Abstract
Our present day understanding of nervous system development is an amalgam of insights gained from studying different aspects and stages of nervous system development in a variety of invertebrate and vertebrate model systems, with each model system making its own distinctive set of contributions. One aspect of nervous system development that has been among the most extensively studied in the nematode Caenorhabditis elegans is the nature of the gene regulatory programs that specify hardwired, terminal cellular identities. I first summarize a number of maps (anatomical, functional, and molecular) that describe the terminal identity of individual neurons in the C. elegans nervous system. I then provide a comprehensive summary of regulatory factors that specify terminal identities in the nervous system, synthesizing these past studies into a regulatory map of cellular identities in the C. elegans nervous system. This map shows that for three quarters of all neurons in the C. elegans nervous system, regulatory factors that control terminal identity features are known. In‐depth studies of specific neuron types have revealed that regulatory factors rarely act alone, but rather act cooperatively in neuron‐type specific combinations. In most cases examined so far, distinct, biochemically unlinked terminal identity features are coregulated via cooperatively acting transcription factors, termed terminal selectors, but there are also cases in which distinct identity features are controlled in a piecemeal fashion by independent regulatory inputs. The regulatory map also illustrates that identity‐defining transcription factors are reemployed in distinct combinations in different neuron types. However, the same transcription factor can drive terminal differentiation in neurons that are unrelated by lineage, unrelated by function, connectivity and neurotransmitter deployment. Lastly, the regulatory map illustrates the preponderance of homeodomain transcription factors in the control of terminal identities, suggesting that these factors have ancient, phylogenetically conserved roles in controlling terminal neuronal differentiation in the nervous system. WIREs Dev Biol 2016, 5:474–498. doi: 10.1002/wdev.233
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
When Sydney Brenner established Caenorhabditis elegans as a model system his vision was to first draw a series of maps—anatomical, lineage and functional maps—that describe this simple metazoan organism and to then use these maps as a starting point for microbial‐style genetic analysis.1, 2 His explicit focus was on the nervous system and his stated goal was to define the genetic programs that instruct the development and function of this most complex of all tissue types.1, 2 And, indeed, we have come a long way. Based on Brenner's initial efforts, no other nervous system is as extensively characterized on so many different levels—anatomically, functionally, and developmentally—as that of C. elegans.3
The nervous system of C. elegans is composed of 302 neurons in the hermaphrodite and 385 in the male. The position and shape of each neuron has been precisely described and the connectivity of neurons has been elucidated, both in the hermaphrodite and male (Figures 1(a)–(c)).6, 9 In addition to these anatomical maps, easily accessible at www.wormatlas.org,3 there is a comprehensive lineage map that describes the developmental history of every single cell of the animal (Figure 1(d)).10, 11 Both the anatomical and lineage maps have been used with tremendous success to identify mutants in which neuronal anatomy or lineages are abnormal. Among the major findings of initial genetic screens were the identification of molecular cues involved in axon pathfinding (e.g., the unc‐6/Netrin system),12, 13 heterochronic timer systems involved in neuroblast patterning (involving the first miRNAs to be discovered, lin‐4 and let‐7 14, 15) and factors involved in synaptic specificity (unc‐4 homeobox gene).16
Figure 1.
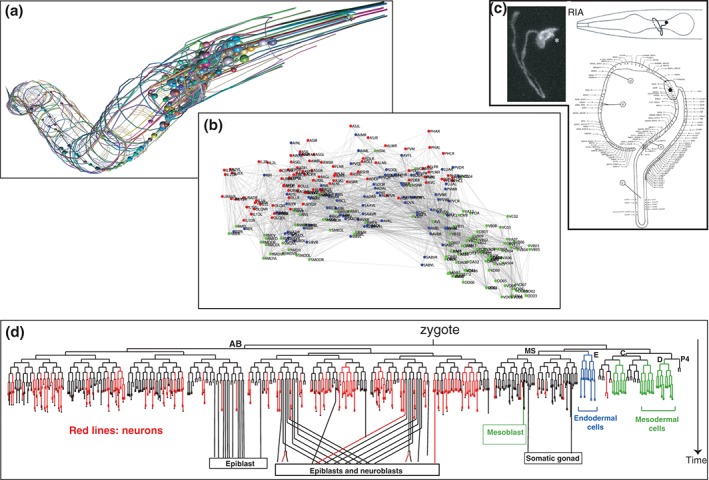
An anatomical and lineage map of the C. elegans nervous system. (a) Illustrations of the entire nervous system and fascicles, kindly provided by Openworm.org. (b) Connectome. (Adapted from Ref 4.) (c) One example of a single neuron type, the glutamatergic RIA interneurons, labeled with gfp (Adapted from Ref 5.) and its synaptic connectivity. (Adapted from Ref 6.) (d) Lineage of cells generated in the embryo. Red lines indicate neurons.
Another map established over the years can be termed a functional map of the nervous system. This map—which is still incomplete but nevertheless remarkably advanced—is based mostly on assessing the behavioral consequences of killing individual neurons through laser ablation. Such microsurgical approaches have revealed functions for almost all sensory neurons, most motor neurons and more than half of all interneurons (Table 1). For example, laser ablation of sensory neurons with specific microtubular structures identified their roles as touch receptor neurons,29 ablation of amphid wing neurons revealed their role in olfaction,33 or ablation of the AIY interneurons revealed their function in thermosensory information processing.26 One of the fascinating themes that emerged over the years is the multifunctionality of many neurons (which is not comprehensively displayed in Table 1). For example, many sensory neurons are polymodal, detecting a wide range of distinct sensory modalities.36, 38, 55
Table 1.
Functional Map of the Extrapharyngeal Nervous System
| Sensory Neuron | Function | Reference | Interneuron | Function | Reference | Motor neuron | Function | Reference |
|---|---|---|---|---|---|---|---|---|
| ADE | Mechanosensory | 17, 18 | ADA | AS | ||||
| ADF | Dauer formation | 19 | AIA | Processing sensory information | 20 | AVL | Defecation | 21 |
| ADL | Olfaction (repulsive) | 22 | AIB | Reversal, pausing, integrating sensory information | 20, 23, 24 | DA | Locomotion | 25 |
| AFD | Thermosensation | 26 | AIM | Sexual attraction | DB | Locomotion | 25 | |
| ALA | Sleep, mechanosensory | 27, 28 | AIN | DD | Locomotion | 21 | ||
| ALM | Mechanosensory | 29 | AIY | Locomotion, integrating sensory information | 26, 30 | DVB | Defecation | 21 |
| ALN | AIZ | Locomotion, processing sensory information | 26, 30 | HSN | Egg‐laying | |||
| AQR | Aggregation, O2 sensation | 31, 32 | AUA | Aggregation | 31 | PDA | ||
| ASE | Gustation | 33 | AVA | Command interneurons | 25, 34 | PDB | ||
| ASG | Gustation | 19, 33 | AVB | Command interneurons | 25, 34 | RIM | Reversal behavior, feeding circuit | 23, 35 |
| ASH | Mechanosensory, gustation | 36 | AVD | Command interneurons | 25, 34 | RMD | Head movement, foraging | 23, 37 |
| ASI | Dauer formation, feeding state, chemosensory, thermosensory | 19, 33, 38 | AVE | Command interneurons | 25, 34 | RME | Head movement | 21 |
| ASJ | Dauer formation and recovery | 19 | AVF | Egg‐laying/locomotion | 39 | RMF | ||
| ASK | Gustation | 33 | AVG | Pioneer/guidepost | 40 | RMG | Aggregation | 41 |
| AVM | Mechanosensory | 29 | AVH | RMH | ||||
| AWA | Olfaction | 33 | AVJ | SAB | Head movement | 42 | ||
| AWB | Olfaction | 33 | AVK | SIA | Guidepost | 43 | ||
| AWC | Olfaction, thermosensation | 33, 44 | BDU | Mechanosensory | 45 | SIB | Guidepost | 43 |
| BAG | CO2 sensation | 46 | CAN | Fluid homeostasis | SMB | Sinusoidal locomotion | 23 | |
| CEP | Mechanosensory | 17, 18 | DVA | Stretch reception | 47 | SMD | Omega turns | 23 |
| FLP | Mechanosensory | 36 | DVC | VA | Locomotion | 25 | ||
| IL1 | Head bends, foraging | 37 | LUA | Pausing | VB | Locomotion | 25 | |
| IL2 | Nictation behavior | 48 | PVC | Command interneurons | 25 | VC | Egg‐laying | 49 |
| OLL | Pathogen avoidance behavior | 50 | PVN | VD | Locomotion | 21 | ||
| OLQ | Mechanosensory, head bends, foraging | 36, 37 | PVP | |||||
| PDE | Mechanosensory | 17, 18 | PVQ | |||||
| PHA | Gustation, mechanosensory | 45, 51 | PVR | |||||
| PHB | Gustation, mechanosensory | 45, 51 | PVT | Pioneer/guidepost | 52 | |||
| PHC | Heat avoidance | 45, 53 | PVW | |||||
| PLM | Mechanosensory | 29 | RIA | Thermosensory processing, quiescence | 26, 54 | |||
| PLN | RIB | Reversal behavior | 23, 30 | |||||
| PQR | Aggregation, O2 sensation | 31, 32 | RIC | Feeding circuit | 35 | |||
| PVD | Mechanosensory, thermosensory | 55 | RID | |||||
| PVM | Mechanosensory | 29 | RIF | Dwelling | 56 | |||
| URA | RIG | |||||||
| URB | RIH | Processes nose touch | 57 | |||||
| URX | O2 sensation | 31, 32 | RIP | Coupling touch and pharyngeal pumping | 25 | |||
| URY | RIR | |||||||
| RIS | Quiescence | 58 | ||||||
| RIV | Omega turns | 23 | ||||||
| SAA | ||||||||
| SDQ | ||||||||
| 38 | 42 | 24 |
One of many values of these functional maps became apparent when mutant animals were identified from genetic screens that precisely phenocopy the ablation of specific neurons. This led to the identification of, e.g., sensory receptors that act in functionally defined sensory neurons (e.g., touch receptor proteins61 and olfactory receptors62) or transcription factors that were found to be required for the development of specific neuron types (e.g., the mec‐3 transcription factor, whose loss results in mechanosensory defects similar to those observed upon ablation of touch sensory neurons63; the che‐1 transcription factor whose loss results in chemotaxis defects similar to those observed upon laser ablation of the ASE gustatory neurons64; or the ttx‐1 and ttx‐3 transcription factors whose losses results in thermotaxis defects similar to those observed upon ablation of the AFD or AIY interneurons65, 66).
With the advent of a number of molecular tools, yet another type of map has gained successively more prominence in the past two decades, a molecular map of the nervous system (Figure 2). In this review, I shall describe molecular maps of the C. elegans nervous system and summarize how important they have been to understand the acquisition of terminal neuronal identity features, allowing us to now establish a conglomerate regulatory map of the nervous system.
Figure 2.
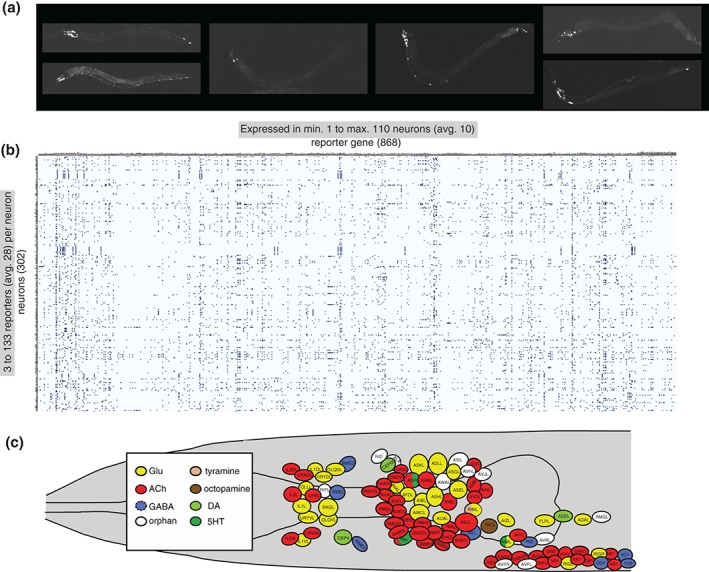
Molecular maps of the C. elegans nervous system. (a) Examples of transgenic worms expressing gfp reporter transgenes. (Reprinted with permission from Ref 7. Copyright 2004) (b) Manually curated expression patterns of transgenes throughout the C. elegans nervous system, kindly extracted from www.wormbase.org by Daniela Raciti and Wen Chen and organized by Lori Glenwinkel. Note that these transgenes may not display the complete expression pattern of the respective gene, but they nevertheless serve as invaluable readouts of the regulatory state of a neuron. (c) Schematic map of neurotransmitter usage in the C. elegans head. (Reprinted with permission from Ref 8. Copyright 2015)
MOLECULAR MAPS OF THE C. ELEGANS NERVOUS SYSTEM
Ever since reporter gene technology was introduced into C. elegans, several thousand reporter lines have been generated to monitor the expression of specific genes (examples are shown in Figure 2(a)). In many cases, reporter genes were generated for genes with suspected functions in the nervous system, such as sensory receptors,22, 67 neuropeptides,68, 69 ion channels,70 neurotransmitter receptors,71 or putative neuronal adhesion/recognition molecules.52 As expected, most of these reporter constructs showed neuron‐type specific expression profiles. In addition, the reporter‐based expression analysis of the many genes characterized for various phenotypes within or outside the nervous system yielded a tremendous number of additional reagents to monitor neuron‐type specific gene expression. Since expression patterns are commonly analyzed in the context of a mature nervous system (larval or early adult stages), in which the identity of neurons can be most readily defined, there has been an inadvertent bias toward the characterization of expression patterns in postmitotic, mature neurons. Even though the precise sites of cellular expression of many reporter constructs (particularly those resulting from large scale expression screens conducted in Vancouver72) have not been determined, the site of expression of 868 reporter genes within the nervous system have been analyzed with single neuron resolution by the C. elegans community, creating more than 260,000 data points (868 × 302). These data points can be extracted from Wormbase and are displayed in Figure 2(b). On average, each of these 868 reporters is expressed in 10 neurons (with a range of 1–110 neurons; very few reporter genes are expressed exclusively in single neuron classes and those that are encode mostly sensory receptor proteins). And, on average, any given neuron type is associated with the expression of 28 distinct reporter genes (ranging from at least 3 to as many as 133). The extent to which each neuron type can be associated with molecular markers is unprecedented in other models, including flies and mice. While gene expression atlases have been generated in these models,73, 74 they usually suffer from a lack of cellular resolution owing to the much more complex cellular anatomy of the fly and mouse nervous systems.
It is important to point out that these molecular maps describe regulatory endpoints, i.e., terminal effector genes that are continuously expressed throughout the life of the neuron and that most often code for proteins that define the mature, functional and anatomical properties of a postmitotic neuron type (e.g., ion channels, sensory receptors, neurotransmitter receptors, and neuropeptides). Equally important, owing to their unsystematic generation by many different labs for many different purposes, these transgenic markers provide a relatively unbiased snapshot of functionally unrelated identity features of a mature neuron.
AN (ALMOST COMPREHENSIVE) NEUROTRANSMITTER MAP
One specific subtype of molecular brain maps is a neurotransmitter map. Neurotransmitter maps are invaluable complements to any anatomical map (specifically, a connectome) as they inform us about the nature of information flow in the nervous system. Like any other organism, the C. elegans nervous system utilizes a plethora of distinct neurotransmitter systems, from fast‐acting neurotransmitters (GABA, glutamate, and acetylcholine) to aminergic neurotransmitters (dopamine, serotonin, octopamine, and tyramine) to more than 200 modulatory neuropeptides.75 Given the diversity of neurotransmitter systems, the neurotransmitter identity of a neuron is a key molecular identity feature of a neuron. While the identity of GABAergic and aminergic neurons has been known for a while (which amount to ~10% of all neuron classes76, 77), it was only the recent analysis of glutamatergic and cholinergic neurons that brought the coverage of neurotransmitter identities to about 90% of all neurons in the nervous system. Cholinergic and glutamatergic neurotransmitter identities were inferred through the systematic expression analysis of vesicular transporters for acetylcholine (ACh) and glutamate (Glu), demonstrating that these two transmitters are the most broadly used neurotransmitters in the C. elegans nervous system (Figure 2(c); ACh: 52 of all 118 neuron classes; Glu 38 of all 118 neuron classes).8, 78 Notable features of the C. elegans neurotransmitter map include the broad usage of ACh and Glu in different neuron types of the nervous system (each utilized by sensory, inter, and motor neurons)8, 78 and its association with specific circuitry (e.g., the ventral nerve cord motor neuron circuit, composed of motor neurons and innervating command interneurons is cholinergic8). Notably, from a developmental standpoint, neurons that share the same neurotransmitter show no apparent lineage relationships (Figure 3).
Figure 3.
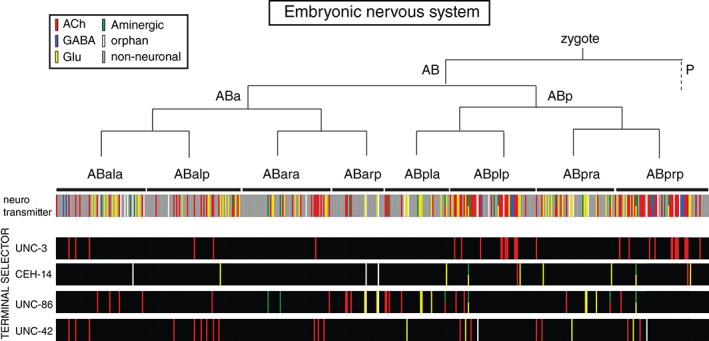
Lack of correlation of lineage with neurotransmitter identity or transcription factor expression. Each bar represents an individual cell. See text for references.
A REGULATORY MAP OF THE NERVOUS SYSTEM
Much like the anatomical, lineage, and functional maps of the worm nervous system, available molecular maps have served as powerful starting points for incisive genetic loss of function analysis. Specifically, reporter genes that are expressed in particular neuron types have offered the opportunity to engage in a bottom–up analysis which seeks to define the regulatory factors that control these terminal neuronal identity features. This type of analysis permitted a significant expansion of the phenotypic analysis of some classical transcription factor mutants. For example, genetic elimination of the LIM homeobox gene mec‐3 phenocopies the loss of mechanosensory neuron function,63 loss of the LIM homeobox gene ttx‐3 phenocopies the loss of AIY neuron function (in both cases, animals show a characteristic cryophilic thermotaxis phenotype),65 and loss of the zinc (Zn) finger transcription factor che‐1 phenocopies the chemotaxis defects of ASE sensory neuron ablation.64 However, it is a priori not clear whether such phenocopying is the result of the respective transcription factor merely regulating one or a few function‐defining genes (e.g., a chemoreceptor protein and/or mechanoreceptor channel) or whether these factors have much more profound effects on the identity of the respective neuron. The ability to phenotype the loss‐of‐function for a regulatory factor of course very much depends on the availability of descriptors of neuronal identity features. The existence of a relatively fine‐grained molecular phenotypic space of individual neuron type (in the form of marker transgenes) has traditionally set C. elegans apart from other systems in which the precise impact of a regulatory pathway on the identity of a neuron often cannot be assessed with the same level of precision.
In principle, the identification of genes controlling terminal identity features for specific neurons can be expected to address the following questions: (1) are the multiple molecular features that define a specific neuron type independently regulated by distinct regulatory factors? or are they coregulated? (2) are there recurrent themes in which the identity of distinct neuron types are controlled? (3) are neurons that share specific functional features, e.g., sensory neurons, or synaptically connected neurons, controlled by the same regulatory factors?
These questions have been extensively addressed over the past decade, either by (1) directly screening for mutants in which specific molecular markers show defective expression; (2) by examination of reporter gene expression in candidate mutants (i.e., loss‐of‐function mutations in transcription factors expressed in a specific neuron type); or (3) by the analysis of mutants retrieved from behavioral screens (the above‐mentioned classic behavioral mutants che‐1, ttx‐3, mec‐3, unc‐3, etc.). The results of dozens of reports ordered by neuron class (sensory, inter, and motor neurons) are summarized in the regulatory maps shown in Figures 4, 5, 6. In sum, transcriptional regulators have been identified that control the differentiation of 76 of the 104 C. elegans extrapharyngeal neuron classes (73%) which amounts to 217 of all the 282 extrapharyngeal neurons (77%). Each of these transcription factors is a terminal regulator since their expression persists throughout the life of the neuron and in the few cases explicitly examined, they have been found to also be continuously required to maintain the differentiated state (earlier, transiently acting factors are not considered here).137
Figure 4.
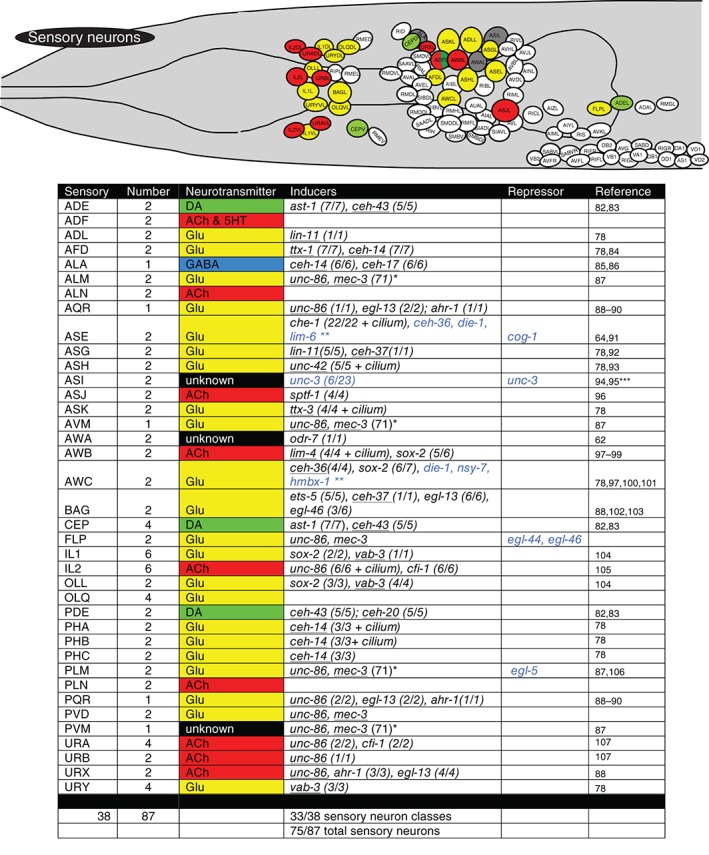
Terminal regulators of sensory neurons. Schematic worm with only sensory neurons colored and tabular summary of terminal regulators of neuronal identity. Color code of neurons in the schematic refer to neurotransmitter identity as show in the tabular summary. ‘Terminal regulators’ refers to the key property of these transcription factors: they are expressed in mature neurons throughout their lifetime, likely a reflection of their continuous role in maintaining the differentiated, terminal state. Early or transiently acting regulators are not shown. Only extrapharyngeal neurons are shown. Black font: most/all tested markers affected (terminal selector). Blue font: only subsets of markers affected. (x/y) indicates x markers out of y markers tested show defective expression. Homeodomain transcription factors are underlined. * indicates that in the case of the touch neurons, expression profiling has identified at least 71 mec‐3‐dependent genes. unc‐86 is known to regulate mec‐3 expression and cooperate with mec‐3 to control touch neuron‐expressed genes.79 ** indicates that the blue labeled factors control only a small subset of left/right asymmetrically expressed chemoreceptors,80, 81 but not bilateral identity of these neurons. *** indicates our own unpublished data. ‘Cilium’ indicates that the respective regulators also control cilium structure, as assessed by dye‐filling defects observed in the respective mutants.
Figure 5.
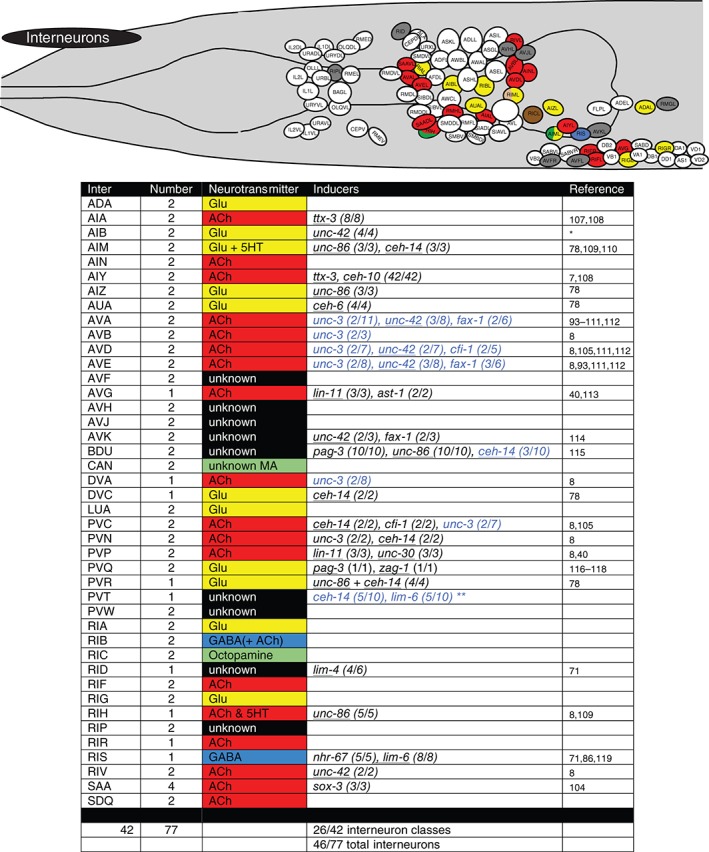
Terminal regulators of interneurons. Schematic worm with only interneurons colored and tabular summary of terminal regulators of neuronal identity. See legend to Figure 4 for an explanation of colors and numbers. * indicates our own unpublished data. ** lim‐6 and ceh‐14 regulate, in conjunction with the lin‐14 heterochronic gene, temporally controlled but not continuously expressed genes in PVT.120
Figure 6.
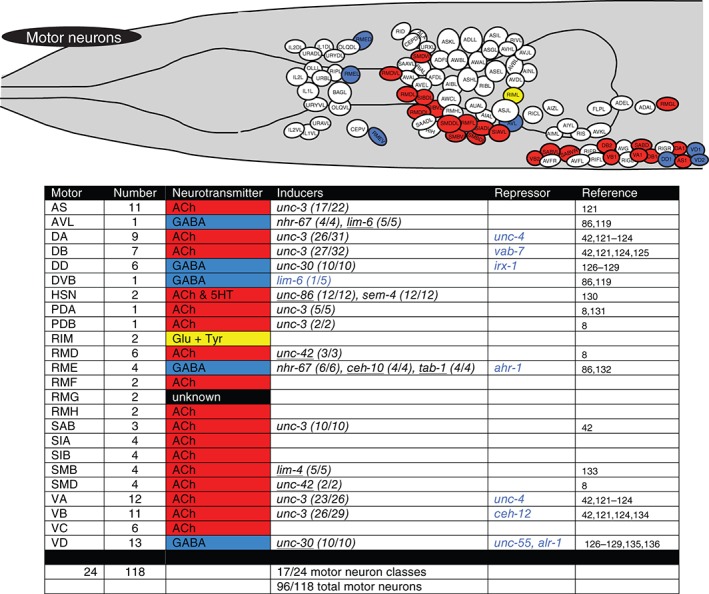
Terminal regulators of motor neurons. Schematic worm with only motor neurons colored and tabular summary of terminal regulators of neuronal identity. See legend to Figure 4 for an explanation of colors and numbers.
Coregulation Versus Piecemeal Regulation
Coming back to the question posed above—what do the transcription factors that control terminal identity features of a neuron exactly do? As discussed already above, a substantial phenotypic space can be probed for most individual neuron types—from anatomy, to connectivity, to molecular markers. It is particularly informative to consider the large panel of molecular markers, i.e., the molecular maps described above (and illustrated in Figure 2), which describe a host of biochemically linked features (e.g., enzymes that act in a pathway to synthesize a neurotransmitter), but more importantly, also biochemically unlinked molecular features of a neuron (as illustrated schematically in Figure 7(a)). The unlinked nature of many of these molecular markers makes it a priori entirely plausible to hypothesize that distinct biochemical features are organized into distinct regulons, i.e., are controlled via distinct sets of transcription factors. Notably, however, this does not appear to be a predominant theme. On the contrary, the in‐depth analysis of the differentiation programs from a number of isolated neuron types—the light touch sensory neurons, GABAergic neurons, cholinergic AIY interneurons, glutamatergic ASE sensory neurons or cholinergic ventral nerve cord motor neurons—reveal that scores of molecular markers that are coexpressed by any of these specific neuron types are coregulated by transcription factors expressed in these neurons (examples shown in Figure 7).7, 87, 91, 121, 126 In each of these cases, the binding sites for the respective transcription factors are known and in each of these cases, the regulation of the terminal identity markers is thought to be largely direct (Figure 7). As expected from the profound molecular defects of these neurons, the neurons are not functional and display various anatomical defects, including axonal outgrowth defects and synaptic connectivity defects. Yet in all cases examined, the neurons are still generated and even still express pan‐neuronal features. In other words, the adoption of pan‐neuronal identity can be genetically separated from the adoption of neuron type‐specific identity. The above mentioned transcription factors have been called ‘Terminal Selectors’ due to the profound impact they have on neuronal identity. The salient features of terminal selectors are summarized in Box 1 (recently reviewed in Ref 139).
Figure 7.
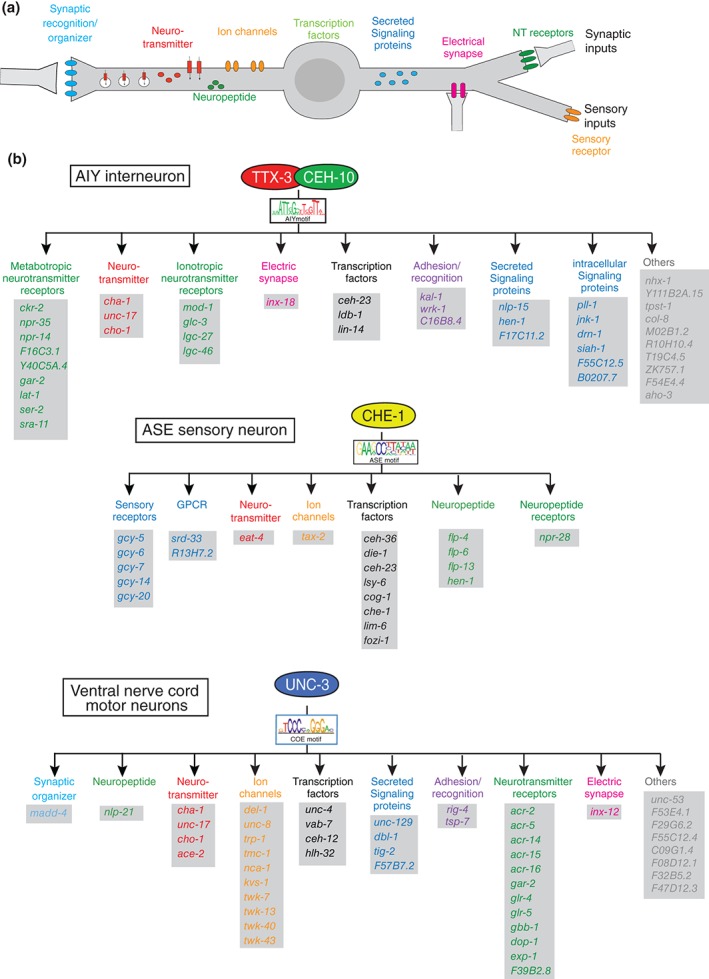
Terminal selector regulons. (a) Schematic illustration of terminal identity features that are controlled by terminal selector‐type transcription factors. These identity features are continuously expressed throughout the life of a neuron. (b) Three examples of terminal selector regulons. All genes shown here were shown to be direct targets of the indicated terminal selectors.7, 121, 138 In addition to the genes shown here, scores of additional genes have been identified as being expressed in the respective neuron types and containing binding sites for the respective regulators, but these additional genes have not been validated for terminal selector‐dependence.
BOX 1. TERMINAL SELECTORS OF NEURONAL IDENTITY.
Terminal selectors are transcription factors that induce expression of the terminally differentiated properties of mature neuron types.139 These factors work mostly in combinations, e.g. as heterodimers that cooperatively bind DNA7, 140 to directly control the expression of effector genes that define the functional properties of a neuron type. Terminal selectors are continuously expressed throughout the life of a neuron and are required to not only induce but also maintain the differentiated state of a neuron, a feat achieved mostly through autoregulation (Box 2).137 Terminal selectors are not only required to induce specific differentiation programs, but are also sufficient to do so, albeit only in some cellular contexts.7, 115, 141 This context dependency is likely dictated by the need of proper cofactors,115 but also by a chromatin environment that may be refractory to terminal selector activity.141
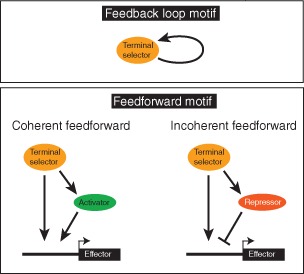
The vast majority of terminal selector targets are, by definition, terminal effector/structural genes, i.e., genes encoding for proteins involved in defining the neuron‐type specific structural and functional features of a neuron (Figure 7). This sets terminal selectors apart from earlier acting transcription factors whose primary role is to trigger ensuing waves of regulatory factors (these factors are not covered here; see another review that considers these earlier acting factors145). Nevertheless, some terminal selector targets encode for regulatory factors that control specific subroutines, likely in conjunction with the upstream terminal selector. For example, the ttx‐3/ceh‐10 terminal selector complex directly activates the expression of the homeobox gene ceh‐23 in the AIY interneurons and these factors together control expression of the G‐protein‐coupled receptor sra‐11. 108 This regulatory architecture is a classic example of a feedforward regulatory motif and is found in other neuron types as well (Box 2).115, 121, 142, 146
BOX 2. REGULATORY NETWORK MOTIFS.
Three types of network motifs142 employed in the context of neuronal specification and terminal selector function in C. elegans. The autoregulatory feedback loop is employed to ensure maintained terminal selector expression and, hence, maintained terminal effector gene expression.137, 143 Feedforward loops involve intervening transcription factors. Coherent feedforward loops have been found in other systems to filter transient input signals,142 but even though frequently observed,108, 115 their function in terminal selector‐mediated gene expression control remains to be shown. Incoherent feedforward loops occur in the context of C. elegans motor neuron class specification or lateralization of gustatory neuron function,121, 144 as discussed in the text.
Since the in‐depth analysis of a number of select cases mentioned above (Figure 7), coregulation of structural genes has been examined in many different neuron types, albeit most often in less depth (summarized in Figures 4, 5, 6). Much of this analysis was performed in the context of mapping glutamatergic and cholinergic neurotransmitter identity throughout the nervous system.8, 78 With this neurotransmitter map in hand, a host of regulatory factors expressed in either glutamatergic or cholinergic neurons were analyzed for how their genetic removal affects the acquisition of the respective neurotransmitter identity. In those cases where an effect was observed, it was tested whether other, unlinked biochemical identity feature are also affected. In most cases, it was found that a factor that regulates neurotransmitter identity also affects the expression of other identity features.8, 78 In a number of cases, the substantial number of markers tested (>20; Figures 4, 5, 6) make a compelling case that factors that regulate neurotransmitter identity regulate large sets of genes expressed in the terminally differentiated neuron, i.e., that many distinct identity features of a terminally differentiated neuron are coregulated. Even in cases where the sample size of examined marker is relatively small, it appears reasonable to extrapolate the effect to the many other genes expressed in the terminally differentiated neuron.
Even though the concept of coregulation appears to apply broadly, the cholinergic command interneurons, a group of interconnected neurons that innervate ventral nerve cord motor neurons, present a notable exception. In these neurons, the unc‐3 transcription factor, which coregulates many, independent terminal identity features (including neurotransmitter identity) in ventral nerve cord motor neurons, appears to only control the acquisition of cholinergic neurotransmitter identity for all command interneurons.8 More than a dozen additional markers expressed in select subsets of command interneurons are not affected in unc‐3 mutants. The additional identity markers (a good number of them encoding for ionotropic Glu receptors) are in turn controlled in a cell‐type specific, piecemeal manner by the unc‐42 homeobox gene, the fax‐1 nuclear hormone receptor, or the cfi‐1 ARID‐type transcription factor.93, 105, 111, 112 Hence, it appears that in some cases, neuronal identity is controlled in a piecemeal manner through parallel acting identity regulators. However, at this point we cannot exclude that the above mentioned factors (unc‐3, unc‐42, and fax‐1) are mere subroutine regulators that operate in the context of aforementioned feedforward motifs, downstream of as yet unknown terminal selectors for command interneuron identity.
Other Features of the Regulatory Map
Two other themes of the regulatory map are striking. First, transcription factors are used over and over again in distinct neuron types. Remarkably, four transcription factors are required to specify the identity of 50 distinct neuron classes, almost half of all neuron types in the C. elegans nervous system (unc‐3: 16 neuron classes; unc‐86: 14 neuron classes; ceh‐14: 10 neuron classes; unc‐42: 10 neuron classes). Note that these four transcription factors are deeply conserved (unc‐3 = EBF/Collier; unc‐86 = Brn3; ceh‐14 = Lhx3/4, unc‐42 = Prop1). In each neuron class, these factors appear to act in combination with other factors and this combinatorial code appears to provide specificity. This combinatorial coding scheme is displayed in Figure 8. For example, the POU homeobox gene unc‐86 acts as a regulator of terminal identity in 14 distinct neuron classes and it appears to operate in conjunction with specific cofactors in distinct neuron classes, such as the LIM homeobox gene mec‐3 in touch sensory neurons,140 the ceh‐14 LIM homeobox gene in a number of distinct interneurons,78 the pag‐3 Zn finger transcription factors in one specific interneuron type,115 or the ttx‐3 LIM homeobox gene in a neurosecretory sensory neuron.107 When reused, the same transcription factor may not necessarily perform as a terminal selector: it can work as a terminal selector in one neuron type or as a subroutine regulator in another. For example, ceh‐14 operates as a terminal selector in 10 different neuron types,78, 84, 85 but operates only as a subroutine regulator (controlling neuropeptide expression) in the BDU neurons.115
Figure 8.
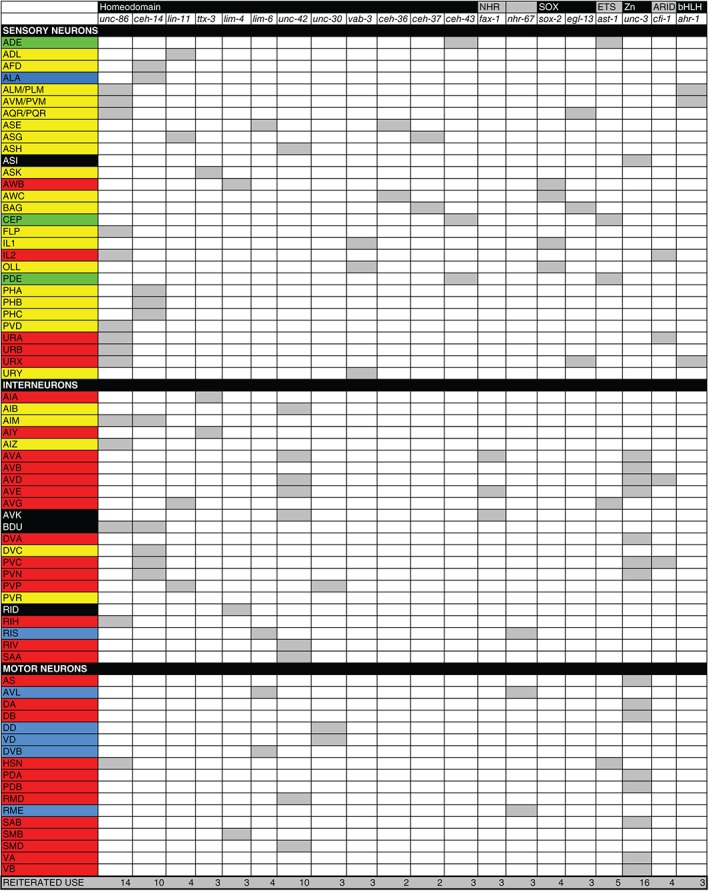
Redeployment of regulators of terminal neuronal identity in distinct neuron types. Data are extracted from Figure 4, 5, 6. Coloring scheme indicates neurotransmitter identity: yellow = Glu, red = ACh, green = aminergic, black = unknown. Only transcription factors that operate in >1 class are shown
Another theme is the striking preponderance of employment of homeodomain transcription factors. While these transcription factors make up only ~10% of all transcription factors in the C. elegans genome, a vast number of identified terminal regulators are homeodomain transcription factors and those that are not most frequently collaborate with a homeodomain transcription factor (homeodomain transcription factors are underlined in Figures 4, 5, 6). Reusage of the same transcription factors, as well as the preponderance of homeodomain transcription factors may indicate the evolutionary history of cell types in the nervous system. Homeodomain proteins may have had very early roles in specifying ancestral neuron types.
Are there other obvious, underlying themes or logical patterns by which transcription factors are used and reused? If one considers the lineage history of each individual neuron type, no obvious patterns emerge. First, if one just considers terminal identity features, it is already evident that the, e.g., neurotransmitter identity does not obviously correlate with lineage (Figure 3). Second, transcription factor expression profiles also show a lack of correlation with lineage. This is illustrated by mapping the expression pattern of four transcription factors that control the terminal identity of at least 50 distinct neuron types on the lineage diagram (Figure 3). The color‐coding in this illustration also shows that neurotransmitter identity generally does not correlate with transcription factor usage; i.e., the same transcription factor can operate in neurons of different neurotransmitter identities. The only exception is the unc‐3 transcription factor, which represents a remarkable case of a transcription factors not only being committed to defining only cholinergic neuron identity, but doing so in the context of a specific circuit, the VNC motor circuit.8
DIVERSIFICATION BY REPRESSORS
As summarized in Figures 4, 5, 6 not every factor that controls terminal neuron differentiation is an inducer, i.e., a likely activator of some, many, or all terminal features of a neuron. In fact, one of the earliest identified regulatory factors in C. elegans is the unc‐4 homeobox gene.16 unc‐4 mutants were initially characterized as synaptic specificity mutants, in which the synaptic input to a specific motor neuron subtype (VA) is altered in a manner that resembles that of another motor neuron subtype (VB).16 unc‐4 has since been shown to repress not just VB‐specific synaptic wiring patterns in the VA neurons but other identity features of the VB neurons as well.122 Additional repressor proteins (e.g., cog‐1 or vab‐7) have since been identified whose loss results in de‐repression or entire switches of neuronal identity programs.125, 148 The repressor nature of proteins like UNC‐4 or COG‐1 is indicated by their proven or probable direct association with the corepressor protein UNC‐37 (orthologous to Drosophila Groucho), which recruits histone deacetylases to repress gene expression.148, 149
Intriguingly, the repressor proteins mentioned above exert their negative effect (either directly or indirectly) on terminal effector genes that are themselves direct targets of terminal selectors. For example, the acetylcholine receptor subunit acr‐5 is expressed in B‐type motor neurons and is a direct target of unc‐3 in these neurons.121 In A‐type motor neurons, however, acr‐5 is repressed by unc‐4, and unc‐4 is itself a target of unc‐3. 121, 122 unc‐3 therefore ensures that one of its direct targets is only expressed in a subtype of motor neurons, by inducing the expression of a subtype‐specific repressor, unc‐4. This regulatory configuration constitutes an incoherent feedforward motif (Box 2).142
The principle of modifying a terminal selector‐induced differentiation program through selective repression in neuronal subtypes has been observed in another intriguing neurobiological context, namely the development of left/right asymmetric neuronal subtype specification. The left and right ASE neurons are the two main gustatory neurons in the worm and their terminal differentiation program is directly controlled by the che‐1 terminal selector.80 While the majority of terminal features of ASE are expressed in both ASE neurons, a subset of putative chemoreceptor proteins, encoded by the gcy genes, are expressed exclusively in the left or the right ASE neuron. gcy genes are also direct targets of che‐1 but the ability of che‐1 to activate gcy genes is restricted by ASEL‐ or ASER‐subtype specific regulatory factors.144 Two of these repressors are the miRNA lsy‐6 (ASEL‐specific) and its target, the homeobox gene cog‐1 (ASER‐specific). Both lsy‐6 and cog‐1 are also direct targets of che‐1. 150, 151
The overall logic of incoherent feedforward regulation is to diversify a ground state into various substates. In an evolutionary context, one could envision an ancestral state in which multiple neurons were induced by the same transcription factor; the recruitment of a repressor in subsets of these neurons may have then diversified the spectrum of targets of the original inducer.
REGULATION OF PANNEURONAL FEATURES
One aspect of neuronal differentiation that has been remarkably unexplored in most model organisms is the acquisition of panneuronal features. A recent study addressed this problem in some depth, using a panel of genes that are expressed in all neurons throughout the nervous system (mostly, but not exclusively, genes involved in the synaptic vesicle cycle).152 Some surprising themes emerged from the analysis of the regulation of more than a dozen pan‐neuronal genes. To appreciate these themes it is important to first consider the cis‐regulatory architecture of genes that are expressed in a neuron‐type specific manner. As schematically indicated in Figure 9, cis‐regulatory control regions of neuron‐type specific genes are organized in a modular manner, composed of response elements to neuron‐type specific terminal selectors. For example, the choline reuptake transporter cho‐1 (involved in clearance of the breakdown product of ACh at cholinergic synapses) contains separable modules with validated binding sites for TTX‐3/CEH‐10, required and sufficient for expression in the cholinergic AIY interneurons and a module with a binding site for UNC‐3, which is required and sufficient for expression of cho‐1 in ventral nerve cord motor neurons.121 Mutation of these sites in the context of fosmid‐based reporters indicates the requirement of these single sites for expression in specific neuron types. This theme reiterates over many of the other neuron‐type specific terminal effector genes tested. The cis‐regulatory control regions of pan‐neuronal genes are organized in a strikingly distinct manner, characterized by redundant, independently acting regulatory inputs (Figure 9).152 Viewed from an evolutionary perspective, this regulatory architecture indicates that pan‐neuronal genes have accumulated responsiveness to many different regulatory factors expressed in a terminally differentiated neuron, perhaps with the purpose of ensuring robustness of gene expression.
Figure 9.
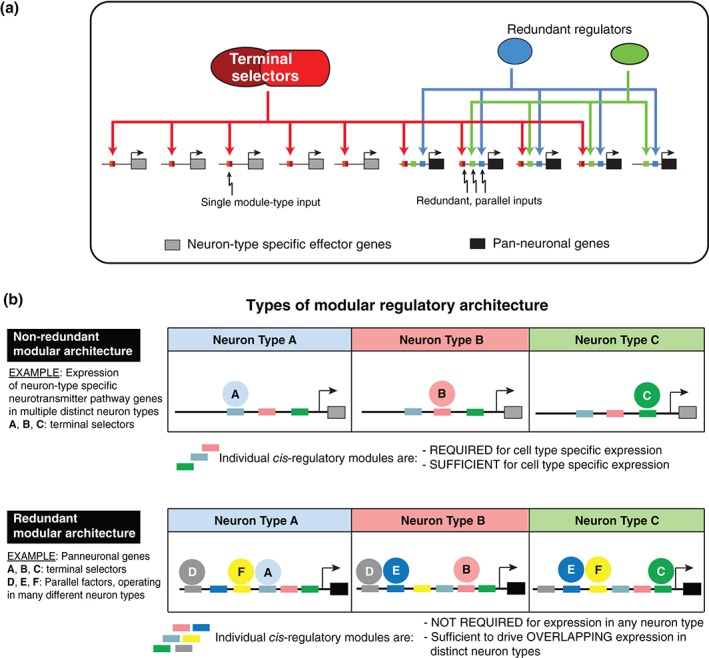
Regulation of pan‐neuronal identity. (Reprinted with permission from 152. Copyright 2015)
HOW IS THE REGULATORY MAP ESTABLISHED?
While terminal differentiation programs have been described in many neuron types throughout the C. elegans nervous system, much less is known about earlier neuronal patterning events in C. elegans. One obvious question is how the expression of terminal selector‐type transcription factors is restricted to specific cell types and therefore, how the regulatory map described above is established. An earlier review has covered a few aspects of neuronal lineage control in C. elegans. 145 I will focus here instead on three aspects of early neuronal patterning, some of which directly relating to the control of terminal selectors.
The Binary Wnt System
In the 1990s, Ralf Schnabel, Jim Priess and others discovered an intriguing binary patterning system that operates during C. elegans embryogenesis.153, 154 A noncanonical Wnt signaling system results in an anterior/posterior difference in the activity of transcriptional outputs of the Wnt system.155 This system has subsequently been explicitly shown to also be involved late in embryonic nervous system patterning, directly regulating the expression of terminal selectors of neuronal identity.156 This work, previously reviewed in Ref 157 provided the first link of terminal patterning of neuronal identity to earlier patterning events, demonstrating a transition from transient regulatory states to terminal, maintained regulatory states. Recent work describes how this system operates in distinct neuroblasts.158
Because of its binary nature, the anterior/posterior patterning system can obviously not act alone to specify the complex expression patterns of terminal selectors. The Wnt output regulators must interact with other transcription factors. The above‐mentioned studies identified several such factors as being involved in defining terminal selector expression.156, 158 The multiplicity of regulatory inputs into terminal selector loci has led to the suggestion of an hourglass regulatory architecture, in which terminal selectors are nodes that integrate various transient regulatory inputs into a stable, maintained regulatory state.159
Differences and Similarities in Early Patterning—The bHLH and SoxB/C Cases
Specific types of basic helix–loop–helix transcription factors operate as proneural genes throughout the animal kingdom and they have also been identified in C. elegans. For example, in one case, a proneural gene hlh‐14 controls the neuronal (vs hypodermal) identity of a lineage that produces the gustatory ASE neurons.160 As expected, hlh‐14 is expressed earlier in the lineage than the terminal selector of ASE identity (the che‐1 transcription factor), but its expression persists long enough to make it conceivable that hlh‐14 may be involved in directly controlling che‐1 expression, perhaps in conjunction with the nuclear hormone receptor nhr‐67, another upstream regulator of che‐1 expression.132 Similarly, the postembryonically generated touch receptor neurons AVM and PVM fail to be generated in animals lacking the proneural gene lin‐32/Atonal,161 which may be involved in directly or indirectly activating expression of the terminal selector of AVM/PVM identity, unc‐86. 162 Whether proneural bHLH genes are required for neuronal fate induction in all neurons of the nervous system (a notion that is surprisingly unexplored in other systems) is currently being investigated.
A recent study shows that one aspect of early neuronal patterning in C. elegans displays a remarkable difference to early neuronal patterning in other organisms.104 The HMG box transcription factor SOX‐2, a member of the SoxB subfamily, is deeply conserved sequence‐wise throughout the animal kingdom. Its function has been addressed in a wide range of animals from deuterostomes (chordates, echinoderms, and hemichordates) and protostomes (Ecdysozoa and Lophotrochozoa) to even earlier diverging animal lineages such as cnidarians. In all these cases, sox‐2 was shown to be expressed in neuronal precursors (upstream of the above‐mentioned proneural bHLH genes) and whenever functional evidence was available, a role for sox‐2 in nervous system development was elucidated.163 Curiously, sox‐2 function in C. elegans is different. sox‐2 is not broadly expressed in neuronal precursors and sox‐2 has no function in embryonic neurogenesis.104 Again in contrast to other organisms, the SoxC‐type sox‐3 gene also has no role in embryonic neurogenesis.104 C. elegans sox‐2 appears to rather be involved in driving terminal differentiation of select neuron types, as well as in the specification of a subset of postembryonic neuroblast.97, 104 The absence of a neurogenic sox‐2 function in C. elegans may relate to what sox‐2 does in other organisms—maintaining the pro‐neuronal developmental potential of neuroepithelial cells during phases of cell proliferation in the developing nervous system.163 Such expansion may not occur in C. elegans embryogenesis.
Redundancy of Early Regulators
It is remarkable how few regulators of early neuronal patterning have been retrieved through genetic mutant screens in C. elegans. At first sight, a relatively trivial explanation could be that essential pleiotropic functions of regulatory factors may hinder retrieval from genetic screens. However, hypomorphic alleles of essential early patterning genes or even regulatory null alleles (i.e., loss of a gene only in specific cells due to mutations in cis‐regulatory elements) have been retrieved in screens for terminal differentiation mutants. For example, a mutant allele of the essential Distalless‐like homeobox gene ceh‐43, retrieved from a genetic screen, disrupts dopaminergic neuron differentiation and is defined by a deletion of a distal enhancer required for ceh‐43 expression in dopaminergic neurons.82 Also, RNAi‐based screens have been conducted for early regulators of ASE lineage specification and differentiation, recovering little more than the above‐mentioned, and somewhat expected proneural bHLH factor for this lineage, hlh‐14. 160 One very intriguing possibility for the poor retrieval rate of early patterning genes is suggested by an older study164 and a more recent study.165 Priess and colleagues found that early patterning in the ABa lineage, which generates about half of the C. elegans nervous system, is controlled by two redundantly acting T‐box genes, tbx‐37 and tbx‐38. 164 Both genes are close paralogs and both need to be genetically eliminated to observe patterning defects. Notably, T‐box genes expanded significantly in C. elegans, and many come in closely paralogous pairs.166 It will be most interesting to determine whether it is a common theme that T‐box factors act redundantly during early patterning decisions.
An intriguing, more recent example of redundancy shows that two unrelated homeobox genes, the Pitx‐type homeobox gene unc‐30 and the Otx‐type homeobox gene ceh‐36 act redundantly in early neuronal lineage decisions.165 Both genes act as terminal selectors in defined neuronal cell types,100, 126 but apparently also moonlight as patterning factors at earlier time points in lineages where their expression overlaps. It is tantalizing to think that early patterning in C. elegans is highly buffered through redundantly acting factors.
CONCLUDING REMARKS
More than half a century after Sydney Brenner first formulated his C. elegans research program,1, 2 it is deeply satisfying to see how the C. elegans field has moved toward fulfilling Sydney Brenner's vision of using the tools of the microbial genetic trade, in combination with his visionary map making efforts, to understand how a nervous system develops. One reason this research program has been particularly successful in the nervous system is the lack of pleiotropies associated with regulators of terminal neuronal identity. In fact, such lack of pleiotropies was recognized by Brenner very early in his analysis of behavioral mutants by electron micrographical analysis.167 Molecular, functional and expression studies subsequently revealed that most transcription factors that are components of the regulatory map are exclusively expressed in the nervous system, i.e., they do not have what could be essential functions elsewhere which could have prevented their retrieval from forward genetic screens. Within the nervous system many of these factors also appear to be committed to controlling terminal differentiation, without being involved in earlier patterning events. One exception to this notion are the unc‐30 and ceh‐36 terminal selectors, which, apart from their terminal selector function in distinct neuron types, have redundant function in controlling earlier lineage decisions.165 These findings illustrate the importance of not solely relying on forward genetic screens, but also taking into account expression patterns and hence possibly redundant gene functions.
One major future challenge lies in connecting terminal regulators of neuronal identity to early lineage patterning events. Using automated lineage tracing in combination with RNAi‐mediated gene knockdown, Du et al. have recently identified a large number of genes with roles in early lineage specification.168 How these genes are coupled to the induction of expression of terminal selectors requires future exploration.
In addition to studying early neuronal patterning events, a great number of questions remain at the level of terminal differentiation programs. For example, the nature of the combinatorial codes of transcription factors that control the differentiated state should be more extensively characterized, perhaps with a specific focus on the ~100 homeobox genes encoded by the C. elegans genome. Importantly, we need a much more comprehensive understanding of the extent to which terminal differentiation programs are controlled in a coregulatory manner by terminal selector‐type transcription factors versus being regulated in a piecemeal manner by distinct cohorts of transcription factors. One important step in this direction needs to be a systematic transcriptome analysis of each and every neuron in the worm. This is now technically feasible and it is easy to predict that the worm, after having been the first organism with a lineage, connectome, and genome, will also be the first with a complete expression atlas with single‐cell resolution. The regulatory programs that control panneuronal genes (e.g. synaptic vesicle machinery) also need to be better understood. The plasticity of the terminally differentiated state will also need to be examined on a much more detailed level. For example, how do postmitotic, embryonically generated neurons alter their differentiated state at defined postembryonic time points? How does circuit activity impinge on the differentiated state of specific neuronal circuit components? How does the sexual identity of the organism impinge on neuronal differentiation? Many of these questions can be addressed by continuing to pursue Sydney Brenner's vision, building higher resolution and now multidimensional maps (gene expression maps at distinct time points, under distinct external conditions), and by then exploiting the unique strengths of the C. elegans model system to genetically dissect these molecular maps through classic mutant analysis.
ACKNOWLEDGMENTS
I thank Daniela Raciti and Wen Chen at Wormbase for extracting expression data from Wormbase, Lori Glenwinkel for building the resulting molecular map, Nuria Flames and Marie Gendrel for sharing unpublished data, Ev Yemini and Nuria Flames for comments on the manuscript. Stephen Larsen at Openworm and Chris Gove at WormBase kindly provided the image in Figure 1(a). Work in my laboratory is funded by the NIH and HHMI.
Conflict of interest: The author has declared no conflicts of interest for this article.
REFERENCES
- 1. Brenner S. The genetics of Caenorhabditis elegans . Genetics 1974, 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenner S. Foreword In: Wood WB, ed. The Nematode Caenorhabditis elegans. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 3. Hall DH, Altun Z. C. Elegans Atlas. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 4. Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol 2011, 7:e1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Margeta MA, Wang GJ, Shen K. Clathrin adaptor AP‐1 complex excludes multiple postsynaptic receptors from axons in C. elegans . Proc Natl Acad Sci USA 2009, 106:1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans . Philos Trans R Soc Lond B Biol Sci 1986, 314:1–340. [DOI] [PubMed] [Google Scholar]
- 7. Wenick AS, Hobert O. Genomic cis‐regulatory architecture and trans‐acting regulators of a single interneuron‐specific gene battery in C. elegans . Dev Cell 2004, 6:757–770. [DOI] [PubMed] [Google Scholar]
- 8. Pereira L, Kratsios P, Serrano‐Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, et al. A cellular and regulatory map of the cholinergic nervous system of C. elegans Elife 2015, 4. pii: e12432. doi: 10.7554/eLife.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. The connectome of a decision‐making neural network. Science 2012, 337:437–444. [DOI] [PubMed] [Google Scholar]
- 10. Sulston JE, Horvitz HR. Post‐embryonic cell lineages of the nematode, Caenorhabditis elegans . Dev Biol 1977, 56:110–156. [DOI] [PubMed] [Google Scholar]
- 11. Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans . Dev Biol 1983, 100:64–119. [DOI] [PubMed] [Google Scholar]
- 12. Hedgecock EM, Culotti JG, Hall DH. The unc‐5, unc‐6, and unc‐40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans . Neuron 1990, 4:61–85. [DOI] [PubMed] [Google Scholar]
- 13. Wadsworth WG, Bhatt H, Hedgecock EM. Neuroglia and pioneer neurons express UNC‐6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron 1996, 16:35–46. [DOI] [PubMed] [Google Scholar]
- 14. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993, 75:843–854. [DOI] [PubMed] [Google Scholar]
- 15. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21‐nucleotide let‐7 RNA regulates developmental timing in Caenorhabditis elegans . Nature 2000, 403:901–906. [DOI] [PubMed] [Google Scholar]
- 16. Miller DM, Shen MM, Shamu CE, Bürglin TR, Ruvkun G, Dubois ML, Ghee M, Wilson L. C. elegans unc‐4 gene encodes a homeodomain protein that determines the pattern of synaptic input to specific motor neurons. Nature 1992, 355:841–845. [DOI] [PubMed] [Google Scholar]
- 17. Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 2000, 26:619–631. [DOI] [PubMed] [Google Scholar]
- 18. Hills T, Brockie PJ, Maricq AV. Dopamine and glutamate control area‐restricted search behavior in Caenorhabditis elegans . J Neurosci 2004, 24:1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans . Science 1991, 251:1243–1246. [DOI] [PubMed] [Google Scholar]
- 20. Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans . Nature 2007, 450:63–70. [DOI] [PubMed] [Google Scholar]
- 21. McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans . Nature 1993, 364:337–341. [DOI] [PubMed] [Google Scholar]
- 22. Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans . Cell 1995, 83:207–218. [DOI] [PubMed] [Google Scholar]
- 23. Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans . Proc Natl Acad Sci USA 2005, 102:3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bendena WG, Boudreau JR, Papanicolaou T, Maltby M, Tobe SS, Chin‐Sang ID. A Caenorhabditis elegans allatostatin/galanin‐like receptor NPR‐9 inhibits local search behavior in response to feeding cues. Proc Natl Acad Sci USA 2008, 105:1339–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans . J Neurosci 1985, 5:956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans . Nature 1995, 376:344–348. [DOI] [PubMed] [Google Scholar]
- 27. Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans . Nat Neurosci 2007, 10:1300–1307. [DOI] [PubMed] [Google Scholar]
- 28. Sanders J, Nagy S, Fetterman G, Wright C, Treinin M, Biron D. The Caenorhabditis elegans interneuron ALA is (also) a high‐threshold mechanosensor. BMC Neurosci 2013, 14:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans . Dev Biol 1981, 82:358–370. [DOI] [PubMed] [Google Scholar]
- 30. Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans . J Neurobiol 2003, 56:178–197. [DOI] [PubMed] [Google Scholar]
- 31. Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans . Nature 2002, 419:925–929. [DOI] [PubMed] [Google Scholar]
- 32. Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 2004, 430:317–322. [DOI] [PubMed] [Google Scholar]
- 33. Bargmann CI, Hartwieg E, Horvitz HR. Odorant‐selective genes and neurons mediate olfaction in C. elegans . Cell 1993, 74:515–527. [DOI] [PubMed] [Google Scholar]
- 34. Piggott BJ, Liu J, Feng Z, Wescott SA, Xu XZ. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans . Cell 2011, 147:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greer ER, Perez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab 2008, 8:118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans . Proc Natl Acad Sci USA 1993, 90:2227–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR‐1 glutamate receptor. Nature 1995, 378:82–85. [DOI] [PubMed] [Google Scholar]
- 38. Beverly M, Anbil S, Sengupta P. Degeneracy and neuromodulation among thermosensory neurons contribute to robust thermosensory behaviors in Caenorhabditis elegans . J Neurosci 2011, 31:11718–11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hardaker LA, Singer E, Kerr R, Zhou G, Schafer WR. Serotonin modulates locomotory behavior and coordinates egg‐laying and movement in Caenorhabditis elegans . J Neurobiol 2001, 49:303–313. [DOI] [PubMed] [Google Scholar]
- 40. Hutter H. Extracellular cues and pioneers act together to guide axons in the ventral cord of C. elegans . Development 2003, 130:5307–5318. [DOI] [PubMed] [Google Scholar]
- 41. Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub‐and‐spoke circuit drives pheromone attraction and social behaviour in C. elegans . Nature 2009, 458:1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kratsios P, Pinan‐Lucarré B, Kerk SY, Weinreb A, Bessereau JL, Hobert O. Transcriptional coordination of synaptogenesis and neurotransmitter signaling. Curr Biol 2015, 25:1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kennerdell JR, Fetter RD, Bargmann CI. Wnt‐Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans . Development 2009, 136:3801–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biron D, Wasserman S, Thomas JH, Samuel AD, Sengupta P. An olfactory neuron responds stochastically to temperature and modulates Caenorhabditis elegans thermotactic behavior. Proc Natl Acad Sci USA 2008, 105:11002–11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li W, Kang L, Piggott BJ, Feng Z, Xu XZ. The neural circuits and sensory channels mediating harsh touch sensation in Caenorhabditis elegans . Nat Commun 2011, 2:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans . Proc Natl Acad Sci USA 2008, 105:8038–8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li W, Feng Z, Sternberg PW, Xu XZ. A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature 2006, 440:684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee H, Choi MK, Lee D, Kim HS, Hwang H, Kim H, Park S, Paik YK, Lee J. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat Neurosci 2012, 15:107–112. [DOI] [PubMed] [Google Scholar]
- 49. Waggoner LE, Zhou GT, Schafer RW, Schafer WR. Control of alternative behavioral states by serotonin in Caenorhabditis elegans . Neuron 1998, 21:203–214. [DOI] [PubMed] [Google Scholar]
- 50. Chang HC, Paek J, Kim DH. Natural polymorphisms in C. elegans HECW‐1 E3 ligase affect pathogen avoidance behaviour. Nature 2011, 480:525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hilliard MA, Bargmann CI, Bazzicalupo P. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr Biol 2002, 12:730–734. [DOI] [PubMed] [Google Scholar]
- 52. Aurelio O, Hall DH, Hobert O. Immunoglobulin‐domain proteins required for maintenance of ventral nerve cord organization. Science 2002, 295:686–690. [DOI] [PubMed] [Google Scholar]
- 53. Liu S, Schulze E, Baumeister R. Temperature‐ and touch‐sensitive neurons couple CNG and TRPV channel activities to control heat avoidance in Caenorhabditis elegans . PLoS One 2012, 7:e32360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nelson MD, Trojanowski NF, George‐Raizen JB, Smith CJ, Yu CC, Fang‐Yen C, Raizen DM. The neuropeptide NLP‐22 regulates a sleep‐like state in Caenorhabditis elegans . Nat Commun 2013, 4:2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chatzigeorgiou M, Yoo S, Watson JD, Lee WH, Spencer WC, Kindt KS, Hwang SW, Miller DM 3rd, Treinin M, Driscoll M, et al. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat Neurosci 2010, 13:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, Bargmann CI. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans . Cell 2013, 154:1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chatzigeorgiou M, Schafer WR. Lateral facilitation between primary mechanosensory neurons controls nose touch perception in C. elegans . Neuron 2011, 70:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Turek M, Lewandrowski I, Bringmann H. An AP2 transcription factor is required for a sleep‐active neuron to induce sleep‐like quiescence in C. elegans . Curr Biol 2013, 23:2215–2223. [DOI] [PubMed] [Google Scholar]
- 59. Bargmann CI. Chemosensation in C. elegans . WormBook 2006:1–29. PMID: 18050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Von Stetina SE, Treinin M, Miller DM. The motor circuit. Int Rev Neurobiol 2006, 69:125–167. [DOI] [PubMed] [Google Scholar]
- 61. Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans . Nature 1994, 367:467–470. [DOI] [PubMed] [Google Scholar]
- 62. Sengupta P, Chou JH, Bargmann CI. odr‐10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 1996, 84:899–909. [DOI] [PubMed] [Google Scholar]
- 63. Way JC, Chalfie M. mec‐3, a homeobox‐containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 1988, 54:5–16. [DOI] [PubMed] [Google Scholar]
- 64. Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che‐1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 2003, 130:1215–1224. [DOI] [PubMed] [Google Scholar]
- 65. Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx‐3 LIM homeobox gene. Neuron 1997, 19:345–357. [DOI] [PubMed] [Google Scholar]
- 66. Satterlee JS, Sasakura H, Kuhara A, Berkeley M, Mori I, Sengupta P. Specification of thermosensory neuron fate in C. elegans requires ttx‐1, a homolog of otd/Otx. Neuron 2001, 31:943–956. [DOI] [PubMed] [Google Scholar]
- 67. Ortiz CO, Etchberger JF, Posy SL, Frøkjaer‐Jensen C, Lockery S, Honig B, Hobert O. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor‐type guanylyl cyclases. Genetics 2006, 173:131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide‐like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci USA 2001, 98:14000–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li C, Kim K, Nelson LS. FMRFamide‐related neuropeptide gene family in Caenorhabditis elegans . Brain Res 1999, 848:26–34. [DOI] [PubMed] [Google Scholar]
- 70. Salkoff L, Butler A, Fawcett G, Kunkel M, McArdle C, Paz‐y‐Mino G, Nonet M, Walton N, Wang ZW, Yuan A, et al. Evolution tunes the excitability of individual neurons. Neuroscience 2001, 103:853–859. [DOI] [PubMed] [Google Scholar]
- 71. Tsalik EL, Niacaris T, Wenick AS, Pau K, Avery L, Hobert O. LIM homeobox gene‐dependent expression of biogenic amine receptors in restricted regions of the C. elegans nervous system. Dev Biol 2003, 263:81–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hunt‐Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. High‐throughput in vivo analysis of gene expression in Caenorhabditis elegans . PLoS Biol 2007, 5:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, Dang C. Allen Brain Atlas: an integrated spatio‐temporal portal for exploring the central nervous system. Nucleic Acids Res 2013, 41:D996–D1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4‐driver line resource for Drosophila neurobiology. Cell Rep 2012, 2:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hobert O. The neuronal genome of Caenorhabditis elegans . WormBook 2013:1–106. PMID: 18050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. McIntire SL, Jorgensen E, Horvitz HR. Genes required for GABA function in Caenorhabditis elegans . Nature 1993, 364:334–337. [DOI] [PubMed] [Google Scholar]
- 77. Chase DL, Koelle MR. Biogenic amine neurotransmitters in C. elegans . WormBook 2007:1–15. PMID: 18050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Serrano‐Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 2013, 155:659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Duggan A, Chalfie M. Control of neuronal development in Caenorhabditis elegans . Curr Opin Neurobiol 1995, 5:6–9. [DOI] [PubMed] [Google Scholar]
- 80. Hobert O. Development of left/right asymmetry in the Caenorhabditis elegans nervous system: from zygote to postmitotic neuron. Genesis 2014, 52:528–543. [DOI] [PubMed] [Google Scholar]
- 81. Hsieh YW, Alqadah A, Chuang CF. Asymmetric neural development in the Caenorhabditis elegans olfactory system. Genesis 2014, 52:544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Doitsidou M, Flames N, Topalidou I, Abe N, Felton T, Remesal L, Popovitchenko T, Mann R, Chalfie M, Hobert O. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans . Genes Dev 2013, 27:1391–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature 2009, 458:885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kagoshima H, Kohara Y. Co‐expression of the transcription factors CEH‐14 and TTX‐1 regulates AFD neuron‐specific genes gcy‐8 and gcy‐18 in C. elegans . Dev Biol 2015, 399:325–336. [DOI] [PubMed] [Google Scholar]
- 85. Van Buskirk C, Sternberg PW. Paired and LIM class homeodomain proteins coordinate differentiation of the C. elegans ALA neuron. Development 2010, 137:2065–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gendrel M, Hobert O. Transcription factor combinations specifying the GABAergic nervous system of C. elegans . In preparation.
- 87. Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature 2002, 418:331–335. [DOI] [PubMed] [Google Scholar]
- 88. Gramstrup Petersen J, Rojo Romanos T, Juozaityte V, Redo Riveiro A, Hums I, Traunmüller L, Zimmer M, Pocock R. EGL‐13/SoxD specifies distinct O2 and CO2 sensory neuron fates in Caenorhabditis elegans . PLoS Genet 2013, 9:e1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Qin H, Powell‐Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR‐1, regulates neuronal development. Dev Biol 2004, 270:64–75. [DOI] [PubMed] [Google Scholar]
- 90. Feng G, Yi P, Yang Y, Chai Y, Tian D, Zhu Z, Liu J, Zhou F, Cheng Z, Wang X, et al. Developmental stage‐dependent transcriptional regulatory pathways control neuroblast lineage progression. Development 2013, 140:3838–3847. [DOI] [PubMed] [Google Scholar]
- 91. Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis‐regulatory architecture of a C. elegans gustatory neuron. Genes Dev 2007, 21:1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sarafi‐Reinach TR, Melkman T, Hobert O, Sengupta P. The lin‐11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans . Development 2001, 128:3269–3281. [DOI] [PubMed] [Google Scholar]
- 93. Baran R, Aronoff R, Garriga G. The C. elegans homeodomain gene unc‐42 regulates chemosensory and glutamate receptor expression. Development 1999, 126:2241–2251. [DOI] [PubMed] [Google Scholar]
- 94. Kim K, Colosimo ME, Yeung H, Sengupta P. The UNC‐3 Olf/EBF protein represses alternate neuronal programs to specify chemosensory neuron identity. Dev Biol 2005, 286:136–148. [DOI] [PubMed] [Google Scholar]
- 95. Prasad BC, Ye B, Zackhary R, Schrader K, Seydoux G, Reed RR. unc‐3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 1998, 125:1561–1568. [DOI] [PubMed] [Google Scholar]
- 96. Gonzalez‐Barrios M, Fierro‐González JC, Krpelanova E, Mora‐Lorca JA, Pedrajas JR, Peñate X, Chavez S, Swoboda P, Gert Jansen, Miranda‐Vizuete A. Cis‐ and trans‐regulatory mechanisms of gene expression in the ASJ sensory neuron of Caenorhabditis elegans . Genetics 2015, 200:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Alqadah A, Hsieh YW, Vidal B, Chang C, Hobert O, Chuang CF. Postmitotic diversification of olfactory neuron types is mediated by differential activities of the HMG‐box transcription factor SOX‐2. EMBO J 2015, 34:2574–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sagasti A, Hisamoto N, Hyodo J, Tanaka‐Hino M, Matsumoto K, Bargmann CI. The CaMKII UNC‐43 activates the MAPKKK NSY‐1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 2001, 105:221–232. [DOI] [PubMed] [Google Scholar]
- 99. Nokes EB, Van Der Linden AM, Winslow C, Mukhopadhyay S, Ma K, Sengupta P. Cis‐regulatory mechanisms of gene expression in an olfactory neuron type in Caenorhabditis elegans . Dev Dyn 2009, 238:3080–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans . Dev Cell 2003, 5:621–633. [DOI] [PubMed] [Google Scholar]
- 101. Koga M, Ohshima Y. The C. elegans ceh‐36 gene encodes a putative homemodomain transcription factor involved in chemosensory functions of ASE and AWC neurons. J Mol Biol 2004, 336:579–587. [DOI] [PubMed] [Google Scholar]
- 102. Rojo Romanos T, Petersen JG, Riveiro AR, Pocock R. A novel role for the zinc‐finger transcription factor EGL‐46 in the differentiation of gas‐sensing neurons in Caenorhabditis elegans . Genetics 2015, 199:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Brandt JP, Aziz‐Zaman S, Juozaityte V, Martinez‐Velazquez LA, Petersen JG, Pocock R, Ringstad N. A single gene target of an ETS‐family transcription factor determines neuronal CO2‐chemosensitivity. PLoS One 2012, 7:e34014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Vidal B, Santella A, Serrano‐Saiz E, Bao Z, Chuang CF, Hobert O. C. elegans SoxB genes are dispensable for embryonic neurogenesis but required for terminal differentiation of specific neuron types. Development 2015, 142:2464–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Shaham S, Bargmann CI. Control of neuronal subtype identity by the C. elegans ARID protein CFI‐1. Genes Dev 2002, 16:972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zheng C, Diaz‐Cuadros M, Chalfie M. Hox Genes promote neuronal subtype diversification through posterior induction in Caenorhabditis elegans . Neuron 2015, 88:514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, Maicas M, Flames N, Mann RS, Colón-Ramos DA, et al. The LIM and POU homeobox genes ttx‐3 and unc‐86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 2014, 141:422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Altun‐Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O A regulatory cascade of three homeobox genes, ceh‐10, ttx‐3 and ceh‐23, controls cell fate specification of a defined interneuron class in C. elegans. Development 2001, 128:1951–1969. [DOI] [PubMed] [Google Scholar]
- 109. Sze JY, Zhang S, Li J, Ruvkun G. The C. elegans POU‐domain transcription factor UNC‐86 regulates the tph‐1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 2002, 129:3901–3911. [DOI] [PubMed] [Google Scholar]
- 110. Kage E, Hayashi Y, Takeuchi H, Hirotsu T, Kunitomo H, Inoue T, Arai H, Iino Y, Kubo T. MBR‐1, a novel helix‐turn‐helix transcription factor, is required for pruning excessive neurites in Caenorhabditis elegans . Curr Biol 2005, 15:1554–1559. [DOI] [PubMed] [Google Scholar]
- 111. Wightman B, Ebert B, Carmean N, Weber K, Clever S. The C. elegans nuclear receptor gene fax‐1 and homeobox gene unc‐42 coordinate interneuron identity by regulating the expression of glutamate receptor subunits and other neuron‐specific genes. Dev Biol 2005, 287:74–85. [DOI] [PubMed] [Google Scholar]
- 112. Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC‐42. J Neurosci 2001, 21:1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schmid C, Schwarz V, Hutter H. AST‐1, a novel ETS‐box transcription factor, controls axon guidance and pharynx development in C. elegans . Dev Biol 2006, 293:403–413. [DOI] [PubMed] [Google Scholar]
- 114. Wightman B, Baran R, Garriga G. Genes that guide growth cones along the C. elegans ventral nerve cord. Development 1997, 124:2571–2580. [DOI] [PubMed] [Google Scholar]
- 115. Gordon PM, Hobert O. A competition mechanism for a homeotic neuron identity transformation in C. elegans . Dev Cell 2015, 34:206–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cameron S, Clark SG, McDermott JB, Aamodt E, Horvitz HR. PAG‐3, a Zn‐finger transcription factor, determines neuroblast fate in C. elegans . Development 2002, 129:1763–1774. [DOI] [PubMed] [Google Scholar]
- 117. Clark SG, Chiu C. C. elegans ZAG‐1, a Zn‐finger‐homeodomain protein, regulates axonal development and neuronal differentiation. Development 2003, 130:3781–3794. [DOI] [PubMed] [Google Scholar]
- 118. Wacker I, Schwarz V, Hedgecock EM, Hutter H. zag‐1, a Zn‐finger homeodomain transcription factor controlling neuronal differentiation and axon outgrowth in C. elegans . Development 2003, 130:3795–3805. [DOI] [PubMed] [Google Scholar]
- 119. Hobert O, Tessmar K, Ruvkun G. The Caenorhabditis elegans lim‐6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 1999, 126:1547–1562. [DOI] [PubMed] [Google Scholar]
- 120. Aurelio O, Boulin T, Hobert O. Identification of spatial and temporal cues that regulate postembryonic expression of axon maintenance factors in the C. elegans ventral nerve cord. Development 2003, 130:599–610. [DOI] [PubMed] [Google Scholar]
- 121. Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci 2011, 15:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Winnier AR, Meir JY, Ross JM, Tavernarakis N, Driscoll M, Ishihara T, Katsura I, Miller DM 3rd UNC‐4/UNC‐37‐dependent repression of motor neuron‐specific genes controls synaptic choice in Caenorhabditis elegans . Genes Dev 1999, 13:2774–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lickteig KM, Duerr JS, Frisby DL, Hall DH, Rand JB, Miller DM 3rd Regulation of neurotransmitter vesicles by the homeodomain protein UNC‐4 and its transcriptional corepressor UNC‐37/groucho in Caenorhabditis elegans cholinergic motor neurons. J Neurosci 2001, 21:2001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Prasad B, Karakuzu O, Reed RR, Cameron S. unc‐3‐dependent repression of specific motor neuron fates in Caenorhabditis elegans . Dev Biol 2008, 323:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Esmaeili B, Ross JM, Neades C, Miller DM 3rd, Ahringer J. The C. elegans even‐skipped homologue, vab‐7, specifies DB motoneurone identity and axon trajectory. Development 2002, 129:853–862. [DOI] [PubMed] [Google Scholar]
- 126. Cinar H, Keles S, Jin Y. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans . Curr Biol 2005, 15:340–346. [DOI] [PubMed] [Google Scholar]
- 127. Jin Y, Hoskins R, Horvitz HR. Control of type‐D GABAergic neuron differentiation by C. elegans UNC‐30 homeodomain protein. Nature 1994, 372:780–783. [DOI] [PubMed] [Google Scholar]
- 128. Eastman C, Horvitz HR, Jin Y. Coordinated transcriptional regulation of the unc‐25 glutamic acid decarboxylase and the unc‐47 GABA vesicular transporter by the Caenorhabditis elegans UNC‐30 homeodomain protein. J Neurosci 1999, 19:6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Petersen SC, Watson JD, Richmond JE, Sarov M, Walthall WW, Miller DM 3rd A transcriptional program promotes remodeling of GABAergic synapses in Caenorhabditis elegans . J Neurosci 2011, 31:15362–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lloret C, et al. Deep homology of a genetic programs controlling serotonergic neuron differentiation in nematodes and mammals. In preparation.
- 131. Richard JP, Zuryn S, Fischer N, Pavet V, Vaucamps N, Jarriault S. Direct in vivo cellular reprogramming involves transition through discrete, non‐pluripotent steps. Development 2011, 138:1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sarin S, Antonio C, Tursun B, Hobert O. The C. elegans Tailless/TLX transcription factor nhr‐67 controls neuronal identity and left/right asymmetric fate diversification. Development 2009, 136:2933–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kim J, Yeon J, Choi SK, Huh YH, Fang Z, Park SJ, Kim MO, Ryoo ZY, Kang K, Kweon HS, et al. The evolutionarily conserved LIM homeodomain protein LIM‐4/LHX6 specifies the terminal identity of a cholinergic and peptidergic C. elegans sensory/inter/motor neuron‐type. PLoS Genet 2015, 11:e1005480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Von Stetina SE, Fox RM, Watkins KL, Starich TA, Shaw JE, Miller DM 3rd UNC‐4 represses CEH‐12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans . Genes Dev 2007, 21:332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Melkman T, Sengupta P. Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr‐1. Development 2005, 132:1935–1949. [DOI] [PubMed] [Google Scholar]
- 136. Zhou HM, Walthall WW. UNC‐55, an orphan nuclear hormone receptor, orchestrates synaptic specificity among two classes of motor neurons in Caenorhabditis elegans . J Neurosci 1998, 18:10438–10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nat Neurosci 2014, 17:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Etchberger JF, Hobert O. Vector‐free DNA constructs improve transgene expression in C. elegans . Nat Methods 2008, 5:3. [DOI] [PubMed] [Google Scholar]
- 139. Hobert O. Terminal selectors of neuronal identity. Curr Top Dev Biol 2016, 116:455–475. [DOI] [PubMed] [Google Scholar]
- 140. Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC‐86 and MEC‐3. Science 1993, 261:1324–1328. [DOI] [PubMed] [Google Scholar]
- 141. Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science 2011, 331:304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet 2007, 8:450–461. [DOI] [PubMed] [Google Scholar]
- 143. Hobert O. Maintaining a memory by transcriptional autoregulation. Curr Biol 2011, 21:R146–R147. [DOI] [PubMed] [Google Scholar]
- 144. Etchberger JF, Flowers EB, Poole RJ, Bashllari E, Hobert O. Cis‐regulatory mechanisms of left/right asymmetric neuron‐subtype specification in C. elegans . Development 2009, 136:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hobert O. Neurogenesis in the nematode Caenorhabditis elegans . WormBook 2010:1–24. PMID: 20891032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Johnston RJ Jr, Copeland JW, Fasnacht M, Etchberger JF, Liu J, Honig B, Hobert O. An unusual Zn‐finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans . Development 2006, 133:3317–3328. [DOI] [PubMed] [Google Scholar]
- 147. Caicedo A, Pereira E, Margolskee RF, Roper SD. Role of the G‐protein subunit alpha‐gustducin in taste cell responses to bitter stimuli. J Neurosci 2003, 23:9947–9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Chang S, Johnston RJ Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans . Genes Dev 2003, 17:2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pflugrad A, Meir JY, Barnes TM, Miller DM 3rd. The Groucho‐like transcription factor UNC‐37 functions with the neural specificity gene unc‐4 to govern motor neuron identity in C. elegans . Development 1997, 124:1699–1709. [DOI] [PubMed] [Google Scholar]
- 150. O'Meara MM, Bigelow H, Flibotte S, Etchberger JF, Moerman DG, Hobert O. Cis‐regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog‐1 affect neuronal development. Genetics 2009, 181:1679–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Sarin S, O'Meara MM, Flowers EB, Antonio C, Poole RJ, Didiano D, Johnston RJ Jr, Chang S, Narula S, Hobert O. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 2007, 176:2109–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Stefanakis N, Carrera I, Hobert O. Regulatory logic of pan‐neuronal gene expression in C. elegans . Neuron 2015, 87:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans . Nature 1997, 390:294–298. [DOI] [PubMed] [Google Scholar]
- 154. Lin R, Hill RJ, Priess JR. POP‐1 and anterior‐posterior fate decisions in C. elegans embryos. Cell 1998, 92:229–239. [DOI] [PubMed] [Google Scholar]
- 155. Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta‐catenin. Trends Cell Biol 2007, 17:465–473. [DOI] [PubMed] [Google Scholar]
- 156. Bertrand V, Hobert O. Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans . Dev Cell 2009, 16:563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bertrand V, Hobert O. Lineage programming: navigating through transient regulatory states via binary decisions. Curr Opin Genet Dev 2010, 20:362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Murgan S, Kari W, Rothbächer U, Iché‐Torres M, Mélénec P, Hobert O, Bertrand V. Atypical transcriptional activation by TCF via a Zic transcription factor in C. elegans neuronal precursors. Dev Cell 2015, 33:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol 2011, 27:681–696. [DOI] [PubMed] [Google Scholar]
- 160. Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. A genome‐wide RNAi screen for factors involved in neuronal specification in Caenorhabditis elegans . PLoS Genet 2011, 7:e1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Zhang Y, Emmons SW. Specification of sense‐organ identity by a Caenorhabditis elegans Pax‐6 homologue. Nature 1995, 377:55–59. [DOI] [PubMed] [Google Scholar]
- 162. Baumeister R, Liu Y, Ruvkun G. Lineage‐specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc‐86 during neurogenesis. Genes Dev 1996, 10:1395–1410. [DOI] [PubMed] [Google Scholar]
- 163. Reiprich S, Wegner M. From CNS stem cells to neurons and glia: Sox for everyone. Cell Tissue Res 2015, 359:111–124. [DOI] [PubMed] [Google Scholar]
- 164. Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T‐box transcription factors TBX‐37 and TBX‐38 link GLP‐1/Notch signaling to mesoderm induction in C. elegans embryos. Development 2004, 131:1967–1978. [DOI] [PubMed] [Google Scholar]
- 165. Walton T, Preston E, Nair G, Zacharias AL, Raj A, Murray JI. The Bicoid class homeodomain factors ceh‐36/OTX and unc‐30/PITX cooperate in C. elegans embryonic progenitor cells to regulate robust development. PLoS Genet 2015, 11:e1005003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans . Science 1998, 282:2033–2041. [DOI] [PubMed] [Google Scholar]
- 167. Brenner S. The genetics of behavior. Br Med Bull 1973, 29:269–271. [DOI] [PubMed] [Google Scholar]
- 168. Du Z, Santella A, He F, Shah PK, Kamikawa Y, Bao Z. The regulatory landscape of lineage differentiation in a metazoan embryo. Dev Cell 2015, 34:592–607. [DOI] [PMC free article] [PubMed] [Google Scholar]


