Abstract
Importance
HER2 is an important drug target in breast cancer, where anti-HER2 therapy has been shown to lead to improvements in disease recurrence and overall survival. HER2 status in head and neck squamous cell carcinoma (HNSCC) has not been well studied. Identification of HER2 positive tumors and characterization of response to HER2 therapy could lead to targeted treatment options in HNSCC.
Objective
To identify HER2 aberrations in HNSCCs and investigate potential for HER2 targeted therapy in HNSCCs.
Design, Setting, and Participants
Retrospective case series of patients with laryngeal and oral cavity SCC enrolled in the University of MichiganSPORE. Publically available sequencing data(TCGA) was reviewed to identify additional mutations and overexpression in HER2 in HNSCC. Established HNSCC cell lines were used for follow-up in vitro analysis.
Interventions
Using targeted, amplicon-based sequencing with the Oncomine Cancer Panel, we assessed the copy number and mutation status of commonly altered genes in HNSCCs. Immunohistochemical staining was performed on tissue microarrays of HNSCCs to assess expression of HER2. Western blotting for HNSCC cell line HER2 expression, and cell survival assays after treatment with HER2 inhibitors were performed.
Main Outcomes and Measures
Prevalence of HER2 genetic aberrations and HER2 overexpression in laryngeal and oral cavity squamous cell carcinomas (SCCs). Prevalence of HER2 aberrations in HNSCC in TCGA. HER2 protein expression in HNSCC cell lines. Response of HNSCC cell lines to targeted HER2 inhibitors.
Results
Forty-two laryngeal SCC samples were screened by targeted sequencing, of which 4 were positive for HER2 amplification. Two samples identified with sequencing showed HER2 overexpression on immunohistochemistry. Two of 94 oral cavity SCC samples were positive for HER2 on immunohistochemistry. Analysis of 288 patients from publicly available HNSCC sequencing data revealed 9 amplifications in HER2. Protein expression was variable across HNSCC cell lines, and a subset of these cell lines show responsiveness to anti-HER2 therapy.
Conclusions and Relevance
HER2 aberrations are identified in a subset of HNSCCs. These tumors may be responsive to targeted therapy against HER2. Screening for HER2 aberrations and applying targeted therapy in HER2 positive patients may provide a useful tool for personalized therapy trials, particularly in patients that are refractory to current treatment paradigms.
Keywords: HNSCC, HER2, ERBB2, neu, amplification, mutant
BACKGROUND
Prognosis and cure rates for advanced head and neck squamous cell carcinomas (HNSCC) remains poor, and survival remains unchanged, particularly in laryngeal squamous cell carcinoma (LSCC)1,2. Current treatment options for advanced HNSCC include combination therapies (surgery, radiation, and/or chemotherapy), and have not changed for years. Moreover, limited treatment options exist for recurrent or metastatic disease. New targeted tumor therapy options, while widely employed in other cancer treatments, are limited in HNSCC.
Human epidermal growth factor receptor 2 (HER2, or ERBB2) is a member of the EGFR family of transmembrane receptor tyrosine kinases intricately involved in cell proliferation and growth. HER2 and other receptors in this family (EGFR, ERBB3, and ERBB4) initiate signaling via homo- and heterodimerization, resulting in activation of downstream MAPK, PIK3CA/AKT, and STAT pathways3. Overexpression of HER2 leads to an increased rate of dimerization, particularly with EGFR, and increased downstream signaling for cell growth and proliferation. HER2 has been shown to be amplified in approximately 15–30% of breast cancers and 10–30% of gastric and esophageal cancers4. Additionally, amplifications in HER2 have been identified in bladder, ovarian, endometrial, pancreatic, and non-small cell lung cancers5.
Historically, HER2 amplification portended a worse prognosis in breast cancer patients, with worse overall and recurrence-free survival6. Prognosis in these patients has subsequently improved largely due to the advent of targeted therapy against HER27,8. Currently, small molecule inhibitors or antibodies targeting HER2 are approved for treatment in HER2 positive breast, gastroesophageal, and non-small cell lung cancers5,9–11. To date, there have been few studies fully characterizing HER2 amplifications and HER2 overexpression in HNSCC12–16. Aberrations in HER2 are a potentially attractive targeted therapy for HNSCC given its important interactions with EGFR via heterodimerization, and their common downstream pathways. Thus, identification and characterization of HER2 positive HNSCCs could lead to potential targetable treatment options for subsets of patients with HER2 positive HNSCCs refractory to current standard of care.
METHODS
Tissue Collection
This study was approved by the University of Michigan Institutional Review Board. Forty-two LSCC and 94 oral cavity squamous cell carcinoma (OSCC) tumor specimens were identified from patients enrolled in the University of Michigan Head and Neck Specialized Program of Research Excellence (SPORE). Patients gave written consent and tumor tissue was collected in the SPORE tissue repository. Patient information, including demographic information, treatments rendered, and patient outcomes were recorded. Specific tissue microarrays (TMAs) comprised of LSCC and OSCC specimens were constructed.
Sequencing of Laryngeal Samples
Using targeted, amplicon-based sequencing with the Oncomine Cancer Panel17, we assessed the copy number and mutation status of several common therapeutic targets in our LSCC samples, including HER2, and EGFR. Amplicon based DNA sequencing and data analysis was performed using 40 ng of isolated DNA using the RNA/DNA formalin-fixed paraffin-embedded isolation kit (Qiagen, Valenica, CA) on the Ion Torrent Personal Genome Machine, utilizing the AmpliSeq Comprehensive Cancer Panel as previously described18. Nucleotide variants and indels were identified using the Torrent Variant Caller plugin, annotated using Annovar,19 and filtered to include candidate somatic mutations by removing germline variants and sequencing artifacts using in-house validated pipelines18. Briefly, called variants were first filtered to remove synonymous variants, as well as those without adequate read support. The variants were filtered such that flow-corrected read depths <20, flow-corrected variant allele-containing reads <6, variant allele frequencies <0.10, and skewed variant read support (i.e. the difference in the number of forward vs. reverse reads containing the variant allele) >5-fold were removed from the variant set, as previously described18. Copy number alterations were identified clinically relevant at >|2| copies, as previously described18,20, using normalized, GC content corrected, total read counts per amplicon from each sample divided by those from a composite “normal” sample consisting of multiple single and pooled normal male DNA samples. Gene-level (e.g. HER2) copy number estimates were determined by taking the coverage-weighted mean of the per-probe ratios, with expected error determined by the probe-to-probe variance, as previously described20.
Immunohistochemistry of Laryngeal and Oral Cavity Tissue Microarray Samples
Immunohistochemical staining for HER2 overexpression was performed on LSCC and OSCC TMA samples. Briefly, the formalin-fixed paraffin-embedded sections from the TMAs were heated, and underwent peroxidase blocking. A HER2 rabbit monoclonal antibody (SP3; Cell Marque, Rocklin, CA) was applied at a 1:150 dilution. A FLEX + Rabbit EnVision System (Dako, Carpinteria, CA) was used for staining. The TMAs were then counterstained with Harris Hematoxylin. Protein expression was scored by a board-certified pathologist in a blinded fashion, and samples with 3+ or 4+ scoring were counted as positive samples, in accordance with current national guidelines21. Additionally, staining and scoring for EGFR overexpression was performed in a similar fashion on the TMA samples.
Analysis of Publically Available HNSCC Sequencing Data
The Cancer Genome Atlas (TCGA) has collected mutation and copy number variation data on 288 HNSCC tumors22. Additionally, we evaluated HNSCC exome sequencing data from Stansky et al (74 patients) and Agrawal et al (32 patients)23,24. We screened for prevalence of amplifications and mutations in HER2, EGFR, ERBB3, and ERBB4 in HNSCC patients, and mRNA levels of HER2 and EGFR.
Western Blotting
The University of Michigan has created multiple cell lines from primary HNSCC tumors. Head and neck cancer cell lines from this repository (UM-SCC) were analyzed for HER2 protein expression. We selected 11 cell lines: UM-SCC-1,-2, -14A, -23, -49, -59, -92, -97, -103, -108, and -110. UM-SCC-23 is derived from LSCC, with the remaining cell lines derived from OSCCs. Briefly, cells were harvested and lysed in RIPA buffer. Ten micrograms of each cell harvest were used and standard western blot protocols were followed. Primary antibodies against β-actin (1:1000, Cat. #4970, Cell Signaling Technology, Danvers, MA), EGFR (1:1000, Cat. #TA312545, Origene, Rockville, MD), and HER2 (1:500, Cat. #4290, Cell Signaling) were incubated overnight at 4 C, followed by a goat anti-rabbit HRP (Cat. #111-035-045, Jackson ImmunoResearch, West Grove, PA) secondary antibody at room temperature for one hour. The blots were then visualized with chemilumiescence and imaged.
HER2 Inhibition and Cell Survival Assays
The UM-SCC cell lines listed above were seeded at 2,000 cells per well in a 384-well plate format in DMEM containing 10% FBS using an automated cell sorter. Cells were allowed to attach and return to growth phase for 20–24 hours prior to dosing. HER2 tyrosine kinase inhibitors (AEE788, BMS599626, CP724714, and TAK285) were prepared from a stock concentration of 10 mM in sterile DMSO, and diluted in cell growth media to working concentrations. Each dose was performed in quadruplicate. At the 60-hour time point, 10 μL of 100 μg/mL resazurin in PBS was added to the plates. Plates were read for cell viability at 72 hours after dosing at 540nm/612nm using a BioTek Cytation 3 plate reader. DMSO-only wells were used as the treatment control, and camptothecin was used as a positive control for cell death. Cell proliferation plots were then constructed using Prism software (GraphPad Software, Inc., La Jolla, CA).
RESULTS
HER2 Amplification in Laryngeal Cancer Specimens
Of the 42 samples collected on the laryngeal TMA, 4 (9.5%) were positive for HER2 amplification on sequencing, with log2 estimated copy number amplification ranging from 2.4 to 9.0 (Supplemental Table I; Figure 1). We also screened for other commonly amplified receptor tyrosine kinases in these patients via copy number analysis (Supplemental Table I). One sample had significant amplification of EGFR in addition to HER2. The remaining three samples also had copy number gains in EGFR, but to a lesser degree (<2.0 copy number amplification ratio). Additional highly amplified or deleted genes were identified and recorded (Figure 1). Overall, we identified similar copy number variations in targetable receptor tyrosine kinases (EGFR, HER2, and FGFR1; Supplemental Table II) in our larynx cohort (n = 42) as has been reported in TCGA LSCC specimens (n = 72). We also collected data on mutations in genes frequently mutated in HNSCC or associated with carcinogenesis in the four HER2 amplified LSCC specimens (Supplemental Table III). Notably, missense mutations in NOTCH1 were identified in all four patients with HER2 amplifications. This finding is in concordance with the high frequency of NOTCH1 mutations in HNSCC that has been previously reported23,24.
Figure 1. Copy number analysis of HER2 positive laryngeal cancer samples demonstrates unique profiles.
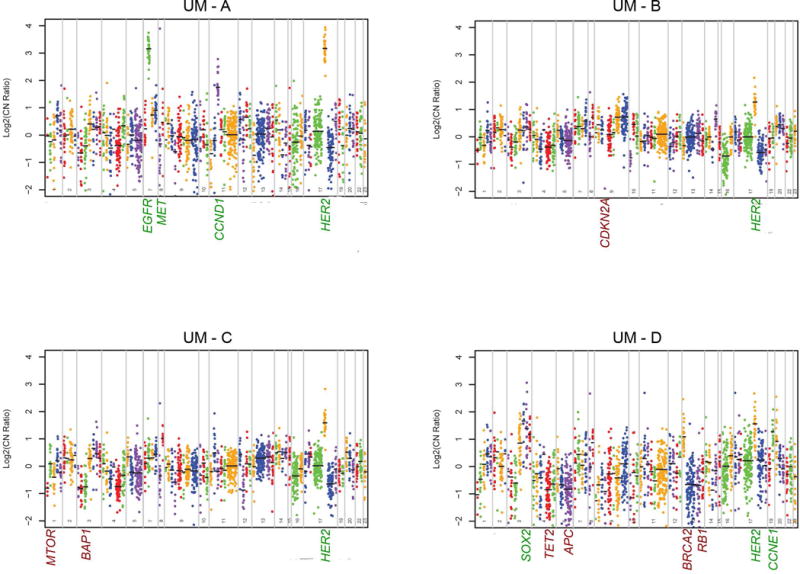
Genes from the oncomine comprehensive cancer panel were assessed for relative copy number by Ion Torrent sequencing. Genes are plotted along the x-axis beginning with chromosome 1 and ending with the X chromosome. Each color represents probe sets for an individual gene and each point represents an individual probe. Only statistically differential genes are highlighted with amplified genes in green text and depleted genes in red text. Log change in copy number is shown on the y-axis.
HER2 Immunohistochemistry of Laryngeal and Oral Cavity Specimens
Two of the 4 LSCC samples identified with sequencing to have amplifications of HER2 were scored to be positive for HER2 overexpression (3+) on immunohistochemistry (Patients 1 and 2; Figure 2A). The remaining 2 samples that showed amplification on sequencing did not show overexpression of HER2 on immunohistochemistry. Two of the OSCC samples demonstrated HER2 overexpression on immunohistochemistry (Patients 3 and 4; Figure 2B). Additionally, we confirmed that the HER2-positive samples stained strongly for EGFR overexpression (3+), as was previously assessed in this cohort25,26.
Figure 2. HER2 immunohistochemistry demonstrates protein overexpression in HNSCC.
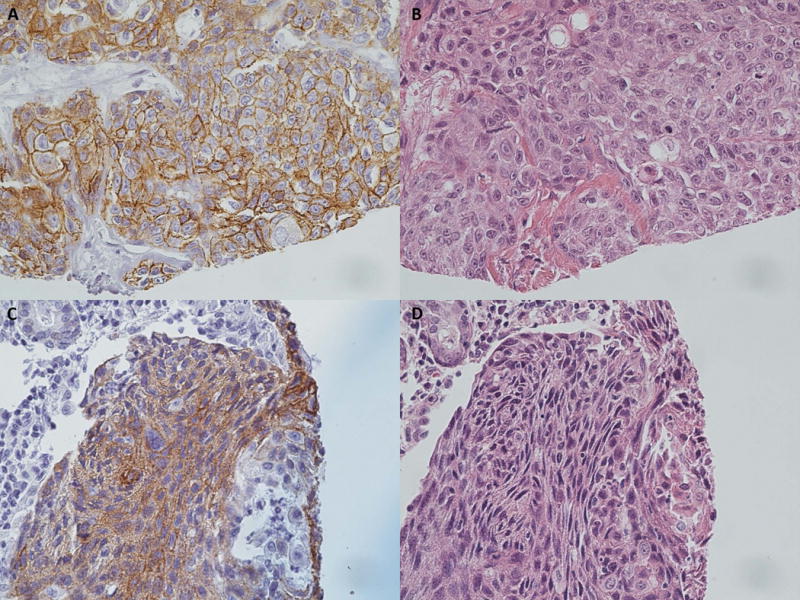
HER2 overexpression is demonstrated in laryngeal SCC (A), and oral cavity SCC (C) samples. Corresponding H & E stains were performed (B, D). Representative images of positive cases were used to demonstrate staining.
HER2 Positive Patient Characteristics
Demographic and treatment information was obtained from the patients whose samples were used for the TMAs (Supplemental Table IV). Clinical information on the four HER2 positive patients on immunohistochemistry was recorded. Patient 1 was a smoker who developed a T2N0M0 SCC of the glottis at age 41, initially treated with radiation. He developed a recurrence 6 years later, but delayed treatment. He eventually underwent salvage total laryngectomy 4 years later, and ultimately died. Patient 2 was a smoker who developed a T4N0M0 SCC of the glottis at age 57, initially treated with a right hemilaryngectomy. He developed a recurrence, and was treated with salvage total laryngectomy and chemoradiation. He is deceased. Patient 3 developed a T2N0M0 SCC of the floor of mouth at age 49, treated with local resection and bilateral selective neck dissections. He is alive and disease-free as of his last office visit 8 years postoperatively. Patient 4 developed a T4N2bM0 SCC of the oral tongue at age 26. She underwent subtotal glossectomy, bilateral selective neck dissections, and free tissue reconstruction. She was unable to complete postoperative chemoradiation due to social issues, and developed a recurrence 7 months later, and eventually underwent chemoradiation, before passing away.
HER2 Amplifications and Mutations in The Cancer Genome Atlas
We next analyzed publicly available HNSCC sequencing data for HER2 alterations22–24. In this data set, 288 samples with documented copy number variations and 385 samples with mutational data were analyzed. We identified 9 (3.1%) HER2 amplifications in HNSCC from these patients, with estimated copy number ranging from 4.1 to 14 (Supplemental Table I, Figure 3A). Additionally, we searched for other HER2 mutations in HNSCC. We identified 11 missense mutations, and 1 nonsense mutation (Figure 3B, D). Seven of the missense mutations were predicted to be possibly or probably damaging to HER2 function based on analysis with the PolyPhen-2 bioinformatic tool27 (Supplemental Table V). We next screened for mutations and amplifications in other EGFR family members (EGFR, ERBB3, ERBB4). Amplifications were identified in EGFR, more frequently than in HER2. No amplifications were seen in ERBB3 or ERBB4. Mutations were identified in EGFR, ERBB3, and ERBB4 (Figure 3B). RNA expression analysis of HER2 and EGFR demonstrated elevated levels of EGFR RNA in a subset of patients with elevated HER2 RNA levels, as well as a subset of patients with elevated EGFR RNA levels in these HNSCC samples (Figure 3C). We identified a HER2 amplified fusion gene product with STARD3, with the first 3 exons from HER2 (noncoding exons), and the remaining exons from STARD3. STARD3 is primarily reported as a cholesterol transport gene in the same chromosomal amplicon as HER2, but it has been implicated in growth and survival of HER2 positive cancers28.
Figure 3. HER2 expression in The Cancer Genome Atlas HNSCC data sets.
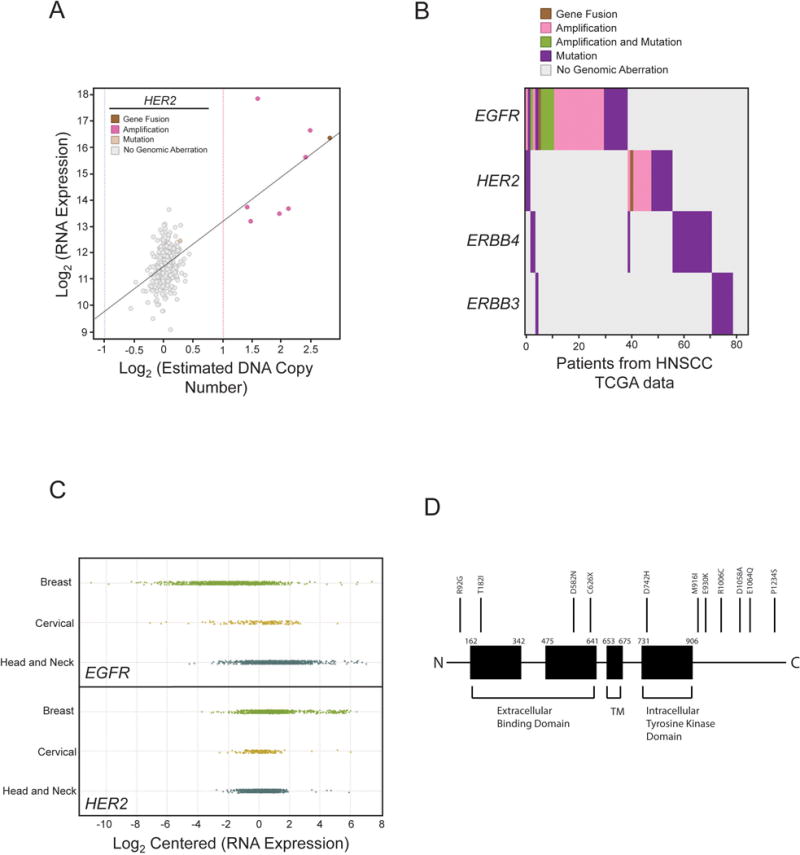
HER2 amplifications are identified in specific HNSCC samples. (A) Using the TCGA data, copy number was plotted along the x-axis on a log scale such that points (which each represent individual patients) to the right of the dotted red line have statistically significant increases in HER2 copy number. RNA overexpression of HER2 is also identified in these HNSCC samples and plotted along the y-axis. (B) Using the TCGA data, we stratified patients along the x-axis by disruptive genomic events in each of the 4 ERBB genes. Color represents the type of genomic aberration as indicated. (C) Comparison of RNA expression of HER2 and EGFR across Breast, Cervical Squamous Cell and Head and Neck Squamous Cell Carcinomas. (D) Mutations in HER2 and other members of the EGFR family are also identified (B, D) and the distribution of mutations in HER2 is shown in the schematic.
HER2 Expression Levels in Head and Neck Squamous Cell Carcinoma Lines
We next assessed for HER2 expression in UM-SCC cell lines. HER2 expression levels were variable across the cell lines. EGFR expression levels were assessed in these cell lines as well, and demonstrated significant variability in expression levels. Notably, UM-SCC-1 had higher HER2 levels in comparison to the other cell lines (Figure 4).
Figure 4. HER2 protein expression in HNSCC cell lines.
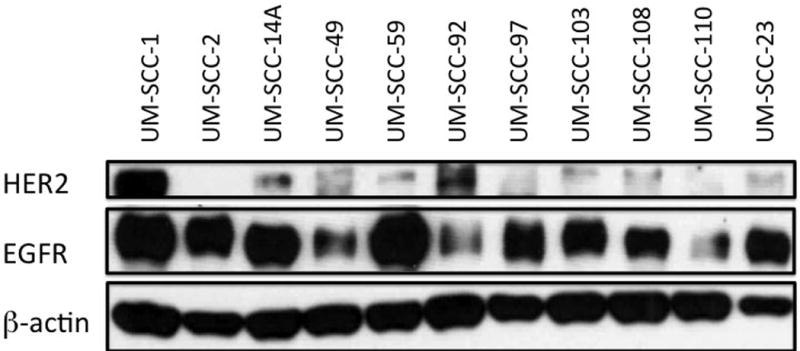
HER2, EGFR levels across multiple OSCC (UM-SCC-1,-2, -14A, -23, -49, -59, -92, -97, -103, -108, -110) and LSCC (UM-SCC-23) cell lines. Overexpression of HER2 is seen in UM-SCC-1 in comparison to the remaining cell lines. β-actin is used to demonstrate overall protein level normalization.
Head and Neck Squamous Cell Carcinoma Line Response to HER2 Inhibitors
We next tested these UM-SCC cell lines for sensitivity to HER2 tyrosine kinase inhibitors currently being assessed in clinical trials. We identified a subset of these cell lines with sensitivity to HER2 inhibition. UM-SCC-1 and UM-SCC-110, and to a lesser extent UM-SCC-23, demonstrated sensitivity to HER2 inhibitors in vitro (AEE788, BMS599626, CP724714, and TAK285; Figure 5A–D; Supplemental Table VI). Notably, CP724714 is a HER2-specific inhibitor, while the other inhibitors do confer some inhibition to EGFR in combination with HER2 inhibition.
Figure 5. Response of HNSCC cell lines to HER2 inhibitors.
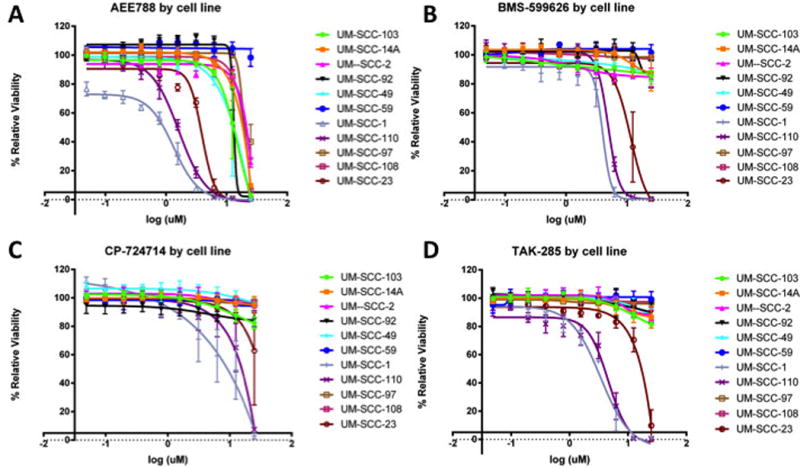
Percent relative viability is plotted on the y-axis, with log10 scales of inhibitor (in μM) on the x-axis. A subset of UM-SCC cell lines demonstrate sensitivity to tyrosine HER2 tyrosine kinase inhibitors. In our panel of cell lines, UM-SCC-1 and -110, and to a lesser extent UM-SCC-23, demonstrate sensitivity to HER2 inhibitors currently in clinical trials (A–D). CP-724714 (C) is a HER2-specific inhibitor, while the remaining inhibitors do have some combination inhibition of EGFR as well. Notably, UM-SCC-1 has significantly higher HER2 expression, and appears to be the most responsive of the UM-SCC cell lines.
DISCUSSION
We are entering into an era with rapid advancements in personalized tumor treatments. Tumor genomes are increasingly being screened, and personalized targeted therapy is actively being investigated for use in clinical care. Currently in HNSCC, targeted therapy has been limited to cetuximab, with early clinical trials testing additional potential drug targets29. In breast and gastro-esophageal cancers, targeted therapy against HER2-amplified tumors has been demonstrated to be successful. The role of HER2 aberrations in HNSCC has not been well investigated. Here we describe amplifications in HER2 and overexpression of HER2 in a subset of LSCC and OSCC specimens. Furthermore, we show in vitro response of some HNSCC cell lines to HER2 inhibitors. Notably, one cell line with significantly increased HER2 expression (UM-SCC-1) demonstrates increased sensitivity to HER2 inhibitors relative to the other UM-SCC lines. Interestingly, another cell line with lower expression levels of HER2 and EGFR (UM-SCC-110) demonstrated some response to HER2 inhibitors in vitro, suggesting targeted inhibitors of HER2 may have broader implications than just HER2-overexpressing tumors.
Targeted next generation sequencing of laryngeal tumor specimens in this study was successful in identifying amplifications in HER2. Multiple proto-oncogenes may be screened simultaneously with this technique, potentially making sequencing a useful high-throughput tool when compared to standard screening assays (in situ hybridization and immunohistochemistry). Of note, we identified four samples positive for HER2 gene amplification on sequencing whereas only two were identified by immunohistochemistry. This can potentially be ascribed to tumor heterogeneity for HER2 expression, which has been well described in literature30,31. It remains unclear whether these HER2 heterogeneous tumors are responsive to anti-HER2 therapy31. Additionally, gene amplification from sequencing studies does not always correlate with RNA and protein expression22. Nevertheless, next-generation sequencing offers a potentially highly sensitive and high-throughput tool for detecting HER2 amplifications in combination with other potentially targetable genes. Confirmation should be performed with established immunohistochemistry protocols.
It is important to clarify the amplification and expression status of additional receptor tyrosine kinases, such as EGFR, in patients with HER2 amplifications. HER2 overexpression is known to cause increased activation of EGFR. Additionally, tumors with EGFR and HER2 amplifications express a more aggressive phenotype and have a worse prognosis32. The two proteins closely interact, forming receptor heterodimers to activate common downstream signaling pathways, and each gene may be integrally involved in development of resistance to targeted treatment against the other. For instance, HER2 amplification has been implicated in acquired resistance to targeted EGFR therapy33,34. Moreover, amplified EGFR status has been shown to lead to decrease response in HER2 positive tumors treated with Herceptin (an anti-HER2 antibody)35. Thus, targeting these patients with dual inhibitor therapy against both EGFR and HER2 may be more effective than with inhibition against either gene alone. In our in vitro data, we confirm that dual inhibitors of EGFR and HER2 are modestly effective in a subset of HNSCC cell lines. Further investigation into combination therapies may provide a means to identify increasingly effective targeted therapy paradigms in HNSCC.
Characterization of point mutations in HER2 may be of interest in HNSCC samples. Patients with nonsense mutations in HER2 will likely be unresponsive to anti-HER2 therapy given the truncated gene product. The response of patients with missense mutations in HER2 is more difficult to predict, as they can be silent, inhibiting or activating mutations36. Additionally, these point mutations could lead to altered interaction and signaling between HER2 and its co-receptors, particularly EGFR. Such mutations would only be identified by sequencing, and would not be picked up by standard immunohistochemistry staining techniques for HER2, thus providing additional benefit for sequencing as a screening tool.
Notably, we have identified a living patient from our cohort who is HER2 positive. This patient is currently free of disease, but should he develop recurrent disease, he could potentially benefit from targeted HER2 therapy as clinical trials are being established to provide targeted therapeutics to recurrent and advanced cancers37. Interestingly, a recent cancer exome sequencing/targeted therapy trial identified a HER2 amplification in a metastatic urothelial carcinoma (another cancer not known to harbor HER2 amplifications)38. Treatment with trastuzumab (an anti-HER2 antibody) in this patient resulted in complete clinical response and regression of liver and lung metastases, providing added rationale for the potential for anti-HER2 therapy in HNSCCs with HER2 aberrations.
Identification of HER2 aberrations in HNSCC is of importance, as these tumors have the potential to be responsive to targeted therapy against HER2. The use of targeted anti-HER2 therapies in HNSCC has not been established, and only recently have agents against HER2 and related targets been expanded to include HNSCC39,40. The potential addition of anti-HER2 drugs may be particularly beneficial for organ preservation protocols for HNSCC, or in patients with advanced, metastatic, or refractory disease. Establishing the role of HER2 amplifications and HER2 overexpression in HNSCC patient response to anti-EGFR therapy will also be important, as resistant patients with HER2 aberrations may be treated successfully with combination therapy. There is potential in the future of HER2 in HNSCC, particularly in personalized medicine trials: screening for HER2 aberrations and targeting HER2 in these patients either as monotherapies or in combination regimens may provide a useful and successful clinical tool for personalized therapy, particularly in patients refractory to current treatment paradigms.
Supplementary Material
Acknowledgments
We would like thank Chia-Jen Liu, MS, Daniel Hovelson, MS, Andi Cani, MS, and Scott Tomlins, MD, PhD from the Department of Pathology, University of Michigan, for technical contributions to NGS and analysis. We thank C.P. for critically reading the manuscript. No compensation was received.
GRANT SUPPORT AND FUNDING
Partial support provided by The University of Michigan Anatomic Pathology Projects Grant.
Dr. A.C.B. is a research fellow funded on an NIH T-32 Training Grant (T32 DC005356). Drs. T.E.C., C.R.B, M.E.S. and J.C.B received funding support from NIH Grant U01DE025184.
Footnotes
FINANCIAL DISCLOSURE/CONFLICT OF INTEREST
The authors have no financial relationships relevant to this article to disclose. There are no conflicts of interest.
PRESENTATION
This study was accepted for presentation as a poster at the 2015 AHNS Translational Research Meeting in Boston, MA, April 21–22, 2015.
AUTHOR CONTRIBUTIONS
Drs. Birkeland and Brenner had full access to data in the study and are responsible for the integrity and accuracy of the data.
Study concept and design: Birkeland, Spector, Brenner
Acquisition, analysis or interpretation of data: Birkeland, Yanik, Tillman, Scott, Foltin, Mann, Michmerhuizen, Ludwig, Sandelski, Komarck, McHugh, Spector, Brenner
Drafting of manuscript: Birkeland, Spector, Brenner
Critical revision of the manuscript for important intellectual content: all authors
Statistical analysis: Birkeland, Spector, Brenner
Obtained funding: Brenner
Administrative, technical, or material support: Carey, Prince, Bradford, McHugh, Spector, Brenner
Study supervision: Carey, Prince, Bradford, Spector, Brenner
References
- 1.Hoffman HT, Porter K, Karnell LH, et al. Laryngeal cancer in the United States: changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116:1–13. doi: 10.1097/01.mlg.0000236095.97947.26. [DOI] [PubMed] [Google Scholar]
- 2.Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncologist. 2010;15:994–1001. doi: 10.1634/theoncologist.2009-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal N, Iqbal N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol Biol Int. 2014;2014:852748. doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan M, Parker BA, Schwab R, et al. HER2 aberrations in cancer: implications for therapy. Cancer Treat Rev. 2014;40:770–80. doi: 10.1016/j.ctrv.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 7.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 8.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–72. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 9.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 10.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 11.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–34. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 12.Falchook GS, Lippman SM, Bastida CC, et al. Human epidermal receptor 2-amplified salivary duct carcinoma: regression with dual human epidermal receptor 2 inhibition and anti-vascular endothelial growth factor combination treatment. Head Neck. 2014;36:E25–7. doi: 10.1002/hed.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Campo JM, Hitt R, Sebastian P, et al. Effects of lapatinib monotherapy: results of a randomised phase II study in therapy-naive patients with locally advanced squamous cell carcinoma of the head and neck. Br J Cancer. 2011;105:618–27. doi: 10.1038/bjc.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nardi V, Sadow PM, Juric D, et al. Detection of novel actionable genetic changes in salivary duct carcinoma helps direct patient treatment. Clin Cancer Res. 2013;19:480–90. doi: 10.1158/1078-0432.CCR-12-1842. [DOI] [PubMed] [Google Scholar]
- 15.Williams MD, Roberts DB, Kies MS, et al. Genetic and expression analysis of HER-2 and EGFR genes in salivary duct carcinoma: empirical and therapeutic significance. Clin Cancer Res. 2010;16:2266–74. doi: 10.1158/1078-0432.CCR-09-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanken H, Gaudin R, Grobe A, et al. Her2 expression and gene amplification is rarely detectable in patients with oral squamous cell carcinomas. J Oral Pathol Med. 2014;43:304–8. doi: 10.1111/jop.12173. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warrick JI, Hovelson DH, Amin A, et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Arch. 2014 doi: 10.1007/s00428-014-1699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grasso C, Butler T, Rhodes K, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff AC, Hammond ME, Kicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford CR, Kumar B, Bellile E, et al. Biomarkers in advanced larynx cancer. Laryngoscope. 2014;124:179–87. doi: 10.1002/lary.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandarana SP, Lee JS, Chanowski EJ, et al. Prevalence and predictive role of p16 and epidermal growth factor receptor in surgically treated oropharyngeal and oral cavity cancer. Head Neck. 2013;35:1083–90. doi: 10.1002/hed.23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahlberg KK, Hongisto V, Edgren H, et al. The HER2 amplicon includes several genes required for the growth and survival of HER2 positive breast cancer cells. Mol Oncol. 2013;7:392–401. doi: 10.1016/j.molonc.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz S, Ang KK, Vermorken J, et al. Targeted therapies for squamous cell carcinoma of the head and neck: current knowledge and future directions. Cancer Treat Rev. 2014;40:390–404. doi: 10.1016/j.ctrv.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Sapino A, Goia M, Recupero D, et al. Current Challenges for HER2 Testing in Diagnostic Pathology: State of the Art and Controversial Issues. Front Oncol. 2013;3:129. doi: 10.3389/fonc.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oakman C, Sapino A, Marchio C, et al. Chemotherapy with or without trastuzumab. Ann Oncol. 2010;21(Suppl 7):vii, 112–9. doi: 10.1093/annonc/mdq283. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch FR, Varella-Garcia M, Cappuzzo F. Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene. 2009;28(Suppl 1):S32–7. doi: 10.1038/onc.2009.199. [DOI] [PubMed] [Google Scholar]
- 33.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–33. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Castanaro C, Luan B, et al. ERBB3/HER2 signaling promotes resistance to EGFR blockade in head and neck and colorectal cancer models. Mol Cancer Ther. 2014;13:1345–55. doi: 10.1158/1535-7163.MCT-13-1033. [DOI] [PubMed] [Google Scholar]
- 35.Diermeier S, Horvath G, Knuechel-Clarke R, et al. Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation. Exp Cell Res. 2005;304:604–19. doi: 10.1016/j.yexcr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–37. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NCI prepares to launch MATCH trial. Cancer DIscov. 2015;5:685. No authors listed. [Google Scholar]
- 38.Beltran H, Eng K, Mosquera JM, et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1:466–74. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garner AP, Bialucha CU, Sprague ER, et al. An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin. Cancer Res. 2013;73:6024–35. doi: 10.1158/0008-5472.CAN-13-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai CJ, Bao R, Tao X, et al. CUDC-101, a multitargeted inhibitor of histone deacetylase, epidermal growth factor receptor, and human epidermal growth factor receptor 2, exerts potent anticancer activity. Cancer Res. 2010;70:3647–56. doi: 10.1158/0008-5472.CAN-09-3360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


