Abstract
The small Maf proteins (sMafs) are basic region leucine zipper (bZIP)-type transcription factors. The basic region of the Maf family is unique among the bZIP factors, and it contributes to the distinct DNA-binding mode of this class of proteins. MafF, MafG and MafK are the three vertebrate sMafs, and no functional differences have been observed among them in terms of their bZIP structures. sMafs form homodimers by themselves, and they form heterodimers with cap ‘n’ collar (CNC) proteins (p45 NF-E2, Nrf1, Nrf2, and Nrf3) and also with Bach proteins (Bach1 and Bach2). Because CNC and Bach proteins cannot bind to DNA as monomers, sMafs are indispensable partners that are required by CNC and Bach proteins to exert their functions. sMafs lack the transcriptional activation domain; hence, their homodimers act as transcriptional repressors. In contrast, sMafs participate in transcriptional activation or repression depending on their heterodimeric partner molecules and context. Mouse genetic analyses have revealed that various biological pathways are under the regulation of CNC-sMaf heterodimers. In this review, we summarize the history and current progress of sMaf studies in relation to their partners.
1. Introduction
It has been over two decades since the small Maf (musculoaponeurotic fibrosarcoma) proteins (sMafs) were isolated (Fujiwara et al., 1993). sMafs are small transcription factors with sizes of approximately 160 amino acids or approximately 18 kDa. sMafs have been diversified into three functionally redundant factors that can form homodimers or heterodimers with various partners. Many lines of inquiry have revealed the importance of sMafs in various biological pathways. Interestingly, the functions of sMaf proteins vary depending on their partners and context. To comprehensively understand sMaf-mediated gene regulation, we and other groups have conducted various molecular and cellular analyses. Here, we summarize the current information related to sMaf functions.
2. Identification of sMaf genes
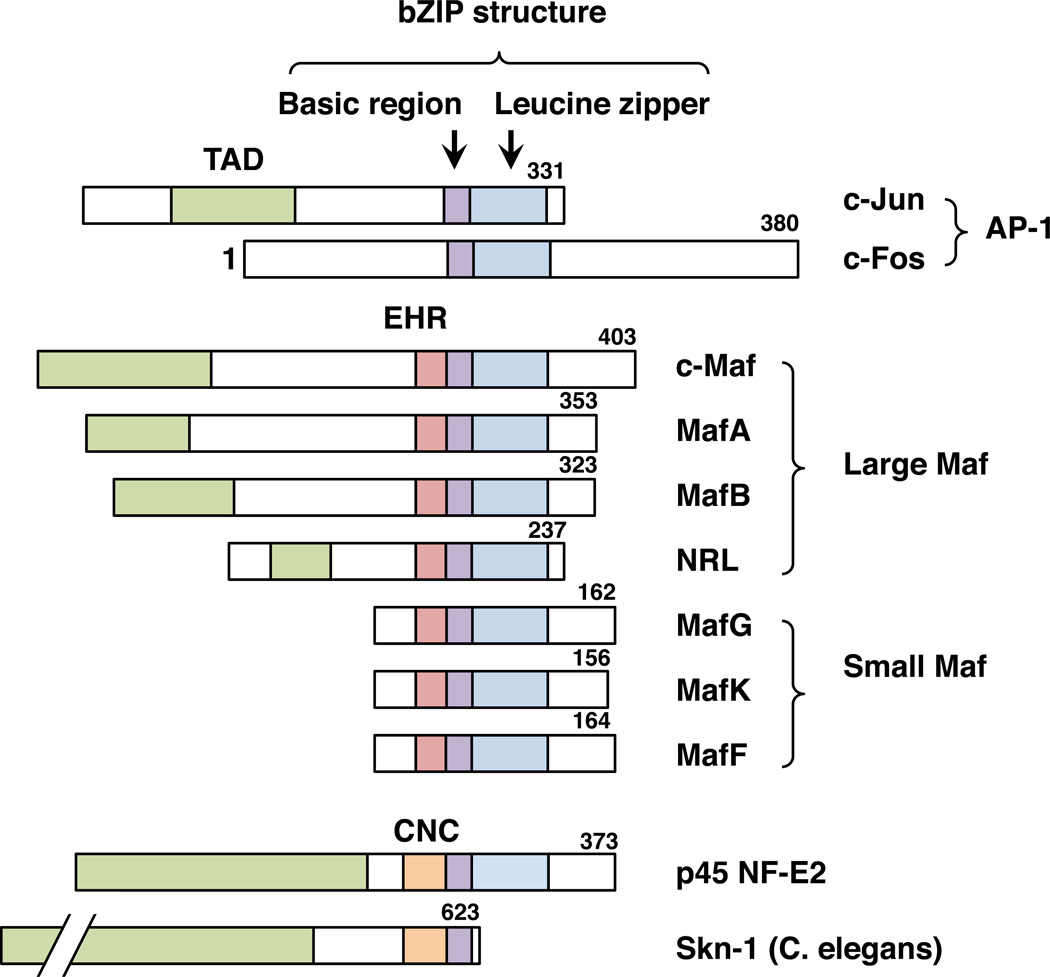
sMafs are basic region leucine zipper (bZIP)-type transcription factors belonging to the Maf family. Although the Maf family and other bZIP-type transcription factors, such as the AP-1 family, share structural similarities, their functions are markedly diversified. The founding member of the Maf family is c-Maf, which was cloned as a cellular counterpart of v-Maf oncogene isolated from avian musculoaponeurotic fibrosarcoma (Nishizawa et al., 1989). Three other members in addition to c-Maf, i.e., MafA, MafB and NRL, constitute the large Maf family (Swaroop et al., 1992; Kataoka et al., 1994; Benkhelifa et al., 1998) (Fig. 1).
Fig. 1.
Structures of the Maf family proteins and other bZIP-type transcription factors. c-Maf, MafA, MafB and NRL constitute the large Maf family. MafG, MafK and MafF constitute the small Maf family. c-Jun and c-Fos are members of the AP-1 family. The protein regions are indicated by different colors: the transcriptional activation domain (TAD; green), extended homology region (EHR; red), CNC domain (orange), basic region (purple) and leucine zipper (blue). The lengths of amino acid sequences for human bZIP factors and C. elegans Skn-1 are provided from the NCBI database (NP_002219, NP_005243, NP_005351, NP_963883, NP_005452, NP_006168.1, NP_002350, NP_002351, NP_001155044, NP_001129495, NP_741404)
Two additional factors, MafK and MafF, were isolated in a low-stringency cDNA library screen using the v-Maf gene (Fujiwara et al., 1993). MafG was later isolated in a separate screening (Kataoka et al., 1995). Though each of the large Maf proteins has a transcriptional activation domain, MafF, MafG and MafK lack any canonical transcriptional activation domains, and based on their small size, they have been classified as the small Maf (sMaf) family (Fig. 1). The official full gene names of MafF, MafG and MafK are “v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F, G, and K”, respectively.
Before the sMaf genes were cloned, a protein complex binding to a specific motif in the human β-globin locus control region (LCR) and other erythroid-related regulatory regions was identified as nuclear factor erythroid 2 (NF-E2) (Mignotte et al., 1989a; Mignotte et al., 1989b; Romeo et al., 1990). Later, a 45 kDa subunit of NF-E2 (p45 NF-E2) was purified and cloned (Andrews et al., 1993a; Ney et al., 1993). The p45 subunit was identified as a bZIP factor, but p45 alone could not bind to the NF-E2-binding motif (Andrews et al., 1993b). NF-E2 was finally identified as a heterodimeric protein complex comprising p45 and another bZIP factor, sMaf (Igarashi et al., 1994). This heterodimer was found to bind to the NF-E2 motif.
MafF, MafG, and MafK are conserved among vertebrates, including human, mouse, rat, chicken and zebrafish. Zebrafish have four sMafs: two MafGs (Mafga and Mafgb); MafK; and MafF, which was originally cloned as MafT (Takagi et al., 2004). In fruit fly, Maf-S was identified as the only sMaf (Veraksa et al., 2000). Therefore, it is assumed that MafF, MafG, and MafK originated from two rounds of whole genome duplication during molecular evolution.
3. Gene structure and expression profile of sMafs
In human and mouse, the genes encoding sMafs (sMaf genes) are composed of three exons. It has been reported that all mouse sMaf genes harbor alternatively used multiple first exons (Motohashi et al., 1996; Onodera et al., 1999; Katsuoka et al., 2005a), and according to the NCBI Reference Sequence (RefSeq) and UCSC EST databases, all human sMaf genes also have multiple alternative first exons. MafF, MafG and MafK are expressed broadly in various tissues, but each sMaf gene has a distinct expression profile (Toki et al., 1997; Onodera et al., 1999). A recent transcriptome sequencing analysis also supports that each sMaf gene exhibits a broad but distinct expression profile (Petryszak et al., 2013). Mechanisms underlying the differential expression of sMaf genes have been well studied in mice, as follows.
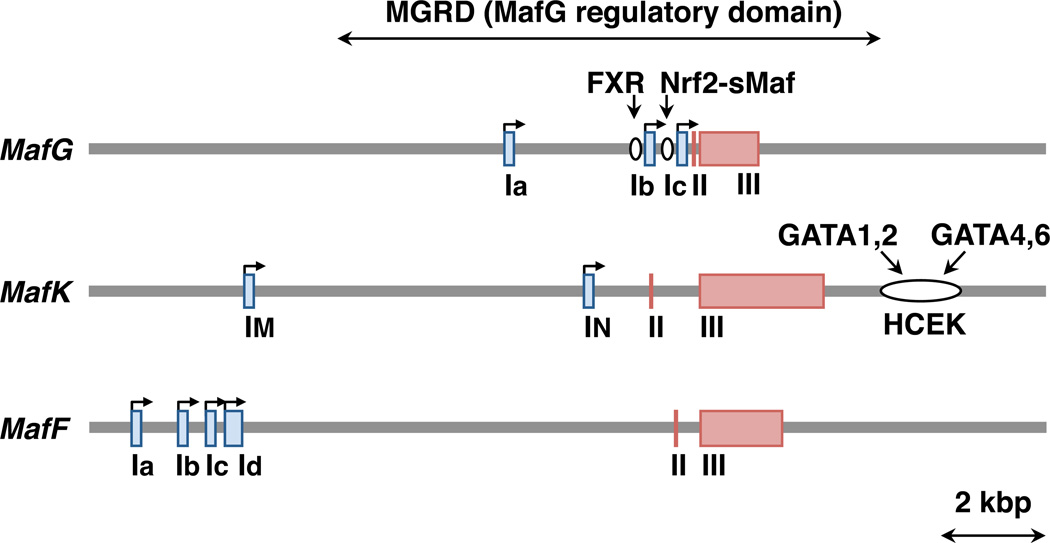
In the mouse MafK gene, two alternative first exons have been identified as follows: the IM exon is broadly utilized, whereas the IN exon is specifically utilized in neural tissues (Fig. 2) (Motohashi et al., 1996; Motohashi et al., 1998). In the 3’ region of MafK, there is an enhancer referred to as Hematopoietic and Cardiac Enhancer for MafK (HCEK), which drives MafK expression in both hematopoietic and cardiac tissues (Katsuoka et al., 2000). Hematopoietic GATA factors (GATA1 or GATA2) and cardiac GATA factors (GATA4 or GATA6) bind to GATA motifs in HCEK and regulate MafK gene expression in specific tissues. It has also been shown that NGF (Torocsik et al., 2002), TGF-β (Okita et al., 2013), AKT (Ro et al., 2010) and Wnt signaling (Wang et al., 2015) influence MafK gene expression.
Fig. 2.
Structures of mouse mafG, mafK, and mafF genomic loci. The first exons are indicated by blue boxes. The other exons are indicated by red boxes. Bent arrows indicate transcription from promoters. The first exons are differentially retained in different splice isoforms. Known cis-regulatory regions, the FXR-binding region, the Nrf2-sMaf-binding region and the hematopoietic and cardiac enhancer for MafK (HCEK) region are indicated by open circles with the names of transcription factors acting through these regions.
In mice, three alternative first exons of MafG have been identified (Ia, Ib and Ic; Fig. 2) (Katsuoka et al., 2005a). In the promoter proximal region of Ic exon, there is an antioxidant response element (ARE) to which the Nrf2-sMaf heterodimer binds (Katsuoka et al., 2005a) (see details in Chapter 5). An important observation is that MafG is induced by Nrf2 inducers, such as electrophilic chemicals, through the ARE (Crawford et al., 1996; Katsuoka et al., 2005a). Certain chemicals have also been reported to induce the expression of the MafK and MafF genes (Moran and Mulcahy, 2002). MafG is also induced by the Farnesoid X receptor (FXR) through a FXR response element (de Aguiar Vallim et al., 2015)(Fig. 2). The three first exons of the MafG gene are clustered in a relatively compact region, and the 6.8 kb upstream region, which includes these three exons and the 3.3 kb downstream region of this gene, are able to recapitulate endogenous mouse MafG expression in transgenic reporter mice in vivo. Therefore, this region has been referred to as the MafG Regulatory Domain (MGRD) (Yamazaki et al., 2012). The MGRD retains stable, strong enhancer activity when employed in mice in vivo (Yamazaki et al., 2012), and it has been used for transgenic overexpression or transgenic complementation rescue analyses (Yamazaki et al., 2012; Hirotsu et al., 2014; Murakami et al., 2014).
Similarly, in mice, the three alternative first exons of MafF have been identified (Ia, Ib and Ic; Fig. 2) (Onodera et al., 1999). Another first exon, which is located downstream of Ic (tentatively referred to here as Id; Fig. 2) is registered in the RefSeq database. Although the RefSeq data show that Ic and Id include alternative start codons that produce alternative protein isoforms, the functional significance of either isoform has not yet been validated. MafF is induced by proinflammatory cytokines in myometrial cells (Massrieh et al., 2006), and it is upregulated in the livers of diet-induced obesity mouse models (Kim et al., 2015).
4. Protein structure of sMafs
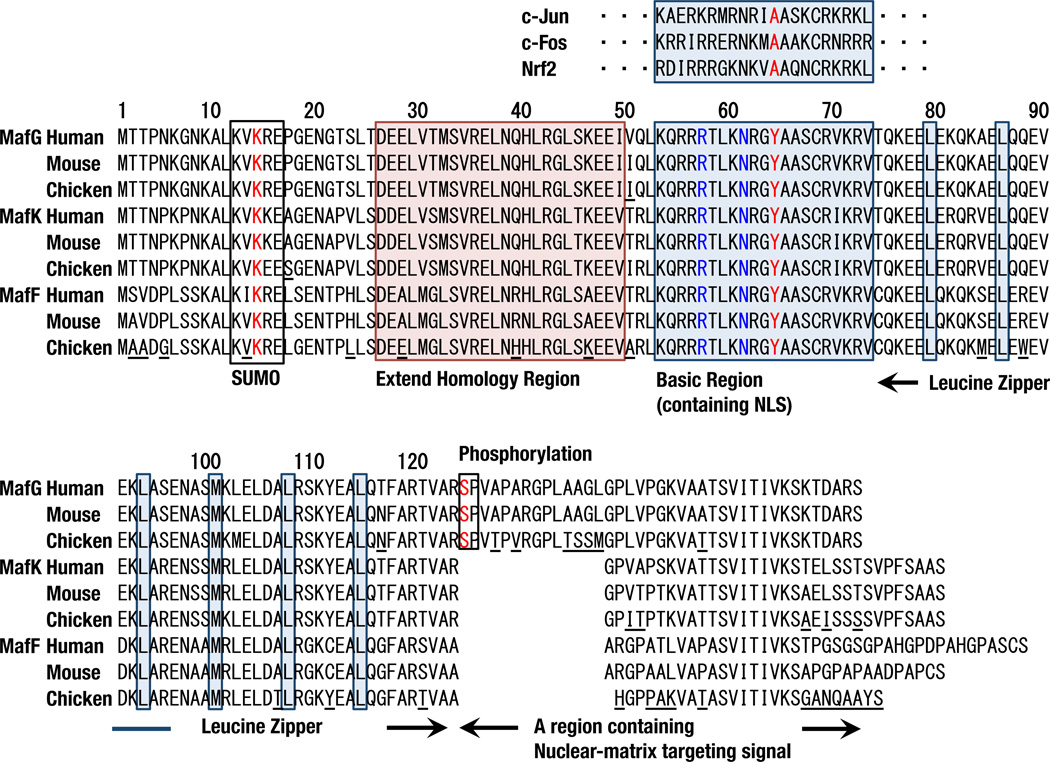
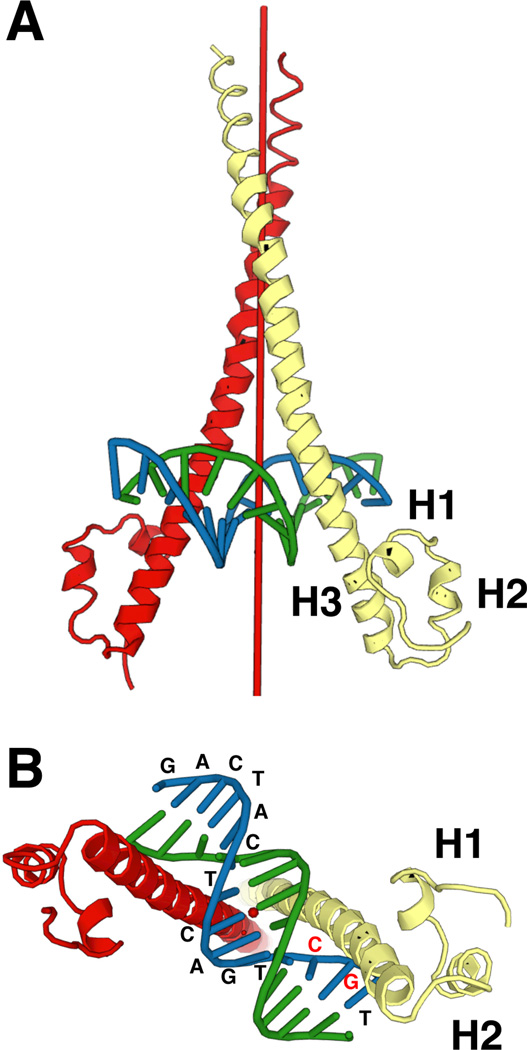
Many domains or motifs have been recognized in the small 18 kDa structures of sMafs (Fig. 3). sMafs harbor a basic region that functions as the DNA-binding motif. The sMafs also harbor a leucine zipper structure that is required for dimer formation with themselves and with other bZIP transcription factors. The elaborate dimer formation involved in the meticulous gene regulation based on this structure is described in Chapter 5. Each sMaf also retains an extended homology region (EHR), which is a domain conserved among the sMaf and large Maf proteins (Kusunoki et al., 2002) that contributes to stable DNA binding. X-ray crystallographic analysis has determined the structure of the MafG-DNA complex (Kurokawa et al., 2009). MafG consists of three helices (H1, H2, and H3). H1, H2, and the beginning of H3 comprise the EHR, and the remaining portion of H3 includes the bZIP region (Fig. 4A). The basic region in H3 fits into the major groove of DNA, and H1 and H2 interact with an N-terminal region of H3 from the outside (Fig. 4B). A tyrosine residue in the basic region of MafG contributes to the unique DNA-binding mode of Maf, as detailed in chapter 5.1.
Fig. 3.
Amino acid sequence alignments of human, mouse and chicken sMafs. SUMO consensus motifs, extended homology regions, basic regions, leucine zipper leucine residues, and phosphorylation sites are indicated by boxes. Amino acid residues not conserved among individual sMafs are underlined in the chicken sequences. MafF is less conserved among species. Tyr64 in red is conserved among basic regions of the small and large Maf families but not in the other bZIP factors. Arg57 and Asn61 in blue are not unique to the Maf family but critical for GC binding.
Fig. 4.
Structure of the MafG homodimer binding to DNA. Two different views (A and B) of the crystal structure of the MafG homodimer and DNA are shown (Protein Data Bank (PDB) Code 3A5T). A symmetry axis is indicated as a red pole in the center of MafG homodimers. Oligonucleotide sequences are shown at each location.
Similar to other bZIP factors, the basic region of sMafs includes a nuclear localization signal (LaCasse et al., 1993). In addition, the C-terminal region of each sMaf contains a nuclear matrix-targeting signal (Motohashi et al., 2011). The C-terminal region is required for localization of the p45 NF-E2-sMaf heterodimer to the subnuclear domain, where proper transcription can be facilitated. Although sMafs are primarily localized in the nucleus, extranuclear localization of sMafs was reported in Hepatitis C virus-replicating cells (Carvajal-Yepes et al., 2011). It has been suggested that extranuclear sMafs can result in mislocalization of Nrf2 and thereby inhibit the induction of Nrf2 target genes.
Post-translational modifications have also been reported for MafG. A sumoylation consensus motif (ΨKXEX) is found in the N-terminal region and is required for sMaf homodimer-mediated repression (Motohashi et al., 2006). In mice, a significant amount of endogenous MafG is conjugated with SUMO-2/3 in bone marrow cells. The C-terminal region of MafG contains an ERK phosphorylation site (Fang et al., 2014), and phosphorylation of this site has been reported to stabilize MafG by inhibiting its ubiquitination.
5. Dimer formation and DNA binding of sMafs
5.1. sMaf Homodimers
An interesting feature of sMafs is that they form homodimers by themselves (Kataoka et al., 1995). Because no functional differences have been reported between the bZIP domains of sMafs, it is assumed that any combination of sMafs can form homodimers (e.g., MafK-MafG and MafK-MafF). The consensus binding motif for sMaf homodimers are palindromic (TGCTGACTCAGCA); this motif is known as the Maf recognition element (MARE) (Fig. 5A). This element is also defined as T-MARE (MARE containing a TRE (TPA-responsive element: TGACTCA or AP-1-binding site) in its core)(Fig. 5A).
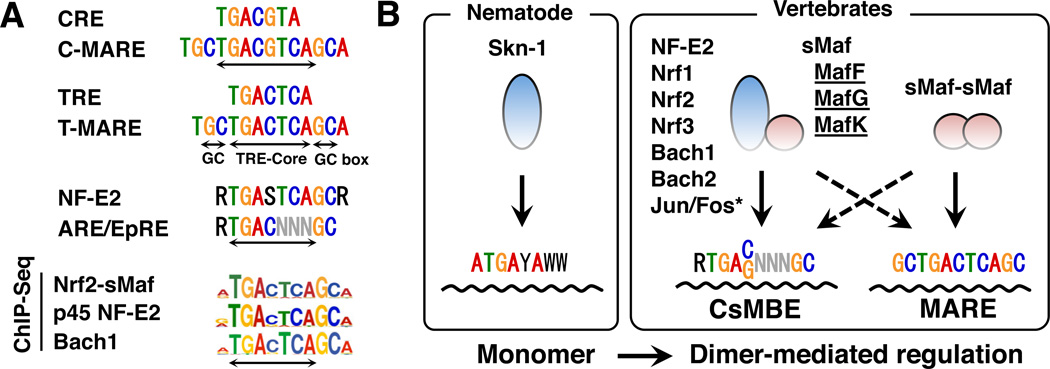
Fig. 5.
Cis-acting consensus sequences recognized by CNC and sMaf. (A) Alignment of the CRE, C-MARE, TRE, T-MARE, NF-E2, and ARE/EpRE motifs. De novo motifs revealed by ChIP-Seq studies (Nrf2, p45NF-E2, and Bach1) are shown below [modified data from (Hirotsu et al., 2012b; Warnatz et al., 2011; Fujita et al., 2013) are shown]. (B) Binding mode of Skn-1, CNC, Bach, and sMaf proteins. Although Skn-1 binds to DNA as a monomer (left box), CNC and Bach proteins bind to DNA as heterodimers with sMafs (right box). Although CNC-sMaf and Bach-sMaf heterodimers preferentially bind to CsMBE, sMaf homodimers preferentially bind to MARE. Asterisk: the exact binding sequences for Jun-sMaf and Fos-sMaf heterodimers have not yet been determined.
An important finding here is that unlike AP-1 factors, the basic regions of Maf factors recognize the flanking GC boxes of MARE in addition to the core region. The basic region of a Maf family protein possesses a specific tyrosine residue (Tyr64, Fig. 3) that is critical for the recognition of the GC box (Kimura et al., 2007) (Fig. 5); this position is usually substituted by alanine residues in other bZIP factors (Fig. 5). A crystallographic analysis determined that Tyr64 does not interact directly with the GC box but affects the side-chain orientation of Arg57 and Asn61 (Fig. 2); these two residues are not unique to sMafs but critical for the recognition of GC (Kurokawa et al., 2009). Although the EHR domain of a sMaf does not contact the DNA directly, the domain supports the recognition of specific DNA sequences by sMafs (Kurokawa et al., 2009). sMafs are known to bind not only to T-MARE but also to palindromic (TGCTGACGTCAGCA) motifs, which are defined as C-MARE (MARE-containing CRE (cAMP-responsive element: TGACGTCA) motif) (Kataoka et al., 1995) (Fig. 5A). However, the binding of sMaf to C-MARE has not been studied extensively.
Because sMafs lack any transcriptional activation domains, sMaf homodimers act as transcriptional repressors (Motohashi et al., 2000). It has been reported that sumoylation at Lysine 14 (K14) within the sMaf sumoylation motif is required for sMaf homodimer-mediated repression (Motohashi et al., 2006). Although the overexpression of wild-type MafG in bone marrow represses its target gene expression, the overexpression of sumoylation-deficient MafG (K14 is mutated to arginine) fails to repress target gene expression. One plausible explanation for this observation is that this is an active repression rather than passive repression, in which the homodimer simply occupies the binding site because a HDAC inhibitor blocks the sMaf homodimer-mediated repression (Motohashi et al., 2006).
5.2 Heterodimers with CNC/Bach transcription factors
sMafs form heterodimers with CNC transcription factors, p45 NF-E2, and NF-E2-related factors (Nrf1, Nrf2 and Nrf3) and with the Bach (BTB and CNC homology) family transcription factors (Bach1 and Bach2), which are distantly related to the CNC family (Fig. 5B) (Igarashi et al., 1994; Oyake et al., 1996; Itoh et al., 1997; Johnsen et al., 1998; Kobayashi et al., 1999). The approved gene symbols for the CNC factors are NFE2, NFE2L1, NFE2L2 and NFE2L3. To avoid any confusion, wewish to emphasize here that NFE2L1 and NFE2L2 (encoding Nrf1 and Nrf2) are different molecules than the nuclear respiratory factors Nrf1 and Nrf2 (Scarpulla, 1997). Because CNC and Bach proteins cannot efficiently bind to DNA by themselves, sMafs are obligatory binding partners. Notably, the CNC homolog in C. elegans, Skn-1, lacks the leucine zipper domain and acts as a monomer (Blackwell et al., 1994) (Fig. 5B). Thus, we surmise that during the course of molecular evolution, the CNC and Bach factors acquired sMafs as indispensable partners to perform higher order transcriptional regulation through the formation of heterodimers (Fig. 5B).
Intriguingly, we can identify several consensus sequences that bind CNC-sMaf heterodimers in various biological contexts. As described above, the original NF-E2 motif “RTGASTCAGCR” was reported as the binding site for the p45 NF-E2-sMaf heterodimer. Another example is the antioxidant/electrophile response element (ARE/EpRE) “RGTGACNNNGC”, which is found in the regulatory regions of genes encoding phase II drug metabolizing enzymes and antioxidant enzymes (Friling et al., 1990; Rushmore et al., 1991). ARE/EpRE is subsequently recognized as the binding site for Nrf2-sMaf heterodimers (Itoh et al., 1997).
Recent ChIP-Seq analyses of the binding sites for the Nrf2-, p45-NF-E2-, and Bach1-sMaf heterodimers revealed that the representative binding motifs for these heterodimers are indeed “RTGACTCAGCA” (Hirotsu et al., 2012b, Fujita et al., 2013, Warnatz et al., 2011) (Fig. 5A). A common feature of these motifs is that the TRE-core is located at the center, with a flanking sequence of R (A or G) on one side and a GC sequence (or GC box) on the other side. The R flanking the TRE core is known to reduce sMaf binding (Kimura et al., 2007). Upon binding of CNC-sMaf heterodimers, CNC factors recognize the R side and sMaf recognizes the GC box side. We previously proposed that the CNC-sMaf heterodimer preferentially binds the CNC-sMaf-binding element (CsMBE) “RTGA(C/G)NNNGC” (Otsuki et al., 2016), thus discriminating this element from the elements that preferentially host Maf homodimers (MARE) (Fig. 5B). Nonetheless, some Nrf2-sMaf-binding sites harbor GC boxes on both sides of the TRE-core, allowing the binding of sMaf homodimers (Fig. 5B) (Otsuki et al., 2016). We surmise that both CNC-sMaf heterodimers and sMaf homodimers participate in the transcriptional regulation of genes through these sites. Indeed, several studies have suggested a switch between CNC-sMaf heterodimers and sMaf homodimers (Motohashi et al., 2000; Yang et al., 2010).
5.3 Heterodimers with other bZIP transcription factors
In addition to CNC and Bach factors, it has been reported that sMafs form heterodimers with other bZIP transcription factors, such as c-Fos and c-Maf (Kataoka et al., 1995; Dhakshinamoorthy et al., 2002). A comprehensive interaction analysis of 49 bZIP transcription factors also showed that sMafs have an affinity to several bZIP factors, including c-Jun and ATF7 (Newman and Keating, 2003). However, the biological significance of these sMaf-containing heterodimers is currently unclear, and further studies are needed to clarify their contribution.
6. In vivo roles of sMafs revealed by mouse genetic studies
To examine the in vivo function of sMafs, MafF, MafG and MafK, knockout mice were generated (Shavit et al., 1998; Onodera et al., 1999; Onodera et al., 2000). Though MafF and MafK knockout mice show no apparent phenotype, MafG knockout mice exhibit mild phenotypes (see chapter 6.1). Therefore, the phenotypic analyses of sMaf double and triple compound knockout mice were conducted to delineate the functions of sMafs in vivo. Because CNC proteins cannot bind to DNA and exert their functions in the absence of sMafs, it seems reasonable to deduce that the phenotypes observed in sMaf mutant mice will also be observed in CNC mutant mice (Fig. 6A). Indeed, available lines of evidence demonstrate that the substantial phenotypes observed in sMaf mutant mice are commonly observed in CNC mutant mice. The phenotypes observed in sMaf mutant mice are summarized in Fig. 6B and discussed below.
Fig. 6.
Genetic analyses of CNC, Bach, and sMaf mutant mice. (A) In the absence of sMaf, CNC and Bach proteins cannot bind to DNA to exert their functions. It is expected that phenotypes observed in CNC and Bach mutant mice are also observed in sMaf mutant mice. (B) Summary of phenotypes observed in sMaf single, double, and triple knockout mutant mice. Note that the phenotype observed in the CNC and Bach mutant mice are mostly observed in sMaf mutant mice depending on the magnitudes of sMaf loss-of-functions. When similar phenotypes are reported in the analyses of CNC or Bach mutant mice, the compositions of CNC-sMaf heterodimers are shown with their illustrations.
6.1. Loss of function of p45 NF-E2-sMaf heterodimers
A founding member of the CNC family of transcription factors is p45 NF-E2, which is specifically expressed in hematopoietic lineages (Andrews et al., 1993a). Indeed, p45 NF-E2 knockout mice show abnormal megakaryocyte differentiation and thrombocytopenia (Shivdasani et al., 1995). In agreement with this observation, MafG knockout mice show similarly abnormal megakaryocyte differentiation and thrombocytopenia (Shavit et al., 1998), whereas MafG−/−::MafK−/− compound mutant mice suffer from much more severe thrombocytopenia (Onodera et al., 2000). A ChIP-Seq analysis of p45 NF-E2-binding sites revealed that p45 NF-E2 binds to a set of genes that are critical for platelet function and production (Fujita et al., 2013). Surprisingly, it has been reported that the overexpression of p45 NF-E2, MafG and MafK can differentiate mouse and human fibroblasts into megakaryocytes (Ono et al., 2012). These results demonstrate that p45 NF-E2-sMaf heterodimers are critical regulators of megakaryocyte differentiation.
It has been reported that p45 NF-E2-sMaf heterodimers regulate β-globin gene expression (Igarashi et al., 1995; Sawado et al., 2001). In the far upstream region of the β-globin gene, there is a LCR composed of several DNase I hypersensitive sites harboring NF-E2 motifs (Raich et al., 1990; Talbot et al., 1990). Overexpression and knockout analyses of p45 NF-E2 using leukemic cell lines showed the involvement of p45 NF-E2-sMaf heterodimers in β-globin gene regulation (Lu et al., 1994; Kotkow and Orkin, 1995). However, mouse genetic studies have failed to show the importance of both p45 NF-E2 and sMafs in β-globin gene regulation (Shivdasani and Orkin, 1995; Onodera et al., 2000), suggesting possible compensation by other CNC factors.
6.2. Loss of function of Nrf1 -sMaf heterodimers
In addition to megakaryocytic phenotypes, MafG knockout and MafG::MafK compound knockout mice exhibit neurological phenotypes. MafG knockout mice show abnormal behavior, such as limb grasping and impaired coordination (Shavit et al., 1998; Onodera et al., 2000). MafG−/−::MafK+/− mice show much more severe neurological phenotypes than MafG single knockout mice, such as ataxia accompanied by progressive neuronal degeneration (Katsuoka et al., 2003). These phenotypes are accelerated in MafG−/−::MafK−/− mice, which cannot survive until weaning (Onodera et al., 2000). Importantly, similar neurological phenotypes were also observed in central nervous system (CNS)-specific Nrf1 knockout mice (Kobayashi et al., 2011; Lee et al., 2011), supporting the notion that Nrf1-sMaf heterodimers are critical for neuronal homeostasis. However, the target genes regulated by Nrf1-sMaf heterodimers in the CNS remain to be fully elucidated.
In addition, conditional liver-specific Nrf1 knockout mice suffer from steatohepatitis (Xu et al., 2005; Ohtsuji et al., 2008). However, because MafG−/−::MafK−/− mice die before weaning (Onodera et al., 2000), this phenotype has not been confirmed in sMaf knockouts. Nrf1 knockout mouse studies have suggested that Nrf1 regulates genes involved in various metabolic pathways, such as fatty acid and amino acid metabolism, as well as proteasome subunit genes (Hirotsu et al., 2012a). These genes are thought to be regulated by Nrf1-sMaf heterodimers, and further studies, such as sMaf conditional targeting analyses, are expected to support this hypothesis.
6.3. Loss of function of Nrf2-sMaf heterodimers
Nrf2 is a master regulator of antioxidant and xenobiotic metabolizing enzyme genes (Itoh et al., 1997). Under normal unstressed conditions, Nrf2 is primarily ubiquitinated by the Keap1 E3-ubiquitin ligase complex and degraded through the proteasome system. For details regarding the degradation mechanisms for Nrf2, please refer to the review papers (Harder et al., 2015; Suzuki and Yamamoto, 2015). In response to oxidative and electrophilic stresses, however, Nrf2 is not ubiquitinated by the Keap1 E3-ubiquitin ligase complex but is instead stabilized and imported into the nucleus (Cullinan et al., 2004; Kobayashi et al., 2004). Nrf2 heterodimerizes with sMafs to activate specific target genes through ARE/EpRE (Friling et al., 1990; Rushmore et al., 1991). Nrf2 knockout mice show impairment in the oxidative/xenobiotic stress response (Itoh et al., 1997). Recent analyses have also demonstrated that Nrf2 regulates genes involved in glucose metabolism and NADPH production (Mitsuishi et al., 2012).
The first genetic evidence supporting the biological significance of Nrf2-sMaf heterodimers was provided by compound mutant analyses of Keap1 and sMaf mice (Motohashi et al., 2004). Keap1 mutant mice show postnatal lethality due to hyperactivation of Nrf2, which results in hyperproliferation of keratinocytes in the esophagus and forestomach (Wakabayashi et al., 2003). These phenotypes are negated not only by the simultaneous disruption of Nrf2 (Wakabayashi et al., 2003) but also by the disruption of MafG and MafF (Motohashi et al., 2004). Conclusive evidence was provided by the genetic analyses of sMaf triple compound mutant mice (Katsuoka et al., 2005b). In embryonic fibroblasts derived from sMaf triple knockout embryos, the induction of Nrf2-regulated cytoprotective genes is severely impaired. Recent ChIP-Seq analyses of Nrf2-MafG-binding sites support the notion that Nrf2-sMaf heterodimers globally contribute to the regulation of antioxidant and metabolic networks (Hirotsu et al., 2012b).
6.4. Loss of function of Bach-sMaf heterodimers
Bach1 is a negative regulator of heme oxygenase-1 (Hmox1) (Oyake et al., 1996; Sun et al., 2002). The Hmox1 gene has two distant enhancers harboring multiple AREs through which Bach1 represses gene transcription. Heme inhibits Bach1 binding to the enhancers, leading to the derepression of the Hmox1 gene (Sun et al., 2002). Bach1 knockout mice show derepression of the Hmox1 gene in various tissues. In agreement with this observation, derepression of the Hmox1 gene is also observed in brains of MafG−/−::MafK+/− mice (Katsuoka et al., 2003) and fibroblasts from sMaf triple knockout mice (Katsuoka et al., 2005b). These genetic data support the role of Bach1 -sMaf heterodimers as negative regulators of the Hmox1 gene.
Similarly, Bach2-sMaf heterodimers are known to negatively regulate the immunoglobulin heavy chain gene through MAREs in the 3’ enhancer region (Muto et al., 1998). Bach2-deficient mice exhibit defects in antibody class switching and somatic hypermutation in B cells (Muto et al., 2004). However, these B-cell phenotypes remain to be examined in murine sMaf mutant lines to demonstrate that sMafs are the indispensable partner molecules of the Bach2 factor.
6.5. Other phenotypes observed in sMaf mutant mice
Lens defects progressively developing into cataracts were reported in MafG−/−::MafK+/− mice (Agrawal et al., 2015). Though microarray analyses revealed several candidate genes that account for lens phenotypes, it is unclear which CNC factors act as partners of sMafs in this context.
sMaf triple knockout mice are embryonic lethal, indicating that sMafs are indispensable for embryo development (Yamazaki et al., 2012). Although mutant embryos grow normally until embryonic day (E) 9.5, the embryos show growth retardation and fetal liver hypoplasia, and they die around E13.5. Apoptotic cells are observed in fetal livers of sMaf triple knockout mice. Because Nrf1::Nrf2 double knockout mice are also embryonic lethal around E13.5 (Leung et al., 2003), the deficiency of both Nrf1-sMaf and Nrf2-sMaf heterodimers may elicit the lethality of sMaf triple knockout mice.
6.6. sMaf functions revealed by overexpression analyses
To examine the functions of sMafs in vivo, several overexpression analyses of sMafs or their mutants have been conducted. The overexpression of dominant negative MafK in pancreatic β-cells promotes MafA binding to MARE in the promoter region of the insulin-2 gene by inhibiting endogenous sMaf binding (Nomoto et al., 2015), which suggests potential competition between sMaf and large Maf proteins for insulin-2 gene regulation. Adenovirus-mediated overexpression of MafG represses Cyp8b1 and several genes involved in bile acid synthesis, suggesting the negative contribution of sMafs to those genes (de Aguiar Vallim et al., 2015).
7. sMaf functions and diseases
As described in chapter 6 and summarized in Table 1, loss-of-function of sMafs results in various disease phenotypes, such as progressive neuronal degeneration, cataracts, thrombocytopenia and embryonic lethality. In addition, many studies have reported relationships between sMAFs and human diseases.
Table 1.
Phenotypes and diseases associated with CNC/Bach-sMaf functions
| Site | Phenotypes and Diseases | References |
|---|---|---|
| CNS | Progressive neuronal degeneration | Mouse Genetics (Nrf1, Kobayashi et al., 2011; Lee et al., 2011) |
| Mouse Genetics (MafG, MafK, Onodera et al., 2000; Katsuoka et al., 2003) | ||
| Eye | Cataract | Mouse Genetics (MafG, MafK, Agrawal et al., 2015) |
| Liver | Steatohepatitis | Mouse Genetics (Nrf1, Ohtsuji et al., 2008) |
| Hepatotoxicity (e.g., Acetaminophen) | Mouse Genetics (Nrf2, Enomoto et al., 2001) | |
| Hematopoietic | Thrombocytopenia | Mouse Genetics (p45 NFE2, Shivdasani et al., 1995) |
| Mouse Genetics (MafG, MafK, Shavit et al., 1998; Onodera et al., 2000) | ||
| Delays in hematopoietic recovery | Mouse Genetics (MafG, Li et al., 2010) | |
| Chronic myeloid leukemia | SNPs (MAFG and MAFF, Martínez- Hernández et al., 2014) | |
| Embryogenesis | Growth Retardation/Lethality | Mouse Genetics (Nrf1, Nrf2, Leung et al., 2003) |
| Mouse Genetics (MafF, MafG MafK Yamazaki et al., 2012) | ||
| Tumorigenesis | Tumor suppression | |
| Mouse Genetics (Nrf2, Iida et al., 2007; Bauer et al., 2011) | ||
| SNPs (NRF2 Suzuki et al., 2013; Review Paper: Cho et al., 2015) | ||
| SNPs (MAFG Wang et al., 2010) | ||
| Tumor progression | Somatic Mutation (NRF2, KEAP1, Review Paper: Cho et al., 2015) | |
| siRNA Knockdown (Bach1, MafG Fang et al., 2014) |
7.1. Involvement of sMafs in disease prevention
Several SNPs in MAFF and MAFG were found to be associated with chronic myeloid leukemia (Martínez-Hernández et al., 2014), and SNPs in MAFG were found to be associated with lung cancers (Wang et al., 2010). In both cases, it was suggested that impairment of Nrf2-sMaf heterodimer function contributes to the disease onsets and/or progressions. The links between the Nrf2-Keap1 system and diseases have been extensively studied. Impaired Nrf2 functions result in the dysregulation of antioxidant and xenobiotic metabolism and an increased susceptibility to various stimuli, including exogenous chemicals (e.g., acetaminophen) and endogenous factors (e.g., electrophilic compounds and reactive oxygen species) (Enomoto et al., 2001; Cho et al., 2015). In addition, several lines of evidence have revealed the contribution of Nrf2 to anti-inflammatory responses. Impaired Nrf2 functions result in prolonged inflammation, which is associated with various disorders, including neurodegeneration and arteriosclerosis (Mimura and Itoh, 2015; Yamazaki et al., 2015). Although direct evidence has not been provided, it is likely that sMAFs are involved in disease prevention together with NRF2.
7.2. sMafs and cancer
The involvement of sMaf in tumor onset and progression is complicated. Because Nrf2-sMaf heterodimers function as critical regulators of antioxidant and xenobiotic metabolism, impairment of the heterodimers increases the risk of various cancers. Conversely, many SNPs in NRF2 and KEPA1 that result in constitutional activation of Nrf2, are observed in many tumors (Suzuki and Yamamoto, 2015). Cancer cells exploit Nrf2-sMaf heterodimers to produce reducing power, which is required for cell survival and cell cycle progression (Mitsuishi et al., 2012).
MafG is involved in aberrant hypermethylation in certain types of cancers. Constitutively activating mutations in the BRAF oncoprotein promote proliferation and malignant transformation (Dhomen and Marais, 2007). Recently, an RNAi screening study reported that the Bach1-MafG heterodimer recruits a chromatin-remodeling factor (CHD8) and a DNA methyl-transferase (DNMT3B) to facilitate the hypermethylation and repression of the mismatch repair gene MLH1 and other tumor suppressor genes harboring CpG island promoters (Fang et al., 2014). Considering these observations, inhibitors for sMafs may be new pharmacological targets for cancer treatments.
8. Conclusion
In summary, accumulating lines of evidence unequivocally demonstrate that sMafs play roles as critical regulatory hubs for the CNC-sMaf transcription factor network. Understanding the cross-talk among sMaf-containing hetero- and homodimers is especially important for answering the question of how higher organisms have adopted the dimer-mediated transcription factor network. Further analyses, including proteome, transcriptome, and cistrome analyses, will provide an in-depth understanding of the cellular dynamics of sMafs and their partners, thus yielding more comprehensive insights into sMafs.
Highlights.
The small Maf (sMaf) proteins are bZIP-type transcription factors.
sMaf proteins form homodimers and heterodimers with CNC and Bach proteins.
sMaf proteins participate in activation or repression depending on their partners.
Various biological pathways are under the regulation of sMaf-containing dimers.
Acknowledgments
This review and the corresponding Gene Wiki article are written as parts of the Gene Wiki Review series, which is a series resulting from a collaboration between the GENE journal and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by the National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. This work was supported, in part, by MEXT/JSPS KAKENHI (24249015, 26111002). The authors thank all researchers and staff who contributed to the research we summarized here.
The corresponding Gene Wiki entry for this review can be found here:
https://en.wikipedia.org/wiki/small_Maf
https://en.wikipedia.org/wiki/MAFF_(gene)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal SA, Anand D, Siddam AD, Kakrana A, Dash S, Scheiblin DA, Dang CA, Terrell AM, Waters SM, Singh A, Motohashi H, Yamamoto M, Lachke SA. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum. Genet. 2015;134:717–735. doi: 10.1007/s00439-015-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993a;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Kotkow KJ, Ney PA, Erdjument-Bromage H, Tempst P, Orkin SH. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc. Natl. Acad. Sci. U.S.A. 1993b;90:11488–11492. doi: 10.1073/pnas.90.24.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AK, Cho HY, Miller-Degraff L, Walker C, Helms K, Fostel J, Yamamoto M, Kleeberger SR. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS One. 2011;6:e26590. doi: 10.1371/journal.pone.0026590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkhelifa S, Provot S, Lecoq O, Pouponnot C, Calothy G, Felder-Schmittbuhl M-P. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene. 1998;17:247–254. doi: 10.1038/sj.onc.1201898. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Carvajal-Yepes M, Himmelsbach K, Schaedler S, Ploen D, Krause J, Ludwig L, Weiss T, Klingel K, Hildt E. Hepatitis C virus impairs the induction of cytoprotective Nrf2 target genes by delocalization of small Maf proteins. J. Biol. Chem. 2011;286:8941–8951. doi: 10.1074/jbc.M110.186684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-Y, Marzec J, Kleeberger SR. Functional polymorphisms in NRF2: implications for human disease. Free Radic. Bio. Med. 2015;88:362–372. doi: 10.1016/j.freeradbiomed.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DR, Leahy KP, Wang Y, Schools GP, Kochheiser JC, Davies KJ. Oxidative stress induces the levels of a MafG homolog in hamster HA-1 cells. Free Radic. Bio. Med. 1996;21:521–525. doi: 10.1016/0891-5849(96)00160-8. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim TQ, Tarling EJ, Ahn H, Hagey LR, Romanoski CE, Lee RG, Graham MJ, Motohashi H, Yamamoto M, Edwards PA. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell Metab. 2015;21:298–310. doi: 10.1016/j.cmet.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakshinamoorthy S, Jaiswal AK. c-Maf negatively regulates ARE-mediated detoxifying enzyme genes expression and anti-oxidant induction. Oncogene. 2002;21:5301–5312. doi: 10.1038/sj.onc.1205642. [DOI] [PubMed] [Google Scholar]
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr. Opin. Genet. Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O’Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG Island Methylator phenotype. Mol. Cell. 2014;55:904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R, Takayama-Tsujimoto M, Satoh H, Gutierrez L, Aburatani H, Fujii S, Sarai A, Bresnick EH, Yamamoto M, Motohashi H. NF-E2 p45 is important for establishing normal function of platelets. Mol. Cell Biol. 2013;33:2659–2670. doi: 10.1128/MCB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara KT, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- Harder B, Jiang T, Wu T, Tao S, de la Vega MR, Tian W, Chapman E, Zhang DD. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem. Soc. Trans. 2015;43:680–686. doi: 10.1042/BST20150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y, Hataya N, Katsuoka F, Yamamoto M. NF-E2-related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1beta genes. Mol. Cell Biol. 2012a;32:2760–2770. doi: 10.1128/MCB.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y, Higashi C, Fukutomi T, Katsuoka F, Tsujita T, Yagishita Y, Matsuyama Y, Motohashi H, Uruno A, Yamamoto M. Transcription factor NF-E2 - related factor 1 impairs glucose metabolism in mice. Genes Cells. 2014;19:650–665. doi: 10.1111/gtc.12165. [DOI] [PubMed] [Google Scholar]
- Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012b;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Itoh K, Motohashi H, Hayashi N, Matuzaki Y, Nakauchi H, Nishizawa M, Yamamoto M. Activity and expression of murine small Maf family protein MafK. J. Biol. Chem. 1995;270:7615–7624. doi: 10.1074/jbc.270.13.7615. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- Iida K, Itoh K, Maher JM, Kumagai Y, Oyasu R, Mori Y, Shimazui T, Akaza H, Yamamoto M. Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis. 2007;28:2398–2403. doi: 10.1093/carcin/bgm146. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Johnsen Ø, Murphy P, Prydz H, Kolstø A-B. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Igarashi K, Itoh K, Fujiwara KT, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Engel JD, Yamamoto M. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J. Biol. Chem. 2005a;280:4483–4490. doi: 10.1074/jbc.M411451200. [DOI] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell Biol. 2005b;25:8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Onodera K, Suwabe N, Engel JD, Yamamoto M. One enhancer mediates mafK transcriptional activation in both hematopoietic and cardiac muscle cells. EMBO J. 2000;19:2980–2991. doi: 10.1093/emboj/19.12.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Tamagawa Y, Kure S, Igarashi K, Engel JD, Yamamoto M. Small Maf compound mutants display central nervous system neuronal degeneration, aberrant transcription, and Bach protein mislocalization coincident with myoclonus and abnormal startle response. Mol. Cell Biol. 2003;23:1163–1174. doi: 10.1128/MCB.23.4.1163-1174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kwon EY, Park S, Kim JR, Choi SW, Choi MS, Kim SJ. Integrative systems analysis of diet-induced obesity identified a critical transition in the transcriptomes of the murine liver and epididymal white adipose tissue. Int. J. Obes. (Lond.) 2015;40:338–345. doi: 10.1038/ijo.2015.147. [DOI] [PubMed] [Google Scholar]
- Kimura M, Yamamoto T, Zhang J, Itoh K, Kyo M, Kamiya T, Aburatani H, Katsuoka F, Kurokawa H, Tanaka T, Motohashi H, Yamamoto M. Molecular basis distinguishing the DNA binding profile of Nrf2-Maf heterodimer from that of Maf homodimer. J. Biol. Chem. 2007;282:33681–33690. doi: 10.1074/jbc.M706863200. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap’n’collar family transcription factor Nrf3. J. Biol. Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Tsukide T, Miyasaka T, Morita T, Mizoroki T, Saito Y, Ihara Y, Takashima A, Noguchi N, Fukamizu A, Hirotsu Y, Ohtsuji M, Katsuoka F, Yamamoto M. Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Genes Cells. 2011;16:692–703. doi: 10.1111/j.1365-2443.2011.01522.x. [DOI] [PubMed] [Google Scholar]
- Kotkow KJ, Orkin SH. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa H, Motohashi H, Sueno S, Kimura M, Takagawa H, Kanno Y, Yamamoto M, Tanaka T. Structural basis of alternative DNA recognition by Maf transcription factors. Mol. Cell Biol. 2009;29:6232–6244. doi: 10.1128/MCB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunoki H, Motohashi H, Katsuoka F, Morohashi A, Yamamoto M, Tanaka T. Solution structure of the DNA-binding domain of MafG. Nat. Struct. Biol. 2002;9:252–256. doi: 10.1038/nsb771. [DOI] [PubMed] [Google Scholar]
- LaCasse E, Lochnan H, Walker P, Lefebvre Y. Identification of binding proteins for nuclear localization signals of the glucocorticoid and thyroid hormone receptors. Endocrinology. 1993;132:1017–1025. doi: 10.1210/endo.132.3.8440170. [DOI] [PubMed] [Google Scholar]
- Lee CS, Lee C, Hu T, Nguyen JM, Zhang J, Martin MV, Vawter MP, Huang EJ, Chan JY. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8408–8413. doi: 10.1073/pnas.1019209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- Li X-M, Hu Z, Zafar A-B, Jorgensen ML, Bungert J, Slayton W. Intrinsic and extrinsic effects of mafG deficiency on hematopoietic recovery following bone marrow transplant. Exp. Hematol. 2010;38:1251–1260. doi: 10.1016/j.exphem.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Lu S-J, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hernández A, Gutierrez-Malacatt H, Carrillo-Sánchez K, Saldaña-Alvarez Y, Rojas-Ochoa A, Crespo-Solis E, Aguayo-González A, Rosas -López A, Ayala-Sanchez JM, Aquino-Ortega X. Small MAF genes variants and chronic myeloid leukemia. Eur. J. Haematol. 2014;92:35–41. doi: 10.1111/ejh.12211. [DOI] [PubMed] [Google Scholar]
- Massrieh W, Derjuga A, Doualla-Bell F, Ku CY, Sanborn BM, Blank V. Regulation of the MAFF transcription factor by proinflammatory cytokines in myometrial cells. Biol. Reprod. 2006;74:699–705. doi: 10.1095/biolreprod.105.045450. [DOI] [PubMed] [Google Scholar]
- Mignotte V, Eleouet JF, Raich N, Romeo P-H. Cis-and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc. Natl. Acad. Sci. U.S.A. 1989a;86:6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignotte V, Wall L, Grosveld F, Romeo P-H. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 1989b;17:37–54. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura J, Itoh K. Role of Nrf2 in the pathogenesis of atherosclerosis. Free Radic. Bio. Med. 2015;88:221–232. doi: 10.1016/j.freeradbiomed.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Moran JA, Mulcahy RT. Differential induction of mafF, mafG and mafK expression by electrophile-response-element activators. Biochem. J. 2002;361:371–377. doi: 10.1042/0264-6021:3610371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Fujita R, Takayama M, Inoue A, Katsuoka F, Bresnick EH, Yamamoto M. Molecular determinants for small Maf protein control of platelet production. Mol. Cell Biol. 2011;31:151–162. doi: 10.1128/MCB.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Igarashi K, Onodera K, Takahashi S, Ohtani H, Nakafuku M, Nishizawa M, Engel JD, Yamamoto M. Mesodermal-vs. neuronal-specific expression of MafK is elicited by different promoters. Genes Cells. 1996;1:223–238. doi: 10.1046/j.1365-2443.1996.d01-230.x. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Miyoshi C, Uchimura Y, Saitoh H, Francastel C, Engel JD, Yamamoto M. MafG sumoylation is required for active transcriptional repression. Mol. Cell Biol. 2006;26:4652–4663. doi: 10.1128/MCB.02193-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–876. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Ohta J, Douglas Engel J, Yamamoto M. A core region of the mafK gene IN promoter directs neurone-specific transcription in vivo. Genes Cells. 1998;3:671–684. doi: 10.1046/j.1365-2443.1998.00222.x. [DOI] [PubMed] [Google Scholar]
- Murakami S, Shimizu R, Romeo PH, Yamamoto M, Motohashi H. Keap1-Nrf2 system regulates cell fate determination of hematopoietic stem cells. Genes Cells. 2014;19:239–253. doi: 10.1111/gtc.12126. [DOI] [PubMed] [Google Scholar]
- Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, Igarashi K. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3’ enhancer. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- Ney PA, Andrews NC, Jane SM, Safer B, Purucker ME, Weremowicz S, Morton CC, Goff SC, Orkin SH, Nienhuis AW. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol. Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto H, Kondo T, Miyoshi H, Nakamura A, Hida Y, Yamashita K, Sharma AJ, Atsumi T. Inhibition of small Maf function in pancreatic beta-cells improves glucose tolerance through the enhancement of insulin gene transcription and insulin secretion. Endocrinology. 2015;156:3570–3580. doi: 10.1210/en.2014-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita Y, Kamoshida A, Suzuki H, Itoh K, Motohashi H, Igarashi K, Yamamoto M, Ogami T, Koinuma D, Kato M. Transforming growth factor-beta induces transcription factors MafK and Bach1 to suppress expression of the heme oxygenase-1 gene. J. Biol. Chem. 2013;288:20658–20667. doi: 10.1074/jbc.M113.450478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Wang Y, Suzuki H, Okamoto S, Ikeda Y, Murata M, Poncz M, Matsubara Y. Induction of functional platelets from mouse and human fibroblasts by p45NF-E2/Maf. Blood. 2012;120:3812–3821. doi: 10.1182/blood-2012-02-413617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera K, Shavit JA, Motohashi H, Katsuoka F, Akasaka JE, Engel JD, Yamamoto M. Characterization of the murine mafF gene. J. Biol. Chem. 1999;274:21162–21169. doi: 10.1074/jbc.274.30.21162. [DOI] [PubMed] [Google Scholar]
- Onodera K, Shavit JA, Motohashi H, Yamamoto M, Engel JD. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J. 2000;19:1335–1345. doi: 10.1093/emboj/19.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic. Bio. Med. 2016;91:45–57. doi: 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N. Expression Atlas update-a database of gene and transcript expression from microarray-and sequencing-based functional genomics experiments. Nucleic Acids Res. 2013;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich N, Enver T, Nakamoto B, Josephson B, Papayannopoulou T, Stamatoyannopoulos G. Autonomous developmental control of human embryonic globin gene switching in transgenic mice. Science. 1990;250:1147–1149. doi: 10.1126/science.2251502. [DOI] [PubMed] [Google Scholar]
- Ro YT, Jang BK, Shin CY, Park EU, Kim CG, Yang SI. Akt regulates the expression of MafK, synaptotagmin I, and syntenin-1, which play roles in neuronal function. J. Biomed. Sci. 2010;17:18. doi: 10.1186/1423-0127-17-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo P-H, Prandini M-H, Joulin V, Mignotte V, Prenant M, Vainchenker W, Uzan G. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990;344:447–449. doi: 10.1038/344447a0. [DOI] [PubMed] [Google Scholar]
- Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- Sawado T, Igarashi K, Groudine M. Activation of β-major globin gene transcription is associated with recruitment of NF-E2 to the β-globin LCR and gene promoter. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory chain expression in mammalian cells. J. Bioenerg. Biomembr. 1997;29:109–119. doi: 10.1023/a:1022681828846. [DOI] [PubMed] [Google Scholar]
- Shavit JA, Motohashi H, Onodera K, Akasaka J, Yamamoto M, Engel JD. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev. 1998;12:2164–2174. doi: 10.1101/gad.12.14.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Orkin SH. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8690–8694. doi: 10.1073/pnas.92.19.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, Orkin SH. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoeitin/MGDF in megakaryocyte development. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shibata T, Takaya K, Shiraishi K, Kohno T, Kunitoh H, Tsuta K, Furuta K, Goto K, Hosoda F. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol. Cell Biol. 2013;33:2402–2412. doi: 10.1128/MCB.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Bio. Med. 2015;88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Xu J, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl. Acad. Sci. U.S.A. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Kobayashi M, Li L, Suzuki T, Nishikawa K, Yamamoto M. MafT, a new member of the small Maf protein family in zebrafish. Biochem. Biophys. Res. Commun. 2004;320:62–69. doi: 10.1016/j.bbrc.2004.05.131. [DOI] [PubMed] [Google Scholar]
- Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human beta-globin dominant control region. EMBO J. 1990;9:2169. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki T, Itoh J, Kitazawa J, Arai K, Hatakeyama K, Akasaka J, Igarashi K, Nomura N, Yokoyama M, Yamamoto M. Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene. 1997;14:1901–1910. doi: 10.1038/sj.onc.1201024. [DOI] [PubMed] [Google Scholar]
- Torocsik B, Angelastro JM, Greene LA. The basic region and leucine zipper transcription factor MafK is a new nerve growth factor-responsive immediate early gene that regulates neurite outgrowth. J. Neurosci. 2002;22:8971–8980. doi: 10.1523/JNEUROSCI.22-20-08971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap ‘n’ collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wang R, Zheng J, Zhang DS, Yang YH, Zhao ZF. Wnt1-induced MAFK expression promotes osteosarcoma cell proliferation. Genet. Mol. Res. 2015;14:7315–7325. doi: 10.4238/2015.July.3.7. [DOI] [PubMed] [Google Scholar]
- Wang X, Chorley BN, Pittman GS, Kleeberger SR, Brothers J, II, Liu G, Spira A, Bell DA. Genetic variation and antioxidant response gene expression in the bronchial airway epithelium of smokers at risk for lung cancer. PLoS One. 2010;5:e11934. doi: 10.1371/journal.pone.0011934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnatz H-J, Schmidt D, Manke T, Piccini I, Sultan M, Borodina T, Balzereit D, Wruck W, Soldatov A, Vingron M. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Biol. Chem. 2011;286:23521–23532. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chen L, Leung L, Yen TB, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Katsuoka F, Motohashi H, Engel JD, Yamamoto M. Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol. Cell Biol. 2012;32:808–816. doi: 10.1128/MCB.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tanji K, Wakabayashi K, Matsuura S, Itoh K. Role of the Keap1/Nrf2 pathway in neurodegenerative diseases. Pathol. Int. 2015;65:210–219. doi: 10.1111/pin.12261. [DOI] [PubMed] [Google Scholar]
- Yang H, Ko K, Xia M, Li TW, Oh P, Li J, Lu SC. Induction of avian musculoaponeurotic fibrosarcoma proteins by toxic bile acid inhibits expression of glutathione synthetic enzymes and contributes to cholestatic liver injury in mice. Hepatology. 2010;51:1291–1301. doi: 10.1002/hep.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]