Abstract
The UbiA superfamily of intramembrane prenyltransferases catalyzes a key biosynthetic step in the production of ubiquinones, menaquinones, plastoquinones, hemes, chlorophylls, vitamin E, and structural lipids. These lipophilic compounds serve as electron and proton carriers for cellular respiration and photosynthesis, as antioxidants to reduce cell damage, and as structural components of microbial cell walls and membranes. This article reviews the biological functions and enzymatic activities of representative members of the superfamily, focusing on the remarkable recent research progress revealing that the UbiA superfamily is centrally implicated in several important physiological processes and human diseases. Because prenyltransferases in this superfamily have distinctive substrate preferences, two recent crystal structures are compared to illuminate the general mechanism for substrate recognition.
Keywords: Prenyltransferases, intramembrane enzymes, UBIAD1, COQ2, quinones, structural lipids
The UbiA superfamily of intramembrane prenyltransferases
Many essential bioactive compounds must be prenylated to become soluble and functional in biological membranes [1–3]. The prenylation reaction, which generates the basic skeleton of these molecules, is catalyzed by intramembrane prenyltransferases, collectively known as the UbiA superfamily. Since the discovery of the prototype UbiA enzyme [4], a number of superfamily members, producing a broad spectrum of lipophilic compounds, have been identified [2,3].
The UbiA superfamily of prenyltransferases (Figure 1) has recently stimulated much scientific interest because this superfamily is involved in a wide variety of biological processes and diseases (Table 1, Key Table). The microbial UbiA and MenA [3], and their respective eukaryotic homologs COQ2 [5,6] and UBIAD1 [7], are key biosynthetic enzymes of ubiquinones and menaquinones, respectively. In humans, mutations in COQ2 cause infantile multisystem disease [8–13]. UBIAD1 maintains vascular homeostasis by promoting endothelial cell survival [14], prevents oxidative damage in cardiovascular tissues [15], and sustains mitochondrial function [16], while dysfunctional UBIAD1 has been linked to cardiovascular degeneration [14,15], Parkinson’s disease [16], Schnyder corneal dystrophy [17,18], and urologic cancers [19]. Other superfamily members include DPPR synthase, which is essential for the cell wall formation in mycobacteria [20]; DGGGP synthase, which generates the distinctive core structure of archaeal membranes [21]; and eukaryotic COX10, which produces hemes for terminal oxidases in the mitochondrial respiration chain [22]. In plants, there are chlorophyll synthase; homogentisate prenyltransferases, which synthesize plastoquinones and vitamin E; and many other prenyltransferases that generate a large variety of secondary metabolites [2,3]. In sum, the UbiA superfamily plays a crucial role in the cellular respiration, photosynthesis, antioxidation, and structural maintenance of almost all living organisms.
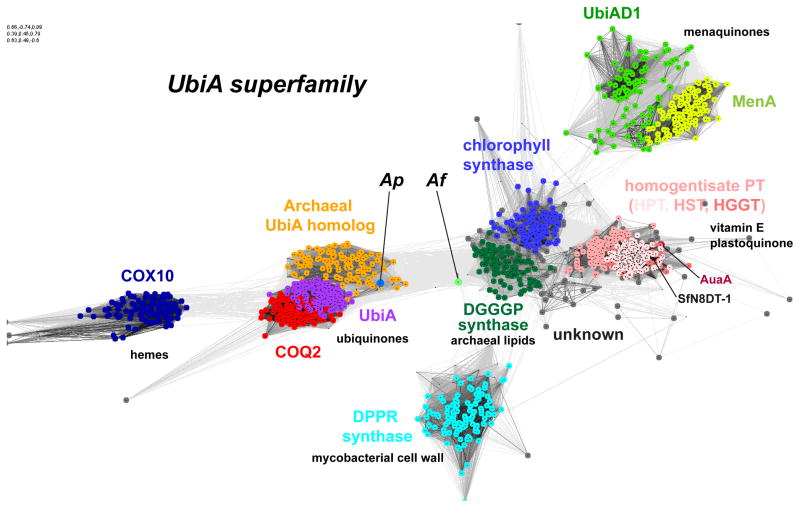
Figure 1. Homology clustering and function of UbiA superfamily prenyltransferases.
Each dot represents a homolog sequence and each grey line shows the PSI-BLAST [95] comparison of two sequences, with darker lines indicating higher similarity (lower E values). Homologs of significant similarity (e.g., all COQ2 proteins) form clusters (shown in same color). The UbiA superfamily includes CoQ2 (colored in red) and UbiA (magenta); UBIAD1 (green) and MenA (yellow green); homogentisate prenyltransferases (HPT, HST, and HGGT in different pink colors); chlorophyll synthase (blue); COX10 (dark blue); DGGGP synthase (dark green); and DPPR synthase (cyan). The Ap homolog represents an archaeal group (orange) close to UbiA and COQ2. The position of the Af homolog (annotated as bacteriochlorophyll synthase in NCBI) is relatively distinct from functionally characterized groups of the superfamily. Other members with unknown functions are shown as grey dots. The sequence clustering was generated by the CLANS (CLuster ANalysis of Sequences) program [96] and PSI-BLAST searches, which were carried out in an NCBI protein database of non-redundant proteins filtered for 70% maximum sequence identity (nr70). The search started from the following sequences: UbiA (NCBI GI: 85676792) and MenA (332345926) from Escherichia coli; the ApUbiA homolog (14601492) from Aeropyrum pernix; COQ2 (14250676), UBIAD1 (7019551), and COX10 (13623563) from Homo sapiens; the Af homolog (499181604) from Archaeoglobus fulgidus; chlorophyll synthases (332645327) and HPT (281193026) from Arabidopsis thaliana; HGGT (526117663) from Vitis vinifera; HST (699979766) from Medicago sativa; DPPR synthase (618730398) from Mycobacterium tuberculosis; DGGGP synthase (2495885) from Methanocaldococcus jannaschii; naringenin 8-dimethylallyltransferase (SfN8DT-1; 403399456) from Sophora flavescens; and AuaA (for aurachins; 133919228) from Stigmatella aurantiaca.
Table 1.
The UbiA superfamily of intramembrane prenyltransferases a.
| Subfamilies | Prenyl acceptors |
Prenyl donors |
End metabolic products |
Biological functions b |
Occurrence | Diseases and applications |
|
|---|---|---|---|---|---|---|---|
| PHB prenyl-transferases | UbiA | PHB | XPP | Ubiquinones | Respiration chain Antioxidant |
Proteobacteria | Chemoenzymatic synthesis |
| CoQ2 | Eukaryotic mitochondria | Infantile multi- system diseases | |||||
| UbiA homolog | Unknown | Likely XPP |
Unknown | Unknown | Many archaea, bacteria | Not studied | |
| LepGT1 | PHB | GPP | Shikonin | Secondary metabolite | L. erythrorhizon | Antibacterial, antiinflammatory | |
| DHNA prenyl-transferases | MenA | DHNA | XPP | Menaquinones Phylloquinones | Respiration, photosynthesis | Most bacteria, archaea | New Mtb drugs |
| UBIAD1 | Menadione Phyllo-quinonec | GGPP | MK-4 | Lipid metabolism b | Human mitochondria | Schnyder corneal dystrophy | |
| Cell signaling b | Human | Urologic cancers | |||||
| Respiration chain | Drosophila mitochondria | Parkinson’s disease | |||||
| Antioxidant | Zebrafish | Cardiovascular degeneration | |||||
| DPPR synthase | pRpp | DP | DPA | Cell wall formation | Coryne-bacterianeae | New Mtb drugs | |
| DGGGP synthase | GGGP | GGPP | Glycerol diether and tetraether lipids | Core lipids of archaeal membranes | Euryarchaeota | Lipid technologies | |
| IPGP c | XPP derived | Thaumarchaeota, crenarchaeota, euryarchaeota | |||||
| Heme O synthase | HOS, COX10 | Protoheme IX (Heme B) | FPP | Heme O, A | Cofactor of terminal oxidases | Eukaryotes | Mitochondrial diseases |
| CyoE, CtaB | Aerobic microbes | ||||||
| Chlorophyll synthase | Chlorophyl-lide | PhyPP, GGPP | Chlorophyll | Photosynthesis | Plants, photo-synthetic bacteria | Agriculture | |
| Homo-gentisate phytyl- and prenyl-transferases | HPT | homogentisic acid | PhyPP | Tocopherols | Antioxidants Nutrients |
Plants Cyanobacteria | Plant metabolic engineering Nutrition |
| HGGT | GGPP | Tocotrienols | |||||
| HST | SPP | Plastoquinones | Photosynthesis Respiration |
||||
| Prenyl-transferases in secondary metabolism (incomplete list) | HlPTs | Acylphloro-glucinols, etc. | DMAPP | Bitter Acid | Phytoalexins, alkaloids, etc. | Hop | Agriculture Natural drugs |
| SfN8DT SfG6DT G4DT |
Genistein, Naringenin, pterocarpan. | Usually DMAPP | Prenylated flavonoids isoflavonoids |
S. flavescens Legume |
|||
| AuaA | MHQ | FPP | Aurachins | S. aurantiaca | |||
| Pyr6 | HPPO | FPP | Pyripyropene A | A. fumigatus | |||
Abbreviations: XPP, isoprenyl diphosphate of various lengths; GGPP, geranylgeranyl diphosphate; FPP, farnesyl diphosphate; PhyPP, phytyl diphosphate; SPP, solanesyl diphosphate; pRpp, phosphoribose diphosphate; DP, decaprenyl phosphate; GGGP, geranylgeranylglyceryl phosphate; IPGP, isoprenylglyceryl phosphate of variable lengths; SfN8DT, naringenin 8-dimethyallyltransferase; G4DT, 4-dimethylallyltransferase; SfG6DT, genistein 6-dimethylallyltransferase; MHQ, 2-methyl-4-hydroxyquinoline. HPPO, 4-hydroxy-6-(3-pyridinyl)-2H-pyran-2-one.
The biological functions are referred to the prenyltransferases, or if unspecified, to the end metabolic products.
The mechanisms remain unclear.
Most UbiA superfamily enzymes catalyze the prenylation of aromatic substrates (Figure 2). The archetypal enzyme, UbiA, fuses an isoprenyl chain to the meta-position of p-hydroxybenzoate (PHB; Figure 2A and Box 1) [4,23]. MenA recognizes 1,4-dihydroxy-2-naphthoic acid (DHNA) and couples prenylation with the decarboxylation of DHNA (Figure 2B) [24]. UBIAD1 is similar to MenA, but probably has a weak activity of cleaving off the phytyl tail of phylloquinone and replacing it with an isoprenyl tail (Figure 2C) [7]. DPPR synthase uses a ribose moiety (Figure 2D) as the prenyl acceptor [20], while for DGGGP synthase the acceptor is a linear substrate (Figure 2E) [25]. COX10 and chlorophyll synthase fuse prenyl or phytyl tails to acceptors that have large porphyrin rings (Figure 2F and 2G) [22,26,27]. Homogentisate phytyl- and prenyl- transferases decarboxylate homogentisic acid and attach different tails (Figure 2H) [28]. Overall, prenyltransferases in the UbiA superfamily can be distinguished by their distinct preferences for structurally diverse substrates. However, it remains unclear how these intramembrane enzymes select their prenyl donor and acceptor substrates. This review therefore discusses the novel structural mechanism of substrate specificity employed by the UbiA superfamily, as well as the biological functions, enzymatic activities, and disease involvement of representative members.
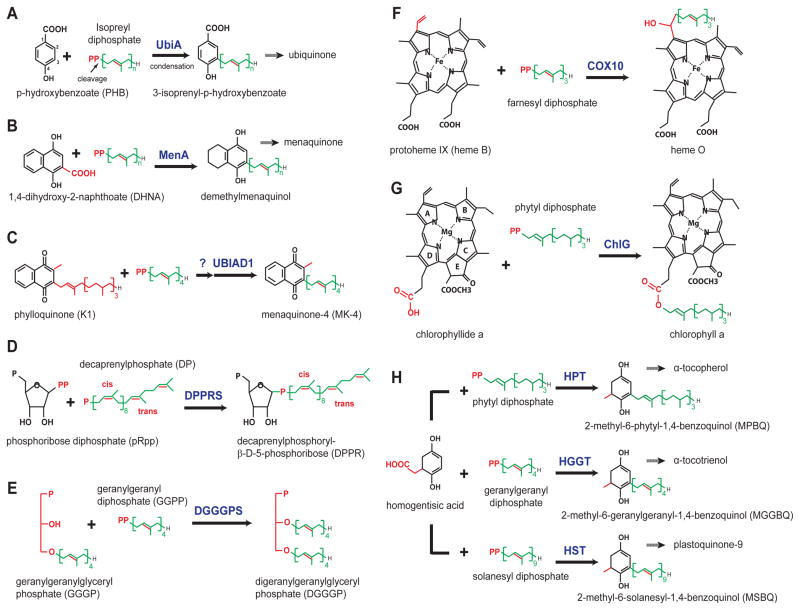
Figure 2. Reactions catalyzed by the UbiA superfamily enzymes.
Key features in different chemical structures are shown in red, and the prenyl or phytyl chains in green. (A) UbiA cleaves the diphosphate group from XPP and fuses it to the meta-position of PHB [4,23]. (B) MenA catalysis involves a decarboxylation step [24]. (C) UBIAD1 catalysis may involve side-chain exchange [7]. (D) DPPR synthase fuses a ribose to a unique prenyl substrate with several cis-bonds [20]. (E) DGGGP synthase fuses two linear substrates [25]. (F–G) COX10 and ChlG prenylate heme and chlorophyl acceptors, respectively, both of which have large porphyrin rings [22,27]. (H) Homogenisate transferases, HPT, HGGT, and HST, use different prenyl or phytyl diphosphates as donor substrates [28].
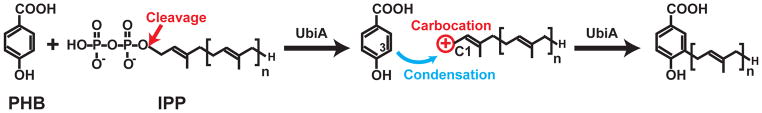
Box 1. A general catalytic mechanism of UbiA superfamily prenytransferases.
UbiA catalysis starts with the cleavage of the diphosphate group from the XPP substrate (Figure I). The cleavage generates a highly reactive carbocation intermediate at the end of the isoprenyl chain. To complete the prenylation reaction, the carbocation reacts regiospecifically at the meta-position of the aromatic PHB substrate to form a C–C bond. In this superfamily, UbiA and MenA recognize XPPs of various chain lengths, whereas several other members are specific to a certain XPP length (Figure 2 and Table 1).
PHB prenyltransferases: COQ2, UbiA and LePGT1
Ubiquinones are electron carriers that are essential for the bacterial and mitochondrial respiratory chains, and also have potent antioxidant activity that protects membranes against lipid peroxidation [1]. Ubiquinones have the ideal chemical property for these functions: the quinone ring allows reversible redox activity, and the isoprenoid side chain confers solubility in the membrane. Fusion of these two chemical groups is an evolutionally conserved and rate-limiting step during ubiquinone biosynthesis [1,23]. The reaction is catalyzed by a PHB prenyltransferase, named UbiA in prokaryotes [4] and COQ2 in eukaryotes [5,6].
COQ2 mutations (Box 2) have been identified as causative for infantile multisystem diseases and nephropathy [8–13]. In these diseases, COQ2 mutations result in primary deficiency of ubiquinone-10 (UQ10; 10 refers to the number of isoprenyl units) [13], which can be alleviated by UQ10 supplementation at an early infantile stage [29]. Fibroblasts from COQ2-mutant patients show mild UQ10 deficiency [30,31]; this medium level of deficiency however generates a maximal response of reactive oxygen species deteriorating to cells [32]. Recently, COQ2 mutations have been suggested to cause multiple system atrophy [33], a progressive neurodegenerative disorder that displays combined symptoms of autonomic dysfunction, parkinsonism, and cerebellar ataxia [34]. Although this proposal remains to be validated [35,36], the identified COQ2 mutants showed lowered enzymatic activity [33], which might in turn increase the oxidative stress in susceptible brain cells [34]. The Coq2 gene is also strongly associated with statin-induced myopathy, a drug side effect in which statin reduces the synthesis of a COQ2 substrate precursor, farnesyl diphosphate, which in turn hinders UQ10 synthesis [37].
Box 2. Structural interpretation of disease-related mutations.
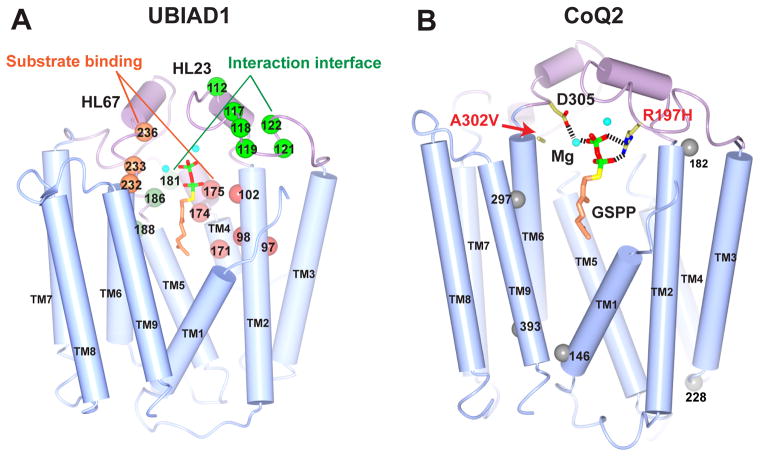
The newly determined archaeal structures offer plausible explanations for disease-linked mutations in eukaryotic UBIAD1 and CoQ2. In UBIAD1, all reported SCD mutations [17,18,52,59] are clustered around the central cavity (Figure I A). As a hot spot for SCD mutations, N102 may be involved in either substrate binding or stabilization of the carbocation intermediate [90,91]. The mutations of R119G, N232S/H, and D236E may also affect the binding of the XPP substrate. A second group of mutations, which includes A97T, G98S. S171P, Y174C, and T175I, is predicted to cluster around the proposed binding pocket for aromatic substrates [90]. Because of their surface-exposed locations (Figure I A), other mutations, including D112G/N, G117R/E, D118G, R119G, L121F, and V122E/G on HL2-3 and K181R, G186R, and L188H on HL4-5, can probably perturb interactions with proteins involved in lipid metabolism [19,52–54].
Several COQ2 mutations cause infantile multisystem diseases and nephropathy [9]. R197H may affect diphosphate binding, and the larger side chain of A302V (Figure I B) may clash with one of the Mg2+ binding sites to interfere with COQ2 catalysis [90]. However, it is unclear how the Y297C [11], N228S, S146N [12], and M182R mutations [13] affect COQ2 activity. In addition, the V393A mutation that potentially associates with multiple system atrophy [33] is not at the central cavity in this COQ2 model.
Escherichia coli UbiA (EcUbiA) is well characterized in biochemistry (Box 1). EcUbiA requires divalent metal ions, preferably magnesium, for the enzymatic activity [4,23]. The apparent Km values of PHB and geranyldiphosphate (GPP) are in the submillimolar range, whereas longer isoprenyldiphosphates (XPP; X stands for various lengths) have higher affinity [23]. EcUbiA can hydrolyze GPP in absence of PHB, which suggests that the diphosphate can be cleaved (Figure 2A) independently of the condensation reaction [38]. The prenylation of PHB is regiospecific and occurs only at the meta-position. However, EcUbiA can accept a variety of PHB derivatives, providing that the benzoate group is retained and that the para-benzene group can function as a donor for electrons and hydrogen bonds [39]. Engineered UbiA is promising for chemoenzymatic applications because it catalyzes regioselective alkylation in water, which is a challenge to conventional organic chemistry [39].
UbiA and COQ2 produce ubiquinones of various lengths that are characteristic of different organisms, with UQ6 in Saccharomyces cerevisiae, UQ8 in E. coli, UQ9 in rodents, UQ9-10 in plants, and UQ10 in humans [40–42]. The UQ chain lengths are determined by the in vivo availability to UbiA and COQ2 of different XPP substrates; UbiA and COQ2 are, however, indiscriminate with respect to chain lengths [43]. There are many examples showing that UbiA-deficient E. coli or COQ2-deficient yeast could be complemented by COQ2 or UbiA from organisms producing different UQs [5,40,44]. Similar chain-length promiscuity was observed in vitro: XPPs varying from 2 to 10 isoprenyl units were substrates for EcUbiA [23], and COQ2 from yeast and plants could also take XPPs of various lengths [44].
As an exception to the chain-length promiscuity, the PHB geranyltransferase from Lithospermum erythrorhizon (LePGT1) uses only GPP as its substrate [45]. Instead of participating in the UQ biosynthesis, LePGT1 is a key enzyme that generates shikonin, a plant secondary metabolite with antiviral activities. When chimeric constructs of EcUbiA and LePGT1 were tested, the N-terminal half of the enzymes influenced the chain-length specificity [46].
UbiAD1 and MenA
UBIAD1 is associated with many physiological processes and diseases, mostly because menaquinones, as ubiquinones, are essential in electron transport and antioxidation. Identification of UBIAD1 as the prenyltransferase for menaquinone-4 (MK4) biosynthesis [7] unravels a long-standing mystery: how vitamin K1, the dietary form of vitamin K, is converted to K2 (i.e. MK4), which is the major storage form in birds and mammals [47–49]. MK4 is required for electron transport in the Drosophila mitochondria. In Drosophila, MK4 is made by the HEIX protein, a UBIAD1 homolog; mutations in the Heix gene cause mitochondrial dysfunction and exacerbate the phenotype of defective Pink1, a mitochondrial kinase involved in human Parkinson’s disease [16]. As to antioxidation, UBIAD1 in zebrafish embryos reduces oxidative stress and cell damage in the heart and blood vessels, thereby maintaining the survival of vascular endothelial cells; loss of UBIAD1 triggers vascular degeneration, cranial hemorrhages, and cardiovascular failure [14,15]. Embryonic lethality is also observed for Ubiad1-deficient mice [50]. Therefore, UBIAD1 may play a general role in embryonic development in many species.
In humans, Ubiad1 is the causative gene of Schnyder corneal dystrophy (SCD), a rare autosomal eye disease characterized by the deposition of cholesterol and phosopholipids in the cornea [17,18]. The majority of older SCD patients require corneal transplants due to progressive loss of visual acuity [51]. SCD-causative Ubiad1 mutations (Box 2) not only reduce the MK-4 synthesis [52], but also interfere with the binding of UBIAD1 to proteins involved in cholesterol and triglyceride metabolism [19,53]. Several proteins in this pathway, ApoE [54], TBL2 [19,53], SOAT1, and HMGCR [52], were found to interact with UBIAD1. In particular, SCD mutations in UBIAD1 make the UBIAD1-HMGCR complex less dissociable, which may prevent the degradation of HMGCR and result in cholesterol accumulation [55]. Therefore, UBIAD1 may have important functions other than MK4 biosynthesis [50].
Human Ubiad1 is also referred as Tere1, the transitional epithelial response gene. In bladder and prostate cancers [56,57], Tere1 expression is substantially reduced, which in turn may activate a signaling pathway that induces cancer cell proliferation [58]. Conversely, Tere1 overexpression inhibits the growth and proliferation of cell lines derived from these urological cancers [56,57]. Tere1, or Ubiad1, therefore seems also to be implicated in cell growth and signaling [58].
In the particular cell types used in the studies here reviewed, UBIAD1 protein occupies various subcellular locations and exhibits distinct functional roles. In the endoplasmic reticulum (ER) of human bone cells [7], UBIAD1 shows a weak activity of side-chain exchange (Figure 2C). UBIAD1 in human endothelial cells participates in the Golgi synthesis of UQ10, which in turn protects the cells from oxidative stress through nitric oxide signaling [15]. How UBIAD1, a MenA homolog (Figure 1), is linked to UQ10 synthesis remains to be elucidated (M. M. Santoro, personal communication). In Drosophila, MK4 supplementation rescues severe mitochrondial deficiency induced by Heix mutations, suggesting that this UBIAD1 homolog is essential for producing MK4 to support mitochondrial electron transport [16]. In bladder cancer cell lines, UBIAD1/TERE1 synthesizes MK4 in mitochondria but also shows non-mitochondrial localization in the ER and Golgi [53]. By contrast, UBIAD1 in corneal keratocytes localizes to mitochondria but not to the ER [59]. Overall, different subcellular localizations of UBIAD1 may reflect its various functions in the respiratory chain, lipid metabolism, or antioxidation in different cell types.
Menaquinones are the quinones most widely used by microbes in their respiratory and photosynthetic electron transport chains [1]. Facultative bacteria such as E. coli use both ubiquionone and menaquinone; these quinones serve as electron carriers under aerobic and anaerobic conditions, respectively. Conversely, gram-positive bacteria, including Mycobacterium tuberculosis (Mtb) and other pathogens, use only menaquinones for electron transport [1]. Because menaquinone is essential to gram-positive bacteria, recent studies of MenA [60–62], the prenyltransferase (Figure 2B) for menaquinone biosynthesis [24], have focused on exploring MenA as an antibiotic target in Mtb and other gram-positive pathogenic bacteria. MenA inhibitors showed significant and specific activity against nonreplicating Mtb, which is particularly interesting because current first-line drugs are ineffective against latent Mtb infection [62].
Other UbiA superfamily members
DPPR synthase
Mycobacteria and other Corynebacterianeae share a characteristic, highly impermeable cell wall, in which arabinogalactan covalently tethers an inner peptidoglycan layer to an outer lipid-rich mycolic acid layer. The DPPR synthase generates an arabinogalactan precursor (Figure 2D) through catalyzing the first committed step in a decaprenylphosphoryl-D-arabinose (DPA) pathway [20]. DPPR synthase is required for the survival of Mtb [63]; disruption of this gene results in the loss of cell wall arabinan and associated mycolic acids [64]. Among genes in the DPA pathway, knockdown mutants of DPPR synthase and DprE show the strongest phenotype in Mtb [63]; DprE is the target of benzothiazinones, the so-called “magic drugs” for treating tuberculosis that have much higher efficacy than first-line drugs targeting the mycobacterial cell wall [65]. Therefore, DPPR synthase is a promising drug target that catalyzes the fusion of two specific substrates (Figure 2D) not found in humans [20,66].
DGGGP synthase
The core structure of archaeal membranes is made of glycerol diether and tetraether lipids; this is a major evolutionary feature that distinguishes archaea from bacteria and eukaryotes [21]. DGGGP synthase generates the skeleton of diether lipid [25] in an unsaturated form (Figure 2E), which is the common precursor to most diether lipids (archaeols) and tetraether lipids (caldarchaeols) in archaea. The prenyl acceptor in DGGGP synthase is a linear compound, which is an exception to the ring structures used by other prenyltransferases in the superfamily (Figure 2). To produce the macro-circular tetraether lipids, DGGGP synthase has been proposed in certain archaea species to accommodate longer DGGGP derivatives containing ring structures and ether-linked diphosphate groups [67]. DGGGP synthase’s atypical substrate specificity may be the key to understanding the biogenesis of the unique archaeal membrane lipids.
Heme O synthase (protoheme IX farnesyltransferase)
Heme A is the prosthetic group of the terminal heme-copper oxidases (e.g., the cytochrome c oxidase in mitochondria, also known as complex IV) that reduces oxygen to water in the respiration chain [22,26]. Heme A is generated by the sequential actions of heme O synthase (HOS or COX10), which attaches a farnesyl tail to protoheme IX (Figure 2F), and heme A synthase (HAS or COX15), which converts heme O to heme A in a two-step reaction. In bacteria, heme O is often directly used as a cofactor for terminal oxidases [22]. Heme O synthase has been identified and characterized in organisms throughout the tree of life, including CyoE in E. coli [68], CtaB [69] in Bacillus subtilis, and COX10 in yeasts and humans [22]. Human COX10 mutations cause mitochondrial diseases such as Leigh Syndrome [70,71] and have been suggested as a risk factor in Alzheimer disease [72].
Heme A synthesis is likely coordinated with the biogenesis of complex IV, because free hemes are cytotoxic and mitochondria lacks a heme degradation pathway [73]. A core subunit of complex IV, COX1, is stabilized by the fanesyl tail of heme A [74]. Through heme A biogenesis, COX10 may participate in the posttranslational maturation of COX1 [75].
Chlorophyll synthase
As the terminal enzyme for chlorophyll biosynthesis, chlorophyll synthase (ChlG) esterifies chlorophyllide with geranylgeranyl or phytyl diphosphate (Figure 2G) [27]. Like the COX system [26], ChlG is involved in the cotranslational insertion of chlorophyll into key proteins of photosystems (PS) I and II [76]. The attachment of chlorophylls to these nascent apoproteins assists their proper folding inside membranes and prevents the accumulation of free phototoxic chlorophylls [77]. The chlorophyll-binding PSI/II subunits accumulate only when the chlorophyll synthesis is carried via ChlG, suggesting a direct transfer of chlorophyll from ChlG to nascent apoproteins [78]. In addition, ChlG is involved in the feedback control of the entire chlorophyll biosynthesis pathway, in which reduced ChlG activity leads to the down regulation of metabolic flux [79]. Biochemically, the orientation of the chlorophyllide binding has been deduced from modified compounds, with rings C, D, and E (Figure 2G) binding to the interior of ChlG and rings A and B at the enzyme surface [27].
Homogentisate phytyl- and prenyl- transferases
In photosynthetic organisms, the branching point leading to the biosynthesis of tocopherols, tocotrienols, and plastoquinones is marked by homogentisate phytyltransferase (HPT), geranylgeranyl transferase (HGGT), and solanesyltransferase (HST), respectively (Figure 2H) [28]. Plastoquinones transfer electrons from photosystem II to the cytochrome b6f complex and shuttle protons across the photosynthetic membrane. Tocopherols and tocotrienols are a group of eight potent lipid-soluble antioxidants collectively known as vitamin E [80]. As critical enzymes in vitamin E biogenesis, HPT and HGGT have been a focus for metabolic engineering to improve the vitamin E content in transgenic plants [81].
Biochemically, homogentisate transferases differ in the specificity of their donor-substrate preferences (Figure 2H). HGGT and HPT have a strong preference for geranylgeranyl and phytyl diphosphate, respectively [82]. HST prefers a longer substrate, solanesyl diphosphate; when short XPPs are provided, the HST catalyzed prenylation is largely decoupled from decarboxylation, which suggests that short XPPs bind and react differently in HST [83].
Intramembrane prenyltransferases involved in secondary metabolism
Plants make numerous kinds of prenylated secondary metabolites, including flavonoids, phenylpropanoids, and coumarins, which have various biological functions with applications in agriculture and medicine [3,84]. Examples include bitter acids in hops, which give flavor to beers [85], and phytoalexins, which appear in both legumes [86] and Sophora flavescens [87]. In bacteria, aurachins are potent inhibitors of the respiration chain [88]. The fungal pyripyropene A is a promising cholesterol-control drug [89]. Intramembrane prenyltransferases have been identified in all these cases, but there are still many genes of unknown function (Figure 1), and together they could generate a large variety of secondary metabolites in many organisms [2].
Among this functional group of enzymes, naringenin 8-dimethylallyltransferase and genistein 6-dimethylallyltransferase recognize flavanone and isoflavone, respectively [84]. Chimeric proteins have been generated to show that the fifth predicted transmembrane helix of these enzymes determines the specificity for these two closely related aromatic substrates [87].
Structural insights into the UbiA superfamily
Crystal structures of archaeal homologs
Crystal structures have recently been determined [90,91] for archaeal homologs from two hyperthermophilic species, Aeropyrum pernix (Ap) and Archaeoglobus fulgidus (Af). On the basis of sequence clustering (Figure 1) and alignment (Figure 3A), the Ap homolog is close to UbiA and COQ2, whereas the Af homolog seems to be relatively distant from known members of the superfamily. The enzymatic activities of these archaeal homologs are unknown, as are their cognate native substrates. Despite the uncertainties, both crystal structures were determined with and without short XPP substrates bound. At 2.4Å resolution, the Af structure is of particularly high accuracy, and shows confirmed positions of bound Mg2+ ions in the active site. Together, these structures significantly advanced understanding of the enzymatic mechanism used by this superfamily of enzymes.
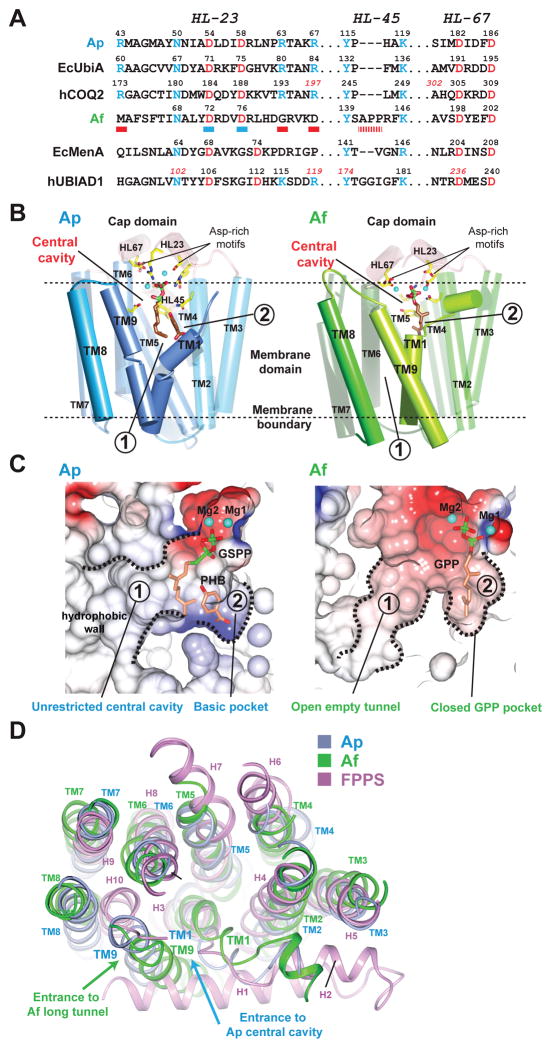
Figure 3. Sequence and structural comparison of the Ap and Af homologs illuminates the mechanism for substrate recognition.
(A) Alignment of conserved sequence motifs. The Ap homolog retains key residues (two Asp-rich motifs colored in red and other residues in blue) in UbiA/COQ2. The Af homolog lacks some conserved residues (red bars underneath the sequence) found in UbiA/COQ2 and differs from MenA/UBIAD1 [91] in the first Asp-rich motif (blue bars; DX3D in Af vs. DX5D in MenA and UBIAD1). (B) Crystal structures of the archaeal homologs. The Ap (PDB: 4OD5) [90] and Af (PDB: 4TQ3) [91] structures are shown in the same orientation, with the cap domain colored in pink and the TM regions of Ap and Af in blue and green, respectively. Locations of the two substrate-binding pockets in each enzyme are indicated by circled numbers. The positions of TM1 and TM9 (with highlighted colors) are notably different in Ap and Af. This difference creates alternative openings of pocket ➀ to lipids (see C), which is between TM9 and TM1 in Ap and between TM9 and TM8 in Af. (C) Substrate-binding pockets in electrostatic surface representation. In Ap structure, the central cavity is directly opened to lipid without restriction; in Af structure, the GPP substrate binds in a small pocket that restricts the chain length. (D) Structure comparison. The Ap (blue), Af (green), and FPPS (PDB: 1RQI; purple) [97] structures are superimposed.
The overall structure of the Ap UbiA homolog [90] contains nine transmembrane helices (TM) that form a U-shaped architecture with a large central cavity (Figure 3B, left). An extramembrane cap domain is formed over the central cavity and contains most of the conserved residues (Figure 3A), including two Asp-rich motifs that are essential for the activity of various enzymes in the superfamily [45,46,66,88,90–92]. The non-cleavable XPP analog, geranyl thiolodiphosphate (GSPP), binds in the central cavity via an extensive interaction network of conserved residues. Both Asp-rich motifs coordinate Mg2+ ions that engage the diphosphate group of GSPP. The large central cavity also contains a hydrophobic bottom wall and a small basic pocket, which were proposed to bind the isoprenyl chain and an aromatic substrate, respectively [90]. For EcUbiA and LepGT1, the affinity for PHB changes when residues at this basic pocket are mutated [46,90]. Location of this putative binding pocket is also consistent with the finding that residues in TM5 determine the specificity for aromatic substrates [87]. The central cavity opens laterally to the lipid bilayer (Figure 3B and 3C), creating a unique passage to the active site that may facilitate the binding and release of long-chain substrates and products in membranes, respectively. This unrestricted binding chamber also explains the chain-length promiscuity of UbiA, which accepts a variety of XPP lengths to generate UQ6-10 in different species.
The overall architecture of the Af homolog [91] is similar to Ap (Figure 3B, right). However, the two substrate-binding pockets are significantly different (Figure 3C). In the Af structure, GPP or dimethylallyl diphosphate (DMAPP) binds to a small pocket, which is restricted by the protein backbone (Figure 3C, right) and is not capable of accommodating longer XPPs. Af also has a significantly longer tunnel coming off the active site, which has been proposed to bind longer XPPs [91]. However, the prenyl-acceptor substrate of Af should occupy one of these two binding sites; therefore a more plausible scenario is that the small pocket and long tunnel bind two linear substrates, as in the case of DGGGP synthase (Figure 2E). With a membrane opening, the long tunnel in Af is generated by the larger spacing between TM9 and TM8 (Figures 3B, 3C, and 3D). This is accompanied by another change in the Af structure: TM9 and TM1 are brought close together, thereby blocking the direct opening of the Af central cavity to lipids. Because the Ap and Af homologs appear to have distinct substrates and functions (Figure 1), these structural variations imply that the different positions of TM9 relative to TM1 and TM8 might define the “gates” to control substrate specificity; this may be a general mechanism used by enzymes in the superfamily. Different spacing between these transmembrane helices, and size and charge changes in critical residues, together may create large variations in the two substrate-binding pockets, thereby allowing different prenyltransferases to use remarkably distinctive substrates (Figure 2). Conversely, XPP is a donor substrate used by most of the superfamily members (except DPPR synthase; Figure 2D), therefore structural features that recognize the disphosphate group of XPP (Figure 3B) are likely to be similar. Future structural studies may also reveal how these prenyltransferases can carry out different reactions, such as the coupling of decarboxylation with prenylation (Figures 2B and 2H).
Comparison to soluble prenyltransferases
Intramembrane aromatic prenyltransferases (APT) share no sequence or structure similarity to soluble APTs, which have a β-barrel structure [93]. While soluble APTs are involved in secondary metabolic pathways, most intramembrane APTs participate in primary metabolism [2]. The prenylation of aromatic compounds is essentially a Friedel-Crafts alkylation, in which the reactive cation generated from XPP cleavage must be shielded from water [2]. In soluble APTs, the shielding is achieved by a barrel of β-sheets. In intramembrane APTs, a deep lipophilic pocket surrounded by transmembrane helices may play a similar shielding role.
Interestingly, there is considerable structural similarity between intramembrane APTs and the soluble trans-prenyltransferses that catalyze the elongation of isoprenyl chains. The farnesyl diphosphate synthase (FPPS), which adopts a typical isoprenyl synthase fold [94], has seven helices (H3–H9) that are superimposable to TM1-7 in the intramembrane APTs (Figure 3D). FPPS also contains two Asp-rich motifs and shares about 12% sequence identity with EcUbiA. Although there is no direct evidence of an evolutionary path for these APTs, it is interesting to postulate how a common ancestor could diverge into soluble and membrane bound enzymes to perform different but essential cellular functions.
Concluding Remarks and Future Perspectives
Understanding of both structural and functional aspects of the UbiA superfamily is increasing. It will be particularly interesting to explore UBIAD1’s roles in mitochondrial electron transport, embryonic development, and vascular homeostasis (see Outstanding Questions). In addition, it remains to be elucidated how UBIAD1 affects lipid metabolism in SCD and urological cancers. The microbial enzymes of particular biological and medical significance include MenA, which is a close homolog to UBIAD1; DGGGP synthase, which generates characteristic archaeal lipids; and DPPR synthase, which is essential for the integrity of mycobacterial cell wall. Finally, structural information is highly desired for a biochemically tractable, active enzyme to illuminate how the specificity of aromatic substrates is achieved in this superfamily.
Outstanding Questions Box.
Does UBIAD1 generally function in a tissue-specific manner?
A discernible phenotype of UBIAD1 mutations in humans is Schnyder corneal dystrophy, which is characterized by the deposition of cholesterol and phosopholipids. UBIAD1 has association with several lipid-metabolism proteins, but it remains unclear why dramatic lipid deposition only occurs in the cornea.
Are the cardiovascular and neurological roles of UBIAD1, which were discovered in animal studies, conserved in humans?
While UBIAD1 is linked to cardiovascular degeneration in zebrafish and neurological degeneration in Drosophila, these physiological roles, and the related cellular functions, remain to be demonstrated in humans.
Is there a missing enzyme involved in vitamin K turnover?
A second enzyme may cleave off the side chain of phylloquinone (Figure 2c) and generate menadione as the substrate for UBIAD1.
Is COQ2 a genetic risk factor for multiple system atrophy (MSA)?
Establishing this link requires extended analysis of different populations and MSA subtypes.
Does DGGGP synthase produce the tetraether lipids in Archaea?
These macro-circular lipids, which form unique monolayer membranes, were thought to be generated by a novel enzyme that catalyzed a highly unusual chemistry: a head-to-head reaction between non-activated phytyl chains. To circumvent this problem, it has been proposed that DGGGP synthase can directly produce the tetraether lipids by accommodating longer precursors.
What is the structural basis of substrate specificity and promiscuity in the UbiA superfamily?
Prenyltransferases in this superfamily are characterized by their preferences for diverse substrates. Understanding the structural differences that dictate these preferences is the key to understand the catalytic mechanisms of these enzymes, which can be subsequently engineered for chemoenzymatic synthesis.
Figure I. Scheme of UbiA catalysis.
UbiA is a representative member of the superfamily.
Figure I. Disease-related mutations in UBIAD1 and COQ2.
The homology models are generated by PHYRE2 [98] using the Ap structure (PDB: 4OD5) as template. (A) SCD mutations in UBIAD1. Mutations around the two putative substrate-binding pockets are represented by orange spheres, and mutations at a putative partner-interaction interface in green. (B) COQ2 mutations identified in infantile multisystem diseases. Residues potentially involved in substrate binding are shown in sticks and mutations of unknown mechanism in grey spheres.
Trends.
Emerging data reveal that the UbiA superfamily of prenyltransferases, including UBIAD1, COQ2, DPPR synthase and DGGGP synthase, is involved in a wide variety of biological processes and diseases.
UBIAD1 catalyzes the vitamin K turnover, prevents oxidative damage in cardiovascular tissues, sustains mitochondrial function, and plays a role in lipid metabolism. Dysfunctional UBIAD1 is linked to cardiovascular degeneration, Parkinson’s disease, and Schnyder corneal dystrophy.
DPPR synthase is required for the formation of M. tuberculosis cell wall and thus is a promising target for anti-tuberculosis drugs.
DGGGP synthase is proposed to synthesize the diether and tetraether membrane lipids, key evolutionary features of archaea.
Acknowledgments
I thank T. Ellenberger, P. Yuan, and Y. Yang for critical reading of the manuscript. W. L. is supported by a R01 (HL121718) from the National Heart, Lung, and Blood Institute, a grant-in-aid (14GRNT20310017) from the American Heart Association, and a scholar award from the American Society of Hematology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta. 2010;1797:1587–605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Bonitz T, et al. Evolutionary relationships of microbial aromatic prenyltransferases. PLoS One. 2011;6:e27336. doi: 10.1371/journal.pone.0027336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heide L. Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol. 2009;13:171–9. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Young IG, et al. Biochemical and genetic studies on ubiquinone biosynthesis in Escherichia coli K-12:4-hydroxybenzoate octaprenyltransferase. J Bacteriol. 1972;110:18–25. doi: 10.1128/jb.110.1.18-25.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsgren M, et al. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem J. 2004;382:519–26. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashby MN, et al. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J Biol Chem. 1992;267:4128–36. [PubMed] [Google Scholar]
- 7.Nakagawa K, et al. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature. 2010;468:117–21. doi: 10.1038/nature09464. [DOI] [PubMed] [Google Scholar]
- 8.Mollet J, et al. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J Clin Invest. 2007;117:765–72. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakobs BSB, et al. A novel mutation in COQ2 leading to fatal infantile multisystem disease. J Neurol Sci. 2013;326:24–8. doi: 10.1016/j.jns.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Scalais E, et al. Early myoclonic epilepsy, hypertrophic cardiomyopathy and subsequently a nephrotic syndrome in a patient with CoQ10 deficiency caused by mutations in para-hydroxybenzoate-polyprenyl transferase (COQ2) Eur J Paediatr Neurol. 2013;17:625–630. doi: 10.1016/j.ejpn.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Quinzii C, et al. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78:345–9. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diomedi-Camassei F, et al. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol. 2007;18:2773–80. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- 13.Desbats MA, et al. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency. J Inherit Metab Dis. 2014;10:145–156. doi: 10.1007/s10545-014-9749-9. [DOI] [PubMed] [Google Scholar]
- 14.Hegarty J, et al. UBIAD1-mediated vitamin K2 synthesis is required for vascular endothelial cell survival and development. Development. 2013;140:1713–9. doi: 10.1242/dev.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mugoni V, et al. Ubiad1 Is an Antioxidant Enzyme that Regulates eNOS Activity by CoQ10 Synthesis. Cell. 2013;152:504–18. doi: 10.1016/j.cell.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vos M, et al. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science. 2012;336:1306–10. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- 17.Orr A, et al. Mutations in the UBIAD1 gene, encoding a potential prenyltransferase, are causal for Schnyder crystalline corneal dystrophy. PLoS One. 2007;2:e685. doi: 10.1371/journal.pone.0000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss JS, et al. Mutations in the UBIAD1 gene on chromosome short arm 1, region 36, cause Schnyder crystalline corneal dystrophy. Invest Ophthalmol Vis Sci. 2007;48:5007–12. doi: 10.1167/iovs.07-0845. [DOI] [PubMed] [Google Scholar]
- 19.Fredericks WJ, et al. The bladder tumor suppressor protein TERE1 (UBIAD1) modulates cell cholesterol: implications for tumor progression. DNA Cell Biol. 2011;30:851–64. doi: 10.1089/dna.2011.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, et al. Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-{alpha}-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. J Biol Chem. 2005;280:24539–43. doi: 10.1074/jbc.M504068200. [DOI] [PubMed] [Google Scholar]
- 21.Jain S, et al. Biosynthesis of archaeal membrane ether lipids. Front Microbiol. 2014;5:641. doi: 10.3389/fmicb.2014.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, et al. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta. 2012;1823:1604–1616. doi: 10.1016/j.bbamcr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melzer M, Heide L. Characterization of polyprenyldiphosphate: 4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim Biophys Acta. 1994;1212:93–102. doi: 10.1016/0005-2760(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 24.Suvarna K, et al. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J Bacteriol. 1998;180:2782–7. doi: 10.1128/jb.180.10.2782-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, Poulter CD. Biosynthesis of archaebacterial ether lipids. Formation of ether linkages by prenyltransferases. J Am Chem Soc. 1993;115:1270–1277. [Google Scholar]
- 26.Hederstedt L. Heme A biosynthesis. Biochim Biophys Acta. 2012;1817:920–927. doi: 10.1016/j.bbabio.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 27.Rüdiger W, et al. Enzymes of the last steps of chlorophyll biosynthesis: Modification of the substrate structure helps to understand the topology of the active centers. Biochemistry. 2005;44:10864–10872. doi: 10.1021/bi0504198. [DOI] [PubMed] [Google Scholar]
- 28.Schledz M, et al. A novel phytyltransferase from Synechocystis sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Lett. 2001;499:15–20. doi: 10.1016/s0014-5793(01)02508-x. [DOI] [PubMed] [Google Scholar]
- 29.Montini G, et al. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med. 2008;358:2849–50. doi: 10.1056/NEJMc0800582. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Martin JM, et al. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum Mol Genet. 2007;16:1091–1097. doi: 10.1093/hmg/ddm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinzii CM, et al. Respiratory chain dysfunction and oxidative stress correlate with severity of primary CoQ10 deficiency. FASEB J. 2008;22:1874–85. doi: 10.1096/fj.07-100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinzii CM, et al. Effects of inhibiting CoQ10 biosynthesis with 4-nitrobenzoate in human fibroblasts. PLoS One. 2012;7:e30606. doi: 10.1371/journal.pone.0030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuji S. Mutations in COQ2 in familial and sporadic multiple-system atrophy. N Engl J Med. 2013;369:233–44. doi: 10.1056/NEJMoa1212115. [DOI] [PubMed] [Google Scholar]
- 34.Bleasel JM, et al. Lipid dysfunction and pathogenesis of multiple system atrophy. Acta Neuropathol Commun. 2014;2:15. doi: 10.1186/2051-5960-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon BS, et al. Mutant COQ2 in multiple-system atrophy. N Engl J Med. 2014;371:80. doi: 10.1056/NEJMc1311763. [DOI] [PubMed] [Google Scholar]
- 36.Ogaki K, et al. Analysis of COQ2 gene in multiple system atrophy. Mol Neurodegener. 2014;9:44. doi: 10.1186/1750-1326-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Needham M, Mastaglia FL. Statin myotoxicity: A review of genetic susceptibility factors. Neuromuscul Disord. 2014;24:4–15. doi: 10.1016/j.nmd.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Bräuer L, et al. A structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. Chembiochem. 2008;9:982–92. doi: 10.1002/cbic.200700575. [DOI] [PubMed] [Google Scholar]
- 39.Wessjohann L, Sontag B. Prenylation of Benzoic Acid Derivatives Catalyzed by a Transferase from Escherichia coli Overproduction: Method Development and Substrate Specificity. Angew Chemie Int Ed English. 1996;35:1697–1699. [Google Scholar]
- 40.Suzuki K, et al. Evidence that Escherichia coli ubiA product is a functional homolog of yeast COQ2, and the regulation of ubiA gene expression. Biosci Biotechnol Biochem. 1994;58:1814–9. doi: 10.1271/bbb.58.1814. [DOI] [PubMed] [Google Scholar]
- 41.Swiezewska E, et al. Biosynthesis of ubiquinone and plastoquinone in the endoplasmic reticulum-Golgi membranes of spinach leaves. J Biol Chem. 1993;268:1494–9. [PubMed] [Google Scholar]
- 42.Kalén a, et al. Nonaprenyl-4-hydroxybenzoate transferase, an enzyme involved in ubiquinone biosynthesis, in the endoplasmic reticulum-Golgi system of rat liver. J Biol Chem. 1990;265:1158–64. [PubMed] [Google Scholar]
- 43.Meganathan R. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol Lett. 2001;203:131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- 44.Ohara K, et al. Functional characterization of OsPPT1, which encodes p-hydroxybenzoate polyprenyltransferase involved in ubiquinone biosynthesis in Oryza sativa. Plant Cell Physiol. 2006;47:581–90. doi: 10.1093/pcp/pcj025. [DOI] [PubMed] [Google Scholar]
- 45.Ohara K, et al. Homogeneous purification and characterization of LePGT1 - A membrane-bound aromatic substrate prenyltransferase involved in secondary metabolism of Lithospermum erythrorhizon. FEBS J. 2013;280:2572–2580. doi: 10.1111/febs.12239. [DOI] [PubMed] [Google Scholar]
- 46.Ohara K, et al. Functional characterization of LePGT1, a membrane-bound prenyltransferase involved in the geranylation of p-hydroxybenzoic acid. Biochem J. 2009;421:231–41. doi: 10.1042/BJ20081968. [DOI] [PubMed] [Google Scholar]
- 47.Okano T, et al. Conversion of phylloquinone (Vitamin K1) into menaquinone-4 (Vitamin K2) in mice: two possible routes for menaquinone-4 accumulation in cerebra of mice. J Biol Chem. 2008;283:11270–11279. doi: 10.1074/jbc.M702971200. [DOI] [PubMed] [Google Scholar]
- 48.Will BH, et al. Comparative metabolism and requirement of vitamin K in chicks and rats. J Nutr. 1992;122:2354–2360. doi: 10.1093/jn/122.12.2354. [DOI] [PubMed] [Google Scholar]
- 49.Thijssen HH, et al. Phylloquinone and menaquinone-4 distribution in rats: synthesis rather than uptake determines menaquinone-4 organ concentrations. J Nutr. 1996;126:537–543. doi: 10.1093/jn/126.2.537. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa K, et al. Vitamin K2 biosynthetic enzyme, UBIAD1 is essential for embryonic development of mice. PLoS One. 2014;9:e104078. doi: 10.1371/journal.pone.0104078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss JS. Visual morbidity in thirty-four families with Schnyder crystalline corneal dystrophy (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2007;105:616–648. [PMC free article] [PubMed] [Google Scholar]
- 52.Nickerson ML, et al. The UBIAD1 prenyltransferase links menaquinone-4 [corrected] synthesis to cholesterol metabolic enzymes. Hum Mutat. 2013;34:317–29. doi: 10.1002/humu.22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fredericks WJ, et al. The TERE1 (UBIAD1) bladder tumor suppressor protein interacts with mitochondrial TBL2: regulation of trans-membrane potential, oxidative stress and SXR signaling to the nucleus. J Cell Biochem. 2013;114:2170–87. doi: 10.1002/jcb.24567. [DOI] [PubMed] [Google Scholar]
- 54.McGarvey TW, et al. An interaction between apolipoprotein E and TERE1 with a possible association with bladder tumor formation. J Cell Biochem. 2005;95:419–28. doi: 10.1002/jcb.20432. [DOI] [PubMed] [Google Scholar]
- 55.Schumacher MM, et al. The prenyltransferase UBIAD1 is the target of geranylgeraniol in degradation of HMG CoA reductase. Elife. 2015;4:1–21. doi: 10.7554/eLife.05560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGarvey TW, et al. Isolation and characterization of the TERE1 gene, a gene down-regulated in transitional cell carcinoma of the bladder. Oncogene. 2001;20:1042–1051. doi: 10.1038/sj.onc.1204143. [DOI] [PubMed] [Google Scholar]
- 57.McGarvey TW, et al. TERE1, a novel gene affecting growth regulation in prostate carcinoma. Prostate. 2003;54:144–155. doi: 10.1002/pros.10174. [DOI] [PubMed] [Google Scholar]
- 58.Xia Y, et al. Down-regulation of TERE1/UBIAD1 activated Ras-MAPK signalling and induced cell proliferation. Cell Biol Int Rep. 2010;17:e00005. doi: 10.1042/CBR20100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nickerson ML, et al. UBIAD1 mutation alters a mitochondrial prenyltransferase to cause Schnyder corneal dystrophy. PLoS One. 2010;5:e10760. doi: 10.1371/journal.pone.0010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li K, et al. Multitarget drug discovery for tuberculosis and other infectious diseases. J Med Chem. 2014;57:3126–3129. doi: 10.1021/jm500131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dhiman RK, et al. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol. 2009;72:85–97. doi: 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Debnath J, et al. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J Med Chem. 2012;55:3739–55. doi: 10.1021/jm201608g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolly GS, et al. Assessing the essentiality of the decaprenyl-phospho-d-arabinofuranose pathway in Mycobacterium tuberculosis using conditional mutants. Mol Microbiol. 2014;92:194–211. doi: 10.1111/mmi.12546. [DOI] [PubMed] [Google Scholar]
- 64.Alderwick LJ, et al. Arabinan-deficient mutants of Corynebacterium glutamicum and the consequent flux in decaprenylmonophosphoryl-D-arabinose metabolism. Glycobiology. 2006;16:1073–1081. doi: 10.1093/glycob/cwl030. [DOI] [PubMed] [Google Scholar]
- 65.Makarov V, et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324:801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang H, et al. Identification of amino acids and domains required for catalytic activity of DPPR synthase, a cell wall biosynthetic enzyme of Mycobacterium tuberculosis. Microbiology. 2008;154:736–43. doi: 10.1099/mic.0.2007/013532-0. [DOI] [PubMed] [Google Scholar]
- 67.Villanueva L, et al. A re-evaluation of the archaeal membrane lipid biosynthetic pathway. Nat Rev Microbiol. 2014;12:438–48. doi: 10.1038/nrmicro3260. [DOI] [PubMed] [Google Scholar]
- 68.Saiki K, et al. In vitro heme O synthesis by the cyoE gene product from Escherichia coli. J Biol Chem. 1993;268:26041–26045. [PubMed] [Google Scholar]
- 69.Mogi T. Over-expression and characterization of Bacillus subtilis heme O synthase. J Biochem. 2009;145:669–675. doi: 10.1093/jb/mvp024. [DOI] [PubMed] [Google Scholar]
- 70.Antonicka H, et al. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum Mol Genet. 2003;12:2693–2702. doi: 10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- 71.Pitceathly RDS, et al. COX10 Mutations Resulting in Complex Multisystem Mitochondrial Disease That Remains Stable Into Adulthood. JAMA Neurol. 2013;70:1556–61. doi: 10.1001/jamaneurol.2013.3242. [DOI] [PubMed] [Google Scholar]
- 72.Vitali M, et al. Analysis of the genes coding for subunit 10 and 15 of cytochrome c oxidase in Alzheimer’s disease. J Neural Transm. 2009;116:1635–1641. doi: 10.1007/s00702-009-0324-8. [DOI] [PubMed] [Google Scholar]
- 73.Khalimonchuk O, et al. Oligomerization of Heme o Synthase in Cytochrome Oxidase Biogenesis Is Mediated by Cytochrome Oxidase Assembly Factor Coa2. J Biol Chem. 2012;287:26715–26726. doi: 10.1074/jbc.M112.377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsukihara T, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 75.Khalimonchuk O, et al. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chidgey JW, et al. A Cyanobacterial Chlorophyll Synthase-HliD Complex Associates with the Ycf39 Protein and the YidC/Alb3 Insertase. Plant Cell. 2014;26:1–14. doi: 10.1105/tpc.114.124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sobotka R. Making proteins green; Biosynthesis of chlorophyll-binding proteins in cyanobacteria. Photosynth Res. 2014;119:223–232. doi: 10.1007/s11120-013-9797-2. [DOI] [PubMed] [Google Scholar]
- 78.Eichacker LA, et al. Stabilization of the chlorophyll binding apoproteins, P700, CP47, CP43, D2, and D1, by synthesis of Zn-pheophytin a in intact etioplasts from barley. FEBS Lett. 1996;395:251–256. doi: 10.1016/0014-5793(96)01026-5. [DOI] [PubMed] [Google Scholar]
- 79.Shalygo N, et al. Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol Biol. 2009;71:425–436. doi: 10.1007/s11103-009-9532-8. [DOI] [PubMed] [Google Scholar]
- 80.Schneider C. Chemistry and biology of vitamin E. Mol Nutr Food Res. 2005;49:7–30. doi: 10.1002/mnfr.200400049. [DOI] [PubMed] [Google Scholar]
- 81.Cahoon EB, et al. Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol. 2003;21:1082–7. doi: 10.1038/nbt853. [DOI] [PubMed] [Google Scholar]
- 82.Yang W, et al. Vitamin E biosynthesis: functional characterization of the monocot homogentisate geranylgeranyl transferase. Plant J. 2011;65:206–17. doi: 10.1111/j.1365-313X.2010.04417.x. [DOI] [PubMed] [Google Scholar]
- 83.Sadre R, et al. Catalytic reactions of the homogentisate prenyl transferase involved in plastoquinone-9 biosynthesis. J Biol Chem. 2010;285:18191–18198. doi: 10.1074/jbc.M110.117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yazaki K, et al. Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry. 2009;70:1739–1745. doi: 10.1016/j.phytochem.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 85.Li H, et al. A heteromeric membrane-bound prenyltransferase complex from Humulus lupulus catalyzes three sequential aromatic prenylations in the bitter acid pathway. Plant Physiol. 2015;167:650–659. doi: 10.1104/pp.114.253682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen G, et al. Characterization of an Isoflavonoid-Specific Prenyltransferase from Lupinus albus. Plant Physiol. 2012;159:70–80. doi: 10.1104/pp.112.195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sasaki K, et al. Molecular characterization of a membrane-bound prenyltransferase specific for isoflavone from Sophora flavescens. J Biol Chem. 2011;286:24125–24134. doi: 10.1074/jbc.M111.244426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stec E, Li SM. Mutagenesis and biochemical studies on AuaA confirmed the importance of the two conserved aspartate-rich motifs and suggested difference in the amino acids for substrate binding in membrane-bound prenyltransferases. Arch Microbiol. 2012;194:589–595. doi: 10.1007/s00203-012-0795-0. [DOI] [PubMed] [Google Scholar]
- 89.Itoh T, et al. Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat Chem. 2010;2:858–864. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 90.Cheng W, Li W. Structural Insights into Ubiquinone Biosynthesis in Membranes. Science. 2014;343:878–81. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang H, et al. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol. 2014;12:e1001911. doi: 10.1371/journal.pbio.1001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saiki K, et al. Identification of the functional domains in heme O synthase. Site-directed mutagenesis studies on the cyoE gene of the cytochrome bo operon in Escherichia coli. J Biol Chem. 1993;268:26927–26934. [PubMed] [Google Scholar]
- 93.Kuzuyama T, et al. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–7. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wallrapp FH, et al. Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily. Proc Natl Acad Sci U S A. 2013;110:E1196–202. doi: 10.1073/pnas.1300632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 97.Hosfield DJ, et al. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J Biol Chem. 2004;279:8526–9. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 98.Kelley LA, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]