Abstract
Muscle hypertrophy occurs when the rate of protein synthesis exceeds the rate of degradation. A main factor determining the rate of protein synthesis is ribosome abundance or translational capacity. The production of ribosomes is a primary determinant of translational capacity. Based on studies from our laboratory, we propose the novel hypothesis that ribosome biogenesis is necessary for skeletal muscle hypertrophy.
Keywords: skeletal muscle, hypertrophy, ribosome, translational capacity, protein synthesis
Introduction
Adult skeletal muscle as an organ is amazingly plastic. Muscle robustly changes its size, composition and metabolic profile in order to optimally adapt to environmental and physiological demands (4). In response to resistance exercise training, muscle progressively accumulates protein mass leading to hypertrophy of individual myofibers. Conversely, under chronic conditions of energy deprivation, unloading, and/or disease, muscle undergoes a relatively rapid atrophy resulting from a loss in myofiber cross-sectional area. A critical determinant of muscle mass is the ratio of protein synthesis to protein breakdown; however, commitment to protein synthesis may be the preferred target of regulation in all but emergency situations (22).
Protein synthesis does not happen without ribosomes, which are nature’s nano machines that convert mRNA genetic code into polypeptide chains. Changes to these protein factories during periods of muscle growth are key to understanding the mechanisms of skeletal muscle plasticity. Specifically, during muscle hypertrophy, the amount of protein synthesis per unit RNA (translation efficiency) and the total ribosomal content (translation capacity) both increase within muscle fibers (2, 20). As one would imagine, these two processes are intimately connected and, as has been reported in the heart, there is an emerging recognition that the initial increase in translational efficiency in response to a growth stimulus is required for the subsequent increase in translational capacity that ultimately leads to the protein accretion that underlies skeletal muscle hypertrophy (16).
In this review, we first provide a brief description of the ribosome and the regulation of ribosome biogenesis, followed by a discussion of our work supporting the main thesis and then finish with a review of studies from other investigators in the field that provide additional compelling evidence in support of our hypothesis that ribosome biogenesis is necessary for skeletal muscle hypertrophy.
Ribosome Biogenesis
The functionally mature human ribosome (80S) is a 4.3 megadalton ribonucleoprotein complex composed of two subunits. The large (60S) subunit contains three (28S, 5S, and 5.8S) ribosomal RNAs (rRNAs) and 47 ribosomal proteins (RPLs). The small (40S) subunit contains one (18S) rRNA molecule and 33 ribosomal proteins (RPSs). Three of the four rRNA molecules (28S, 5.8S, and 18S) are transcribed together as a single precursor rRNA molecule called the 47S pre-rRNA. The 47S pre-rRNA is transcribed by the dedicated RNA polymerase I (Pol I) from ribosomal DNA sequences (rDNA). The human genome contains hundreds of copies of tandem repeats of rDNA sequences distributed across five chromosomes. The rDNA clusters are organized into characteristic regions within the nucleus known as the nucleolus (12).
Cleavage of the 47S pre-rRNA by processing factors and small nucleolar ribonucleoproteins (snoRNPs) leads to the formation of the 28S, 5.8S, and 18S rRNA molecules required for small and large subunit assembly. The only rRNA molecule not transcribed in the nucleolus is the 5S rRNA, which is transcribed by RNA polymerase III (Pol III). 5S rRNA molecules are transported into the nucleolus after synthesis in order to be assembled with the 28S and the 5.8S rRNAs to form the large subunit. Pol III is also responsible for transcribing transfer RNAs (tRNAs), which are transported out of the nucleus, loaded with amino acids, and used for polypeptide elongation (12).
The various ribosomal proteins are encoded by messenger RNAs (mRNAs), which are transcribed by RNA polymerase II (Pol II). Ribosomal protein mRNAs are transported out of the nucleus and translated into protein in the cytoplasm. mRNAs encoding assembly factors and transport proteins are also transcribed by Pol II and transported into the cytoplasm for translation. Ribosomal proteins and protein-processing factors are subsequently imported back into the nucleolus to be assembled onto the small and large subunits. Small subunit ribosomal proteins (RPSs) assemble onto the 18S rRNA scaffold and large subunit ribosomal proteins (RPLs) assemble onto the combined 28S, 5S, and 5.8S rRNA scaffold. Following assembly, the 40S and 60S subunits are exported out into the cytoplasm, where they bind their respective co-factors. The 40S and 60S subunits do not combine and mature into the functional 80S ribosome until translation initiation occurs on mRNA molecules (12).
As can be seen from the previous discussion, ribosome biogenesis is a highly complex process with numerous points of regulation. Despite this complexity, the general consensus is that the primary point of regulation in ribosome biogenesis is transcription of 47S pre-rRNA by Pol I and, in particular, the initiation of transcription (19). The first step in 47S pre-rRNA transcription is the formation of the pre-initiation complex (PIC) by the binding of upstream binding factor (UBF) proteins at the rDNA promoter which, in turn, recruits selectivity factor 1 (SL-1; also known as TIF-1B) complex. Ribosomal DNA transcription is initiated with the recruitment of Pol I to the PIC via Rrn3 (also known as TIF-1A) which serves as a bridge between SL-1 and Pol1 (19). The formation and activation of this Pol I “regulon” is regulated by the extracellular signal-regulated kinas (ERK) and mechanistic target of rapamycin (mTOR) pathways via UBF and Rrn3 phosphorylation (19). Activation of mTOR complex 1 (mTORC1) stimulates the UBF-mediated transcription of rRNAs as well as the 5′-terminal oligopyrimidine (TOP)-mediated transcription of mRNAs encoding ribosomal proteins (24, 31). Accordingly, blocking mTORC1 activity has been shown to inhibit the translation of the numerous 5′-TOP mRNAs encoding ribosomal proteins and impair the formation of Pol I transcription complex in the promoter region of the rRNA gene (15, 17). In addition to these signaling pathways, numerous studies have collectively provided evidence that the proto-oncogene c-Myc has a central role in the control of ribosome biogenesis through its regulation of all three RNA polymerases and, by extension, their respective gene products such as Pol I regulon components, ribosomal proteins and 5S rRNA (32).
Origin of our hypothesis
In an effort to identify new genes that have a critical role in skeletal muscle growth our laboratory has performed a series of transcriptome analyses over the last few years (7, 8, 18). In addition to identifying a small set of conserved genes that were up-regulated during hypertrophy and re-growth following atrophy, we found there appeared to be a positive relationship between the change in total RNA content of the muscle and the magnitude of muscle growth (7, 8). During hypertrophic growth, the 62% increase in skeletal muscle mass was associated with a 2.5-fold increase in RNA content whereas RNA abundance remained stable throughout a period of muscle re-growth in which mass increased by a modest 15% (7). This apparent relationship between RNA content and muscle growth was further bolstered by our more recent study comparing the hypertrophic response of young and old mice (18). Following two weeks of synergist ablation, the plantaris muscle of 24 month old mice showed a significantly blunted hypertrophic response compared to 5 month old mice that was associated with a 50% increase (compared to a 2.5-fold increase in young mice) in the RNA content of the muscle (18). Knowing that total RNA can serve as a proxy of ribosome content, this finding suggested the ability of old muscle to produce new ribosomes in response to a growth stimulus was impaired and, moreover, might be the underlying cause for the decreased hypertrophy. In supported of this notion, we reported the increase in rDNA transcription during hypertrophy was significantly less in old muscle compared to young muscle; after 3 days of synergist ablation, 45S pre-rRNA expression increased 3-fold in the plantaris muscle of young mice and only 1.7-fold in old mice (18). Further, following seven days of synergist ablation, 45S pre-rRNA expression continued to be elevated 3-fold in plantaris muscle of young mice but returned to baseline levels in old mice. Collectively, these data provided the impetus for proposing the hypothesis that ribosome biogenesis is necessary for skeletal muscle hypertrophy. In the following sections, we will present evidence from colleagues in the field, as well as studies on cardiac hypertrophy, that support our hypothesis.
Ribosome biogenesis in cardiac hypertrophy
What evidence is there for the necessity of ribosome biogenesis in skeletal muscle hypertrophy? While most of the evidence supporting this hypothesis has only emerged in the last few years, the cardiac field has a long history investigating the importance of ribosome biogenesis in cardiac hypertrophy. In reviewing the literature, Hannan and coworkers concluded that increased translational capacity was required for cardiac hypertrophy and that increased translational efficiency alone was not sufficient to promote cardiac hypertrophy (16). This conclusion was best exemplified by a study from Brandenburger and colleagues showing that ribosome biogenesis was required for cardiomyocyte hypertrophy and that UBF was essential in mediating this process (6).
Ribosome biogenesis in skeletal muscle hypertrophy
As mentioned above, total RNA can be used as a measure of ribosome content given that ~85% of the RNA in the cell is rRNA; thus, an increase in RNA content is considered to be indicative of an increase in ribosome biogenesis (39). This convention is supported by our work and others showing that increases in RNA content during muscle hypertrophy are associated with an increase rDNA transcription as reflected by higher 45S pre-rRNA expression (18, 33). In the following sections, we will take advantage of this convention to explore what is known about the relationship between ribosome biogenesis and skeletal muscle hypertrophy.
Skeletal muscle hypertrophy during post-natal development
The rapid growth that occurs during post-natal development is no better characterized than in skeletal muscle. The increase in skeletal muscle size during post-natal development is primarily the result of cell hypertrophy with possibly a small contribution from hyperplasia, an increase in cell number (38). This hypertrophic growth was beautifully described by White and colleagues who reported a 7.6-fold increase in myofiber cross-sectional area between post-natal day 7 (P7) and adulthood (P56) in the mouse extensor digitorum longus muscle with no change in fiber number; remarkably, more than 40% of this hypertrophic growth happened between P7 and P21 (36). A rich body of early work clearly showed that post-natal skeletal muscle hypertrophy was associated with a significant increase in RNA abundance and that undernutrition, in the form of a low-protein diet, negatively impacted both growth and RNA content of the muscle (38). More recently, Fiorotto and coworkers provided a detailed description of the effect of undernutrition on post-natal muscle protein metabolism and growth (11). The recovery of skeletal mass following undernutrition was dependent upon the restoration of translational capacity which coincided with a 2-fold increase in UBF expression (11). This study also showed that during normal maturation there was a progressive decline in both ribosome abundance and UBF expression of the muscle, consistent with the decrease in the fractional growth rate (11). These studies collectively demonstrate the existence of a strong connection between post-natal skeletal muscle hypertrophy and ribosome content that is sensitive to nutrition.
Hypertrophy in adult skeletal muscle
In addition to our studies, numerous animal (1, 14, 15, 23, 26, 33, 35), human (3, 9, 10, 25, 27, 29, 30) and in vitro (24) studies have provided further evidence that skeletal muscle hypertrophy is associated with an increase in ribosome biogenesis. The following section will provide a brief overview of these studies with an emphasis on those findings that support our hypothesis that ribosome biogenesis is necessary for skeletal muscle hypertrophy.
One of the first studies to really investigate the importance of ribosome biogenesis and skeletal muscle hypertrophy was a study by Nader and colleagues (24). Using an in vitro model of hypertrophy, the authors showed a ~70% increase in rRNA 48 hr post-serum stimulation that was associated with increased UBF availability as the result of cyclin D1-dependent activation of CDK4 phosphorylation of Rb (24). When mTORC1 activity was inhibited by rapamycin during serum stimulation, the subsequent increases in UBF availability and rRNA abundance were not observed which, in turn, was associated with a complete loss of myotube hypertrophy (24). This is a critical finding as it suggests that the ribosomal pool is limiting and must be expanded to support myotube hypertrophy. In agreement with this finding, Stec and coworkers reported that the specific inhibition of Pol I activity by CX-5461 treatment prevented serum-stimulated increase in total RNA as well as myotube hypertrophy (29). Moving into the mouse, the Nader laboratory next investigated Pol1 regulon expression during the initial phase of hypertrophy induced by synergist ablation (33). After three days of synergist ablation, there was a 2-fold increase in total RNA concentration as the result of a 3-fold increase in 45S pre-rRNA expression. The increase in rRNA expression was paralleled by ~2.5- to 6-fold increase in the transcript levels of Pol I regulon components such as Polr1b, Rrn3 (TIF-1A), Polr1e (PAF53), Ttf1 and Taf1c as well as UBF mRNA and protein. Consistent with the up-regulation of the Pol I regulon, chromatin immunoprecipitation assay showed enhanced binding of Pol I and UBF at the rDNA promoter. The findings from these studies provide clear evidence that in response to a hypertrophic stimulus, there is a dramatic increase in translational capacity of the muscle as the result of Pol 1-dependent transcription of rDNA.
In an effort to clarify how increases in translational efficiency and capacity are related to the degree of muscle hypertrophy, Nakada and coworkers modified the synergist ablation model in such a way as to produce four different levels of hypertrophy (26). After five days of synergist ablation, plantaris muscle weight increased by 8%, 22%, 32% and 45% with rRNA content increasing by 1.8-fold, 2.2-fold and 2.5-fold in only the top three groups, respectively. Further, the increase in translational capacity observed at five days was strongly correlated (r = 0.98) to muscle weight after 14 days of synergist ablation. In contrast, the level of p70S6K phosphorylation, a readout of mTORC1 activation and translation efficiency, showed no further increase above 20% hypertrophy leading the authors to conclude that increased translational capacity was required for robust hypertrophic growth (26).
Although the synergist ablation model of hypertrophy has been very useful in identifying the cellular and molecular mechanisms regulating skeletal muscle hypertrophy, there are concerns that it may not accurately reflect the underlying processes involved in the hypertrophic response following resistance exercise (5, 21). To better mimic resistance exercise, a rodent model of electrical stimulation was developed and successfully used in the first study to report elevated p70S6K phosphorylation following high-force contractions (2, 37). This model of hypertrophy was recently used by West and colleagues to better understand the influence of mTORC1 signaling on translational capacity following an acute bout of resistance exercise (35). Transcription of rDNA was increased between 12 and 36 hr after exercise and preceded the trend toward increased translational capacity. In agreement with Nader and coworkers in vitro findings, inhibition of mTORC1 signaling by rapamycin completely prevented the increase in ribosome biogenesis; however, the increase in c-Myc and UBF phosphorylation following electrical stimulation were rapamycin-insensitive suggesting these two pathways may work together in a temporal fashion to promote ribosome biogenesis. In support of this notion, the Hornberger laboratory reported that the ~3-fold increase in translational capacity following seven days of synergist ablation was not completely blocked by rapamycin treatment but only blunted by 32% (14). Further, the significant increase in both UBF protein and Rb hyperphosphorylation levels at this later time point were insensitive to mTORC1 inhibition. More recently, Goodman and coworkers provided intriguing evidence showing that YAP was capable of driving c-Myc expression in a rapamycin-independent manner (13). Similar to what has been described in cardiac hypertrophy, it appears that mTORC1 regulation of translational efficiency is important in mediating the initial events of ribosome biogenesis and that more long-term, c-Myc regulation of ribosome biogenesis may be necessary to sustain a full hypertrophic response (16). Further, the early activation mTORC1 may also be necessary to insure robust c-Myc protein expression as inhibition of mTORC1 activity by rapamycin has been shown to reduce the amount of c-MYC mRNA associated with the polysome (34). The proposed temporal relationship between translational efficiency and capacity during skeletal muscle hypertrophy is graphically shown in Figure 1.
Figure 1.
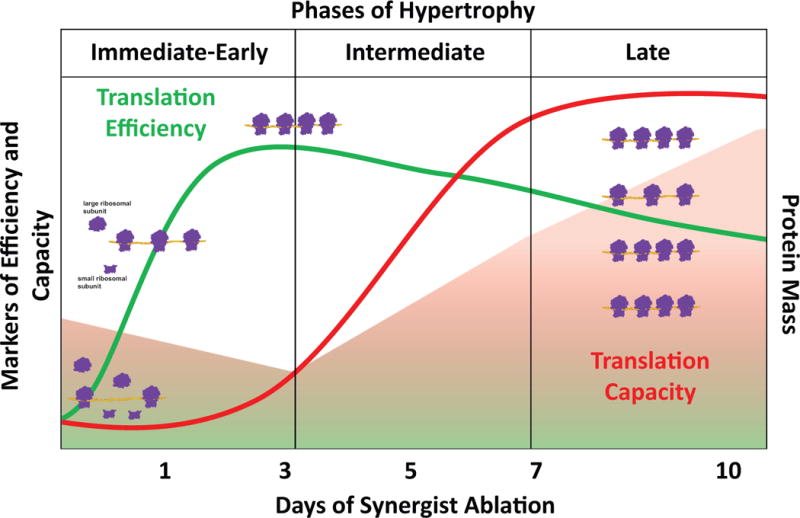
The temporal relationship between translational efficiency and capacity during skeletal muscle hypertrophy. During the initial phase of hypertrophy there is increased translational efficiency resulting in enhanced polysome formation (more ribosomes per mRNA). During the intermediate phase, as translational efficiency begins to return to baseline, translational capcity begins to increase as the result of ribosome biogensis; during this phase protein accretion begins to occur. In the late phase, peak translational capacity (maximum number of ribosomes) is reached which promotes further polysome formation, resulting in significant protein accretion leading to myofiber hypertrophy.
How well do the findings on ribosome biogenesis using animal models of hypertrophy translate into what is observed in humans following an acute bout of resistance exercise (RE) and with RE training? Nader and colleagues conducted a study to determine if the change in gene expression following an acute bout of RE was altered after 12 weeks of RE training (25). Relevant to the current discussion, these investigators found that both c-Myc and 45S pre-rRNA expression were significantly elevated in response to RE regardless of training status; however, in the trained state these genes were less response to RE with 45S pre-RNA expression significantly attenuated by 40% and c-Myc expression slightly, though still significant, above control levels (25). Although these findings are somewhat limited, they do suggest that the response to RE in terms of ribosome biogenesis is conserved between humans and rodents.
Building on the findings of Nader and coworkers, Figueiredo and colleagues performed the first detailed human study investigating the regulation of ribosome biogenesis in response to RE, both acutely and following eight weeks of RE training (9, 25). The total RNA concentration following training tended toward an increase (1.3-fold) but did not achieve statistical significance because of variability between subjects; however, the increase in total RNA from pre-training to post-training was significantly correlated (r = 0.72) to the percent change in the cross-sectional area of the muscle (9). Consistent with this trend, 45S pre-rRNA expression in resting muscle was increased by 2-fold after training and reflected in a 2- to 4-fold increase in mature 18S, 5.8S and 28S rRNAs. The activation of mTORC1 and ERK signaling pathways remained response to exercise independent of training status as well as cyclin D1 and UBF proteins and Rrn3 (TIF-1A) Ser649 phosphorylation (9). Although UBF phosphorylation did change following exercise, UBF Ser388 phosphorylation was ~5-fold higher in the rested state following eight weeks of RE training which the authors speculated was necessary for the increased translational capacity observed in trained muscle.
In a recent follow-up study, Figueiredo and colleagues investigated the impact of different recovery strategies on ribosome biogenesis following an acute bout of RE (10). To provide an important temporal component to the study, muscle biopsies were collected at 2, 24 and 48 hr post-exercise. This time course allowed the authors to capture activation of the upstream p38/MNK/eIF4E signaling cascade as p38Thr180/Tyr182, MNK1Thr197 and eIF4ESer209 phosphorylation were all significantly elevated at 2 hr post-exercise (10). Furthermore, UBFSer388 phosphorylation, 45S pre-rRNA and cMyc expression were significantly increased at both the 24 hr and 48 hr post-exercise time point (10). Collectively, these data provide convincing evidence that ribosome biogenesis is activated in response to a single bout of RE in humans.
Although the studies by Figueiredo and coworkers clearly showed that ribosome biogenesis occurs following RE, the findings of their studies do not speak to whether or not ribosome biogenesis is necessary for skeletal muscle hypertrophy in humans. A study by the Bamman laboratory reported that in older individuals (age 60–75 y) only extreme responders (to 4 weeks of RE) showed a significant increase in total RNA (+26%) and rRNA content (+40%) which was associated with a ~350% increase in c-Myc protein (29). These findings were not completely unexpected given this group’s earlier study showing blunted ribosome biogenesis in older individuals following a bout of RE, in agreement with Kirby and colleagues study comparing the hypertrophic response of young and old mice (18, 30). A curious difference not observed in older mice was the significantly greater ribosome content of skeletal muscle from older humans, presumably as a result of higher 45S pre-rRNA expression (18, 30). The reason why older human skeletal muscle has more ribosomes remains unknown but the authors speculate it may reflect some type of compensatory mechanism for impaired translational efficiency known to occur with aging (30). Although these findings do not definitively demonstrate that skeletal muscle hypertrophy requires ribosome biogenesis they do show that the hypertrophic response is enhanced by increases in translational capacity.
An unanswered question is if the reported increase in rRNA abundance following resistance exercise truly results in the production of more functional ribosomes? While this questions remains to be answered, Srivastava and others have reported that changes in total RNA during skeletal muscle maturation were strongly correlated with the level of polysome formation suggesting that the increase in rRNA observed with hypertrophy is indicative of more functionally active ribosomes (28, 38). This issue takes on greater importance given that skeletal muscle from older humans was shown to have a higher concentration of total RNA compared to young skeletal muscle, begging the question as to whether or not this actually indicates a higher translation capacity (30).
Summary
The results of studies from our laboratory, as well as other investigators in the field, have clearly shown ribosome biogenesis is increased in response to resistance exercise and that this response is conserved across species and affected in a similar manner with aging. Moreover, there is a strong correlation between the level of ribosome biogenesis and the magnitude of hypertrophy that is also conserved in both humans and rodents. While the findings of these studies collectively make a strong argument for the necessity of ribosome biogenesis in skeletal muscle hypertrophy, they remain descriptive in nature. The challenge for the field moving forward will be to carry out in vivo loss- and gain-of-function experiments with components of the Pol I regulon and master regulators (such as c-Myc) to rigorously test the hypothesis that ribosome biogenesis is necessary for skeletal muscle hypertrophy. The results of such experiments are expected to show that both translational efficiency and capacity serve critical roles in the regulation of skeletal muscle hypertrophy.
Key Points.
Skeletal muscle hypertrophy requires an increase in the rate of protein synthesis.
One way in which the rate of protein synthesis can be enhanced is by increasing the translational capacity of the muscle through ribosome biogenesis.
Evidence from both human and rodent studies reveals that the magnitude of muscle hypertrophy is highly correlated to the increase in translational capacity of the muscle, highlighting the importance of ribosome biogenesis.
Acknowledgments
THE authors apologize to colleagues in the field whose work was unable to be cited because of space limitations.
Funding: This work was supported in part by grants (AR061939 and AR064896) from the National Institutes of Health to J.J.M.
Footnotes
Disclosure: The authors declare no conflicts of interest.
References
- 1.Adams GR, Haddad F, Baldwin KM. Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. Journal of applied physiology. 1999;87(5):1705–12. doi: 10.1152/jappl.1999.87.5.1705. [DOI] [PubMed] [Google Scholar]
- 2.Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. The American journal of physiology. 1999;276(1 Pt 1):C120–7. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- 3.Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. Journal of applied physiology. 2005;98(2):482–8. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- 4.Blaauw B, Schiaffino S, Reggiani C. Mechanisms modulating skeletal muscle phenotype. Comprehensive Physiology. 2013;3(4):1645–87. doi: 10.1002/cphy.c130009. [DOI] [PubMed] [Google Scholar]
- 5.Bodine SC, Stitt TN, Gonzalez M, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature cell biology. 2001;3(11):1014–9. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 6.Brandenburger Y, Jenkins A, Autelitano DJ, Hannan RD. Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2001;15(11):2051–3. doi: 10.1096/fj.01-0853fje. [DOI] [PubMed] [Google Scholar]
- 7.Chaillou T, Jackson JR, England JH, et al. Identification of a conserved set of upregulated genes in mouse skeletal muscle hypertrophy and regrowth. Journal of applied physiology. 2015;118(1):86–97. doi: 10.1152/japplphysiol.00351.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. Journal of applied physiology. 2013;115(7):1065–74. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron-Smith D, Blazevich AJ. Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. American journal of physiology. Endocrinology and metabolism. 2015;309(1):E72–83. doi: 10.1152/ajpendo.00050.2015. [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo VC, Roberts LA, Markworth JF, et al. Impact of resistance exercise on ribosome biogenesis is acutely regulated by post-exercise recovery strategies. Physiological reports. 2016;4(2) doi: 10.14814/phy2.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiorotto ML, Davis TA, Sosa HA, Villegas-Montoya C, Estrada I, Fleischmann R. Ribosome abundance regulates the recovery of skeletal muscle protein mass upon recuperation from postnatal undernutrition in mice. The Journal of physiology. 2014;592(23):5269–86. doi: 10.1113/jphysiol.2014.279067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 13.Goodman CA, Dietz JM, Jacobs BL, McNally RM, You JS, Hornberger TA. Yes-Associated Protein is up-regulated by mechanical overload and is sufficient to induce skeletal muscle hypertrophy. FEBS letters. 2015;589(13):1491–7. doi: 10.1016/j.febslet.2015.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman CA, Frey JW, Mabrey DM, et al. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. The Journal of physiology. 2011;589(Pt 22):5485–501. doi: 10.1113/jphysiol.2011.218255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman CA, Mabrey DM, Frey JW, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25(3):1028–39. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannan RD, Jenkins A, Jenkins AK, Brandenburger Y. Cardiac hypertrophy: a matter of translation. Clinical and experimental pharmacology & physiology. 2003;30(8):517–27. doi: 10.1046/j.1440-1681.2003.03873.x. [DOI] [PubMed] [Google Scholar]
- 17.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(10):4441–5. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirby TJ, Lee JD, England JH, Chaillou T, Esser KA, McCarthy JJ. Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. Journal of applied physiology. 2015;119(4):321–7. doi: 10.1152/japplphysiol.00296.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene. 2015;556(1):27–34. doi: 10.1016/j.gene.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. Journal of applied physiology. 2007;102(1):306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy JJ, Mula J, Miyazaki M, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138(17):3657–66. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millward DJ, Garlick PJ, Nnanyelugo DO, Waterlow JC. The relative importance of muscle protein synthesis and breakdown in the regulation of muscle mass. The Biochemical journal. 1976;156(1):185–8. doi: 10.1042/bj1560185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazaki M, McCarthy JJ, Fedele MJ, Esser KA. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. The Journal of physiology. 2011;589(Pt 7):1831–46. doi: 10.1113/jphysiol.2011.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. American journal of physiology. Cell physiology. 2005;289(6):C1457–65. doi: 10.1152/ajpcell.00165.2005. [DOI] [PubMed] [Google Scholar]
- 25.Nader GA, von Walden F, Liu C, et al. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. Journal of applied physiology. 2014;116(6):693–702. doi: 10.1152/japplphysiol.01366.2013. [DOI] [PubMed] [Google Scholar]
- 26.Nakada S, Ogasawara R, Kawada S, Maekawa T, Ishii N. Correlation between Ribosome Biogenesis and the Magnitude of Hypertrophy in Overloaded Skeletal Muscle. PloS one. 2016;11(1):e0147284. doi: 10.1371/journal.pone.0147284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts LA, Raastad T, Markworth JF, et al. Post-exercise cold water immersion attenuates acute anabolic signalling and long-term adaptations in muscle to strength training. The Journal of physiology. 2015;593(18):4285–301. doi: 10.1113/JP270570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava U. Polyribosome concentration of mouse skeletal muscle as a function of age. Archives of biochemistry and biophysics. 1969;130(1):129–39. doi: 10.1016/0003-9861(69)90018-6. [DOI] [PubMed] [Google Scholar]
- 29.Stec MJ, Kelly NA, Many GM, Windham ST, Tuggle SC, Bamman MM. Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. American journal of physiology. Endocrinology and metabolism. 2016 doi: 10.1152/ajpendo.00486.2015. ajpendo 00486 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stec MJ, Mayhew DL, Bamman MM. The effects of age and resistance loading on skeletal muscle ribosome biogenesis. Journal of applied physiology. 2015;119(8):851–7. doi: 10.1152/japplphysiol.00489.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature reviews. Cancer. 2010;10(4):301–9. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 33.von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. American journal of physiology. Cell physiology. 2012;302(10):C1523–30. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- 34.Wall M, Poortinga G, Hannan KM, Pearson RB, Hannan RD, McArthur GA. Translational control of c-MYC by rapamycin promotes terminal myeloid differentiation. Blood. 2008;112(6):2305–17. doi: 10.1182/blood-2007-09-111856. [DOI] [PubMed] [Google Scholar]
- 35.West DW, Baehr LM, Marcotte GR, et al. Acute resistance exercise activates rapamycin-sensitive and -insensitive mechanisms that control translational activity and capacity in skeletal muscle. The Journal of physiology. 2016;594(2):453–68. doi: 10.1113/JP271365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC developmental biology. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong TS, Booth FW. Protein metabolism in rat gastrocnemius muscle after stimulated chronic concentric exercise. Journal of applied physiology. 1990;69(5):1709–17. doi: 10.1152/jappl.1990.69.5.1709. [DOI] [PubMed] [Google Scholar]
- 38.Young VR. Regulation of protein synthesis and skeletal muscle growth. Journal of animal science. 1974;38(5):1054–70. doi: 10.2527/jas1974.3851054x. [DOI] [PubMed] [Google Scholar]
- 39.Zak R, Rabinowitz M, Platt C. Ribonucleic acids associated with myofibrils. Biochemistry. 1967;6(8):2493–9. doi: 10.1021/bi00860a028. [DOI] [PubMed] [Google Scholar]


