB. Angew Chem, Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Franc G, Kakkar AK. Chem Soc Rev. 2010;39:1536–1544. doi: 10.1039/b913281n. [DOI] [PubMed] [Google Scholar]
- 24.Lallana E, Sousa-Herves A, Fernandez-Trillo F, Riguera R, Fernandez-Megia E. Pharm Res. 2011:1–34. doi: 10.1007/s11095-011-0568-5. [DOI] [PubMed] [Google Scholar]
- 25.Khanna K, Varshney S, Kakkar A. Macromolecules. 2010;43:5688–5698. [Google Scholar]
- 26.Hourani R, Jain M, Maysinger D, Kakkar A. Chem–Eur J. 2010;16:6164–6168. doi: 10.1002/chem.201000241. [DOI] [PubMed] [Google Scholar]
- 27.Gao H, Matyjaszewski K. J Am Chem Soc. 2007;129:6633–6639. doi: 10.1021/ja0711617. [DOI] [PubMed] [Google Scholar]
- 28.Hansen CM. J Paint Technol. 1967;39:104–17. [Google Scholar]
- 29.Mahmud A, Patel S, Molavi O, Choi P, Samuel J, Lavasanifar A. Biomacromolecules. 2009;10:471–478. doi: 10.1021/bm800846a. [DOI] [PubMed] [Google Scholar]
- 30.Yan J, Ye Z, Chen M, Liu Z, Xiao Y, Zhang Y, Zhou Y, Tan W, Lang M. Biomacromolecules. 2011;12:2562–2572. doi: 10.1021/bm200375x. [DOI] [PubMed] [Google Scholar]
- 31.Danquah M, Fujiwara T, Mahato RI. Biomaterials. 2010;31:2358–2370. doi: 10.1016/j.biomaterials.2009.11.081. [DOI] [PubMed] [Google Scholar]
- 32.Hourani R, Sharma A, Kakkar A. Tetrahedron Lett. 2010;51:3792–3795. [Google Scholar]
- 33.Kim B-S, Park SW, Hammond PT. ACS Nano. 2008;2:386–392. doi: 10.1021/nn700408z. [DOI] [PubMed] [Google Scholar]
- 34.Wei X, Gong C, Gou M, Fu S, Guo Q, Shi S, Luo F, Guo G, Qiu L, Qian Z. Int J Pharm. 2009;381:1–18. doi: 10.1016/j.ijpharm.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 35.Murphy MP, Smith RAJ. Adv Drug Delivery Rev. 2000;41:235–250. doi: 10.1016/s0169-409x(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 36.Richter A, Olbrich C, Krause M, Kissel T. Int J Pharm. 2010;389:244–253. doi: 10.1016/j.ijpharm.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, Zhu J, Chen T, Lin S, Cai C, Zhang L, Zhuang Y, Wang X-S. Biomaterials. 2009;30:108–117. doi: 10.1016/j.biomaterials.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Boddapati SV, Tongcharoensirikul P, Hanson RN, D’Souza GGM, Torchilin VP, Weissig V. J Liposome Res. 2005;15:49–58. doi: 10.1081/lpr-64958. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Li CX, Li XC, Yuan Z, An YL, He BL. J Appl Polym Sci. 2001;80:1976–1982. [Google Scholar]
- 40.Nederberg F, Appel E, Tan JPK, Kim SH, Fukushima K, Sly J, Miller RD, Waymouth RM, Yang YY, Hedrick JL. Biomacromolecules. 2009;10:1460–1468. doi: 10.1021/bm900056g. [DOI] [PubMed] [Google Scholar]
- 41.Stojkovic M, Westesen K, Zakhartchenko V, Stojkovic P, Boxhammer K, Wolf E. Biol Reprod. 1999;61:541–547. doi: 10.1095/biolreprod61.2.541. [DOI] [PubMed] [Google Scholar]
- 42.Ankola DD, Viswanad B, Bhardwaj V, Ramarao P, Kumar MNVR. Eur J Pharm Biopharm. 2007;67:361–369. doi: 10.1016/j.ejpb.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Kwon SS, Nam YS, Lee JS, Ku BS, Han SH, Lee JY, Chang IS. Colloids Surf, A. 2002;210:95–104. [Google Scholar]
- 44.Amorim CdM, Couto AG, Netz DJA, de Freitas RA, Bresolin TMB. Nanomed Nanotech Biol Med. 2010;6:745–752. doi: 10.1016/j.nano.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Dwan’Isa JPL, Rouxhet L, Preat V, Brewster ME, Arien A. Pharmazie. 2007;62:499–504. [PubMed] [Google Scholar]
- 46.Patel SK, Lavasanifar A, Choi P. Biomacromolecules. 2009;10:2584–2591. doi: 10.1021/bm900512h. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Xiao Y, Allen C. J Pharm Sci. 2004;93:132–143. doi: 10.1002/jps.10533. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Li Y, Liu L, Sun Q, Shuai X, Zhu W, Chen Y. Biomacromolecules. 2010;11:1331–1338. doi: 10.1021/bm100116g. [DOI] [PubMed] [Google Scholar]
- 49.Swarnakar NK, Jain AK, Singh RP, Godugu C, Das M, Jain S. Biomaterials. 2011;32:6860–6874. doi: 10.1016/j.biomaterials.2011.05.079. [DOI] [PubMed] [Google Scholar]
- 50.Hsu C-H, Cui Z, Mumper RJ, Jay M. AAPS PharmSciTech. 2003;4:269–280. doi: 10.1208/pt040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y, Zhang Y, Fan Z, Li S. J Polym Sci, Part B: Polym Phys. 2010;48:286–293. [Google Scholar]
- 52.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 53.Jeong Y-I, Cheon J-B, Kim S-H, Nah J-W, Lee Y-M, Sung Y-K, Akaike T, Cho C-S. J Controlled Release. 1998;51:169–178. doi: 10.1016/s0168-3659(97)00163-6. [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Zhu B, Dong T, Pan P, Shuai X, Inoue Y. Macromol Biosci. 2008;8:1116–1125. doi: 10.1002/mabi.200800085. [DOI] [PubMed] [Google Scholar]
- 55.Francis MF, Lavoie L, Winnik FM, Leroux J-C. Eur J Pharm Biopharm. 2003;56:337–346. doi: 10.1016/s0939-6411(03)00111-5. [DOI] [PubMed] [Google Scholar]
- 56.Nagarajan R, Ganesh K. Macromolecules. 1989;22:4312–4325. [Google Scholar]
- 57.Bule MV, Singhal RS, Kennedy JF. Carbohydr Polym. 2010;82:1290–1296. [Google Scholar]
- 58.McCarthy S, Somayajulu M, Sikorska M, Borowy-Borowski H, Pandey S. Toxicol Appl Pharmacol. 2004;201:21–31. doi: 10.1016/j.taap.2004.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.