Abstract
Studying gene-environment (GxE) interactions is important, as they extend our knowledge of the genetic architecture of complex traits and may help to identify novel variants not detected via analysis of main-effects alone. The main statistical framework for studying GxE interactions uses a single regression model that includes both the genetic main and GxE interaction effects (the ‘joint’ framework). The alternative ‘stratified’ framework combines results from genetic main-effect analyses carried out separately within the exposed and unexposed groups. Although there have been several investigations using theory and simulation, an empirical comparison of the two frameworks is lacking. Here, we compare the two frameworks using results from GWAS of systolic blood pressure for 3.2 million low frequency and 6.5 million common variants across 20 cohorts of European ancestry, comprising 79,731 individuals. Our cohorts have sample sizes ranging from 456 to 22,983 and include both family-based and population-based samples. In cohort-specific analyses, the two frameworks provided similar inference for population-based cohorts. The agreement was reduced for family-based cohorts. In meta-analyses, agreement between the two frameworks was less than that observed in cohort-specific analyses, despite the increased sample size. In meta-analyses, agreement depended on 1) the minor allele frequency, 2) inclusion of family-based cohorts in meta-analysis, and 3) filtering scheme. The stratified framework appears to approximate the joint framework well only for common variants in population-based cohorts. We conclude that the joint framework is the preferred approach and should be used to control false positives when dealing with low frequency variants and/or family-based cohorts.
Introduction
Genome-wide association studies (GWAS) and subsequent meta-analyses have successfully identified hundreds of genetic variants associated with many disease traits (http://www.genome.gov), accelerating the progress in the genetic dissection of complex human traits. Meta-analysis has become a key component of GWAS to increase sample sizes and therefore power [de Bakker, et al. 2008; Evangelou and Ioannidis 2013], and most new discoveries are now driven by large-scale consortia such as the CHARGE (Cohorts for Heart and Aging Research in Genetic Epidemiology) [Psaty, et al. 2009], GIANT (Genetic Investigation of Anthropometric Traits) [Shungin, et al. 2015; Willer, et al. 2009], ICBP (International Consortium of Blood Pressure) [International Consortium for Blood Pressure Genome-Wide Association, et al. 2011], and MAGIC (the Meta-Analyses of Glucose and Insulin-related traits Consortium) [Prokopenko, et al. 2009]. The identified genetic variants, however, typically have small effects, explaining only a small part of the heritability of most complex traits [Manolio, et al. 2009].
Studying gene-environment (GxE) interactions is becoming popular as it can potentially identify novel genetic variants not detected via main-effects analysis alone [Manning, et al. 2012], extend our knowledge of the genetic architecture of complex traits [Hunter 2005], and enable “profiling” of individuals at high risk for disease [Le Marchand and Wilkens 2008; Thomas 2010]. Meta-analysis is more critical for analysis of GxE interactions, as identifying GxE interactions requires even larger sample sizes than those needed to identify genetic main effects [Thomas 2010]. The main statistical framework for the analysis of GxE interactions is using a single regression model that includes both genetic main and GxE interaction effects; we call this the ‘joint’ framework. Under this framework, one can use the traditional 1 degree of freedom (DF) test of the interaction effect or a 2 DF test that jointly tests for both the genetic main and interaction effects [Kraft, et al. 2007]. The 2 DF test has been shown to be particularly useful to identify variants with low main effect and moderate interaction effects, as such variants would be difficult to detect when using either a marginal genetic main effect or the aforementioned 1DF interaction test [Kraft, et al. 2007]. Meta-analysis approaches for the 2 DF test have been developed by Manning et al [Manning, et al. 2011], and by combining data from 52 studies and accounting for body mass index as a possible interaction variable, MAGIC identified multiple novel loci associated with fasting insulin levels [Manning, et al. 2012].
For dichotomous exposure variables, such as yes/no status of smoking or drinking, another framework has emerged, which we call the ‘stratified’ framework. Under this framework, samples are stratified into two groups: the exposed and unexposed groups. Genetic main-effect analysis is performed separately in each stratum. These stratum-specific genetic effects are subsequently combined to perform a 1DF test [Randall, et al. 2013] or a 2DF test [Aschard, et al. 2010]. Although the stratified framework approximates the joint framework, because main-effect models are readily available in many software packages, it is easier to implement in a large-scale consortium setting. Indeed, the stratified framework has been used in several projects of the GIANT consortium including a recent publication [Shungin, et al. 2015].
As of today, there is no clear consensus on which framework (joint versus stratified) should be preferred. Several papers compared specific aspects of each approach. This includes simulation-based studies demonstrating power comparisons [Magi, et al. 2010; Manning, et al. 2011], theoretical work demonstrating close equivalence (in large samples) between statistical tests from the two frameworks [Aschard, et al. 2010; Magi, et al. 2010], and power computations [Behrens, et al. 2011]. However, no empirical comparison using real data has been performed so far. As part of the CHARGE Gene-Lifestyle Interactions Working Group, we performed GWAS of systolic blood pressure for 3.2 million low frequency variants (with 1% ≤ MAF < 5%) and 6.5 million common variants (with MAF ≥ 5%), imputed using reference haplotypes from the 1000 Genomes Project [1000 Genomes Project Consortium, et al. 2012], across 20 cohorts of European ancestry. Using this unique resource we provide a comparison of the two frameworks in several ways. First, to explore the role of the total sample size on the extent of agreement between the two frameworks; second, to understand the impact of unequal sample size between the two (exposed and unexposed) strata, using ‘current-smoking’ status, which leads to highly unequal sample sizes in the two strata, and ‘ever-smoking’ status, which leads to similar sample sizes in the two strata; third, to understand the impact of meta-analysis, by comparing cohort-specific GWAS results and results from meta-analysis; and fourth, to understand the impact of family-based cohorts on meta-analysis by comparing meta-analysis results from1) population-based cohorts only, 2) family-based cohorts only, and 3) all cohorts.
Methods
Study Samples, Genotype and Phenotype Data
We used data from 20 studies with participants of European ancestry. Table 1 summarizes these studies; a detailed description is provided in the Supplemental Materials. Each study obtained informed consent from participants and approval from the appropriate institutional review boards. Genotyping was performed using Illumina (San Diego, CA, USA) or Affymetrix (Santa Clara, CA, USA) genotyping arrays. To infer genotypes for single nucleotide polymorphisms (SNPs), short insertions and deletions (indels), and larger deletions that were not genotyped directly on the genotyping arrays but are available from the 1000 Genomes Project [1000 Genomes Project Consortium, et al. 2012], each study performed imputation using MACH [Li, et al. 2010], Minimac [Howie, et al. 2012], IMPUTE2 [Howie, et al. 2009], or BEAGLE [Browning and Browning 2009] software. For imputation, all studies used the 1000 Genomes Project Phase I Integrated Release Version 3 Haplotypes (2010–11 data freeze, 2012-03-14 haplotypes), which contain haplotypes of 1,092 individuals of all ethnic backgrounds. Information on genotype and imputation for each study is presented in Table S1.
Table 1.
The 20 Participating Cohorts of European Ancestry and their Sample Sizes. Cohorts are divided into two groups (population-based and family-based) and ordered with respect to sample size within each group.
| Cohort | Sample Size | CurSmk | EverSmk | |||||
|---|---|---|---|---|---|---|---|---|
| Yes | No | % Yes | Yes | No | % Yes | |||
| Population | CROATIA-Korcula | 456 | 112 | 344 | 24.6% | 237 | 219 | 52.0% |
| CROATIA-Vis | 483 | 141 | 342 | 29.2% | 277 | 206 | 57.3% | |
| BioMe | 1,480 | 134 | 1,346 | 9.1% | 441 | 1,039 | 29.8% | |
| CARDIA | 1,649 | 406 | 1,243 | 24.6% | 693 | 956 | 42.0% | |
| HealthABC | 1,662 | 106 | 1,556 | 6.4% | 951 | 711 | 57.2% | |
| RS2 | 1,998 | 408 | 1,590 | 20.4% | 1,410 | 588 | 70.6% | |
| AGES | 2,410 | 345 | 2,065 | 14.3% | 1,440 | 970 | 59.8% | |
| MESA | 2,591 | 298 | 2,293 | 11.5% | 1,447 | 1,144 | 55.8% | |
| RS3 | 2,966 | 673 | 2,293 | 22.7% | 2,031 | 935 | 68.5% | |
| CHS | 2,975 | 357 | 2,618 | 12.0% | 1,595 | 1,380 | 53.6% | |
| RS1 | 4,991 | 1,162 | 3,829 | 23.3% | 3,317 | 1,674 | 66.5% | |
| GS:SFHS* | 6,439 | 994 | 5,445 | 15.4% | 3,133 | 3,306 | 48.7% | |
| ARIC | 9,465 | 2,339 | 7,126 | 24.7% | 5,685 | 3,780 | 60.1% | |
| WGHS | 22,983 | 2,680 | 20,303 | 11.7% | 11,284 | 11,699 | 49.1% | |
|
| ||||||||
| Family | HERITAGE | 499 | 75 | 424 | 15.0% | 191 | 308 | 38.3% |
| GENOA | 1,064 | 169 | 895 | 15.9% | 535 | 529 | 50.3% | |
| HyperGEN | 1,251 | 114 | 1,137 | 9.1% | 424 | 827 | 33.9% | |
| ERF | 2,491 | 984 | 1,507 | 39.5% | 1,721 | 770 | 69.1% | |
| FamHS | 3,683 | 523 | 3,160 | 14.2% | 1,668 | 2,015 | 45.3% | |
| FHS | 8,195 | 2,520 | 5,675 | 30.8% | 4,281 | 3,914 | 52.2% | |
|
| ||||||||
| Total | 79,731 | 14,540 | 65,182 | 18.2% | 42,761 | 36,970 | 53.6% | |
Abbreviations: BioMe, Biobank of Institute for Personalized Medicine at Mount Sinai; CARDIA, Coronary Artery Risk Development in Young Adults; HealthABC, Health, Aging, and Body Composition study; RS2, Rotterdam Study cohort 2; AGES, Age Gene Environment Susceptibility Study; MESA, Multi-Ethnic Study of Atherosclerosis; RS3, Rotterdam Study cohort 3; CHS, Cardiovascular Health Study; RS1, Rotterdam Study cohort 1; GS:SFHS, Generation Scotland Scottish Family Health Study; ARIC, Atherosclerosis Risk in Communities; WGHS, Women’s Genome Health Study; HERITAGE, Health, Risk Factors, Exercise Training and Genetics; GENOA, Genetic Epidemiology Network of Arteriopathy; HyperGEN, Hypertension Genetic Epidemiology Network; ERF, Erasmus Rucphen Family study; FamHS, Family Heart Study; FHS, Framingham Heart Study
For this manuscript, GS:SFHS, although a family-based study, removed related individuals using IBS values calculated from genetic data.
In total, 79,731 subjects between 18 and 80 years of age with genotype, phenotype and covariate information were available in this analysis. Resting SBP was measured on an mmHg scale. For subjects taking antihypertensive or BP lowering medications, the SBP value was adjusted by adding 15 mmHg [Newton-Cheh, et al. 2009; Tobin, et al. 2005]. This medication-adjusted SBP variable is approximately normally distributed. In addition, to reduce the effect of possible outliers, winsorising has been applied for this SBP value that is more than 6 standard deviations away from the mean. Two smoking exposure variables were considered: ‘current smoking’ status (CurSmk), defined as being a smoker at the time of the blood pressure measurements, and ‘ever smoking’ status (EverSmk), defined as being a smoker at the time of the measurement or else being a former smoker. If subjects had partially missing data for SBP, smoking variable, and any covariates, they were excluded from analysis.
Cohort-specific GWAS Analysis
For the ‘joint’ framework, a regression model including both genetic main and GxE interaction effects
| (Equation 1) |
was applied to the entire sample. Y is the medication-adjusted SBP value, E is the smoking variable (with 0/1 coding for the absence/presence of the smoking exposure), G is the dosage of the imputed genetic variant coded additively (from 0 to 2), and C is the vector of all other covariates, which include age, sex, field center (for multi-center studies), principal components (to account for population stratification and admixture) and additional cohort-specific covariates (if any). Each study conducted GWAS analysis and provided the genetic main effect βG and the interaction effect βGE and their 2×2 robust covariance matrix. For the 1 DF test, we used a Wald test statistic that approximately follows a chi-squared distribution with 1 DF under H0: βGE = 0. Similarly for the 2 DF test, we used a Wald test statistic, which approximately follows a chi-squared distribution with 2 DF under H0: βG = βGE = 0.
For the ‘stratified’ framework, analyses of the genetic main-effect regression models
| (Equation 2) |
were applied separately to the E = 0 unexposed group and to the E = 1 exposed group. Note that C is the same vector of the covariates as used in Equation (1). Each study conducted GWAS analysis and provided the stratum-specific effects and their robust standard errors (SE). Robust covariance matrices and robust SEs were sought as a safeguard against mis-specification of the mean model [Tchetgen Tchetgen and Kraft 2011; Voorman, et al. 2011]. To obtain robust covariance matrices and robust SEs, studies of unrelated subjects used either the R package sandwich [Zeileis 2006] or ProbABEL [Aulchenko, et al. 2010]. To account for relatedness in families, four family studies used the generalized estimating equations (GEE) approach, treating each family as a cluster, with the R packages geepack [Halekoh, et al. 2006]. The remaining two studies used the linear mixed effect model approach with a random polygenic component (for which the covariance matrix depends on the kinship matrix) with GenABEL [Aulchenko, et al. 2007] or R (Table S1).
For the 1 DF test in the stratified framework, we used the approach of Randall et al [Randall, et al. 2013], who define
| (Equation 3) |
where and are stratum-specific genetic effects; and are their respective robust standard errors; and r is the Spearman rank correlation coefficient between and , calculated from the genome-wide results. The statistic Zdiff approximately follows a standard normal distribution under H0: βGE = 0. For the 2DF test in the stratified framework, we used the test proposed by Aschard et al [Aschard, et al. 2010]:
| (Equation 4) |
which approximately follows a 2 DF chi-squared distribution under H0: βG = βGE = 0 when the two strata are independent. Note that the 1DF test includes the correlation term “r” to correct for any relatedness between E = 1 and E = 0 strata, whereas such correction is not available for the 2 DF test. Both tests in the stratified framework were computed using the R package EasyStrata [Winkler, et al. 2015].
Meta-analysis of GWAS Results
Variants with minor allele frequency (MAF) below 1% were excluded from each cohort-specific analysis. Extensive quality control (QC) using the R package EasyQC [Winkler, et al. 2014] was performed for all cohort-specific GWAS results. In meta-analysis, to exclude unstable cohort-specific results that reflect small sample size and low MAF, variants were excluded based on the minor allele count (MAC). In the joint framework, variants were included in the meta-analysis if MAC0 (= 2 * MAFE0 * NE0) ≥ 10, (with MAFE0 and sample size NE0 for E=0 stratum) and MAC1 (= 2 * MAFE1 * NE1) ≥ 10. In the stratified framework, we considered two filtering schemes (schemes A and B). Scheme A applied the MAC filter in each stratum separately: variants with MAC0 ≥ 10 (regardless of MAC1 values) were included in the meta-analysis for E = 0 and variants with MAC1 ≥ 10 were included in the meta-analysis for E = 1. Scheme B applied the same filter as the joint framework in both strata (E = 0 and E = 1). Variants were further excluded if imputation quality measure < 0.5. This value of 0.5 was used regardless of the software used for imputations, because imputation quality measures are shown to be similar across imputation software (Supplementary Information S3 through S5 from Marchini and Howie [Marchini and Howie 2010]).
To compare the two frameworks when using meta-analysis, we first performed meta-analysis using the 1 DF and 2 DF tests in each framework. For the 1 DF test in the joint framework, inverse-variance weighted meta-analysis was performed on the cohort-specific interaction effects βGE, using METAL [Willer, et al. 2010]. For the 2 DF test, the joint meta-analysis of Manning et al [Manning, et al. 2011] was performed using the cohort-specific βG, βGE, and their corresponding robust covariance matrix. In the stratified framework, meta-analysis was performed separately within each stratum using METAL. These stratum-specific meta-analysis results for and were subsequently combined to perform the 1DF test (Equation 3) and the 2DF test (Equation 4) using EasyStrata [Winkler, et al. 2015]. During meta-analysis, genomic control correction [Devlin and Roeder 1999] was applied to cohort-specific GWAS results if their genomic control lambda value was greater than 1. After meta-analysis was performed, a variant was excluded if the overall sample size, i.e. the sample size combined across multiple cohorts, for the variant was below 2,000.
Cohort-specific Results
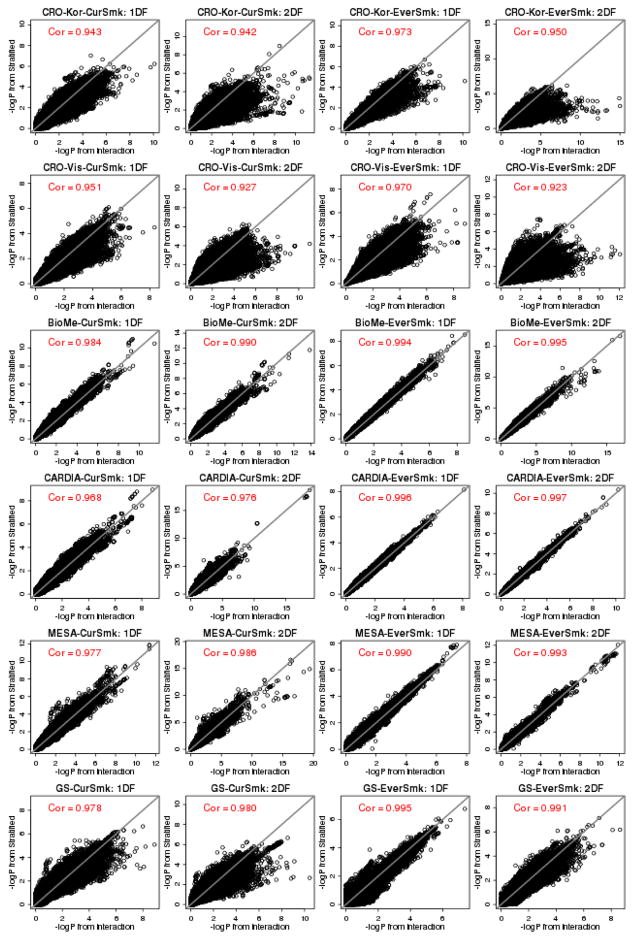
To compare the performance of the two frameworks for all cohort-specific GWAS results, we made scatterplots of −log10P values obtained from the joint framework (x-axis) and the stratified framework (y-axis) using both the 1 DF interaction and 2 DF joint tests (Figures 1, 2, and Figure S1); correlation is shown in Table 2. Cohort-level comparison was restricted to variants with MAC0 ≥ 10 and MAC1 ≥ 10. The genomic control lambda values of cohort-specific GWAS results ranged from 0.98 to 1.15 (Table S2).
Figure 1.
Scatterplots of cohort-level −log10(p) values for the 6 select population-based cohorts showing the weakest correlations. Each point shows −log10(p) value from the joint framework (x-axis) and the stratified framework (y-axis) at a variant. Cohorts are ordered with respect to sample sizes (shown in Table 1). The remaining 8 population-based cohorts that had correlation over 0.99, which are shown in Figure S1.
Figure 2.
Scatterplots of cohort-level −log10(p) values for the 6 family-based cohorts. Each point shows −log10(p) value from the joint framework (x-axis) and the stratified framework (y-axis) at a variant. Cohorts are ordered with respect to sample size (shown in Table 1).
Table 2.
Correlation between the two frameworks for cohort-specific GWAS results. Scatterplots are shown in Figures 1, 2, and S1.
| Cohort | CurSmk | EverSmk | |||
|---|---|---|---|---|---|
| 1DF | 2DF | 1DF | 2DF | ||
| Population | CROATIA-Korcula | 0.943 | 0.942 | 0.973 | 0.950 |
| CRO-Vis | 0.951 | 0.927 | 0.970 | 0.923 | |
| BioMe | 0.984 | 0.990 | 0.994 | 0.995 | |
| CARDIA | 0.968 | 0.976 | 0.996 | 0.997 | |
| HealthABC | 0.993 | 0.994 | 0.998 | 0.998 | |
| RS2 | 0.992 | 0.994 | 0.999 | 0.999 | |
| AGES | 0.997 | 0.998 | 0.999 | 0.999 | |
| MESA | 0.977 | 0.986 | 0.990 | 0.993 | |
| RS3 | 0.998 | 0.999 | 1.000 | 1.000 | |
| CHS | 0.991 | 0.994 | 0.999 | 0.999 | |
| RS1 | 0.996 | 0.998 | 0.996 | 0.997 | |
| GS:SFHS | 0.978 | 0.980 | 0.995 | 0.991 | |
| ARIC | 0.992 | 0.994 | 0.992 | 0.993 | |
| WGHS | 0.999 | 1.000 | 1.000 | 1.000 | |
|
| |||||
| Family | HERITAGE | 0.762 | 0.819 | 0.886 | 0.902 |
| GENOA | 0.998 | 0.998 | 0.992 | 0.992 | |
| HyperGEN | 0.885 | 0.921 | 0.935 | 0.942 | |
| ERF | 0.973 | 0.979 | 0.974 | 0.979 | |
| FamHS | 0.926 | 0.950 | 0.960 | 0.968 | |
| FHS | 0.935 | 0.951 | 0.939 | 0.951 | |
Impact of imbalance in exposure groups
Within each cohort, the number of current smokers is smaller than the number of non-smokers, with percentages of current smokers ranging from 6% to 39% of the cohort sample. When considering ever-smoking instead, the two strata are much more balanced, with percentages of ever smokers ranging from 29% to 70% within each cohort. When all cohorts are combined, current smokers are 18.2% of the entire sample, whereas ever smokers are 53.6% (Table 1).
For both tests and for almost all studies, we observed a higher correlation of the −log10P values between the two frameworks for EverSmk compared to CurSmk. The impact of unequal sample sizes in the two strata can be seen from cohorts with small sample sizes. For example, for CROATIA-Korcula study (N = 456; 25% CurSmk; 52% EverSmk), the smallest population-based cohort, the correlation between the two frameworks for the 1 DF test was 0.94 and 0.97 for CurSmk and EverSmk, respectively (the first row in Figure 1). The scatterplot exhibited many variants that are away from the diagonal line, showing weak agreement. The joint framework had higher genomic control values for this cohort (and the CROATIA-Korcula cohort) (Table S2). However, this pattern was not consistent across cohorts, as the stratified framework had higher genomic control values than the joint framework for several other cohorts.
Sample size for asymptotic equivalence
For population-based cohorts, correlation of −log10P values between the two frameworks generally increased with sample sizes. Out of 14 population-based cohorts, 8 cohorts had excellent agreement between the two frameworks showing correlations over 0.99 for both tests and for both smoking measures (Figure S1): the sample size of these population-based cohorts ranges from 1,663 to 22,983. For the Women’s Genome Health Study (WGHS, N=22,983, 11.7% CurSmk; 49.1% EverSmk), the largest cohort, both frameworks provided almost identical −log10P values, demonstrating the asymptotic equivalence (the last row in Figure S1).
Family-based cohorts
For family-based cohorts, we found less agreement between the two frameworks. For Health, Risk Factors, Exercise Training and Genetics (HERITAGE; N=499; 15% CurSmk; 38% EverSmk), the smallest family-based cohort, the correlation between the two frameworks for the 1 DF test was 0.78 and 0.88 for CurSmk and EverSmk, respectively (the first row in Figure 2). In contrast to population-based cohorts, agreement between the two frameworks did not increase with their sample sizes for family-based cohorts. Out of 6 family-based cohorts, only one cohort GENOA (N = 1,064; 16% CurSmk; 50% EverSmk) showed correlations over 0.99 for both tests and for both smoking measures (Figure 2). The Framingham Heart Study (FHS; N=8,195; 31% CurSmk; 52% EverSmk) is the largest family-based cohort, but the correlation between the two frameworks for the 1 DF test was only 0.94 for both smoking measures (the last row in Figure 2). These correlations were less than those found for the smallest population-based cohort CROATIA-Korcula (N=456).
The complexity of pedigree structure may have a greater impact on the agreement between the two frameworks than sample sizes alone. The GENOA cohort consists of mostly sibling pairs without parents and therefore has the simplest pedigree structure. FamHS, HERITAGE and HyperGEN cohorts have mostly nuclear families. Two remaining cohorts ERF and FHS consist of multi-generation families and therefore have more complex pedigree structures. In family-based cohorts, in particular with large extended pedigrees, most families often are split into the two strata under the stratified framework (making the strata non-independent). Note that the 1 DF test in the stratified framework includes the Spearman rank correlation coefficient between stratum-specific genetic effects to correct for any relatedness between E = 1 and E = 0 strata in Equation (3). Indeed, we observed higher Spearman rank correlation between stratum-specific effects with family-based cohorts (Table 3), ranging from 0.000 to 0.016 with population-based cohorts, and from 0.017 to 0.105 with family-based cohorts. Although the 2 DF test in the stratified framework does not take account for such potential relatedness across strata, correlation between two frameworks for the 2 DF test was generally higher than correlation for the 1 DF test.
Table 3.
Spearman rank correlation coefficients between the two stratum-specific genetic effects calculated from the genome-wide results used for the 1 DF test in the stratified framework
| Cohort | CurSmk | EverSmk | |
|---|---|---|---|
| Cohort-level for population-based cohorts | CROATIA-Korcula | 0.000 | −0.003 |
| CROATIA-Vis | 0.014 | 0.005 | |
| BioMe | 0.001 | 0.002 | |
| CARDIA | 0.000 | −0.002 | |
| HealthABC | 0.007 | 0.010 | |
| RS2 | 0.003 | −0.001 | |
| AGES | 0.016 | 0.014 | |
| MESA | 0.012 | 0.044 | |
| RS3 | 0.006 | 0.006 | |
| CHS | 0.001 | 0.004 | |
| RS1 | 0.013 | 0.005 | |
| GS:SFHS | 0.003 | 0.006 | |
| ARIC | 0.012 | 0.012 | |
| WGHS | 0.014 | 0.027 | |
|
| |||
| Cohort-level for family-based cohorts | HERITAGE | 0.105 | 0.076 |
| GENOA | 0.017 | 0.030 | |
| HyperGEN | 0.052 | 0.093 | |
| ERF | 0.053 | 0.066 | |
| FamHS | 0.071 | 0.078 | |
| FHS | 0.091 | 0.112 | |
|
| |||
| Meta-level | Population-based cohorts | 0.034 | 0.045 |
| Family-based cohorts | 0.090 | 0.095 | |
| All cohorts | 0.055 | 0.065 | |
Meta-analysis Results
Meta-analysis was performed under three scenarios: 1) using 14 population-based cohorts, 2) using 6 family-based cohorts and 3) using all 20 cohorts. For each scenario, meta-analysis was performed once for the joint framework and twice (using two filtering schemes) for the stratified framework. Figure 4 shows the agreement between the two frameworks when the stratified framework used a filtering scheme A. Figure 5 shows the agreement when the stratified framework used scheme B. Correlation is shown in Table 4. We observed that scheme B improved the agreement between the two frameworks.
Figure 4.
Scatterplots of meta-level −log10(p) values using a scheme A in the stratified framework. The joint framework used a filtering scheme B. Row 1 shows results for meta-analysis including the 14 population-based cohorts, row 2 for meta-analysis including the 6 family-based cohorts, and row 3 for meta-analysis including all 20 cohorts.
Figure 5.
Scatterplots of meta-level −log10(p) values using a scheme B in the stratified framework. The joint framework used a filtering scheme B. Row 1 shows results for meta-analysis including the 14 population-based cohorts, row 2 for meta-analysis including the 6 family-based cohorts, and row 3 for meta-analysis including all 20 cohorts.
Table 4.
Correlation between the two frameworks for meta-analysis results. Scatterplots are shown in Figures 4 and 5.
| Stratified Framework with | Meta-analysis with | CurSmk | EverSmk | ||
|---|---|---|---|---|---|
| 1DF | 2DF | 1DF | 2DF | ||
| Scheme A | Population cohorts | 0.942 | 0.970 | 0.950 | 0.982 |
| Family cohorts | 0.860 | 0.893 | 0.889 | 0.924 | |
| All cohorts | 0.904 | 0.947 | 0.927 | 0.965 | |
|
| |||||
| Scheme B | Population cohorts | 0.957 | 0.990 | 0.965 | 0.995 |
| Family cohorts | 0.882 | 0.946 | 0.905 | 0.950 | |
| All cohorts | 0.923 | 0.98 | 0.948 | 0.985 | |
Filtering schemes
Each cohort contributed more variants to meta-analysis with filtering scheme A (applying the MAC filter separately to each stratum) (Table S3). This is more noticeable in cohorts with small sample sizes with CurSmk variable because of the unbalanced sample sizes between the two strata. For example, the CROATIA-Korcula cohort contributed 8.46 million variants to E=0 stratum meta-analysis but 6.641 million variants to E=1 meta-analysis under scheme A. The difference (roughly 1.82 million) corresponding to the number of variants with MAC0 ≥ 10 and MAC1 < 10 arose from highly unbalanced sample sizes in the two strata. Under scheme B (applying the same filter to both strata in the stratified framework and in the joint framework), a smaller number of variants (6.640 million for CROATIA-Korcula) were contributed to the meta-analysis as variants needed to have MAC0 ≥ 10 and MAC1 ≥ 10.
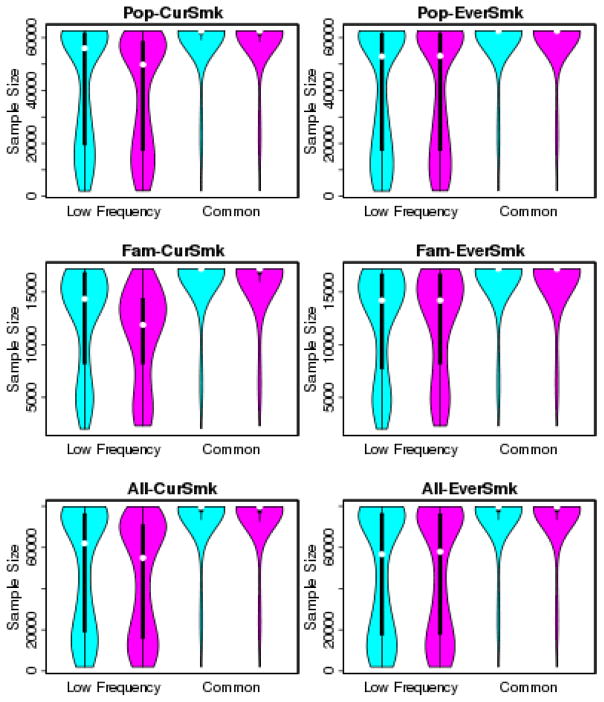
The final number of variants resulting from meta-analysis was slightly larger under scheme A (9.76 million variants under scheme A vs. 9.68 million variants under scheme B in meta-analysis combining all cohorts for CurSmk, Table S4). The difference was mostly from low frequency variants (with 1% ≤ MAF < 5%) (3.2 million variants under scheme A vs. 3.1 million under scheme B); there were 6.5 million common variants (with MAF ≥ 5%) under both schemes. Because each cohort contributed more variants under scheme A, there were more cohorts contributing to each variant, resulting in larger sample sizes under scheme A. The difference in the overall sample size, the sample size combined across multiple cohorts, was more notable for low frequency variants and for CurSmk (Figure 3).
Figure 3.
Violin plots of sample sizes arising from meta-analysis under two filtering schemes. Cyan color under scheme A (a stratum-specific filter) and magenta color under scheme B. Row 1 shows results for meta-analysis including the 14 population-based cohorts (with total sample size 62,548), row 2 for meta-analysis including the 6 family-based cohorts (with sample size 17,183), and row 3 for meta-analysis including all 20 cohorts (with total sample size 79,731).
In meta-analysis, the stratified framework had higher genomic control lambda values for the 1 DF test, regardless of filtering schemes. The Spearman rank correlation between stratum-specific effects for the 1 DF test was also slightly increased (0.034) after meta-analysis of population-based cohorts (Table 3). The lambda values for the 2 DF test were generally similar between the two frameworks (Table S5).
Population-based vs. family-based results
Regardless of the schemes (A and B), we found a surprising reduction of agreement between the two frameworks in meta-analysis compared to cohort-specific analyses. For meta-analysis combining 14 population-based cohorts (with a total sample size of 62,548), the correlation between the two frameworks for the 1 DF test was 0.94 and 0.96 with the use of schemes A and B in the stratified framework, respectively, for CurSmk (the top left in Figures 4 and 5). Note that we had found higher correlations on the cohort-level: for population-based cohorts, about 80% of sample (49,450 subjects) was from the 8 cohorts that had correlation over 0.999 between the two frameworks for the 1 DF test and for CurSmk. Compared to the 1 DF test, using the 2 DF test generally increased the significance of p-values, possibly reflecting true main effect associations that are missed by the 1 DF tests. The 2 DF test also had higher correlation between the two frameworks compared to the 1 DF test.
When meta-analysis included family-based cohorts, the level of agreement became even less. For meta-analysis combining the 6 family-based cohorts (with sample size 17,183), the correlation between the two frameworks for the 1 DF test was 0.86 and 0.88 with the use of schemes A and B in the stratified framework, respectively, for CurSmk (the middle left in Figures 4 and 5). Again, these correlation values on the meta-level were lower than those observed on the cohort-level. Furthermore, there were a noticeable number of variants that had highly discrepant p-values between two frameworks using the 2 DF test with the use of the scheme A in the stratified framework (the middle second and fourth columns in Figure 4).
When all 20 cohorts were combined (with total sample size 79,731), the correlation was approximately the average of the two values for the two meta-analysis results (population-based and family-based). With the use of scheme A in the stratified framework, the scatterplot for the 2 DF test still included those variants with highly discrepant p-values between two frameworks (the last row of Figure 4).
Low-frequency vs common variants
To examine how the concordance between the two frameworks depends on the MAF, we generated two scatterplots for each scatterplot in Figures 4 and 5, one including about 3 million low frequency variants (with MAF < 5%) and another including 6.5 million common variants (with MAF ≥ 5%). The filtering scheme in the stratified framework had a larger impact on the concordance of low frequency variants (Supplemental Figures 2 and 4). For common variants, the two schemes for the stratified framework provided almost identical performance, providing similar agreement between the two frameworks (Supplemental Figures 3 and 5). Moreover, when meta-analysis included family-based cohorts (rows 2 and 3 of Figure 4), those variants that showed highly discrepant p-values between the two frameworks were all low frequency variants (Figure S2).
To further understand this discrepancy for low frequency variants, we examined the variants from the meta-analysis of the 6 family-based cohorts for the CurSmk measure (the middle second in Figure 4). The three selected variants are presented in Table 5. For all variants, meta-analysis for the E=1 stratum is identical regardless of filtering schemes. The difference came from meta-analysis of the E = 0 stratum. For example, with the first variant (2:48619812, MAF = 1.2%), the meta-analysis for E = 0 stratum used 3 cohorts under scheme A but one cohort (FamHS) under scheme B. When two remaining cohorts were included, the final 2 DF p-values were changed dramatically. The second variant shared this feature although all 6 cohorts contributed to the scheme A meta-analysis for E=0 stratum. However, the 2 DF p-values for both schemes were similar for the third variant. It appears that the use of the generalized estimating equations (GEE) approach for the analysis of the family-based cohorts may lead to spurious results for low frequency variants. This finding is consistent with the recent publication [Sitlani, et al. 2015]. The variants that showed highly discrepant p-values from meta-analysis combining all cohorts (the third row in Figure 4) also shared this feature.
Table 5.
Comparison of schemes A and B for family-based meta-analysis for CurSmk at select variants
| Marker | Level | Type | N | MAC | MAC | Effect | StdErr | P | Stratified | Interaction |
|---|---|---|---|---|---|---|---|---|---|---|
| E=0 | E=0 | E=1 | E=0 | E=0 | E=0 | 2DF P | 2DF P | |||
| 2:48619812 (MAF=1.2%) | Meta | Scheme A | 4,479 | 12.8 | 1.0 | 7.8E-41 | 8.5E-40 | |||
| Meta | Scheme B | 3,160 | 0.6 | 2.7 | 0.83 | 0.63 | 0.89 | |||
| Cohort | FamHS | 3,160 | 77.7 | 13.7 | 0.6 | 2.6 | 0.83 | 0.60 | 0.89 | |
| Cohort | GENOA | 895 | 29.9 | <10 | 3.2 | 3.1 | 0.30 | |||
| Cohort | HERITAGE | 424 | 10.3 | <10 | 15.8 | 1.0 | 1.2E-54 | |||
|
| ||||||||||
| 6:142093034 (MAF=1.7%) | Meta | Scheme A | 12,798 | 4.7 | 0.5 | 3.8E-23 | 2.2E-22 | |||
| Meta | Scheme B | 10,342 | −0.1 | 1.0 | 0.89 | 0.47 | 0.61 | |||
| Cohort | ERF | 1,507 | 66.9 | 52.7 | 1.2 | 2.1 | 0.58 | 0.68 | 0.78 | |
| Cohort | FamHS | 3,160 | 140.3 | 25.2 | 0.2 | 1.6 | 0.88 | 0.98 | 0.89 | |
| Cohort | FHS | 5,675 | 141.8 | 63.0 | −0.5 | 1.5 | 0.74 | 0.47 | 0.5 | |
| Cohort | GENOA | 895 | 43.8 | <10 | −4.0 | 2.6 | 0.12 | 0.15 | 0.12 | |
| Cohort | HERITAGE | 424 | 19.1 | <10 | −6.3 | 0.5 | 1.9E-33 | |||
| Cohort | HyperGEN | 1137 | 30.5 | <10 | 0.3 | 4.4 | 0.95 | |||
|
| ||||||||||
| 12:5679139 (MAF=1.3%) | Meta | Scheme A | 4,479 | 1.9 | 2.1 | 0.37 | 1.8E-09 | |||
| Meta | Scheme B | 3,160 | 4.1 | 2.8 | 0.15 | 5.0E-09 | 1.9E-11 | |||
| Cohort | FamHS | 3,160 | 84.1 | 12.5 | −4.1 | 2.8 | 0.14 | 6.3E-10 | 3.8E-12 | |
| Cohort | GENOA | 895 | 33.5 | <10 | 3.1 | 3.7 | 0.41 | 0.2 | 0.21 | |
| Cohort | HERITAGE | 424 | 10.5 | <10 | −3.6 | 4.8 | 0.46 | 0.59 | 2.4E-08 | |
Discussion
Gene-environment interactions play important roles in the pathobiology of disease traits, improving our understanding about which combinations of genes and environments may be predisposed to unfavorable health outcomes. Modeling gene-lifestyle interactions may discover more trait loci through context dependent (or “refined”) main effects as well as true interactions. To actively investigate the role of such interactions on cardiovascular traits, we have established a Gene-Lifestyle Interactions Working Group within the CHARGE Consortium. The working group includes over 50 cohorts from around the world, spanning four race/ethnic groups (European, African, Hispanic, and Asian ancestry). This offers us an opportunity to compare and contrast two analysis frameworks for studying gene-environment interactions.
Using actual results from 20 cohorts of European ancestry, we empirically compared the two frameworks. In cohort-specific analyses, we observed that agreement between the two frameworks were generally good and depended on 1) balance between sample sizes of the two strata, 2) total sample size, and 3) whether the cohort is population-based or family-based. In meta-analyses, agreement between the two frameworks was less than that observed in cohort-specific analyses, despite the increased sample size. In meta-analyses, agreement depended on 1) the minor allele frequency, 2) inclusion of family-based cohorts in meta-analysis, and 3) filtering scheme. The discrepancy was more notable for low frequency variants.
The joint framework that considers the genetic main and interaction effects jointly in a single linear model has been the main statistical approach for studying interactions. It utilizes the entire sample and works well whether environmental exposures are categorical or continuous. The stratified framework has emerged because main-effect models are readily available in many software packages and easier to implement in a large-scale consortium setting. However, the stratified framework, appropriate for population-based cohorts, was developed to approximate the joint framework. Our findings from cohort-specific results support the equivalence between the two frameworks for population-based cohorts. For family-based cohorts, however, we found less agreement between the two frameworks. Most family-based cohorts, in particular large extended pedigrees, include both exposed and unexposed members within each family. The stratified framework is unable to fully account for family structures across strata. The Spearman rank correlation coefficient in the 1 DF test may partly correct for any correlation between the strata (that may arise from family data). In contrast, the 2 DF test does not take into account any relatedness across the strata: the null distribution of the 2 DF test holds when the exposed and unexposed groups are independent. We observed that the stratified framework was less suitable for approximating the joint framework for family studies with complex pedigree structures (such as the Framingham heart study).
To increase the sample sizes, most large scale consortia include both population-based and family-based studies. It is also becoming standard to perform analysis of low frequency variants imputed using the 1000 Genomes project. In our meta-analysis, we had about 3 million low frequency variants. However, with inclusion of family-based studies in meta-analysis, disagreement between the two frameworks was more pronounced for low frequency variants. With the use of stratum-specific filters, we observed less agreement and a notable number of variants that had highly discrepant p-values between the two frameworks, where 20% of subjects were from family-based cohorts. If the stratified framework is already in use, then using a consistent filter for both strata may improve the agreement, thereby providing a similar inference as the joint framework.
To our knowledge, this is the first report comparing the joint and stratified frameworks using real data. The stratified framework appears to approximate the joint framework well only for common variants in population-based cohorts. We conclude that the joint framework is the preferred approach and should be used to control false positives when dealing with low frequency variants and/or family-based cohorts. As our findings were based on an empirical evaluation using one phenotype, they may not be generalized to all situations. Even though we focused on a continuous outcome, the methods are generally applicable to dichotomous outcomes under the logistic regression framework [Aschard, et al. 2010; Magi, et al. 2010]. With dichotomous outcomes, we expect similar conclusion but may require more stringent MAC thresholds to produce valid logistic regression results [Ma, et al. 2013]. A more comprehensive investigation covering the various scenarios with both continuous and dichotomous outcomes, among others, would strengthen our findings.
Supplementary Material
Acknowledgments
We thank anonymous reviewers for their constructive and insightful comments. The work was partly supported by R01HL118305 and K25HL121091 from the National Heart, Lung, and Blood Institute (NHLBI), national Institutes of Health (NIH). Study-specific acknowledgments are included in the Supplemental Materials.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Hancock DB, London SJ, Kraft P. Genome-wide meta-analysis of joint tests for genetic and gene-environment interaction effects. Hum Hered. 2010;70(4):292–300. doi: 10.1159/000323318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens G, Winkler TW, Gorski M, Leitzmann MF, Heid IM. To stratify or not to stratify: power considerations for population-based genome-wide association studies of quantitative traits. Genet Epidemiol. 2011;35(8):867–79. doi: 10.1002/gepi.20637. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17(R2):R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14(6):379–89. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- Halekoh U, Hojsgaard S, Yan J. The R Package geepack for Generalized Estimating Equations. Journal of Statistical Software. 2006;15(2):1–11. [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–9. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- Le Marchand L, Wilkens LR. Design considerations for genomic association studies: importance of gene-environment interactions. Cancer Epidemiol Biomarkers Prev. 2008;17(2):263–7. doi: 10.1158/1055-9965.EPI-07-0402. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Blackwell T, Boehnke M, Scott LJ, Go TDi. Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37(6):539–50. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34(8):846–53. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AK, LaValley M, Liu CT, Rice K, An P, Liu Y, Miljkovic I, Rasmussen-Torvik L, Harris TB, Province MA, et al. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP x environment regression coefficients. Genet Epidemiol. 2011;35(1):11–8. doi: 10.1002/gepi.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41(6):666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall JC, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL, Kilpelainen TO, Esko T, Magi R, Li S, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9(6):e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitlani CM, Rice KM, Lumley T, McKnight B, Cupples LA, Avery CL, Noordam R, Stricker BH, Whitsel EA, Psaty BM. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Stat Med. 2015;34(1):118–30. doi: 10.1002/sim.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchetgen Tchetgen EJ, Kraft P. On the robustness of tests of genetic associations incorporating gene-environment interaction when the environmental exposure is misspecified. Epidemiology. 2011;22(2):257–61. doi: 10.1097/EDE.0b013e31820877c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. Gene--environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11(4):259–72. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24(19):2911–35. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- Voorman A, Lumley T, McKnight B, Rice K. Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS One. 2011;6(5):e19416. doi: 10.1371/journal.pone.0019416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, Ferreira T, Fall T, Graff M, Justice AE, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192–212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Kutalik Z, Gorski M, Lottaz C, Kronenberg F, Heid IM. EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics. 2015;31(2):259–61. doi: 10.1093/bioinformatics/btu621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A. Object-oriented computation of sandwich estimators. Journal of Statistical Software. 2006;16(9) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.