Abstract
Objective
Influenza A virus causes acute respiratory infections that induce annual epidemics and occasional pandemics. While a number of studies indicated that the virus-induced intracellular signaling events are important in combating influenza virus infection, the mechanism how specific molecule plays a critical role among various intracellular signaling events remains unknown. Raf/MEK/ERK cascade is one of the key signaling pathways during influenza virus infection, and the Sprouty-related Ena/VASP homology 1-domain-containing protein (Spred) has recently been identified as a negative regulator of Raf-dependent ERK activation. Here we examined the role of Raf/MEK/ERK cascade through Spred in influenza A (H1N1) viral infection, because the expression of Spred-2 was significantly enhanced in human influenza viral-induced pneumonia autopsy samples.
Design
Prospective animal trial.
Setting
Research laboratory
Subjects
Wild-type (WT) and Spred-2 knockout (KO) mice inoculated with H1N1
Intervention
WT or Spred-2 KO mice were infected by intranasal inoculation of H1N1(A/PR/8). An equal volume of PBS was inoculated intranasally into mock-infected mice.
Measurements and Main Results
H1N1 infection of Spred-2 KO mice led to higher mortality with greater viral load, excessive inflammation, and enhanced cytokine production compared with WT mice. Administration of MEK-inhibitor, U0126, improved mortality and reduced both viral load and cytokine levels. Moreover, bone marrow chimeras indicated that H1N1-induced lung pathology was most severe when Spred-2 expression was lacking in nonimmune cell populations. Furthermore, microarray analysis revealed knockdown of Spred-2 led to enhanced phosphatidylinositol 3-kinase (PI3K) signaling pathway, resulting that viral clearance was regulated by Spred-2 expression through the PI3K signaling pathway in murine lung epithelial cells.
Conclusions
These data support an important function of Spred-2 in controlling influenza virus-induced pneumonia and viral replication. Spred-2 may be a novel therapeutic target for controlling the immune response against influenza H1N1 virus infection.
Keywords: influenza virus, Sprouty-related Ena/VASP homology 1-domain-containing protein-2, mitogen-activated protein kinase, phosphatidylinositol 3-kinase, mouse, cytokine
INTRODUCTION
Influenza A (H1N1) viral infections contribute to the annual mortality associated with acute viral pneumonia and have been identified as the etiologic agents for historic global pandemics (1–3), the most severe of which on record occurred in 1918. More recently in 2009, the currently circulating H1N1 virus resulted in a worldwide outbreak (4, 5). Influenza A viruses are members of the enveloped Orthomyxoviridae family. The virus genome consists of eight single-stranded, negative sense RNA segments, which encode up to 12 viral proteins. The three subunits (PB1, PB2, and PA) of the viral polymerase and the nucleoprotein (NP) together with the viral RNA form the replication and transcription active ribonucleoprotein (RNP) complex (3, 6).
Influenza virus infection occurs by a multiple step pathway, including endocytosis, replication, nuclear export and finally, budding from the cell membrane. Epithelial cells of the upper respiratory tract are the primary target of influenza virus infection. Viral infection is initially sensed by the host innate immune system, triggering a rapid antiviral response involving various cell signaling cascades. This eventually leads to an anti-viral immune response characterized by production of cytokines and chemokines such as type-I IFN, CXCL1 (KC) and CCL2 (MCP-1) (7, 8). The Raf/MEK/ERK cascade, which belongs to the mitogen-activated protein (MAP) kinase cascade family and has an important role in cell growth, differentiation and survival, as well as cytokine/chemokine production, is one of the key regulators of influenza virus infection (9–12). Several studies have demonstrated that influenza virus activates several signaling cascades, including the Raf/MEK/ERK-, JNK- and p38-pathways (13, 14). Such activation plays a critical role in virus production and RNP export from the nucleus during influenza virus infection (15–17).
Recently, the Spred (Sprouty-related Ena/VASP homology 1 domain-containing) proteins have been implicated as general inhibitors of Raf/MEK/ERK signaling (18). There are three Spreds (Spred-1, Spred-2, and Spred-3), which bind to Ras and Raf, thereby suppressing activation of Raf (18, 19). Spred-2 is ubiquitinously expressed in various tissues including lung, whereas Spred-1 and Spred-3 are preferentially expressed in the brain and cerebellum (20, 21). However, the role of Spred proteins in influenza virus infection remains completely unknown. Animal models of influenza are essential to research efforts aimed at understanding the viral and host factors that contribute to the disease and transmission outcomes of influenza virus infection in humans. In various laboratory animal models, mice are the most widely used animal models for influenza virus research, and offer a system in which the host response to infection can be studied in depth coupled with ability to manipulate mice genetically (22). In the present study, we have focused on Spred-2 and investigated the physiopathological role of this protein in influenza virus infection using Spred-2 KO mice. We demonstrate that Spred-2 plays an indispensable role in the regulation of influenza virus replication and the ensuing inflammatory response.
MATERIALS AND METHODS
Ethics statement
The Animal Care and Use Committee at Okayama University approved all animal experiments conducted in this study, and all methods were carried out based on the Policy on the Care and Use of Laboratory Animals, Okayama University. Human autopsy lung samples from patients who died following confirmed H1N1 influenza infection at the University of Michigan Health Systems (UMHS: Ann Arbor, MI) were collected as previously described (23). Ethical Committee at Okayama University approved the use of all human autopsy samples and waived the need for consent.
Human autopsy sample
Three age and sex matched non-influenza-related autopsy cases were selected as controls. Blocks of formalin-fixed, paraffin-embedded lung tissue were used for all studies, including RNA isolation and immunohistochemistry.
Mice
Age (8–12 week) and sex matched mice were used in all experiments. Spred-2 KO mice backcrossed into the C57BL/6 background were kindly provided by Dr. Akihiko Yoshimura (Keio University, Japan) (24). C57BL/6 mice were used as WT mice. These mice were housed in the specific-pathogen-free animal facility at 25°C with a 12-hr light/dark cycle, in the Department of Animal Resources at Okayama University.
Reagents
Rat mAbs specific for mouse CD3 (17A2), CD4 (L3T4), CD8 (53–6.7), CD11b (M1/70), CD16/32 (2.4G2), CD45 (30-F11), CD69 (H1.2F3), Gr-1 (RB6-8C5), and NK1.1 (PK136) were purchased from BD Pharmingen (San Jose, CA). Rat Anti-F4/80 (BM8) mAb was purchased from BioLegend (San Diego, CA). Antibodies to p44/42 MAPK (Erk1/2; 137F5), phospho-p44/42 MAPK Thr202/Thr204 (Erk1/2; D13.14.4E), SAPK/JNK (56G8), phospho-SAPK/JNK Thr183/Thr185 (81E11), p38 MAPK, phospho-p38 MAPK Thr180/Thr182 (D3F9), Akt (C67E7), phospho-Akt Thr308 (244F9), and GAPDH (14C10) were purchased from Cell Signaling Technology (Danvers, MA). U0126 and LY294402 were from Calbiochem (Darmstadt, Germany) and Cell Signaling Technology, respectively. The mouse cell line MLE-12 was purchased from the American Type Culture Collection (Manassas, VA).
Virus infection and sampling
Influenza virus A/Puerto Rico/8/34 (H1N1) was used throughout the study. Mice were infected by intranasal inoculation of 100 plaque forming units (PFU) of virus in 25 µl of PBS. An equal volume of PBS was inoculated intranasally into mock- infected mice (25). In some experiments, mice were treated intranasally with 20 µl of U0126 (200 µg) or control DMSO on days 0, 2, and 4 of viral challenge. Lungs were harvested at the indicated time after influenza infection. The left lobe of the lung was used for histological assessment, and each right lobe was homogenized for the analysis of mRNA, protein, flow cytometry, and virus infectious titer. Lung homogenates were serially diluted with Eagle’s Minimum Essential Medium medium (Sigma-Aldrich, St. Louis, MO) and virus infectious titers were measured using the 50% tissue culture infectious doses (TCID50) assay based on cytopathic effect as previously described (7).
Short-interfering (si) RNA assay
A total of 1.0 × 106 MLE-12 cells were transfected with 2 µg of a mixture of Spred2-specific, or non-targeting control siRNAs (Thermo Scientific, Yokohama, Japan), using mouse macrophage nucleofector kit (Lonza, Cologne, Germany) according to the manufacturer’s instructions and plated in a 24-well plate (7). Cells were used at 24 hours post transfection.
Reverse transcription and real-time quantitative PCR analysis
Total RNA was isolated from cultured cells and whole lungs using High Pure RNA Isolation and High Pure RNA Tissue kits (Roche Applied Science, Penzberg, Germany), respectively. RNA from human autopsy samples was isolated using an RNeasy FFPE kit (Qiagen, Germantown, MD). Total RNA was extracted, and 1µg of total RNA was reverse-transcribed to cDNA according to the procedure previously described. Real-time quantitative PCR analysis was performed using StepOne with Taqman PCR master mix (Applied Biosystems, Foster City, CA). The thermal cycling conditions included 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s and 55°C for 1.5 min for denaturing and annealing, respectively. Quantification of the genes of interests were normalized to GAPDH and expressed as fold increases over the negative control for each treatment at each time point, as previously described (7).
Microarray Analysis
For cRNA amplification and labeling, total RNA was amplified and labeled with Cyanine 3 (Cy3) using Agilent Low Input Quick Amp Labeling Kit, one-color (Agilent Technologies, Santa Clara, CA) following the manufacturer's instructions. Briefly, 100ng of total RNA was reversed transcribed to double-strand cDNA using a poly dT-T7 promoter primer. Primer, template RNA and quality-control transcripts of known concentration and quality were first denatured at 65°C for 10 min and incubated for 2 hours at 40°C with 5× first strand Buffer, 0.1 M DTT, 10 mM dNTP mix, and AffinityScript RNase Block Mix. The AffinityScript enzyme was inactivated at 70°C for 15 min. cDNA products were then used as templates for in vitro transcription to generate fluorescent cRNA. cDNA products were mixed with a transcription master mix in the presence of T7 RNA polymerase and Cy3 labeled-CTP and incubated at 40°C for 2 hours. Labeled cRNAs were purified using RNeasy mini spin columns (Qiagen) and eluted in 30 µl of nuclease-free water. After amplification and labeling, cRNA quantity and cyanine incorporation were determined using a Nanodrop ND-1000 spectrophotometer and an Agilent Bioanalyzer. Next for each hybridization, 1.65 µg of Cy3 labeled cDNA were hybridized at 65°C for 17 hours to an Agilent SurePrint G3 Mouse GE 8×60K Microarray (Design ID: 028005). After washing, microarrays were scanned using an Agilent DNA microarray scanner. Intensity values of each scanned feature were then quantified using Agilent feature extraction software version 10.7.3.1, which performs background subtractions. We only used features which were flagged as no errors (present flags) and excluded features which were not positive, not significant, not uniform, not above background, saturated and population outliers (marginal and absent flags). Normalization was performed using Agilent GeneSpring GX version 11.0.2. (per chip: normalization to 75 percentile shift; per gene: normalization to median of all samples). A total of 55,681 probes were present on the Agilent SurePrint G3 Mouse GE 8×60K Microarray (Design ID: 028005) without control probes.
Western blotting
MLE-12 cells or lung samples were lysed in lysis buffer (Cell Signaling Technology), briefly sonicated, kept on ice for 30 minutes, and centrifuged at 15,000 rpm for 10 minutes. The supernatant was collected and stored at −80°C until required. Total protein concentration of the samples was measured by Bradford protein assay (Bio-Rad Laboratories Inc., Hercules, CA). Equal concentrations and amounts (15–30 µg) of cell lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Invitrogen, Carlsbad, CA). The proteins were then transferred onto a nitrocellulose membrane. After the overnight incubation with appropriate primary antibody, the membrane was counter-stained with horseradish peroxidase-conjugated rabbit or mouse IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and visualized with enhanced chemiluminescence detection reagents (GE Healthcare, Little Chalfont, UK). Quantifications of intensity were calculated by Image J software.
Measurement of cytokines
Murine cytokines were measured using a standard sandwich ELISA, as per the manufacturer’s protocol (R&D Systems, Minneapolis, MN). Antibodies and recombinant chemokines, CCL2 and CXCL1, were purchased from R&D Systems. The ELISAs employed in this study did not cross-react with other murine cytokines available. ELISAs for IFN-α and IFN-β levels were purchased from PBL Interferon Source. For cytokine measurement in the lung, lung tissues were homogenized in PBS containing 0.1% TritonX-100 and complete protease inhibitor (Roche Applied Science) then centrifuged to obtain cleared supernatants. The cytokine levels in lung homogenates were normalized to the protein present in cell-free preparations of each sample measured by the Bradford assay, as described previously (7).
Flow cytometry
Flow cytometric analyses of lung cells were performed as previously described. In brief, whole lungs were minced and digested with 0.02% (w/v) collagenase and 0.0005% (w/v) DNase I in HBSS containing 20mM HEPES, pH 7.4, with a gentleMACS Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions, to obtain a single-cell suspension. The cells were stained with the indicated antibodies after 10 minutes of pre-incubation with CD16/CD32 antibody (Fc block) and fixed overnight with 1% formalin. Cells were analyzed using a MACSQuant Analyzer (Miltenyi Biotec), and data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR).
Histological assessment
After BAL lavage, some of the lungs from WT and Spred-2 KO mice were instilled with 4% buffered paraformaldehyde, removed, and fixed in the same solution. After paraffin embedding, sections for microscopy (4-µm slices) were stained with H&E. An index of pathologic changes in coded H&E slides was established by scoring the inflammatory cell infiltrates around airways and vessels for severity (0, normal; 1, <3 cells thick; 2, 4—10 cells thick; 3, >10 cells thick) and overall extent (0, normal; 1, <25% of sample; 2, 25—50%; 3, 51—75%; 4, >75%). The index was calculated by multiplying severity by extent, with a maximal possible score of 12 (26). Immunostaining was carried out using the Histofine Simple Stain MAX-PO (Nichirei Biosciences Inc, Tokyo, Japan), according to the manufacturer’s instructions. In brief, sections (4 µm slices) were treated with 0.3% H2O2 in methanol and then incubated with anti-Spred-2 antibodies (2G11, Sigma-Aldrich) (for human tissue) or anti-phospho-ERK (D13.14.4E) (for human and murine tissue) overnight at 4°C. Sections were rinsed and incubated with peroxidase-labeled polymer at room temperature for 30 minutes. As a chromogen, diaminobenzidine (DAKO, Carpinteria, CA) was used. For immunofluorescent analysis, MLE-12 cells were grown on 8-well chamber slides (Thermo Fisher Scientific Inc.) in complete DMEM (supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin). After incubation with H1N1 virus, MLE-12 cells were washed with PBS, then fixed in ice-cold acetone, incubated with anti-influenza A virus nucleoprotein (NP) antibody (AA5H) (Abcam, Cambridge, UK), followed by the addition of Alexa568-labeled anti-mouse IgG (Invitrogen). Finally, the sections were analyzed by Olympus confocal microscope system (Olympus, Tokyo, Japan).
Bone marrow chimeras
Recipient WT C57BL/6 and Spred-2 KO mice received two 4.50 Gy X-ray total body irradiation separated by 3 hours. Donor bone marrow was isolated from femurs and tibias of WT and Spred-2 KO mice. Two to 4 hours following the last irradiation dose, 2 × 106 bone marrow cells in 100 µl PBS were administered via tail vein injection. The following bone marrow chimeras were created (donor → host) (27): WT → WT, Spred-2 KO → WT, Spred-2 KO → Spred-2 KO, and WT → Spred-2 KO. 8 weeks after reconstitution, bone marrow chimeras were infected with H1N1.
Statistics
Statistical significance was evaluated by ANOVA. All data were expressed as mean ± SEM. Differences of p < 0.05 were considered significant. All statistical calculations were performed using GraphPad Prism 4.0 (GraphPad Software, San Diego, CA).
RESULTS
Spred-2 expression is enhanced in the lungs during H1N1 infection from both human autopsy and mouse samples
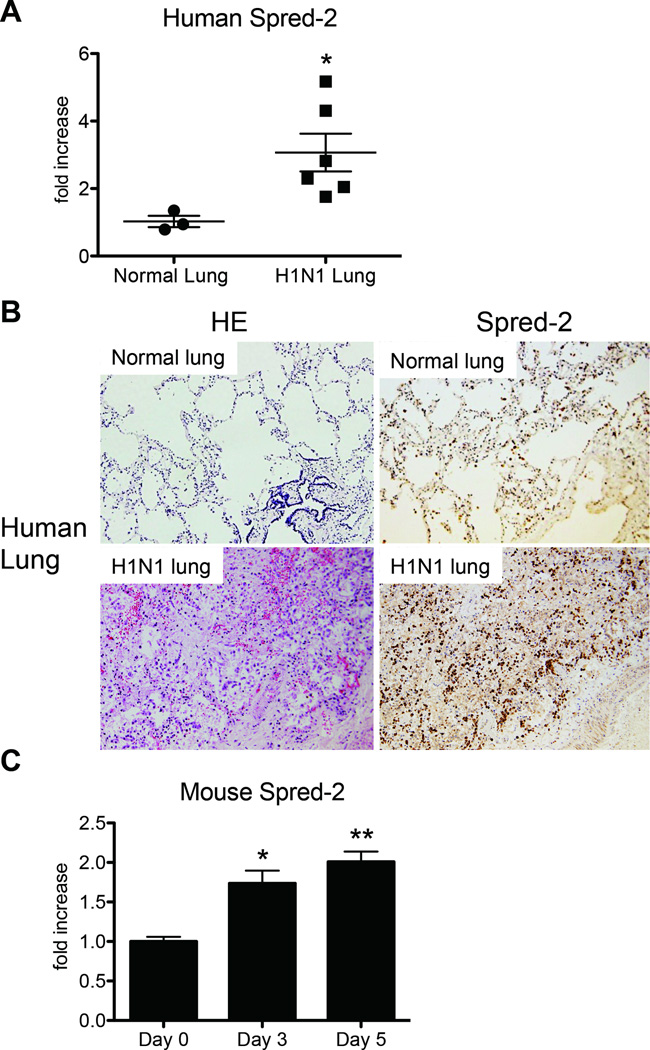
We first assessed the expression of Spred-2 in paraffin-embedded autopsy lung tissue from patients succumbing to H1N1 infection. Microscopic autopsy findings were described in a previous report (23). Compared with non-influenza-related autopsy samples, expression of Spred-2 was significantly higher in humanlung samples from H1N1-related deaths (Fig. 1A). This result was also supported by immunohistochemical analysis (Fig. 1B), which shows enhanced staining of Spred-2 in H1N1 infected humanlung tissue. Moreover, intranasal infection of wild-type (WT) mice with H1N1 virus showed a significant increase of Spred-2 gene expression at both day 3 and day 5 post-infection (Fig. 1C).
Figure 1. H1N1-infected lungs exhibited increased expression of Spred-2.
(A) The expression of Spred-2 in paraffin-embedded autopsy lung samples from 3 non-H1N1 and 6 H1N1-related deaths were evaluated by quantitative real-time PCR. *p < 0.05 compared with normal lung group. (B) Representative immunohistochemical examinations of human lung autopsy samples showed enhanced Spred-2 expression in the lungs following H1N1-related death. (C) WT mice were inoculated intranasally with H1N1 at 1 × 102 pfu. Quantitative real-time PCR was performed to determine transcript level of Spred-2 at days 0, 3 and 5 after inoculation of influenza virus.*p < 0.05, ** p < 0.01 compared with Day 0. Data shown indicate mean ± SEM and are from a representative experiment of 3 independent experiments. Each time point represents at least 5 mice per group.
Spred-2 KO mice are susceptible to influenza A virus (H1N1) infection
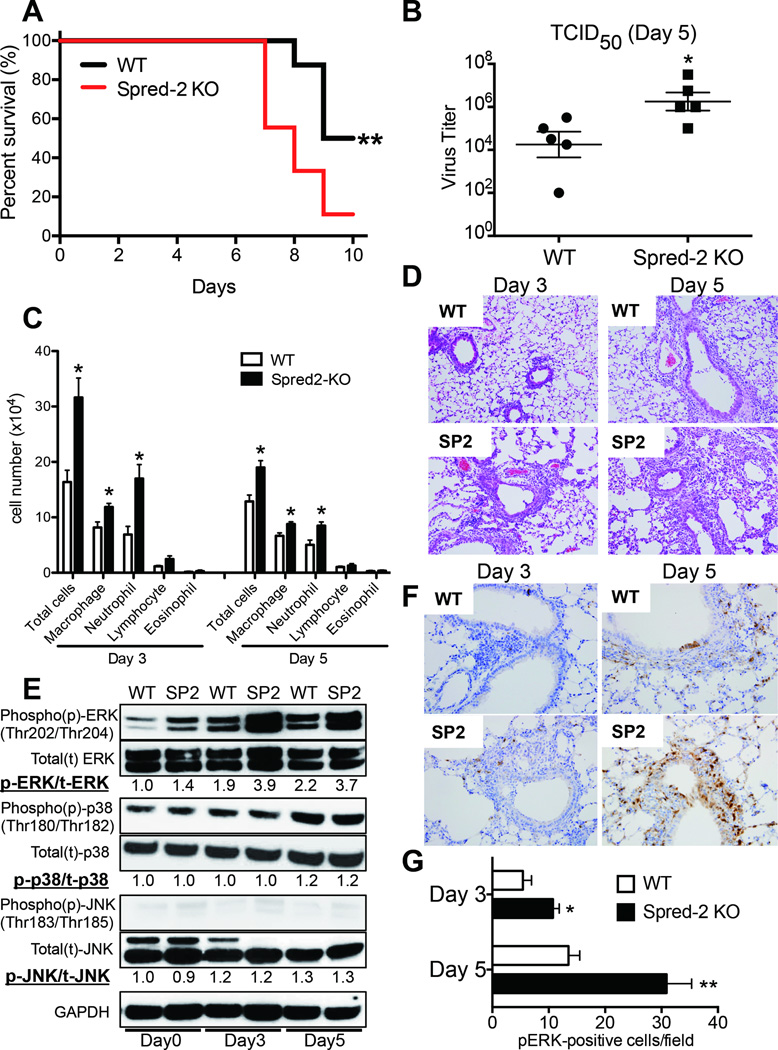
To directly examine the importance of Spred-2 during influenza virus infection, we examined whether Spred-2 is correlated with the pathogenesis of influenza virus infection. First, we monitored the survival of WT and Spred-2 KO mice following H1N1 infection. We confirmed that the absence of Spred-2 led to increased mortality after viral challenge when compared with WT mice (Fig. 2A). In agreement with the mortality data, Spred-2 KO mice also showed a significantly greater viral load, measured by 50% tissue culture infective dose (TCID50) at day 5 post infection (Fig. 2B). Additionally, the cellular appearance of the bronchoalveolar lavage (BAL) demonstrated an increased number of total cells, macrophages, and neutrophils (Fig. 2C). These findings were confirmed in lung histology studies at both day 3 and day 5 post infection, showing a significant increase in lung inflammation in Spred-2 KO mice, as compared with WT mice (Fig. 2D). Thus, we used this model of infection to examine the contribution of the MEK-ERK pathway, which is downstream of Raf activation. Following H1N1 infection, ERK was phosphorylated in both WT and Spred-2 KO mice. In addition, ERK phosphorylation was enhanced in Spred-2 KO mice compared with WT mice (Day 3: 2.0 fold, Day 5: 1.7 fold). However, despite enhanced ERK phosphorylation in Spred-2-deficient mice, there is no difference in p38 and JNK between WT and Spred-2 KO mice (Fig. 2E). Moreover, immunohistochemical analysis of samples from Spred-2 KO mice showed significantly increased number of phosphorylated ERK, especially in bronchial epithelial and inflammatory cells, compared with WT mice (Fig.2F and 2G).
Figure 2. Spred-2 deficiency abrogates survival rate, viral load and lung pathology.
(A) Survival rate of WT and Spred-2 KO mice after intranasal injection with H1N1 at 1 × 102 pfu. Results are expressed as the percentage of survival from 16 individual mice per group. **p < 0.01 compared with WT mice.(B) Viral load in WT and Spred-2 KO mice at day 5 after intranasal injection with H1N1 (1 × 102 pfu). *p < 0.05 compared with H1N1 injection group treated with WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. Each time point indicates 5 mice per group. (C) Cellular analysis of BAL from WT and Spred-2 KO mice at days 3 and 5 after intranasal injection with H1N1. *p < 0.05 compared with WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. Each time point indicates 5 mice per group. (D) Histological appearance of lungs isolated from WT and Spred-2 KO mice at days 3 and 5 after intranasal injection with H1N1. Shown are representative sections from 1 mouse of 5 per group. H&E staining. Original magnification, ×100. (E) Lungs from WT and Spred-2 KO mice were harvested at day 0, 3, and 5 after H1N1 challenge and extracts were immunoblotted with phosphorylated (p)-ERK, ERK, p-p38, p38, p-JNK, JNK, or GAPDH Abs. GAPDH was used as a loading control. Experiment was repeated three times. A representative gel images and the value of phosphorylation compared with lung from WT mice (Day 0) are shown. (F) Immunohistochemical examination showed increased number of phosphorylated ERK-positive cells in Spred-2 KO mice. Shown are representative sections from 1 mouse of 5 per group. Original magnification, ×200. (G) Average number of pERK-positive cells per each field at x200 magnification, 10 fields per lung, 5 lungs per group. *p < 0.05, **p < 0.01 compared with WT mice. SP2 = Spred-2 KO
Spred-2 KO mice display an enhanced immune response during influenza virus infection
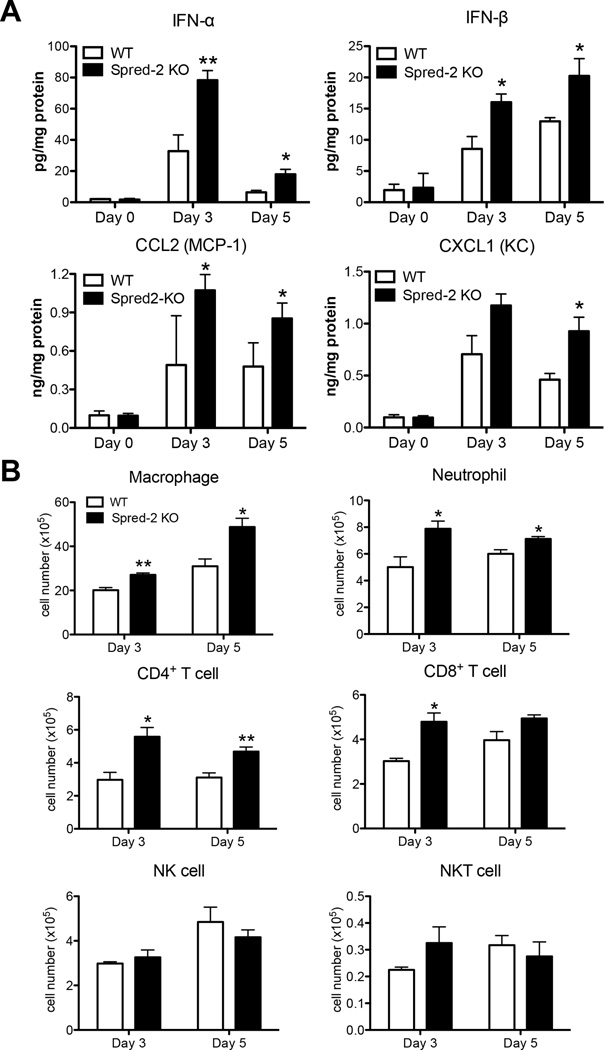
To help elucidate the mechanism underlying the increased mortality and severe inflammation seen in Spred-2 KO mice, we examined the cytokine and chemokine profile in whole lungs during H1N1 challenge. Type-I IFN protein expression was significantly higher in H1N1-infected whole lung homogenates from Spred-2 KO mice compared with lungs from WT mice (Fig. 3A). Additionally, H1N1-infected whole lungs from Spred-2 KO mice had significantly higher protein levels of CCL2 and CXCL1, molecules that play a critical role in the recruitment of monocytes/macrophages and neutrophils into inflammatory lesions (Fig. 3A). In agreement with the chemokine profile observed in whole lungs, flow cytometric analysis demonstrated increased macrophage (WT vs. Spred-2 KO; Day 3: 20.2 ± 1.2 vs. 27.1 ± 0.8, p<0.01, Day 5: 31.0 ± 3.3 vs. 48.8 ± 4.0, p<0.05) and neutrophil (WT vs. Spred-2 KO; Day 3: 5.0 ± 0.8 vs. 7.9 ± 0.6, p<0.05, Day 5: 6.0 ± 0.3 vs. 7.1 ± 0.2, p<0.05) recruitment during H1N1 infection in Spred-2 KO mice (Fig. 3B). Moreover, the numbers of both CD4+(WT vs. Spred-2 KO; Day 3: 3.0 ± 0.5 vs. 5.6 ± 0.6, p<0.05, Day 5: 3.1 ± 0.3 vs. 4.7 ± 0.3, p<0.01) and CD8+ (WT vs. Spred-2 KO; Day 3: 3.0 ± 0.1 vs. 4.8 ± 0.6, p<0.05, Day 5: 4.0 ± 0.4 vs. 4.9 ± 0.2) T cells were significantly higher in Spred-2 KO mice, whereas no significant differences were found in the number of NK cells (WT vs. Spred-2 KO; Day 3: 3.0 ± 0.1 vs. 3.3 ± 0.3, Day 5: 4.9 ± 0.7 vs. 4.2 ± 0.3) and NKT cells (WT vs. Spred-2 KO; Day 3: 0.23 ± 0.01 vs. 0.33 ± 0.06, Day 5: 0.32 ± 0.04 vs. 0.28 ± 0.06) between both mouse strains (Fig. 3B).
Figure 3. Deficiency of Spred-2 modulates immune response during influenza virus infection.
(A) Protein levels of cytokines and chemokines in whole lungs from WT and Spred-2 KO mice at days 3 and 5 after influenza virus infection, determined by ELISA. (B) Flow cytometry analysis of lung macrophage (CD11b+F4/80+), neutrophil (Gr-1high), CD4+ T cell (CD3+CD4+), CD8+ T cell (CD3+CD8+), NK cell (CD3−NK1.1+), and NKT cell (CD3+NK1.1+) isolated from influenza challenged WT and Spred-2 KO mice at days 3 and 5.*p < 0.05, **p < 0.01 compared with WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 3 independent experiments. Each point had 5 mice per group.
Inhibition of the Raf/MEK/ERK signaling cascade leads to impaired pathogenesis of influenza virus infection
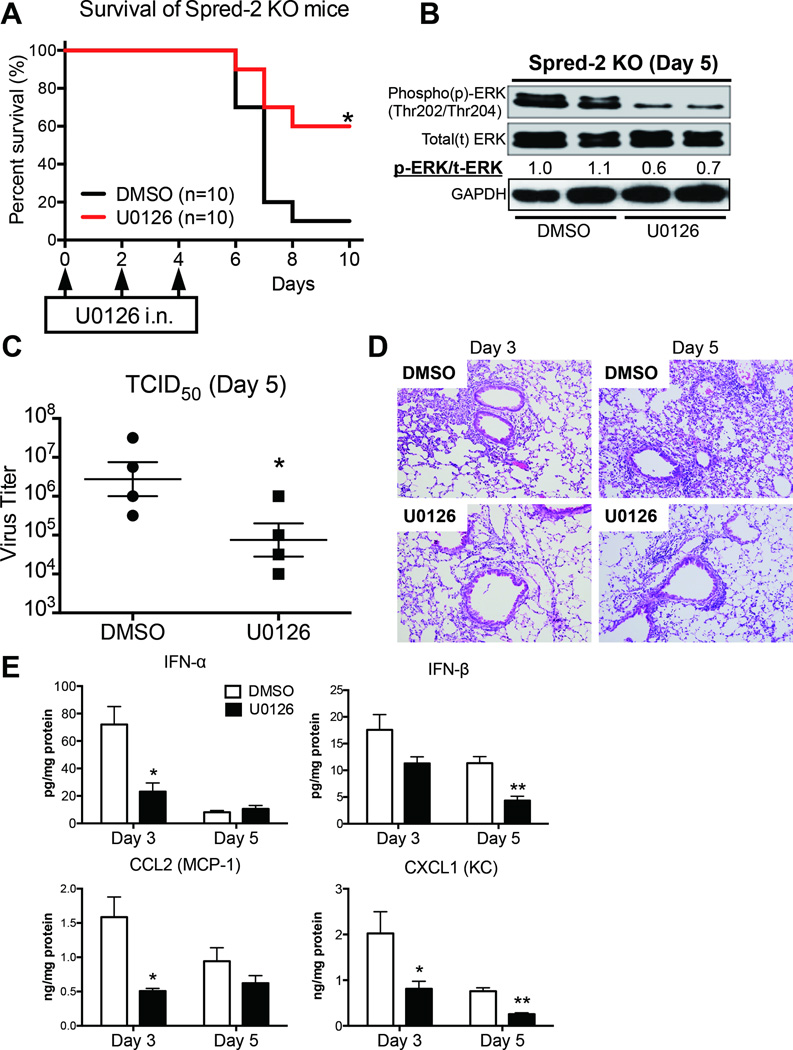
To directly test whether the Raf/MEK/ERK signaling pathway correlates with abrogated pathogenesis of influenza virus infection in Spred-2 KO mice, we used the specific MEK inhibitor U0126. Intranasal administration of U0126 in Spred-2 KO mice during influenza infection led to lower mortality with decreased expression of phosphorylated-ERK (0.6–0.7 fold) in the lungs compared with the control DMSO-treated group (Fig.4A and 4B). Furthermore, measurement of TCID50 indicated significantly lower viral load in the lungs of mice receiving U0126 compared with DMSO controls at day 5 post-infection (Fig. 4C). Additionally, histological appearance demonstrated that the U0126-treated group displayed reduced inflammation in the lungs (Fig. 4D). We also found significantly impaired production of IFN-α (WT vs. Spred-2 KO; Day 3: 72.0 ± 13.1 vs. 23.1 ± 6.4 p<0.05, Day 5: 8.09 ± 1.3 vs. 10.5 ± 2.5), IFN-β (WT vs. Spred-2 KO; Day 3: 17.6 ± 2.8 vs. 11.3 ± 1.2, Day 5: 11.3 ± 1.2 vs. 4.35 ± 0.8, p<0.01), CCL2 (WT vs. Spred-2 KO; Day 3: 1.59 ± 0.30 vs. 0.51 ± 0.04, p<0.05, Day 5: 0.94 ± 0.20 vs. 0.62 ± 0.11), and CXCL1 (WT vs. Spred-2 KO; Day 3: 2.02 ± 0.48 vs. 0.81 ± 0.17, p<0.05, Day 5: 0.76 ± 0.08 vs. 0.26 ± 0.03, p<0.01), compared with the control treated group (Fig. 4E). Conversely, U0126 treatment of WT mice led to no significant reduction in mortality (data not shown).
Figure 4. Blocking of MEK signaling improves survival rate, viral load, and lung inflammation.
U0126 (200 µg; 20 µl volume) was intranasally administrated to mice at days 0, 2 and 4 after influenza virus challenge. DMSO in 20 µl was used as a control for U0126. (A) Survival rate of Spred-2 KO mice treated with either control DMSO or U0126 after intranasal injection with 1 × 102 pfu H1N1. *p < 0.05 compared with WT mice. n=10 per group. (B) Lungs from Spred-2 KO mice treated with either control DMSO or U0126 after intranasal injection with 1 × 102 pfu H1N1 were harvested at day 5 after H1N1 challenge and extracts were immunoblotted with the indicated antibodies. Data are expressed from a representation of two independent experiments. A representative gel images and the value of phosphorylation compared with lung from DMSO-treated mice are shown. (C) Viral load in Spred-2 KO mice treated with control DMSO or U0126 at day 5 after influenza virus infection, measured by TCID50. *p < 0.05 compared with WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. Each group had 5 mice per group. (D) Representative histological appearance of lungs isolated from Spred-2 KO mice treated with either control DMSO or U0126 after intranasal injection with 1 × 102 pfu H1N1, harvested at day 5 after H1N1 infection. H&E staining. Original magnification, ×40; ×200. (E) Protein level of cytokines and chemokines from whole lungs was measured by ELISA.*p < 0.05, **p < 0.01 compared with WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 3 independent experiments. Each group had 5 mice per group.
Differential regulation of inflammation and viral clearance by Spred-2 in immune and nonimmune cell populations
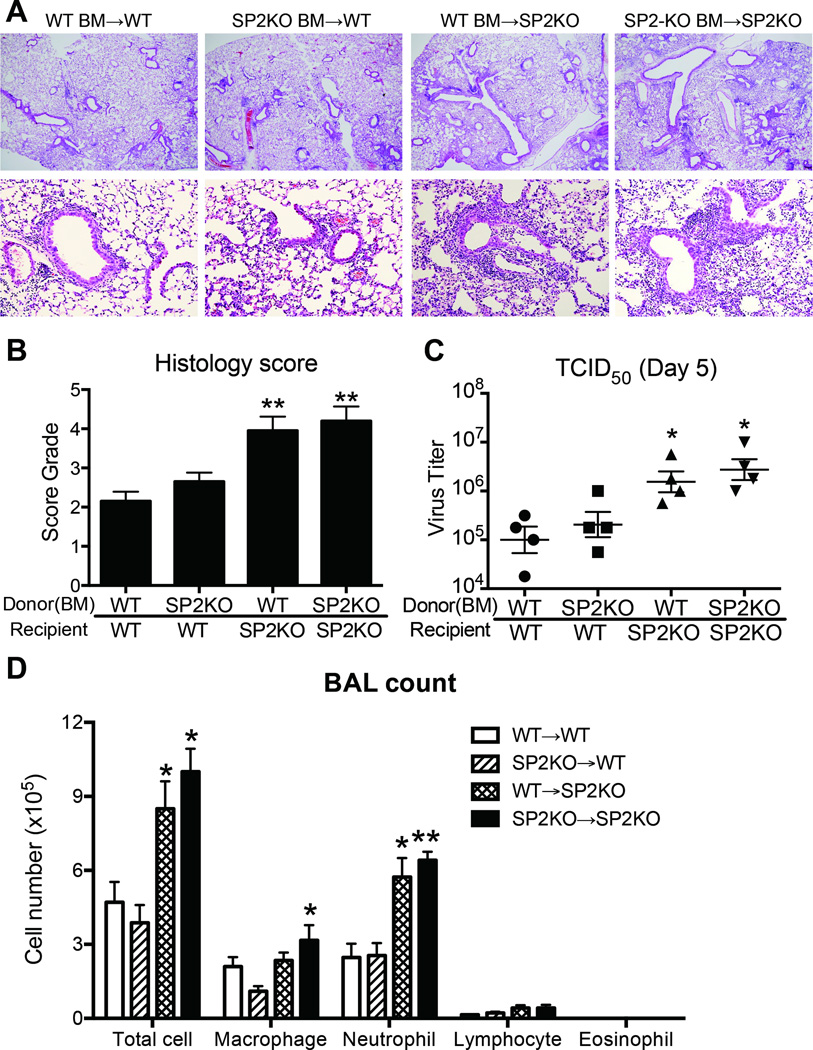
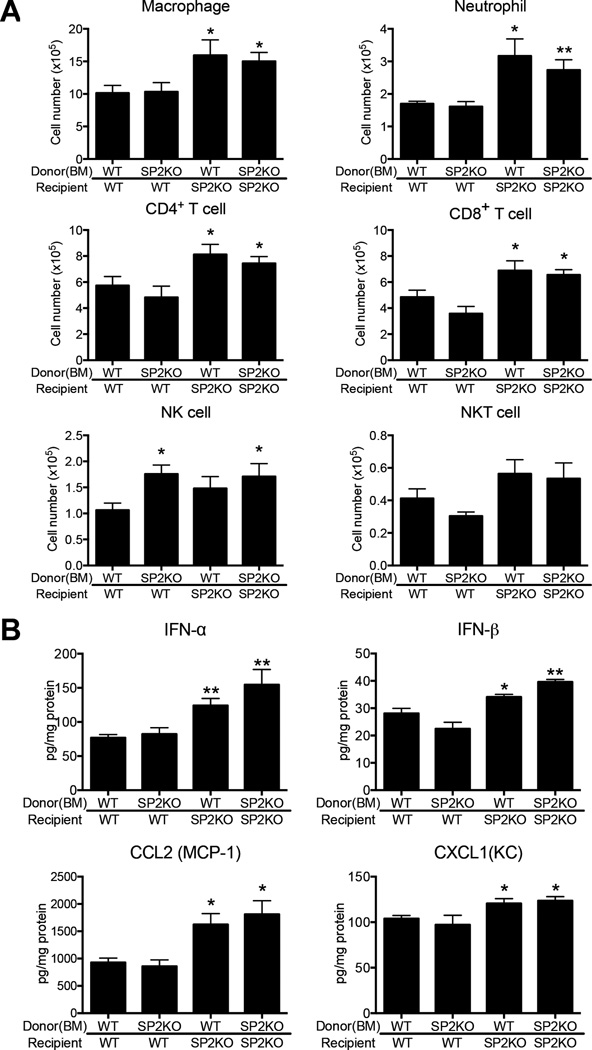
To directly examine which cell types contribute to the pathologic discrepancies between WT and Spred-2 KO mice, we next created bone marrow (BM) chimeras. This allowed us to address the question of whether presence of Spred-2 in either the immune or the nonimmune compartment is crucial to control of H1N1-induced inflammation and viral clearance. Whereas H1N1-induced inflammation was limited in WT chimeras receiving either WT or KO BM, histological inflammation and score was more severe in KO chimeras regardless of bone marrow origin (Fig.5A and 5B), suggesting that Spred-2 expression in nonimmune cells, most likely lung epithelial cells, plays a critical role in regulating influenza virus infection in these animals. Accordingly, TCID50 indicated higher viral load in recipient Spred-2 KO mice compared with WT chimeras (Fig. 5C). Spred-2 KO mice also showed significantly increased leukocyte infiltration, including increased neutrophil infiltration in BAL, regardless of donor BM origin, compared with recipient animals on the WT background (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; Total cell: 4.7 ± 0.8, 3.9 ± 0.7, 8.5 ± 1.1: p<0.05, vs. 10.3 ± 1.2: p<0.05, Macrophage: 2.1 ± 0.4, 1.1 ± 0.3, 2.4 ± 1.1, vs. 3.2 ± 0.3: p<0.05, Neutrophil: 2.5 ± 0.6, 2.6 ± 0.5, 5.7 ± 0.9: p<0.05, vs. 6.4 ± 0.4: p<0.01, Lymphocyte: 0.15 ± 0.03, 0.22 ± 0.05, 0.37 ± 0.12, vs. 0.42 ± 0.16, Eosinophil: not detected)(Fig. 5D). Additionally, flow cytometric analysis of lung cells demonstrated that the numbers of macrophages (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 11.7 ± 1.3, 10.3 ± 1.1, 16.0 ± 2.1: p<0.05, vs. 15.0 ± 1.4: p<0.05), neutrophils (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 1.7 ± 0.1, 1.6 ± 0.2, 3.2 ± 0.5: p<0.05, vs. 2.7 ± 0.3: p<0.01), CD4+(WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 5.7 ± 0.7, 4.8 ± 0.9, 8.1 ± 0.8: p<0.05, vs. 7.4 ± 0.5: p<0.05) and CD8+ T cells (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 4.8 ± 0.5, 3.6 ± 0.5, 6.9 ± 0.8: p<0.05, vs. 6.6 ± 0.4: p<0.05) were significantly higher during influenza virus infection in recipient Spred-2 KO mice, compared with chimeras on the WT background, regardless of donor BM origin (Fig. 6A). On the other hand, the number of NK cells was significantly increased when donor was Spred-2 KO mice (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 1.1 ± 0.1, 1.8 ± 0.2: p<0.05, 1.5 ± 0.2, vs. 1.7 ± 0.3: p<0.05), while there was no significant difference in that of NKT cells (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 0.40 ± 0.06, 0.31 ± 0.03, 0.56 ± 0.09, vs. 0.53 ± 0.10)(Fig. 6A). We further demonstrated that lung protein levels of IFN-α (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 76.9 ± 4.6, 82.3 ± 9.3, 124.0 ± 10.4: p<0.01, vs. 154.6 ± 22.3: p<0.01), IFN-β (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 28.1 ± 1.5, 22.5 ± 2.4, 34.1 ± 1.0: p<0.05, vs. 39.6 ± 0.9: p<0.01), CCL2 (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 927.5 ± 82.0, 858.6 ± 117.0, 1623 ± 199.8: p<0.05, vs. 1811 ± 248.4: p<0.05) and CXCL1 (WT BM→WT, SP2KO BM→WT, WT BM→SP2KO, vs. SP2KO BM→SP2KO; 103.9 ± 3.4, 97.1 ± 10.5, 120.6 ± 5.2: p<0.05, vs. 123.5 ± 4.5: p<0.05) in recipient KO mice were significantly higher when compared with WT animals, with genotype of donor BM having no significant effect (Fig. 6B). Taken together, these data indicate that Spred-2 expression must occur in the non-hematopoietic compartment to exert its protective effect.
Figure 5. Spred-2 in nonimmune cells plays critical role against influenza virus infection.
The following bone marrow chimeras were created (donor → host): WT → WT, Spred-2 KO → WT, Spred-2 KO → Spred-2 KO, and WT → Spred-2 KO. 8 weeks after reconstitution, bone marrow chimeras were intranasally infected with influenza virus (1 × 102 pfu), and then sacrificed at 5 days post-infection. (A) Representative photomicrographs of hematoxylin-stained lung sections of WT → WT, KO → WT, KO → KO, and WT → KO chimeras at day 5 post-H1N1 infection. Original magnification, ×40; ×200. (B) Quantitative analysis of histopathologic changes using a scoring system detailed in the methods. (C) Viral load in BM-chimera mice measured by TCID50. (D) Absolute numbers of inflammatory leukocytes in the BAL fluid of BM-chimera mice. SP2KO = Spred-2 KO. *p < 0.05, **p < 0.01 compared with when both donors and recipients were from WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. Each group had 4–6 mice per group.
Figure 6. Differential immune response by Spred-2 expression between immune and nonimmune cells during influenza virus infection.
(A) FACS analysis of lung macrophage (CD11b+F4/80+), neutrophil (Gr-1high), CD4+ T cell (CD3+CD4+), CD8+ T cell (CD3+CD8+), NK cell (CD3−NK1.1+), and NKT cell (CD3+NK1.1+) isolated from influenza challenged BM-chimera mice at day 5. (B) Protein levels of cytokines and chemokines from whole lungs in BM-chimera mice were measured using ELISA. SP2KO = Spred-2 KO. *p < 0.05, **p < 0.01 compared with when both donors and recipients were from WT mice. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. Each group had 4–6 mice per group.
Spred-2 regulates influenza virus replication in pulmonary epithelial cells
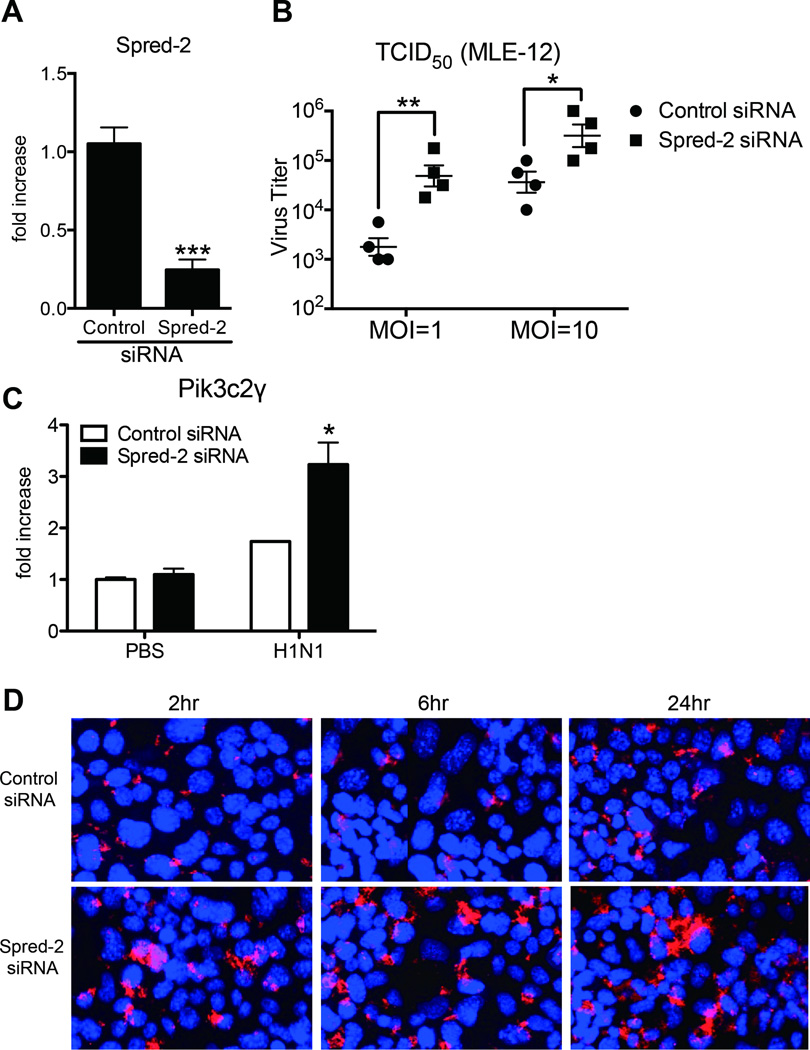
Our results so far suggest nonimmune cells in the lungs play a key role in regulating both viral titers and inflammation. To determine whether influenza virus infection is regulated by Spred-2 in pulmonary epithelial cells, we knocked down Spred-2 gene expression in the mouse pulmonary epithelial cell line, MLE12, and examined signal transduction activity, production of type-I IFNs and viral titer. First, we examined the efficiency of gene knockdown of Spred-2. The gene expression of Spred-2 was significantly and 1/4–1/5 down-regulated by Spred-2 siRNA compared with control siRNA (Fig. 7A). Next, viral load was tested. Viral load measured by TCID50 was significantly higher in Spred-2 siRNA-treated MLE-12 cells compared with the control siRNA treatment (Fig. 7B). To further investigate the role of Spred-2 in epithelial cells during influenza virus infection, we performed microarray analysis to compare gene expression between control and Spred-2 siRNA-transfected MLE-12 cells following H1N1 infection. The analysis revealed that transfection with Spred-2 siRNA resulted in up-regulation of 106 genes with more than a 2-fold induction, while 62 genes were down-regulated by more than half compared with control siRNA-transfected cells (Supplemental Table 1). Most of the up-regulated genes are known to be involved in the induction of inflammation and host immune responses to influenza virus. Importantly, phosphatidylinositol 3-kinase: PI3K, C2 domain containing, gamma polypeptide (Pik3c2γ) was increased in Spred-2 siRNA-transfected MLE-12. This gene encodes the catalytic subunit of PI3Kγ, a subtype of PI3K, and is involved in influenza virus entry (28). Upregulation of Pik3c2γ was verified by real-time PCR, which indicated that Pik3c2γ expression was enhanced following H1N1 infection, and knocking down Spred-2 expression in infected MLE-12 cells induced significantly higher expression of Pik3c2γ compared with control siRNA-transfected cells (Fig. 7C). Furthermore, we performed confocal immunofluorescent analysis to identify influenza virus entry between control and Spred2 siRNA-treated MLE-12 following influenza virus infection, demonstrating that knocking down the Spred-2 gene facilitated nuclear export of RNPs in MLE-12 cells at 2 hours post-infection, while transfection with control siRNA resulted in retention of viral RNPs in the nucleus at 6 hours post-infection (Fig. 7D). Interestingly, knocking down of the Spred-2 gene resulted in enhanced cytoplasmic nucleoprotein staining 24 hours after influenza virus infection (Fig. 7D).
Figure 7. Spred-2 gene knock down induced aggravated influenza virus replication.
(A) Gene expression of Spred-2 from MLE-12 cells following control or Spred-2 siRNA-transfection. ***p < 0.001 compared with control siRNA-transfected MLE-12. Data shown indicate mean ± SEM and are combined from 3 independent experiments. (B) Control siRNA- or Spred-2 siRNA-transfected MLE-12 cells were infected with H1N1 (multiplicity of infection [MOI] = 1 or 10) for 10 hours. The viral load of infectious particles produced from these infections were determined using TCID50. *p < 0.05, **p < 0.01 compared with control siRNA-transfected MLE-12. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. (C) Gene expression of Pik3c2γ following H1N1 stimulation for 24 hours in MLE-12 transfected with control siRNA or Spred-2 siRNA. *p < 0.05 compared with control siRNA-transfected MLE-12. Data shown indicate mean ± SEM and are combined from 3 independent experiments. (D) MLE-12 cells transfected with control siRNA or Spred-2 siRNA were infected with H1N1 (MOI = 10) for 2, 6 or 24 hours and then analyzed using anti-NP specific antibody in immunofluorescence assay and confocal laser microscopy. Red staining indicates influenza virus NP. Blue staining indicates nuclear staining, DAPI. Original magnification, ×400. Data are a representative experiment of 3 independent experiments.
Spred-2 regulates influenza virus infection through PI3K as well as the Raf/MEK/ERK signaling pathway
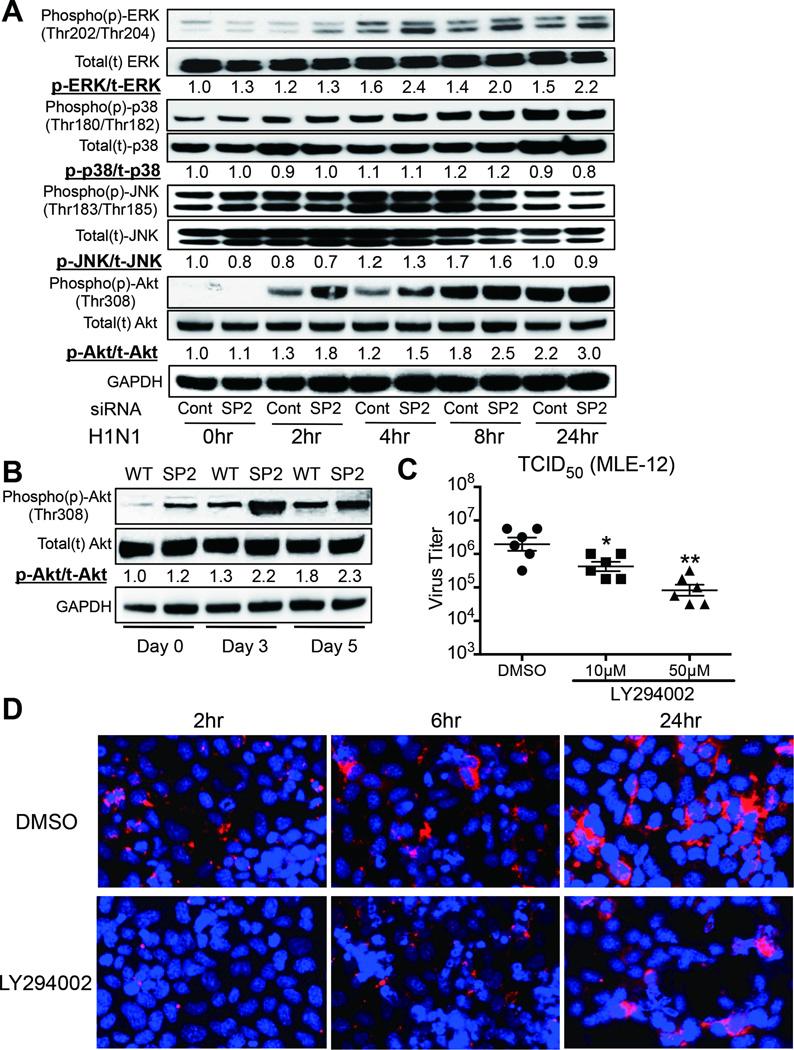
We further assessed the signal transduction activity of both Raf/MEK/ERK and PI3K signaling pathway. H1N1 infection induced an enhancement of ERK activation, but had little effect on JNK- and p38-activation in MLE-12 cells relative to the control (Fig. 8A). Furthermore, ERK-activation was further enhanced in Spred-2 siRNA-treated MLE-12 cells compared with control siRNA treatment. There was no significant difference in JNK- and p38-activation between control siRNA- and Spred-2 siRNA-treated MLE-12 cells. Infection with H1N1 resulted in Akt phosphorylation, a commonly used marker of PI3K activation, and knocking down of Spred-2 expression led to a further increase in p-Akt induction (1.3–1.4 fold) (Fig. 8A). We also confirmed this finding in vivo H1N1 infection of Spred-2 KO mice resulted in higher p-Akt induction compared with WT mice at day 3 (1.7 fold) and day 5 (1.3 fold) post infection (Fig. 8B). Moreover, we demonstrated that treatment of MLE-12 cells with the PI3K inhibitor, LY294002, significantly reduced the viral load compared with control DMSO treatment (Fig. 8C). Furthermore, the previously observed reduction in virus replication under the treatment of LY294002, along with the retarded nuclear export of viral RNPs in the late stages of viral replication and decreased cytoplasmic nucleoprotein staining were also apparent under confocal laser microscopy in LY294002 treated MLE-12 cells infected with H1N1 (Fig. 8D).
Figure 8. Spred-2 influences activation of both Raf/MEK/ERK and PI3K signaling.
(A) Control siRNA- or Spred-2 siRNA-transfected MLE-12 cells infected with H1N1 at indicated times were subjected to immunoblotted with phosphorylated (p)-ERK, ERK, p-p38, p38, p-JNK, JNK, p-Akt, Akt, or GAPDH Abs. GAPDH was used as a loading control. Experiment was repeated three times. A representative gel images and the value of phosphorylation compared with control siRNA-treated (0hr) are shown. (B) Western blotting analysis of total or phosphorylated Akt from H1N1-infected whole lungs in WT or Spred-2 KO mice at 0, 3, and 5 days post-infection. GAPDH was used as a loading control. Experiment was repeated three times. A representative gel images and the value of phosphorylation compared with lungs from WT mice (Day 0) are shown. (C) MLE-12 cells were infected with H1N1 (MOI = 10) treated with control DMSO or LY294002 (10 µM or 50 µM) for 10 hours. Viral load was measured by TCID50. *p < 0.05, **p < 0.01 compared with MLE-12 cells treated with DMSO. Data shown indicate mean ± SEM and are from a representative experiment of 2 independent experiments. (D) Confocal immunofluorescent examination of MLE-12 treated with control DMSO or LY294002 following stimulation with influenza virus for 2, 6, or 24 hours (MOI = 10). Red staining indicates influenza virus NP. Blue staining indicates DAPI. Original magnification, ×400. SP2KO = Spred-2 KO. Data are a representative experiment of 2 independent experiments.
DISCUSSION
The Raf/MEK/ERK cascade is the prototype MAP kinase signaling cascade and plays an important role in cell growth, differentiation, survival, and immune responses (17, 29). Influenza A viruses are ubiquitous pathogens, causing acute respiratory disease in humans and various animal species, induce signal activation through MAP kinase cascades in infected host cells (1, 2, 11). The Spred proteins bind to Ras and Raf, leading to suppression of Raf activation and subsequent inhibition of the Raf/MEK/ERK signaling pathway, thereby inhibiting cellular responses (18). In the present study, we have focused on Spred-2 in an influenza A virus (H1N1) infectious model and demonstrated for the first time that Spred-2 protein expression represents a host protective factor in influenza A virus infection.
The innate immune response is the host’s first defense against the invading influenza virus. Infection of cells with influenza A virus leads to biphasic activation of the Raf/MEK/ERK cascade (30). Once initiated, proinflammatory cytokines and chemokines are released, causing macrophages and neutrophils to migrate to the source of infection (31). We first demonstrated that Spred-2 expression level in the lungs was increased following H1N1 infection in both human and mouse samples. Especially in human samples, Spred-2 was detected in both epithelial and inflammatory cells. Using Spred-2 KO mice, we demonstrated that Spred-2 deficiency during influenza virus infection led to impaired survival, increased virus titer and exacerbated inflammatory status, accompanied by profound ERK activation. MAP kinase cascades also involve not only ERK- but also JNK- and p38-activation, but Spred-2 deficiency correlated solely with ERK activation during influenza virus infection. There was no difference in p38 and JNK activation between WT and Spred-2 KO mice. Moreover, Spred-2 KO mice treated with U0126, an inhibitor of the Raf/MEK/ERK signaling pathway (32, 33), demonstrated improved survival, reduced viral replication and limited lung inflammation with decreased inflammatory cytokine/chemokine levels. Our data agrees with previous studies that indicate a requirement for Raf/MEK/ERK activation in efficient influenza virus replication and cytokine production (15–17, 30). Our findings also indicate that regulation of Raf/MEK/ERK activation by Spred-2 is critical for a protective immune response against influenza virus.
Among the cytokines and chemokines induced during the innate immune response, activation of the type-I IFNs, IFN-α and -β, is the one of the most powerful defense mechanism against influenza viral replication and spread (30). Type-I IFNs also affect many functions resulting in further recruitment of inflammatory cells (34, 35). Additionally, chemokines CXCL1 and CCL2 play roles in the recruitment of neutrophils and macrophages, respectively (36), which represent the dominant leukocyte subtypes recruited to the lung during influenza virus infection (37, 38). This recruitment process is markedly augmented in Spred-2 KO mice, but can be limited by U0126 treatment. Exacerbated recruitment of inflammatory cells into the lungs results in excessive tissue damage, and may be related to higher mortality. It has been previously reported that blocking expression of CXCR2, the receptor for CXCL1, resulted in a reduction of neutrophil influx with prolonged host survival during influenza virus infection (39). Additionally, CCR2 deficiency, a major receptor for CCL2, leads to a milder inflammatory response with reduced lung pathology and increased survival rates as a result of defective macrophage recruitment (40). The above published reports agree with our findings, which show that higher CXCL1 and CCL2 levels in the lungs of Spred-2 KO mice may correlate with both enhanced neutrophil and macrophage migration into lungs and impaired survival rate.
Epithelial cells in the lung play a vital role in both lung immunity and influenza virus replication. Immune cells also have a key role in regulating and modulating the immune response during influenza virus infection, and immunohistochemical analysis of human H1N1-infected lungs strong staining of Spred-2 in both parenchymal cells and inflammatory cells. However, our findings from chimeric mice indicated that Spred-2 expression in epithelial cells, the first infectious site of influenza virus, is critically involved in the regulation of influenza virus replication and amelioration of lung inflammation. Conversely, cells of the hematopoietic compartment, including macrophages, failed to influence the outcome of infection in Spred-2 deficient recipient mice, despite macrophages being known to play a critical role against influenza virus infection (7). We performed additional experiments, which showed there was no significant difference in cytokine (IFN-α and IFN-β) and chemokine (CCL2 and CXCL1) production following H1N1 stimulation between WT and Spred-2 KO mouse using either BM-derived macrophages or alveolar macrophages (data not shown). These data suggest that impairment of influenza virus replication in epithelial cells might enhance pro-inflammatory cytokine and chemokine production, known as the cytokine storm. This uncontrolled cascade of cytokine production has been correlated with severe pneumonia and abrogated survival following influenza virus infection (12, 41). Moreover, the enhanced cytokine and chemokine production observed in epithelial cells may augment inflammatory responses induced by leukocytes, such as macrophages and neutrophils.
Activation of the Raf/MEK/ERK signaling pathway by influenza viruses is required for the efficient export of RNPs from the nucleus into the cytoplasm (15, 17, 42). We showed that Spred-2 knock down in lung epithelial cells enhanced ERK activation and nuclear export of viral RNPs in the late stage of the replication cycle compared with control conditions. Viral load was significantly elevated in the absence of Spred-2. Interestingly, Spred-2 deletion conditions in MLE-12 cells indicated enhanced influenza virus uptake in the early stages of the replication cycle. We further demonstrated that Spred-2 also regulates the PI3K signaling pathway, as the Raf-MEK-ERK and PI3K signaling pathways functionally cross-regulate each other (43, 44). Several studies also report that PI3K signaling regulates influenza virus entry (45–47). Our findings indicated that Spred-2 deletion enhanced the phosphorylation status of Akt, a commonly used marker of PI3K activation. Previous reports support our results showing that PI3Kγ is apparently indispensable for efficient influenza virus entry, and treatment with the PI3K inhibitor LY294002 led to reduced virus titer (28, 48). Thus, our studies show that Spred-2 regulates both ERK and PI3K signaling pathways, either directly or indirectly, and influences influenza virus replication in both early stage (endocytosis) via the PI3K signaling pathway and late stage (nuclear export of RNPs) by the Raf/MEK/ERK signaling pathway. However, ERK may receive multiple potential signals except Ras-Raf-MEK signaling, so the detailed molecular mechanism by which these signaling pathways regulates each stage remains to be elucidated.
In summary, we present a comprehensive in vivo analysis of Spred-2 protection against influenza virus infection. Spred-2 deficiency resulted in impaired survival and exacerbated inflammatory responses, accompanied by enhanced virus replication during influenza virus infection. Expression of Spred-2 in epithelial cells was indispensable for protection against influenza virus. Furthermore, regulation of influenza virus replication by Spred-2 in epithelial cells was influenced by not only the Raf/MEK/ERK but also the PI3K signaling pathways. This study supports the concept that an understanding of the Spred proteins, especially Spred-2, in the immune response to influenza virus may provide mechanistic approaches with clinical applicability.
CONCLUSIONS
Spred-2 KO mice challenged with influenza virus in vivo display higher mortality with greater virus load, excessive inflammation, and enhanced cytokine productions compared with WT mice. In addition, administration of MEK-inhibitor, U0126, improved mortality with lower viral load and decreased cytokine levels. Moreover, bone marrow chimeras indicated that H1N1-induced lung pathogenesis by Spred-2 was dependent on nonimmune cell populations. Similarly, viral clearance was regulated by Spred-2 expression through PI3K signaling pathway in murine lung epithelial cells MLE-12. Together, these results suggest that Spred-2 is a critical regulator in providing an anti-viral response during influenza infection.
Supplementary Material
Acknowledgments
We thank Mr. Hiroyuki Watanabe, Mr. Yasuharu Arashima, Ms. Yuki Nakashima (Department of Pathology and Experimental Medicine, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University), and Ms. Noriko Ouji-Sageshima (Department of Immunology, Nara Medical University) for their technical assistance.
Financial support: This work was supported in part by grants from Ministry of Education, Culture, Sports, Science and Technology, Japan, Young Scientists (B) (25860296), Scientific Research (B) (25293095), Ministry of Health, Labor and Welfare, Japan (20249053), The research grant of Astellas Foundation for research on metabolic disorders, Japan, The Mochida Memorial Foundation for Medical and Pharmaceutical Research, Japan, and The Uehara Memorial Foundation, Japan.
Dr. Morishima’s institution received funding from a Grant from Ministry of Health, Labour and Welfare of Japan and from Okayama University. Dr. Kunkel received support for article research from the National Institutes of Health (NIH).
Footnotes
Copyright form disclosures: Dr. Takahashi disclosed work for hire.
The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Palese P. Influenza: old and new threats. Nat Med. 2004;10(12 Suppl):S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 2.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302(5650):1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 3.Webster RG, Bean WJ, Gorman OT, et al. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao W, Sun W, Qu B, et al. Distinct regulation of host responses by ERK and JNK MAP kinases in swine macrophages infected with pandemic (H1N1) 2009 influenza virus. PLoS One. 2012;7(1):e30328. doi: 10.1371/journal.pone.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Padilla R, de la Rosa-Zamboni D, Ponce de Leon S, et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;361(7):680–689. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- 6.Lamb RA, Takeda M. Death by influenza virus protein. Nat Med. 2001;7(12):1286–1288. doi: 10.1038/nm1201-1286. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Allen RM, Carson WFt, et al. The critical role of Notch ligand Delta-like 1 in the pathogenesis of influenza A virus (H1N1) infection. PLoS Pathog. 2011;7(11):e1002341. doi: 10.1371/journal.ppat.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420(1):1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- 9.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13(9):679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 10.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig S, Planz O, Pleschka S, et al. Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol Med. 2003;9(2):46–52. doi: 10.1016/s1471-4914(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 12.Planz O. Development of cellular signaling pathway inhibitors as new antivirals against influenza. Antiviral Res. 2013;98(3):457–468. doi: 10.1016/j.antiviral.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Kujime K, Hashimoto S, Gon Y, et al. p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J Immunol. 2000;164(6):3222–3228. doi: 10.4049/jimmunol.164.6.3222. [DOI] [PubMed] [Google Scholar]
- 14.Wu W, Booth JL, Duggan ES, et al. Innate immune response to H3N2 and H1N1 influenza virus infection in a human lung organ culture model. Virology. 2010;396(2):178–188. doi: 10.1016/j.virol.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig S, Wolff T, Ehrhardt C, et al. MEK inhibition impairs influenza B virus propagation without emergence of resistant variants. FEBS Lett. 2004;561(1–3):37–43. doi: 10.1016/S0014-5793(04)00108-5. [DOI] [PubMed] [Google Scholar]
- 16.Pinto R, Herold S, Cakarova L, et al. Inhibition of influenza virus-induced NF-kappaB and Raf/MEK/ERK activation can reduce both virus titers and cytokine expression simultaneously in vitro and in vivo. Antiviral Res. 2011;92(1):45–56. doi: 10.1016/j.antiviral.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Pleschka S, Wolff T, Ehrhardt C, et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol. 2001;3(3):301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 18.Wakioka T, Sasaki A, Kato R, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412(6847):647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura A. Regulation of cytokine signaling by the SOCS and Spred family proteins. Keio J Med. 2009;58(2):73–83. doi: 10.2302/kjm.58.73. [DOI] [PubMed] [Google Scholar]
- 20.Engelhardt CM, Bundschu K, Messerschmitt M, et al. Expression and subcellular localization of Spred proteins in mouse and human tissues. Histochem Cell Biol. 2004;122(6):527–538. doi: 10.1007/s00418-004-0725-6. [DOI] [PubMed] [Google Scholar]
- 21.Kato R, Nonami A, Taketomi T, et al. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem Biophys Res Commun. 2003;302(4):767–772. doi: 10.1016/s0006-291x(03)00259-6. [DOI] [PubMed] [Google Scholar]
- 22.Bouvier NM, Lowen AC. Animal models for influenza virus pathogenesis and transmission. Viruses. 2010;2(8):1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harms PW, Schmidt LA, Smith LB, et al. Autopsy findings in eight patients with fatal H1N1 influenza. Am J Clin Pathol. 2010;134(1):27–35. doi: 10.1309/AJCP35KOZSAVNQZW. [DOI] [PubMed] [Google Scholar]
- 24.Wakabayashi H, Ito T, Fushimi S, et al. Spred-2 deficiency exacerbates acetaminophen-induced hepatotoxicity in mice. Clin Immunol. 2012;144(3):272–282. doi: 10.1016/j.clim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Yashiro M, Tsukahara H, Matsukawa A, et al. Redox-active protein thioredoxin-1 administration ameliorates influenza A virus (H1N1)-induced acute lung injury in mice. Crit Care Med. 2013;41(1):171–181. doi: 10.1097/CCM.0b013e3182676352. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Hamada K, Suzaki Y, et al. Subcautaneous vaccination of Mycobacterium bovis Bacillus Calmette-Guerin attenuates allergic inflammation in a murine model of asthma. Allergology International. 2005;54(4):601–609. [Google Scholar]
- 27.Nishimori H, Maeda Y, Teshima T, et al. Synthetic retinoid Am80 ameliorates chronic graft-versus-host disease by down-regulating Th1 and Th17. Blood. 2012;119(1):285–295. doi: 10.1182/blood-2011-01-332478. [DOI] [PubMed] [Google Scholar]
- 28.Fujioka Y, Tsuda M, Hattori T, et al. The Ras-PI3K signaling pathway is involved in clathrin-independent endocytosis and the internalization of influenza viruses. PLoS One. 2011;6(1):e16324. doi: 10.1371/journal.pone.0016324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9(2):180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 30.Droebner K, Pleschka S, Ludwig S, et al. Antiviral activity of the MEK-inhibitor U0126 against pandemic H1N1v and highly pathogenic avian influenza virus in vitro and in vivo. Antiviral Res. 2011;92(2):195–203. doi: 10.1016/j.antiviral.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Ehrhardt C, Seyer R, Hrincius ER, et al. Interplay between influenza A virus and the innate immune signaling. Microbes Infect. 2010;12(1):81–87. doi: 10.1016/j.micinf.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 32.DeSilva DR, Jones EA, Favata MF, et al. Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. J Immunol. 1998;160(9):4175–4181. [PubMed] [Google Scholar]
- 33.Favata MF, Horiuchi KY, Manos EJ, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273(29):18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Sastre A. Antiviral response in pandemic influenza viruses. Emerg Infect Dis. 2006;12(1):44–47. doi: 10.3201/eid1201.051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshmane SL, Kremlev S, Amini S, et al. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannock GA, Paul JA, Barry RD. Immunization against influenza by the ocular route. Vaccine. 1985;3(3 Suppl):277–280. doi: 10.1016/0264-410x(85)90122-7. [DOI] [PubMed] [Google Scholar]
- 38.Wareing MD, Lyon AB, Lu B, et al. Chemokine expression during the development and resolution of a pulmonary leukocyte response to influenza A virus infection in mice. J Leukoc Biol. 2004;76(4):886–895. doi: 10.1189/jlb.1203644. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Lu M, Lau LT, et al. Neutrophils may be a vehicle for viral replication and dissemination in human H5N1 avian influenza. Clin Infect Dis. 2008;47(12):1575–1578. doi: 10.1086/593196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson TC, Beck MA, Kuziel WA, et al. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am J Pathol. 2000;156(6):1951–1959. doi: 10.1016/S0002-9440(10)65068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marjuki H, Yen HL, Franks J, et al. Higher polymerase activity of a human influenza virus enhances activation of the hemagglutinin-induced Raf/MEK/ERK signal cascade. Virol J. 2007;4:134. doi: 10.1186/1743-422X-4-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marjuki H, Gornitzky A, Marathe BM, et al. Influenza A virus-induced early activation of ERK and PI3K mediates V-ATPase-dependent intracellular pH change required for fusion. Cell Microbiol. 2011;13(4):587–601. doi: 10.1111/j.1462-5822.2010.01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36(6):320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehrhardt C, Ludwig S. A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cell Microbiol. 2009;11(6):863–871. doi: 10.1111/j.1462-5822.2009.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehrhardt C, Marjuki H, Wolff T, et al. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 2006;8(8):1336–1348. doi: 10.1111/j.1462-5822.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- 47.Tsutsumi K, Fujioka Y, Tsuda M, et al. Visualization of Ras-PI3K interaction in the endosome using BiFC. Cell Signal. 2009;21(11):1672–1679. doi: 10.1016/j.cellsig.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Maira SM, Finan P, Garcia-Echeverria C. From the bench to the bed side: PI3K pathway inhibitors in clinical development. Curr Top Microbiol Immunol. 2010;347:209–239. doi: 10.1007/82_2010_60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.