Summary
Long non-coding RNAs (lncRNAs) are a diverse and poorly conserved category of transcripts that have expanded greatly in primates, particularly in the brain. We identified a lncRNA, which has acquired 16 microRNA response elements for miR-143-3p in the Catarrhini branch of primates. This lncRNA termed LncND (neuro-development) is expressed in neural progenitor cells and then declines in neurons. Binding and release of miR-143-3p, by LncND, controls the expression of Notch receptors. LncND expression is enriched in radial glia cells (RGCs) in the ventricular and subventricular zones of developing human brain. Down-regulation in neuroblastoma cells reduced cell proliferation and induced neuronal differentiation, an effect phenocopied by miR-143-3p over-expression. Gain-of-function of LncND in developing mouse cortex led to an expansion of PAX6+ RGCs. These findings support role for LncND in miRNA-mediated regulation of Notch signaling within the neural progenitor pool in primates that may have contributed to the expansion of cerebral cortex.
Introduction
Long non-coding RNAs (lncRNAs) have complex and diverse functions in brain development. lncRNAs have relatively low levels of evolutionary conservation with sequence deletions, insertions (Mclean et al., 2003) and accelerated nucleotide substitution (Pollard et al., 2006) at evolutionary divergences. About a third of the lncRNAs are unique to the primate lineage (Derrien et al., 2012), and only ~12% of human lncRNAs appear to be conserved in other vertebrate species (Ulitsky and Bartel, 2013; Cabili et al., 2011). The restricted spatial and temporal expression patterns of many lncRNAs within the brain likely contributes to neuronal diversification in large brain primates (Amaral and Mattick, 2008; Cao et al., 2006; Chodroff et al., 2010; Qureshi et al., 2010) and the specification of individual neuronal subtypes (Mercer et al., 2008).
MicroRNAs (miRNAs) are derived from hairpin precursors that function in association with Argonaute proteins to regulate target genes post-transcriptionally. These ~ 21–23 nucleotide sequences change quite dramatically as cells transition from germ cells to neural stem cells and at all stages of cell differentiation during brain development (reviewed in Fineberg et al., 2009). The many regulatory controls over miRNA levels and cell type specific expression, include the well-known panoply of gene expression mechanisms, e.g. promoters, enhancers and epigenetic modifications; as well as degradation and biogenesis pathways (Ha and Kim, 2014).
An additional source of control over the levels of mature miRNAs is their sequestration and release from binding sites, known as miRNA response elements (MREs) in transcribed pseudogenes, long non-coding RNAs (lncRNAs) and circular RNAs (Cesana et al., 2011; Ebert and Sharp, 2010; Hansen et al., 2013; Kallen et al., 2013; Memczak et al., 2013; Tay et al., 2011, 2014; Wang et al., 2013; Zhang et al., 2013). These natural miRNA-binding platforms known as sponges contain MREs that can relieve mRNA targets from repression or indirectly induce target mRNA repression by release of miRNAs from this reservoir. Natural miRNA sponges impart stability to miRNAs (Bail et al., 2010) by sequence-specifically sequestering miRNAs directed toward specific mRNA targets within Argonaute protein complexes. Short stretches of complementarity to miRNA seeds in regions of relatively unstructured RNA found in lncRNAs could evolve easily (Ebert and Sharp, 2010).
lncRNAs have the potential to sponge miRNAs, and thereby, regulate the expression of mRNAs (Hu et al., 2012; Wang et al., 2013; Wang et al., 2010; Kallen et al., 2013; Tay et al., 2014; Cesana et al., 2011). The first of these was discovered in plants (Franco-Zorrilla et al., 2007) and others have been described during muscle development (Legnini et al., 2014; Cesana et al., 2011; Kallen et al., 2013), and in embryonic stem cells to regulate core pluripotency transcription factors (Wang et al., 2013). The H19 lncRNA has been detected in a complex with miR-17-5p by photo-crosslinking and Argonaute 2 immunopurification (Imig et al., 2014).
We identified and functionally characterized a lncRNA, LncND (lncRNA for Neuronal Development) that is deleted in a region of the genome associated with a human neurodevelopmental disorder. A microdeletion at 2p25.3 includes LncND and 6–7 protein-coding genes (Stevens et al., 2011; Rio et al., 2013; Doco-Fenzy et al., 2014; Bonaglia et al., 2014). Six individuals harboring 2p25.3 terminal or interstitial deletions of different sizes, all including LncND, had intellectual disability (Stevens et al., 2011). A set of monozygotic twins with a karyotype involving this genomic locus had a discordant phenotype: one with a heterozygous deletion of the 2p25.3 region exhibited developmental delay, absence of speech, hyperactivity and the other harbored somatic mosaicism had autism spectrum disorder (Rio et al., 2013). Five patients with a deletion of the subtelomeric region of chromosome 2 (size 2.90–2.97 Mb) had intellectual disability with aggressive outbursts and hyperactivity (Doco-Fenzy et al., 2014). The deleted region included Ysc84-like-1 (SH3YL1), acid phosphatase 1 soluble (ACP1), trans-membrane protein 18 (TMEM18), syntrophin gamma 2 (SNTG2), thyroid peroxidase (TPO), Drosophila peroxidasin homologue (PXDN), myelin transcription factor 1-like (MYT1L) (Figure S1a), and uncharacterized long-non coding RNAs, including LncND. We demonstrated that LncND acts as a miRNA sponge for miR-143-3p and in so doing can regulate the Notch signaling pathway. The knock-down of LncND in neuroblastoma cells repressed NOTCH-1 and NOTCH-2 and differentiated the cells to neurons. RNA-seq analysis indicated a significant number of differentially expressed overlapping genes (including cell-cycle related and neurogenesis genes) after LncND knock-down or miR-143-3p over-expression. Single-cell analysis and in-situ hybridization confirmed the high expression of LncND in radial glia cells in human cortex suggesting its role in maintaining the neural progenitor pool during the expansion of the cerebral cortex by regulating the Notch signaling pathway. This conclusion was supported by a functional assay in which expansion of the radial glial population occurred following in vivo overexpression of LncND.
RESULTS
Characterization of lncRNA with Multiple miRNA Response Elements
Recently, a reference catalog of more than 8000 human lncRNAs was defined from about four billion RNA-seq reads across 24 tissues and cell types (Cabili et al., 2011). A stringent set of 4662 high quality human lncRNA loci (14,353 transcripts) was annotated (Human Body Map LincRNAs, Broad Institute). We focused on brain expressed lncRNAs and selected 358 lncRNA loci (1175 transcripts) that were expressed (fpkm >=1) in two biological replicates from the database. TargetScan (Lewis et al., 2005) was used to predict miRNA binding sites along these full-length lncRNAs. 53 brain expressed miRNAs were selected from profiling 40 normal human tissues (Liang et al., 2007). We postulated that the tendency of an RNA to act as a miRNA sponge will increase with an increasing number of miRNA Response Elements (MREs). Taking these factors into consideration, we identified four lncRNA loci (6 transcripts) with 10 or more MREs for a single miRNA (Figure 1A).
Figure 1. Identification, Characterization and Conservation of LncND transcript.
(A) Computational prediction (using TargetScan) of all the MREs of brain-expressed miRNAs on all the brain-expressed human lncRNAs (from Human Body Map LincRNAs). The graph represents the total lncRNAs having MREs ranging from 1–12. (B) Diagrammatic representation of LncND transcript of 2710 bp length as annotated on UCSC Genome browser showing the MREs located in the 5′ end. Region between 1–1971 bp of LncND transcript is zoomed-in to show the MREs for miR-143-3p, miR-4330, miR-1912 and miR-4286. (C) in-situ hybridization using probes specific for either sense (left) or antisense (right) strand. Brown dots represent the positive signal for LncND and blue staining shows the haematoxylin nuclear stain. Black and red arrows represent the cytoplasmic and nuclear expression of LncND, respectively. The experiment was repeated twice with the same outcome in neural progenitor cells (top) and SHSY5Y cells (bottom). Scale: 50 μm. (D) Multiz Alignment showing the conservation of LncND across some representative species. Red bar represents the 5′ end of human LncND enriched in MREs for several miRNAs. (E) Phylogenetic relationship of LncND among primates. Branch lengths are based on an approximation of the standard likelihood ratio test representing the number of substitutions per site calculated by PhyML. Numbers represented in red are branch support values. The tree was created using phylogeny.fr platform. The numbers of MREs for miR-143-3p are shown in green in parenthesis.
The transcript, TCONS_00003534 transcript (sense strand), has multiple MREs for several miRNAs spread over a relatively short sequence segment. However, with further computational analysis, we observed contradictory information on the directionality of this transcript. RNA-sequencing data from hiPSC-derived neurons (M.A. Lalli., I. Hernandez, and K.S.K., unpublished data) and RNAseq data on the UCSC Genome Browser showed most of the reads derived from the antisense strand, contrary to the annotation in the Human Body Map LincRNAs catalog. The antisense transcript, hereafter referred to as lncRNA for Neuronal Development or LncND, is located on chromosome 2: 663814–666523 (2p25.3 arm of chromosome 2) with seven protein-coding genes in its vicinity (Figure S1A). Several marks of active transcription and conserved regulatory elements are present upstream of the LncND locus (http://genome.ucsc.edu/) (Kent et al., 2002) (Figure S1B). There were multiple MREs for several miRNAs on the antisense strand, among them, miR-143-3p, miR-4286, miR-1912 and miR-4330 (Figure 1B).
To confirm the strand usage of this lncRNA, we performed in situ hybridization (ISH) on undifferentiated human embryonic stem cells (H9 cells) before and after differentiation into neural progenitor cells (NPCs) by dual SMAD inhibition (SMADi) (Chambers et al., 2009). Consistent with our RNAseq data from the hiPSC-derived neurons, only the antisense strand was expressed in NPCs (Figure 1C, top). No signal was detected in undifferentiated H9 cells (Figure S1C). We obtained the same result with the antisense probe in human neuroblastoma cell line, SHSY5Y (Figure 1C, bottom). The signal was localized in the nucleus and the cytoplasm (Figure 1C, black arrows-cytoplasmic and red arrows-nuclear). Therefore, a mis-annotation of the strand usage is present in the Human Body Map LincRNAs catalog, likely due to the use of a sequencing method which lacked strand specificity (Cabili et al., 2011).
Cabili et al., predicted four splice variants of LncND. One of these isoforms (TCONS_00003534) was identified with a small intron (59 bases from 758–816 bp) between two exons (1–757 bp and 817–2651 bp) with a full transcript of 2651 bp. Fragments of the full-length LncND transcript with overlapping regions were successfully amplified from human brain RNA (FirstChoice Human Brain Total RNA from Ambion) using either random hexamers (Figure S1D) or oligo-dT primers (Figure S1E). Therefore, the predicted intron sequence (between 758–816 bp) is part of an exon suggesting that the transcript might be expressed as a single large exon similar to MALAT1 and NEAT1 lncRNAs (Hutchinson et al., 2007; Ji et al., 2003).
LncND is poorly conserved with a primate specific insertion in the 5′ end of LncND RNA (Multiz Alignment, Figure 1D). Interestingly, most of the MREs are located in the primate-specific region (Figure 1D, 1B). The phylogeny tree, created by phylogeny.fr (Dereeper et al., 2008), shows that the LncND sequences among Catarrhini (Old World Monkeys including, Rhesus, Green Monkey, Baboon and Crab-eating Macaque, and Apes including, Chimpanzee, Gorilla, Orangutan and Gibbon and Humans) are closely related, and the MREs for miR-143-3p are less conserved in Platyrrhini (New World Monkeys including, Squirrel monkeys and Marmoset) compared to Catarrhini (Figure 1E).
Expression of LncND During Neuronal Differentiation
To determine the expression of LncND during development, we differentiated H9 hESCs either into neurons through intermediate stages using dual SMAD inhibitors (Chambers et al., 2009) or into mesendoderm using BMP4 and FGF2 (Yu et al., 2011). SMAD inhibition directed the stem cells specifically towards neuroectoderm in which mRNA expression of PAX6, SOX2, NES and HES1, were highly up-regulated at day five of differentiation (Figure S2A, B). Markers for endoderm (GATA6) and mesoderm (Brachyury or the T gene) were low (Ct values lower than 30) (Figure. S2A). With further differentiation to neurons, PAX6 was down-regulated (Figure S2A) and immuno-staining with Tau- and MAP2-specific antibodies became apparent (Figure S2C). LncND expression increased in neural progenitors at day 5 of differentiation and then dropped rapidly in neurons (Figure 2A). We also measured the expression of LncND using a second model of neuronal differentiation (passage through embryoid bodies and neurospheres) and again observed a gradual increase in the expression of LncND till the neurosphere stage, followed by a decrease in its expression in neurons (data not shown). When H9 cells were differentiated into mesendoderm, GATA6 and Brachyury mRNA were highly up-regulated at day 5 (Figure S2D). PAX6 was expressed at very low levels in these cells, consistent with differentiation towards mesendoderm (Figure S2D). LncND expression remained stable at day 2 and down-regulated at day 5 of mesendoderm differentiation (Figure 2B). The expression of LncND in SHSY5Y cells exceeded the levels of neural stem cells at day 5 of differentiation of H9 cells (Figure S2E). Thus, LncND is preferentially elevated in early neural progenitor cells, suggesting its selective role during neuronal differentiation.
Figure 2. Expression of LncND and binding of miR-143-3p to LncND transcript.
(A) H9 cells differentiated into neurons using the dual SMAD inhibition protocol. Expression of LncND at different time points during neuronal differentiation by RT-qPCR. (B) H9 cells differentiated into mesendoderm using BMP4 and FGF2. Expression of LncND during different time points of mesendoderm differentiation. (C) Table showing representative miRNAs with high number of MREs in LncND transcript as predicted by PITA algorithm. (D) Thermodynamic energy prediction for the association of LncND and miR-143-3p by RNAHybrid program. (E) RNA immunoprecipitation with either IgG or Ago2-specific antibody in SHSY5Y cells. Western blot showing Ago2 protein marked by red arrow. Asterisk (*) indicates the heavy and light chain of anti-Ago2 antibody. (F) RNA was extracted from the immunoprecipitates and analyzed by RT-qPCR. The values are represented as fold enrichment compared to the IgG control. GAPDH was used as a negative control and H19 as a positive control. (G) HEK293T cells were treated with miRNA mimics and the luciferase activity of LncND after 72 h was detected. Reduced luciferase activity of LncND was observed in the presence of the miR-143-3p mimic. (H) Increase in the luciferase activity of LncND after the addition of LNA inhibitor for miR-143-3p in SHSY5Y cells. Firefly luciferase activity was normalized to renilla luciferase activity. (I) The endogenous RNA expression of LncND after the overexpression of miR-143-3p in SHSY5Y cells by RT-qPCR. All the experiments (A-B and E-I) were performed in triplicates. Error bars represent standard deviation. p-values indicated were calculated by Student’s t-test (unpaired). ns: p>0.05, *: p≤0.05, **: p≤0.01, ***: p≤0.001, ****: p≤0.0001.
miR-143-3p Binds to LncND
Near-perfect complementarity at the “seed region” from position 2–7 at the 5′end of the miRNA determines target specificity (Friedman et al., 2009; Ellwanger et al., 2011; Betel et al., 2010). PITA algorithm (Kertesz et al., 2007) predicted 16 putative MREs for miR-143-3p at the 5′end of the LncND (Figure 1B). All these sites, except one with a 7mer-1A site, are perfect 6-mer sites together with complementarity at the 3′ end of miRNA (Lewis et al., 2005; Bartel, 2009). A large number of MREs for several other miRNAs were found including miR-4286, miR-1912 and miR-4330 (Figures 2C and 1B). A thermodynamic energy prediction between LncND and miR-143-3p sites indicated highly negative ΔG values for all of its sites (Figure 2D). The other three potential miRNAs also had very low ΔG values (Figure S3).
A miRNA sponge is expected to form a complex with Ago2, a component of the RNA-induced silencing complex (RISC) (Gregory et al., 2005; Meister et al., 2004). LncND was highly enriched (~ 6–7 fold) in Ago2 immunoprecipitation with an Ago2-specific antibody in SHSY5Y cells as compared to the IgG control (Figure 2E, 2F). As a positive control, we observed ~8–9 fold enrichment of H19 lncRNA in Ago2-IP (Figure 2F), which was previously identified as a miRNA sponge enriched in an Ago2-IP (Kallen et al., 2013). GAPDH, used as a negative control, was only ~ 2 fold enriched (Figure 2F). This finding suggests that LncND is associated with the RISC in a setting where it might compete for the binding of miRNAs “shared” with target mRNAs.
To verify a direct interaction between miR-143-3p and LncND, the 5′ end of LncND (1–1511 bp) was cloned in pMIR-Report vector and the luciferase reporter assay was performed in HEK293T cells. When luciferase fused to LncND was expressed in the presence of the miR-143-3p mimic, the luciferase signal was reduced. This finding suggested that miR-143-3p can bind to the 5′ end of LncND with functional consequences (Figure 2G). The LNA-inhibitor for miR-143-3p caused a substantial increase in the luciferase activity of LncND in SHSY5Y cells. (Due to negligible expression of miR-143-3p in HEK293T cells, we used SHYS5Y cells for LNA-inhibition) (Figure 2H). Interestingly, the miR-143-3p mimic did not destroy the endogenous LncND RNA in SHSY5Y cells, suggesting high stability of the LncND-miR-143-3p complex, which is an expected property of a miRNA sponge (Figure 2I). As observed by luciferase activity assay, miR-4330 could also bind to LncND (data not shown), but not miR-4286, nor miR-1912, suggesting that at least two miRNAs could be targeted by LncND. These results suggested that LncND could potentially serve as a miRNA sponge as it binds to miR-143-3p, but is not degraded by the miRNA.
miR-143-3p Targets Notch
High LncND expression in neural progenitors but not in neurons, suggested that LncND could regulate an early neuronal differentiation pathway. Among neurodevelopmental genes NOTCH-1 and NOTCH-2 3′UTR had one MRE for miR-143-3p (Figure 3A). The Notch signaling pathway is involved in differentiation, development, proliferation, cell fate decision, and survival in brain (Shimojo et al., 2008; Artavanis-Tsakonas, 1999; Gaiano and Fishell, 2002; Fox et al., 2008; Louvi and Artavanis-Tsakonas, 2006). miRNA binding sites of the top four miRNAs on the 3′UTR of NOTCH-1 and NOTCH-2 are represented in the table (Figure 2C). To confirm the predicted binding of miR-143-3p to the 3′UTR of NOTCH-1 and NOTCH-2, we transiently expressed miR-143-3p mimic in HEK293T cells or the LNA inhibitor of miR-143-3p in SHSY5Y cells together with the luciferase reporter plasmids for either NOTCH-1 or NOTCH-2 3′UTR. The reduction or increase in the luciferase activity in the presence of miRNA mimic or LNA inhibitor confirmed the potential binding sites for miR-143-3p in the 3′UTR of NOTCH-1 and NOTCH-2 in HEK293T cells (Figure 3B, 3C).
Figure 3. NOTCH-1 and NOTCH-2 are the targets of miR-143-3p.
(A) The binding site of miR-143-3p in NOTCH-1 and -2 3′UTR as predicted by PITA algorithm. (B) Validation of miRNA-binding to the 3′UTR of NOTCH-1 and NOTCH-2 by luciferase activity assay. Reduction in the luciferase activity demonstrated the binding of miR-143-3p to the 3′UTR of NOTCH-1 and NOTCH-2. (C) Increase in the luciferase activity of LncND after the addition of LNA inhibitor for miR-143-3p in SHSY5Y cells. (D) Effect of miRNA overexpression on the endogenous RNA expression of NOTCH-1, NOTCH-2 and LncND in SHSY5Y cells by RT-qPCR. (E) Down-regulation of HES1 and HEY1 mRNA after the overexpression of miR-143-3p in SHSY5Y cells after 48 h. (F) Immuno-blotting (left panel) and quantification (right panel) showing the reduction in the endogenous protein expression of NOTCH-1 and NOTCH-2 at 72 h following the transfection of the miR-143-3p mimic in SHSY5Y cells (G) Immuno-blotting (left panel) and quantification (right panel) showing the increase in NOTCH-1 and NOTCH-2 protein 72 h after LNA inhibition of miR-143-3p in SHSY5Y cells. qPCR results are normalized to HPRT. All experiments were performed at least three times. Error bars represent standard deviation. p-values indicated were calculated by Student’s t-test (unpaired). ns: p>0.05, *: p≤0.05, **: p≤0.01, ***: p≤0.001, ****: p≤0.0001.
Furthermore, the expression of endogenous NOTCH-1 and NOTCH-2 mRNA and protein were significantly down-regulated in the presence of the miR-143-3p mimic (Figure 3D, F). The downstream targets of Notch signaling pathway, HES1 and HEY1 were also down-regulated (Figure 3E), confirming the effect on this pathway by miR-143-3p. Moreover, inhibition of miR-143-3p in SHSY5Y cells, up-regulated the expression of endogenous NOTCH-1 and NOTCH-2 protein (Figure 3G). These results support NOTCH-1 and NOTCH-2 mRNA as miR-143-3p targets, for which LncND may serve as a reservoir. miR-4330 can also down-regulate NOTCH-1 and NOTCH-2 (data not shown); however we did not pursue this further because the expression of this miRNA was very low during the differentiation of stem cells.
Interestingly, like LncND, the miR-143-3p MRE region of NOTCH has undergone active evolutionary change, albeit over a longer time interval than LncND. NOTCH-2 lacks the miR-143-3p MRE in rodents and ferrets; however the MRE is conserved in NOTCH-1 (Figure S4).
LncND Sequesters miR-143-3p to Regulate Notch
Assuming that LncND can compete with the 3′UTR of NOTCH-1 or NOTCH-2 for the binding of miR-143-3p, we would expect that luciferase fused to the 3′ UTR of either NOTCH-1 or NOTCH-2 would show increased activity when LncND is over-expressed. The overexpression or knock-down of LncND, in SHSY5Y cells, in the presence of the 3′UTR of NOTCH-1 and NOTCH-2 fused to luciferase, resulted in up-regulation or down-regulation respectively of the luciferase activity (Figure 4A, 4B). These results suggest that LncND sequesters miR-143-3p to regulate NOTCH-1 and NOTCH-2 expression.
Figure 4. LncND act as a platform to sequester miR-143-3p.
(A) The luciferase activity of NOTCH-1 and NOTCH-2 is up-regulated with the over-expression of LncND in SHSY5Y cells. (B) The luciferase activity of NOTCH-1 and NOTCH-2 is down-regulated with the knock-down of LncND in SHSY5Y cells. Firefly luciferase activity was normalized to renilla luciferase activity in all the experiments. (C) Protein expression analysis of endogenous NOTCH-1 and NOTCH-2 protein with the knock-down of LncND in SHSY5Y cells normalized to beta-actin. Right panel shows quantification from three replicates. (D) Overexpression of LncND in SHSY5Y cells increases the expression of NOTCH-1 and NOTCH-2 endogenous protein levels. Right panel shows quantification from three independent experiments. Error bars represent standard deviation (n=3). p-values indicated were calculated by Student’s t-test (unpaired). ns: p>0.05, *: p≤0.05, **: p≤0.01, ***: p≤0.001, ****: p≤0.0001.
Upon the knock-down of LncND in SHSY5Y cells (~50% knock-down, Figure S5), the protein expression of NOTCH-1 and NOTCH-2 was down-regulated (Figure 4C) whereas no significant change in their mRNA levels was observed (Figure S5). As expected, over-expression of the MRE-containing fragment of LncND (1–1511 bp) in SHSY5Y cells increased the protein levels of endogenous NOTCH-1 and NOTCH-2 (Figure 4D). Taken together, these observations support the role of LncND as a regulator of Notch signaling.
LncND Co-regulatory Networks Direct Cells toward Neuronal Differentiation
LncND appears to sequester miR-143-3p to regulate NOTCH-1 and -2. LncND expression in H9 cells is correlated to NOTCH-1 and NOTCH-2 expression during neuronal differentiation (Figure 5A, left panel). Both increased during early stages of differentiation (day 5), but decreased at day 10 of differentiation and further decreased in neurons (Figure 5A). miR-143-3p had a distinct expression pattern. It rose with differentiation in neuroectoderm and remained elevated (Figure 5A, right panel). This pattern of LncND expression would allow the expression of NOTCH during early neuroectoderm stages despite the increase in miR-143-3p levels at this stage relative to the undifferentiated stem cells. On the other hand, release of miR-143-3p from LncND inhibits NOTCH in late neuroectoderm stages.
Figure 5. LncND knock-down leads to reduced cell proliferation and increased neuronal differentiation of SHSY5Y cells.
(A) H9 cells were differentiated into neurons using dual SMAD inhibitors. RNA from cells was isolated at different time points. Left: Expression of LncND, NOTCH-1 and NOTCH-2 was analyzed by RT-qPCR at different time points during differentiation of H9 cells. Right: miRNA expression during neuronal differentiation of H9 cells was measured using Taqman probes and the values were normalized to U6. (B) Representative differentially expressed genes involved in Notch signaling as identified using Cufflinks 2.1.1 after LncND knock-down (n=3, q-value <= 0.05). (C) Significant correlation and overlap between genes differentially expressed or unchanged after LncND knock-down and miR-143-3p overexpression (p<0.0001, Chi-square test). The numbers in the box represent the number of genes overlapping in each category. (D) Gene ontology (GO) term analysis of overlapping down-regulated genes between si-LncND and miR-143-3p overexpression (left), downregulated genes after LncND knock-down (middle) and miR-143-3p overexpression (right). (E) Immunostaining of SHSY5Y cells treated with control siRNA or siRNAs for LncND and NOTCH-1. Cells were stained for Tau (Green), MAP2 (Red) or Hoechst (Blue) for axons, dendrites and nucleus, respectively, and analyzed by fluorescent microscopy. Scale: 100 μm. (F) Bright-field image of SHSY5Y cells treated with either control siRNA, si-LncND, si-NOTCH-1, control miRNA mimic or miR-143-3p mimic. Knock-down of LncND and NOTCH-1 shows neurite-like processes after 72 h of transfection. Scale: 100 μm. (G) BrdU proliferation assay. SHSY5Y cells were treated with 100 nM of either control siRNA or siRNA for LncND or NOTCH-1 for 72 h. Cells stained with BrdU and 7-AAD were analyzed by flow cytometry. Knock-down of LncND and NOTCH-1 significantly reduced the proliferation of SHSY5Y cells. All the experiments were performed in triplicates. Error bars represent standard deviation. p-values indicated were calculated by Student’s t-test (unpaired). ***: p<0.001, **: p<0.01.
Because inhibition of Notch signaling in neuronal progenitor cells promotes neuronal differentiation (Wen et al., 2009 ; Louvi and Artavanis-Tsakonas, 2006), we investigated the role of LncND in neuronal differentiation by RNAseq. We took advantage of SHSY5Y cells, which can differentiate into neurons in the presence of all-trans retinoic acid (Constantinescu et al., 2007). Undifferentiated SHSY5Y cells were treated with siRNA against LncND or miR-143-3p mimic for three days and the cells were polyA+ RNA-sequenced. Scrambled sequences for siRNA or miRNA mimic were used as negative controls. RNA-seq analysis revealed that 3,680 genes were down-regulated, whereas 3,026 genes were up-regulated in LncND-knock-down cells (Table S2, GEO: GSE73982). On the other hand, 2,389 genes were down-regulated and 1,551 genes were up-regulated after the overexpression of miR-143-3p mimic (Table S2). Among the down-regulated genes after LncND knock-down were those involved in the Notch signaling pathway (Figure 5B). Consistent with the prediction that the LncND knock-down and the miR-143-3p mimic should alter the transcriptome in the same direction, we observed a significant overlap and correlation among the differentially expressed (DE) genes (q-value<=5%) between these two experiments (p value < 0.0001, Chi-square test) (Figure 5C). The 1028 overlapping down-regulated genes contribute significantly to cell cycle processes according to DAVID GO analysis (Figure 5D). Similarly the top downregulated genes (>=1.5 fold) in LncND knock-down cells and miR-143-3p over-expressed cells contribute to cell cycle processes (Figure 5D). Interestingly, among the common set of down-regulated genes (1028 genes), ~45% had a perfect 6-mer match with the miR-143-3p seed region. To place this effect in terms of total down-regulated genes under the two conditions, ~44.6% of genes down-regulated after miR-143-3p overexpression had an MRE for miR-143-3p and ~41.7% of genes down-regulated after LncND knock-down had an MRE for miR-143-3p. In the non-overlapping set only ~ 24% of down-regulated genes had an MRE for miR-143-3p after miR-143-3p overexpression, but not after LncND knock-down and only ~28.2% genes of down-regulated genes had an MRE for miR-143-3p after LncND knock-down but not after miR-143-3p overexpression. Thus, in addition to the high concordance of the overlapping genes when miR-143-3p is over-expressed and LncND is knocked-down, there was also a much lower discordance between the non-overlapping gene sets. The up-regulated genes in both of these experiments were enriched in the terms related to synapses and ion channels (Figure S6A), supporting the idea that miR-143-3p modulates the expression of multiple genes after knock-down of LncND.
LncND Promotes Differentiation of Progenitor Cells
LncND reduction directed cells toward neuronal identity with axon-like and dendrite-like processes (Figure 5E). To support the RNA-seq analysis and confirm the identity of these cells as neurons, we immuno-labeled them for Tau (MAPT) and MAP2 proteins, which are predominantly localized in the axons and dendrites of neurons, respectively (Kosik and Finch, 1987). The undifferentiated SHSY5Y cells express low levels of Tau and MAP2 protein (Figure 5E, top panel), whereas LncND-knock-down cells induced labeling of the neurites with Tau and MAP2 antibodies (Figure 5E, middle panel). The up-regulation of Tau protein is consistent with RNA-seq analysis in the LncND knock-down experiment in which significant up-regulation of MAPT transcript occurred (Table S2). NOTCH-1 knock-down cells (~ 55% knock-down, Figure S6B) phenocopied LncND knock-down (Figure 5E, bottom panel) in SHSY5Y cells, supporting the idea that the neuronal differentiation phenotype is due to a reduction in NOTCH-1 protein after LncND knock-down. Also NES and PAX6 (neural stem cell markers) mRNA expression was down-regulated in LncND- and NOTCH-1 knock-down cells (Figures S6C and S6D). The phenotype was confirmed using a shRNA targeting another region of LncND transcript (Figure S6E). To determine whether this phenotype could be explained by the release of miRNAs bound to LncND, miR-143-3p mimic was over-expressed in SHSY5Y cells. We observed that miR-143-3p over-expression phenocopied LncND-knock-down by inducing cells with axon-like and dendrite-like processes (Figure 5F).
Following LncND knock-down, we observed a reduction in cell density (Figure 5F), consistent with premature differentiation of neural progenitor cells and decreased expression of cell cycle genes among differentially expressed transcripts (Figure 5D). To show the knockdown affected proliferation, we labeled cells with 5-bromo-2′-deoxyuridine (BrdU), a thymidine analog which is incorporated into newly synthesized DNA. SHSY5Y cells, treated either with control siRNA or LncND siRNA, were pulsed with BrdU and then fixed and stained for BrdU- and 7-aminoactinomycin D (7-AAD). ~35-40% cells, treated with control siRNA, were double positive for BrdU and 7-AAD. After three 3 days of siRNA treatment, reduction in LncND expression led to a highly significant decrease (~25%) in cell proliferation, as evident from the decrease in the percentage of double-positive cells (Figure 5G). NOTCH-1 knock-down cells had a comparable reduction of ~22% in cell proliferation (Figure 5G).
Taken together, these findings suggest that low expression of LncND differentiates neural stem cells into neurons, due to the down-regulation of the Notch signaling pathway and several cell cycle-related genes and this effect is phenocopied by miR-143-3p overexpression.
Expression of LncND in Human Cerebral Organoids and Developing Human Cerebral Cortex
The Notch pathway is important for radial glia cell maintenance (Yoon et al., 2008; Gaiano et al., 2000; Shimojo et al., 2008). The Notch signaling effector gene HES1 is highly specific to radial glia in the developing human neocortex, emphasizing the importance of Notch signaling in the maintenance of neural stem cell identity (Hansen et al., 2010). We, therefore, investigated whether LncND might regulate Notch signaling in radial glia cells (RGCs) in human cortex.
The expression of LncND was determined by single-cell RNAseq from three-dimensional cerebral organoids generated from human induced pluripotent stem cells (iPSCs) (Lancaster and Knoblich, 2014) (GEO: GSE74207). The cerebral organoid cultures had distinct germinal zones with SOX2+ cells surrounding a neurotube-like structure (Figure S7A). The presence of radial glia cells was suggested by the immuno-localization of p-vimentin (Figure S7A). The apical localization of neural-specific N-cadherin was also observed (Figure S7B). We captured 68 single-cells on a C1 Single-Cell Auto Prep System (Fluidigm) and sequenced the RNA (Pollen et al., 2014). Out of 61 high quality cells, three expressed LncND (5% cells). The cells were hierarchically clustered according to the expression of the top 200 genes (see Methods). The smallest cluster containing all the LncND expressing cells (LncND cluster) was composed of 8 cells. 1534 genes were up-regulated and 454 genes down-regulated in this cluster as compared to all other cells (Figure S7C). Interestingly, the group of eight cells expressed significantly high levels of known RGC markers (PAX6, FABP7, and NES), Notch pathway genes (Feng et al., 1994; Hutton and Pevny, 2011; Kriegstein and Götz, 2003; Shibata et al., 1997; Götz et al., 1998; Lui et al., 2011) (Figure 6A, LncND expressing cells are indicated in purple bar) and neural progenitor genes including BUB1, MKI67, AURKA, CCNA2, CCNB1, ASPM and DLGAP5 (Figure S7C, Table S3). No genes were differentially expressed between the LncND expressing cells and the other cells within the LncND cluster, suggesting that all 8 cells shared a transcriptional identity. This observation suggests that LncND is present in a relatively uniform population of cells representing RGCs in the human cerebral organoids.
Figure 6. Expression of LncND in neural progenitor cells including radial glia cells in VZ and OSVZ in human neocortex.
(A) Single-cell RNA-seq analysis from human cerebral organoids–Heatmap (Pearson’s correlation), representing the clustering of cells based on the known radial glia cell markers. LncND-expressing cells are represented in purple on top of the heatmap. Genes marked in red box represents example of DE genes between LncND-cluster (marked in black box) and the rest of the cells. (B) Left and middle panel: GW17 primary human neocortex section demonstrating the PAX6+ radial glia cells and nuclear staining with DAPI. Right panel: in-situ hybridization with a probe specific for LncND showing its localization in VZ and OSVZ progenitor cells. Experiment was repeated at least two times. (C) Co-localization of LncND and PAX6+ cells in the OSVZ representing outer radial glia cells (oRG). Red and green boxes of the merged figure are zoomed-in to show the co-localization of LncND and PAX6. Cells were immunostained with PAX6-specific antibody (red) after performing in-situ hybridization with LncND-specific probe (green dots). Experiment was repeated at least two times. Scale: 50μm.
We further investigated the expression domain of LncND during cortical development in vivo in humans by performing ISH in GW (gestational week) 17 cortical slices, corresponding to the peak upper cortical layer neurogenesis period (Workman et al., 2013). ISH with the probe specific for LncND indicated high expression in the ventricular zone (VZ) and inner subventricular zone (ISVZ) and a lower density of label in the outer subventricular zone (OSVZ) (Figure 6B). Only faint labeling was detected in the cortical plate (CP) (Figure 6B). In the OSVZ, LncND positive cells co-localized with PAX6 immuno-labeled cells suggesting that a population of cells expressing LncND are outer RGCs (Hansen et al., 2010; Götz et al., 1998; Fietz et al., 2010) (Figure 6C). This observation is consistent with the single-cell sequencing analysis in which the LncND-expressing cells belong to a cluster of PAX6+ cells (Figure 6A). Taken together, these results suggest that LncND is predominantly expressed in radial glia cells in VZ and OSVZ.
Gain of LncND Function Leads to an Expansion of the Radial Glia Population in the Developing Mouse Cortex
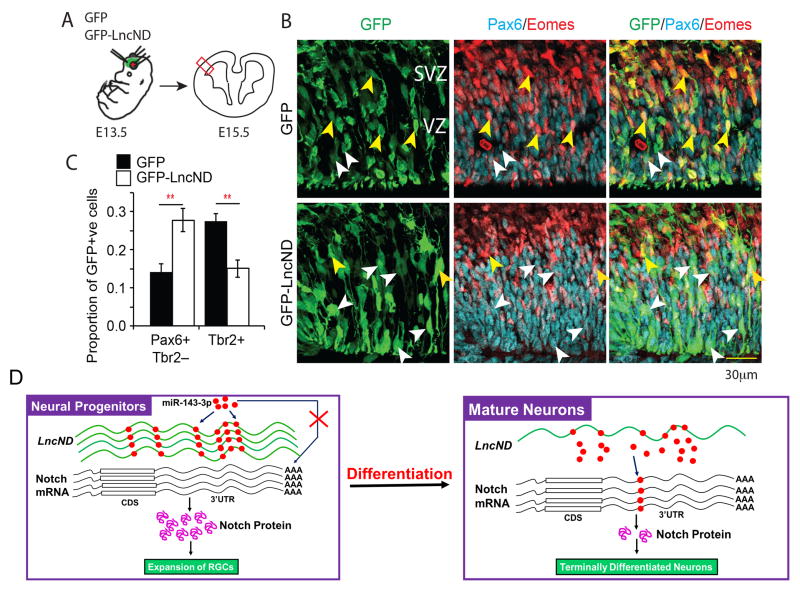
Although LncND is not expressed in mouse, both miR-143-3p and the candidate target mRNAs coding for NOTCH-1 and NOTCH-2 are expressed. Based on the proposed mechanisms of LncND action elucidated through studies in cell lines, we hypothesized that overexpression of LncND would lead to an increase in Notch signaling, which supports the maintenance of radial glia (Gaiano et al., 2000; Shimojo et al., 2008; Yoon et al., 2008). To address this hypothesis, we overexpressed LncND in mouse radial glia by in utero electroporation at E13.5 (Figure 7A) and found that two days later (E15.5), a higher proportion of cells electroporated with the LncND construct expressed the radial glia marker pattern (Pax6+/Tbr2−) compared to control electroporated cells (Figure 7B–C). Furthermore, we detected a corresponding decrease in the proportion of TBR2+ intermediate progenitor cells, whose identity is antagonized by active Notch signaling (Yoon et al., 2008; Shimojo et al., 2008). Together, these findings suggests the role of LncND in the expansion of the pool of cortical radial glia cells in vivo.
Figure 7. Gain of LncND function leads to an expansion of radial glia population in the developing mouse cortex.
(A) Schematic representing experimental design. LncND was overexpressed in developing mouse E13.5 radial glia by in utero electroporation. Brains were harvested at E15.5 and quantification was performed in the lateral cortex (red box, see Methods). (B) Representative images of tissue sections through E15.5 mouse lateral cortex tissue sections immunostained for GFP, radial glia marker Pax6, and intermediate neuronal progenitor cell marker Tbr2. White arrowheads indicate examples of electroporated cells expressing Pax6 but not Tbr2, and yellow arrowheads indicate examples of electroporated cells expressing Tbr2. VZ-ventricular zone, SVZ-subventricular zone. (C) Bar chart represents quantification of the proportion of electroporated cells with the molecular signature of radial glia (Pax6+/Tbr2−) and intermediate progenitors (Tbr2+) in control (filled bars) and LncND-overexpressing conditions (open bars). ** -p<0.01, two tailed Student’s t-test for n=4 biological replicates. (D) Diagrammatic representation of the molecular mechanism of LncND during neuronal development. In neural progenitors, LncND is highly expressed and sequesters miR-143-3p, thus releasing repression on the NOTCH mRNA leading to increased production of NOTCH proteins required for the maintenance of neural progenitors. During differentiation of neural progenitors to neurons, LncND expression decreases, thus releasing miR-143-3p to repress NOTCH mRNA, supporting differentiation. RGCs: Radial glia cells, CDS: Coding DNA sequence.
Discussion
The results presented here describe a novel neurodevelopmental regulatory system mediated by a lncRNA that provides a spatially and temporally specific control element to co-regulate the developmental genes, Notch and miR-143-3p. The features of LncND, which support its function as a platform capable of Ago-mediated miR-143-3p binding to regulate NOTCH-1 and -2 are: (a) Functional sites are likely to be located in regions with little secondary structure. (b) Correlated expression patterns of the lncRNA and its miRNA(s). (c) Cytoplasmic localization of the lncRNA and its miRNA(s). (d) Competition with mRNA targets. (e) Presence in a RNA-induced silencing complex (RISC). (f) Derepression of target genes occur at physiological expression levels. (g) The derepressive effect is attributable to the miRNA binding sites. These features have been attributed to natural sponges (Ebert and Sharp, 2010). Despite the large number of lncRNAs, only a few may have a miRNA sponge-like function according to the above criteria. We found only 4–5 human brain-expressed lncRNAs with more than 10 MREs for at least one brain-expressed miRNA.
We assume the co-localization of LncND with miR-143-3p at the single cell level given that most of the NPCs derived from H9 and RGCs in VZ/OSVZ expressed LncND (Figure 1C and 6B). However, a definitive demonstration of this model will require visualization of LncND and miR-143-3p in the same cell. The 16 MREs for miR-143-3p likely confers efficient binding that does not result in degradation of LncND within a LncND/Ago2 complex. We found novel targets for miR-143-3p: NOTCH-1 and -2. The expression patterns of these RNAs are consistent with LncND serving as a miR-143-3p reservoir early in neurogenesis when Notch expression is critical. Later in neural differentiation when NOTCH expression declines so does LncND, which releases miR-143-3p and, thereby, decreases Notch signaling specifically in LncND-expressing cells. Control mechanisms of this type can smoothen changes in downstream Notch signaling which would otherwise have multiplicative effects over noise in NOTCH levels. The knockdown of LncND promoted differentiation and reduced proliferation, consistent with the known function of NOTCH in neuronal differentiation. However, because LncND has the potential to bind other miRNAs we cannot definitively attribute the LncND knock-down phenotype entirely to miR-143-3p despite the ability of the miR-143-3p mimic to phenocopy LncND knock-down. Nevertheless, the effects we observed of miR-143-3p are also consistent with the known effects of this miRNA in other tissues that include inhibition of proliferation (Kent et al., 2010; Chen et al., 2009; Cordes et al., 2009). These findings suggest a model in which LncND regulates the expression NOTCH during neuronal differentiation by sequestering miR-143-3p in neural precursors (Figure 7D).
Non-coding RNAs have been considered drivers of recent brain expansion (Geschwind & Rakic, 2013; Barry & Mattick, 2012) with ~20% of human lncRNAs restricted in their expression to recent branches along the human lineage and not detectable even in Rhesus (Washietl et al., 2014). Brain expansion during evolution correlates with increased cell diversity. For example, the outer subventricular zone (OSVZ), a germinal zone that expanded greatly in primates (Dehay et al., 2015), has distinctively high expression of miR-143-3p as well as miR-1912 and miR-4330 (Arcila et al., 2014), which also have MREs on LncND. This regulatory circuit appears to have emerged within the primate lineage when MREs for miR-143-3p, an ancient miRNA, evolved as an insertion within the more ancient lncRNA present at this locus. In the absence of the insertion, the lncRNA may have served as a cis control element over the neighboring gene, TMEM18, which mediates the migration of neural stem cells towards glioblastoma cells (Jurvansuu et al., 2008). TMEM18 expression correlated positively with that of LncND upon the knock-down of LncND in SHSY5Y cells (data not shown). Of further relevance to tumor biology is the observation that LncND expression was high in SHSY5Y cells compared to glioblastoma (U251) cells or H9 hESCs (data not shown). This observation is consistent with the origin of neuroblastoma cells from early neural progenitor cells, whereas glioblastoma cells are derived from more downstream lineages in the adult brain when LncND would be expected to decline.
Progenitor cells in the OSVZ, which is thought to have contributed to cortical size and complexity in humans (Hansen et al., 2010) express HES1, a downstream target of the Notch pathway. Inhibition of the Notch signaling pathway has been shown to induce neuronal differentiation of OSVZ radial glia, suggesting that the Notch pathway is necessary for maintaining stemness of human oRG cells (Hansen et al., 2010; Shimojo et al., 2011; Fietz et al., 2010; Lui et al., 2011). In support of this hypothesis that LncND contributes to the maintenance of Notch signaling in human radial glia in VZ and OSVZ, single-cell RNA-Seq of cells captured from an in vitro cerebral organoid model of human cortical development and in-situ hybridization of primary tissue sample revealed LncND expression in radial glia cells. Molecular signatures for human oRG cells during cortical development (Pollen et al., 2015) and a database, CORTECON, that describes changes in RNA expression during cortical development from human embryonic stem cells (van de Leemput et al., 2014) reveal expression of LncND in radial glia cells and neural progenitor cells. Together, our findings suggest a molecular mechanism by which the emergence of LncND in primates and its co-evolution with a highly conserved miRNA-143-3p contributed to the expansion of radial glia in higher primates (Smart et al., 2002; Dehay et al., 2015).
EXPERIMENTAL PROCEDURES
Bioinformatic Analysis for prediction of MREs
miRBase 20 was used to extract the sequences of miRNAs and TargetScan 6.1 version was used for MREs for brain-expressed miRNAs for Figure 1A. For a less stringent analysis (using all miRNAs), PITA algorithm was used to identify perfect complementary pairing between the seed region 2–7 of mature miRNA sequences and the full-length LncND, NOTCH-1 and NOTCH-2. Thermodynamic energy predictions were made using RNAHybrid (Rehmsmeier et al., 2004).
RNA in-situ Hybridization
RNAScope 2.0 Brown Assay (Advanced Cell Diagnostics) localized the expression of sense and antisense strand of LncND with probes to the complementary regions (~1000 bp). Cells were grown in chamber slides and processed according to the manufacturer’s instructions. Hs-PPIB and DapB were used as positive and negative controls, respectively. Images were examined using upright Olympus BX51 microscope.
See the Supplemental Experimental Procedures for further details.
Supplementary Material
Table S2: List of differentially expressed (DE) genes after the knock-down of LncND or overexpression of miR-143-3p mimic.
Table S3: Differentially expressed genes between the cluster of LncND-expressing cells and other cells obtained from single-cell RNA sequencing of cerebral organoids.
Acknowledgments
Mary Raven of the UCSB Microscopy Facility provided assistance. We thank UCSB Laboratory for Stem Cell Biology and Engineering, the Biological Nanostructures Laboratory within the UCSB and UCOP-supported California NanoSystems Institute and Elmer Guzman for help with RNA-sequencing. We thank Jiwon Jang for critically reading the manuscript and providing suggestions, David Bartel for insightful comments, John FredyCastro-Alvarez for technical assistance, Benjamin Purow for providing plasmid constructs for NOTCH and Israel Hernandez for sharing his unpublished data. This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (KSK) and R01 NS075998/NS/NINDS NIH (ARK).
Footnotes
Author Contributions
K.S.K. and N.R. conceived and supervised the project. N.R. designed and performed the experiments. Bioinformatic analysis was performed by H.Z., V.L. and N.R. Cerebral organoids generation and single-cell sequencing was performed by S.E.G. in-utero electroporation experiment was performed by T.J.N. in the lab of A.R.K. Manuscript was prepared by N.R. and K.S.K. with critical suggestions from T.J.N.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Arcila ML, Betizeau M, Cambronne Xa, Guzman E, Doerflinger N, Bouhallier F, Zhou H, Wu B, Rani N, Bassett DS, et al. Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns. Neuron. 2014;81:1255–1262. doi: 10.1016/j.neuron.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science (80- ) 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff La, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Mattick JS. The role of regulatory RNA in cognitive evolution. Trends Cogn Sci. 2012;16:497–503. doi: 10.1016/j.tics.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNA Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Zanini S. A new patient with a terminal de novo 2p25.3 deletion of 1.9 Mb associated with early-onset of obesity, intellectual disabilities and hyperkinetic disorder. Mol Cytogenet. 2014;7:53. doi: 10.1186/1755-8166-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding Rnas in the Mammalian Central Nervous System. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano Ca, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- Chodroff Ra, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnár Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu R, Constantinescu aT, Reichmann H, Janetzky B. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y. J Neural Transm Suppl. 2007:17–28. doi: 10.1007/978-3-211-73574-9_3. [DOI] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H, Kosik KS. The Outer Subventricular Zone and Primate-Specific Cortical Complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- Dereeper a, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer a, Djebali S, Tilgner H, Guernec G, Merkel a, Gonzalez D, Lagarde J, et al. The GENCODE v7 catalogue of human long non-coding RNAs : Analysis of their structure, evolution and expression. 2012:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doco-Fenzy M, Leroy C, Schneider A, Petit F, Delrue MA, Andrieux J, Perrin-Sabourin L, Landais E, Aboura A, Puechberty J, et al. Early-onset obesity and paternal 2pter deletion encompassing the ACP1, TMEM18, and MYT1L genes. Eur J Hum Genet. 2014;22:471–479. doi: 10.1038/ejhg.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp Pa. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwanger DC, Büttner Fa, Mewes HW, Stümpflen V. The sufficient minimal set of miRNA seed types. Bioinformatics. 2011;27:1346–1350. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): A novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Fietz Sa, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs Potentiate Neural Development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Fox V, Gokhale PJ, Walsh JR, Matin M, Jones M, Andrews PW. Cell-cell signaling through NOTCH regulates human embryonic stem cell proliferation. Stem Cells. 2008;26:715–723. doi: 10.1634/stemcells.2007-0368. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352:40–47. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Imig J, Brunschweiger A, Brümmer A, Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M, Hall J. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19–miR-106a interaction. Nat Chem Biol. 2014;11:107–114. doi: 10.1038/nchembio.1713. [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jurvansuu J, Zhao Y, Leung DSY, Boulaire J, Yuan HY, Ahmed S, Wang S. Transmembrane protein 18 enhances the tropism of neural stem cells for glioma cells. Cancer Res. 2008;68:4614–4622. doi: 10.1158/0008-5472.CAN-07-5291. [DOI] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent Oa, Chivukula RR, Mullendore M, Wentzel Ea, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143 / 145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway service Repression of the miR-143 / 145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The Human Genome Browser at UCSC The Human Genome Browser at UCSC. Genome Res. 2002:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Finch Ea. MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: an immunocytochemical study of cultured rat cerebrum. J Neurosci. 1987;7:3142–3153. doi: 10.1523/JNEUROSCI.07-10-03142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein AR, Götz M. Radial glia diversity: A matter of cell fate. Glia. 2003;43:37–43. doi: 10.1002/glia.10250. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Leemput J, Boles NC, Kiehl TR, Corneo B, Lederman P, Menon V, Lee C, Martinez Ra, Levi BP, Thompson CL, et al. CORTECON: A temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83:51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A Feedforward Regulatory Loop between HuR and the Long Noncoding RNA linc-MD1 Controls Early Phases of Myogenesis. Mol Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclean CY, Reno PL, Pollen Aa, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2003;1:1–42. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mercer T, Mercer T, Dinger M, Dinger M, Sunkin S, Sunkin S, Mehler M, Mehler M, Mattick J, Mattick J. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen Aa, Nowakowski TJ, Shuga J, Wang X, Leyrat Aa, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014 doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi Ia, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R, Ho M. Fast and effective prediction of microRNA / target duplexes. 2004 Spring;:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio M, Royer G, Gobin S, de Blois M, Ozilou C, Bernheim a, Nizon M, Munnich a, Bonnefont JP, Romana S, et al. Monozygotic twins discordant for submicroscopic chromosomal anomalies in 2p25.3 region detected by array CGH. Clin Genet. 2013;84:31–36. doi: 10.1111/cge.12036. [DOI] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, Ikenaka K, Wada K, Tanaka K, Inoue Y. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci. 1997;17:9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Dynamic expression of notch signaling genes in neural stem/progenitor cells. Front Neurosci. 2011;5:78. doi: 10.3389/fnins.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IHM, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SJC, van Ravenswaaij-Arts CMa, Janssen JWH, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T, van Rheenen J, Heuts-Vijgen R, Stegmann APa, Smeets EEJGL, et al. MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet A. 2011;155A:2739–2745. doi: 10.1002/ajmg.a.34274. [DOI] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Li H, Liu J. Dynamic signaling for neural stem cell fate determination ND NO ST ND OS NO ST. 2009a:1–11. doi: 10.4161/cam.3.1.7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Li H, Liu J. Dynamic signaling for neural stem cell fate determination. Cell Adh Migr. 2009b;3:107–117. doi: 10.4161/cam.3.1.7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. Modeling Transformations of Neurodevelopmental Sequences across Mammalian Species. J Neurosci. 2013;33:7368–7383. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon M chul, Moon JS, Miyata T, Kong YY. Mind Bomb 1-Expressing Intermediate Progenitors Generate Notch Signaling to Maintain Radial Glial Cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Yu P, Pan G, Yu J, Thomson Ja. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2: List of differentially expressed (DE) genes after the knock-down of LncND or overexpression of miR-143-3p mimic.
Table S3: Differentially expressed genes between the cluster of LncND-expressing cells and other cells obtained from single-cell RNA sequencing of cerebral organoids.