Abstract
Basal subtype breast cancers have a particularly poor prognosis, with high invasiveness and resistance to most targeted therapies. TGFβ and MYC drive central features of basal breast cancer: TGFβ is an autocrine and paracrine signaling factor that drives cell invasion and metastasis, and MYC is a central regulator of cellular proliferation that is upregulated in many cancer types. We show here that genetic or pharmacological inhibition of MYC in MCF10A basal breast cells results in increased sensitivity to TGFβ-stimulated invasion and metastasis, and also show that this signaling loop is dependent on activation of SRC. Analysis of human breast cancer datasets and additional experiments with breast cancer cell lines further suggest the relevance of this signaling loop in basal, but not luminal, breast cancers. Our results imply precaution should be taken when utilizing therapeutic inhibitors of MYC with basal breast cancer patients as this could lead to increased metastasis; however, simultaneous pharmacological inhibition of SRC and MYC for these patients could facilitate the anti-proliferative effects of MYC inhibition while blocking the consequent promotion of metastasis.
Keywords: basal breast cancer, metastasis, TGFβ, MYC, integrins
INTRODUCTION
Breast cancers can be classified into clinically relevant groups with distinct therapeutic responsiveness on the basis of their intrinsic molecular subtypes (1, 2). Human breast cancer cell lines can also be clustered according to these intrinsic molecular subtypes, which are associated with differences in phenotypic and genetic features, as well as differences in responsiveness to drug treatments (1, 3). Both basal subtype breast cancers and basal cell lines are associated with aggressive features, invasiveness, higher grade and poor prognosis, and as basal breast cancers overlap with the triple negative histological phenotype (ER-, PR-, HER2-), there exist few effective targeted therapies for this group of patients (4, 5). Thus, there is a need to better understand the distinct molecular pathways controlling progression of basal breast cancers in order to develop new effective therapeutic approaches.
MYC is a pleiotropic transcription factor that regulates cellular proliferation, growth, differentiation, apoptosis, adhesion, and migration (6). Elevated or deregulated expression of MYC has been found in multiple types of cancer, including breast cancer (7, 8). MYC induces cell proliferation by targeting genes that promote progression of the cell cycle, as well as by inhibiting transcription of growth inhibitory genes (9). It also plays roles in inhibition of apoptosis, induction of angiogenesis and epithelial-mesenchymal transition (EMT) (7). In breast cancer, MYC gene amplification is associated with higher tumor grade, poor clinical prognosis, risk of relapse and ER/PR-negativity (7), and inhibitors of MYC are being evaluated for antitumor activity in clinical trials (10). However, despite its role as a pleiotropic oncogene, a recent study of MYC has shown that it can also behave as a tumor suppressor under certain circumstances through inhibition of cancer cell invasion and metastasis (11).
Integrins are heterodimeric cell membrane proteins that consist of α and β transmembrane subunits; acquisition of malignancy is associated with cancer subtype-dependent increased expression of specific integrins that facilitate cell invasion and metastasis (12). Integrin αvβ3 is often upregulated in breast cancer and has been found to be associated with increased invasion and metastasis (12). A recent study identified a tumor suppressor activity for MYC that was dependent upon specific regulation of integrins αv and β3; specifically, while overexpression of MYC in mammary epithelial cells induced cell proliferation in culture and led to formation of larger tumors in mice, direct inhibition of integrin subunits αv and β3 by MYC was shown to inhibit cell invasion and metastasis (11).
TGFβ is a multi-functional cytokine that can act as a tumor suppressor, inhibiting proliferation and suppressing stromal mitogens, as well as a tumor promoter, allowing evasion of immune surveillance and driving invasion and metastatic colonization (13–15). The consequences of TGFβ signaling are highly dependent on cellular context (16), and it is a prominent player during tumorigenesis (17, 18). Early findings that growth inhibition by TGFβ occurred through transcriptional inhibition of MYC (19) were supported by identification of a SMAD-responsive element in the MYC promoter (20) and demonstration of direct binding of SMAD3 to the MYC promoter (21). As downregulation of MYC also leads to increased breast cancer cell invasion and metastasis, we hypothesized that TGFβ-induced inhibition of proliferation and promotion of invasion could be linked through downregulation of MYC in breast cancer cells.
Here, we show that TGFβ decreases expression of MYC and consequently leads to upregulation of integrin αvβ3, and that this signaling axis can drive invasion and metastasis. We further show that this effect is specific for the basal subtype of breast cancer, and find that MYC knockdown or inhibition can drive the invasive phenotype, but that simultaneous inhibition of SRC can block this effect. Our study reveals a hidden danger of MYC inhibition: while suppressing cancer cell proliferation, these drugs can also stimulate invasion and metastasis of basal subtype breast cancer cells. Importantly, by defining the mechanistic basis for this phenomenon, we have also identified a solution: the simultaneous inhibition of MYC and SRC can block the proliferative capacity of MYC while abrogating its SRC-dependent pro-invasive activity. Based on our findings we propose a novel therapeutic strategy for these patients through simultaneous targeted inhibition of MYC and SRC. This strategy has the potential to block the proliferative capacity of MYC while abrogating the activation of invasion and metastasis mediated through SRC signaling.
METHODS
Cell culture
MCF10A and MCF12A cells were grown in DMEM/F12 (Gibco), supplemented with 5% horse serum (Gibco), 20 ng/ml EGF (Peprotech), 0.5 μg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 10 μg/ml insulin (Sigma) and 100 μg/ml gentamicin (Gibco); and maintained as previously described(22). MCF10CA1h cells were grown in DMEM/F12 (Gibco), supplemented with 5% horse serum (Gibco) and 100 μg/ml gentamicin (Gibco). HCC38, BT-474, BT-549, T47D and MCF7 cells were maintained in RPMI 1640 (Gibco), MDA-MB-231and MDA-MB-468 cells were grown in DMEM (Gibco), and SKBR3 cells in McCoy’s 5A (Gibco), all supplemented with 10% heat inactivated fetal bovine serum (FBS) (Thermo Scientific) and 100 μg/ml gentamicin (Gibco). SUM149 cells were grown in Ham’s F12 medium (Gibco), supplemented with 5% FBS (Thermo Scientific), 5 μg/ml insulin (Sigma), 1 μg/ml hydrocortisone (Sigma), 10 mM HEPES (Gibco), 25 μg/ml gentamicin (Gibco) and 2.5 μg/ml Fungizone (Gibco). SUM149 cells were maintained at 37°C in 10% CO2. All other cell lines were maintained at 37°C in 5% CO2.
293FT cells, used for lentivirus production, were purchased from Invitrogen and maintained in DMEM (Gibco), supplemented with 10% heat inactivated FBS (Thermo Scientific), 1X MEM Non-essential amino acids (Gibco) and 500 μg/ml G418 sulfate (Cellgro). No G418 sulfate was present in the medium used for lentivirus production. Pheonix-A cells, used for retrovirus production, were grown in DMEM (Gibco), supplemented with 10% heat inactivated FBS (Thermo Scientific) and 100 μg/ml gentamicin (Gibco).
For TGFβ treatments recombinant human TGFβ1 (R&D Systems) was used, and unless otherwise indicated, cells were treated with TGFβ at 25 ng/ml for 18 h. For drug treatments following compounds were used: Dasatinib (Selleckchem); PP2 (Calbiochem); c-Myc inhibitor 2 (MI2) (Calbiochem), JQ1 (Cayman Chemical). Dasatinib treatments were performed at 50 nM.
All relevant human cell lines used in experiments were obtained from ATCC, which authenticates using short tandem repeat profiling.
Lentivirus production and shRNA-mediated gene knockdown, and gene expression
Specific gene knockdowns were achieved by lentiviral transduction with Mission shRNAs (Sigma). Third generation lentiviruses were produced using HEK293FT cells. Briefly 6 × 106 cells were plated in 10 cm plates and simultaneously transfected with 3 μg shRNA plasmid and 3 μg of each packaging plasmid: pMD2.G (Addgene, #12259), pRSV-Rev (Addgene, #12253), pMDLg/pRPE (Addgene, #12251) (23); lipofectamine 2000 (Invitrogen) was used for transfection, according to manufacturer’s instructions. Medium was changed after 24 hours and lentivirus containing medium was collected 48 hours post transfection. For lentiviral transduction in a 6-well format, 600 μl viral supernatant, 400 μl regular medium and 6 μg/ml polybrene (Millipore) were used. Following shRNAs were used: MYC NM_002467.3-1661s21c1 (TRC2), targeting sequence 5′-CCAGAGGAGGA ACGAGCTAAA-3′; ITGAV NM_002210.x-2551s1c1 (TRC1/1.5), targeting sequence 5′-CTCTGTTGTATATCCTTCATT-3′; ITGB3 NM_000212.x-2084s1c1, targeting sequence 5′-GTCGTCAGATTCCAGTACTAT-3′ (TRC1/1.5); SRC NM_198291.1-648s21c1 (TRC1/1.5), targeting sequence 5′-GACAGACCTGTCC TTCAAGAA-3. Non target shRNAs were used as a control, specifically SHC002 for TRC1 /1.5 library constructs and SHC202 for TRC2 library constructs, both with the same non-mammalian hairpin sequence 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTT TT-3′. In experiments involving double knockdown with constructs from both TRC1/1.5 and TRC2 libraries, both NTs were used as a control. The same lentivirus production and lentiviral transduction protocols were used for specific gene expression. Refer to ‘Plasmids and cloning’ section for construct details.
For MCF10A cells following antibiotic concentrations were used for selection, where indicated: puromycin 1 μg/ml, neomycin (G418) 200 μg/ml. For MDA-MB231 cells puromycin was used at 1 μg/ml for lentiviral transduction selection and at 0.25 μg/ml for retroviral transduction selection.
Retrovirus production and transduction
Retrovirus for expression of Myc was produced using Pheonix-A cells and pMSCV/puro Myc plasmid (11). Retrovirus with an empty vector was used as a control. Briefly, 3 × 106 cells were plated in 10 cm plates and transfected the following day with 6 μg od plasmid DNA, using Fugene6 transfection reagent (Promega), following manufacturer’s instructions. Retrovirus containing medium was collected 48 hours post transfection. For retroviral transduction of MBA-MB231 and MCF10A cells, 1 × 106 cells were plated in 10 cm plates the day before transduction; 1.8 ml viral supernatant, 4.2 ml regular medium and 10 μg/ml polybrene (Millipore) were used. Medium was changed after 24 hours and antibiotic selection using 0.25 μg/ml puromycin was started 48 hours post transduction.
RNA extraction, cDNA synthesis and real-time quantitative PCR (RT-qPCR) analysis
RNA was isolated using Trizol reagent (Invitrogen), following manufacturer’s instructions. Briefly, the cells were lysed with Trizol, followed by phenol-chloroform phase separation. RNA was precipitated with isopropanol, washed with 75% ethanol and dried RNA pellet was resuspended in DEPC water. RNA from mouse tissue was isolated using RNeasy mini kit (Invitrogen), according to the instruction manual. cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Gene expression levels were assayed by RT-qPCR using 7900 HT FAST Real-Time PCR system, TaqMan Fast Universal Master Mix and TaqMan probes for specific genes (Applied Biosystems): ITGAV Hs00233808_m1, ITGB3 Hs00173978_m1, MYC Hs00153408_m1, SERPINE (PAI-1) Hs01126607_g1, GAPDH Hs99999905_m1. All assays were performed in triplicate and analysis was performed using RQ Manager software (Applied Biosystems) and the 2^−ΔΔCt method to obtain relative quantitation (RQ) values, with GAPDH used as endogenous control.
Plasmids and cloning
Cloning was performed utilizing Gateway cloning approach. MYC was cloned from pMSCV/puro MYC (11). Constitutively active TGFβ Receptor I was cloned from pCMV5 TBRI-HA T204D (Addgene, #19162). For obtaining inserts in an entry vector pENTR/D-TOPO cloning kit with One Shot Mach1-T1 chemically competent E. coli (Invitrogen) was used, following manufacturer’s instructions. PCR of inserts for the TOPO reaction was done using PfuUltra high-fidelity DNA polymerase (Stratagene). Following primers were used: for MYC, forward, 5′-CACCATGCCCCTCAACGTTAGCTT-3′, reverse, 5′-TTACGCACAAGAGTTCCG TAG-3′; for constitutively active TGFβ Receptor I, forward 5′-CACCATGGAG GCGGCGGTCGCTGCTCCGCGTCCCCGGCTGCTCCTCCTC-3′, reverse, 5′-CT AGAGGCTAGCATAATCAGGA-3′; for YFP, forward, 5′-CACCATGGTGAGCAA GGGCGAGGA-3′, reverse, 5′-TTACTTGTACAGCTCGTCCATGCC-3′. After verifying the inserts in entry vectors by sequencing, the LR recombination reaction (defined as the recombination between attL and attR sites) was performed with Gateway LR Clonase II enzyme mix (Invitrogen) and specific destination vectors. MYC was cloned into pLenti CMV Puro DEST (Addgene, #17452) (24) vector, for constitutive expression, and into pLenti CMVtight Neo DEST (Addgene, #26432) vector, for tetracycline-inducible expression. FUW-M2rtTA (Addgene, #20342) plasmid was used to for subsequent tetracycline-inducible expression experiments. Constitutively active TGFβ Receptor I, as well as YFP, were cloned into pLenti 6.3/V5-DEST (Invitrogen). One Shot Stbl3 chemically competent E. coli (Invitrogen) were used for transformation and plasmid amplification after the LR reaction. All final insert sequences were verified by sequencing.
Luciferase assay
For luciferase assays, Dual-Luciferase Reporter Assay System (Promega) was used, according to manufacturer’s instructions. Briefly, 48 hours post-transfection the cells, grown in 6-well plates, were lysed by scraping in 250 μl passive lysis buffer. The lysates were cleared by brief centrifugation and 10 μl of lysate was used per measurement. Luminescence measurements were performed in Veritas microplate luminometer (Turner Biosystems), with a 2-second pre-measurement delay and a 10-second signal integration time. The Firefly luciferase activity was normalized to either Renilla luciferase activity. All samples were assayed in triplicates.
Immunoblot analysis
Cells were lysed by scraping in lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA in water) with added Halt phosphatase inhibitor cocktail (Thermo Scientific) and Halt protease inhibitor cocktail (Thermo Scientific). Lysates were cleared by centrifugation at 13 000 rpm × 10 min × 4°C. Protein concentrations were determined using bicinchonic acid (BCA) protein assay kit (Pierce), following manufacturer’s instructions, and using SpectraMax M5 microplate reader (Molecular Devices). Equal protein amounts were loaded onto Mini-Protean TGX 4–20% gradient gel and SDS-PAGE electrophoresis was performed at 150 V. Transfer was done at 100 V for 90 min. using Immobilon-P PVDF membrane with pore size 0.45 μm (Millipore). Membrane was blocked in 5% non-fat dry milk in TBS+1% Tween20 for 90 min. and probed with primary antibody overnight at 4°C. Following primary antibodies were used: c-Myc (9E10) mouse monoclonal (Santa Cruz, #sc-40) 1:250; integrin αv mouse monoclonal (BD Biosciences #611012) 1:5000; integrin β3 mouse monoclonal (BD Biosciences #611140) 1:200; Src rabbit monoclonal (Cell Signaling, #2123) 1:2000; Phospho-Src (Tyr416) rabbit polyclonal (Cell Signaling, #2101) 1:1000; PAI-1 mouse monoclonal (American Diagnostica, #380) 1:200; HA-tag (HRP-conjugated) mouse monoclonal (Cell Signaling #2999) 1:1000. Secondary antibody incubations were performed for 1 hour at room temperature, using: goat anti-mouse IgG (H+L) cross adsorbed secondary antibody at 1:5000 dilution (Thermo Scientific, #31432) or goat anti-rabbit (Invitrogen #656120) 1:7000. GAPDH was used as loading control probing with GAPDH rabbit monoclonal (Meridian, #H86504M) 1:500 000 for 15 min., followed by incubation in secondary goat anti-mouse antibody 1:10 000 for 30 min., both at room temperature. All antibody incubations were performed in blocking buffer. All washes were performed in TBS+1% Tween20. Either Amersham ECL Plus western blotting detection reagent (GE Healthcare) or Clarity western ECL substrate (Bio-Rad) was used for detection. Blue Devil film (Genesee Scientific) was developed in KODAK X-OMAT 2000A (Kodak).
Immunofluorescence
Cells were fixed in 3% paraformaldehyde in PBS for 30 min., permeabilized with 0.2% Triton-X100 in PBS for 5 min., and blocked in 5% non-fat dry milk in PBS for 10 min; all done at room temperature. Washes in between were done with 10mM glycine in PBS. Incubation with primary antibodies was performed overnight at 4°C, followed by secondary antibody incubation for 30 min. at room temperature, both in blocking buffer. Nuclei were stained with Hoechst 33342 (Invitrogen, #H3570) 1:10 000 for 10 min. at room temperature. The following primary antibodies and dilutions were used: anti- vinculin mouse monoclonal antibody (Sigma, #V9131) 1:100, anti-β-tubulin mouse monoclonal (Sigma, #T4026) 1:200. The following secondary antibodies were used: goat anti-mouse Alexa Fluor 488 (Invitrogen, #A11017) 1:200, goat anti-mouse Alexa Fluor 546 (Invitrogen, #A11030) 1:500. Aqua Polymount (Polysciences) was used to mount coverslips on glass slides. Images were acquired using Olympus IX71 microscope, equipped with Olympus LUCPLanFLN objectives (20X NA 0.45, 40X NA 0.6, 60X NA 0.7) and a QuantiFire XI camera (Optronics).
Proliferation assays
Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen, #C10337) was used to directly detect proliferation by immunofluorescence of newly synthesized DNA. Briefly, cells were incubated with 10 μM EdU (5-ethynyl-2′-deoxyuridine) for 2 hours to allow EdU incorporation into the DNA, followed by fixation in 3% paraformaldehyde in PBS for 15 min. and permeabilization in 0.5% Triton-X100 for 20 min. at room temperature. EdU detection was performed by incubation in Click-iT reaction cocktail containing for 30 min at room temperature. Nuclei were stained with Hoechst 33342 (Invitrogen, #H3570) 1:10 000 for 10 min. at room temperature. Images were acquired using Olympus IX71 microscope, an Olympus LUCPLanFLN 20X NA 0.45 objective and a QuantiFire XI camera (Optronics). Proliferation was determined as percentage of EdU positive cells among Hoechst stained cells.
Invasion assay
Cell culture inserts (8 μm pore size, 24-well format) (BD Biosciences) were coated with 50 μg/well growth factor-reduced Matrigel (BD Biosciences) diluted with plain DMEM/F12 medium to a total volume of 100 μl. The invasion chambers were placed in a cell culture incubator for 3 hours, after which 750 μl of complete MCF10A growth medium was added to the bottom wells. 5 × 104 MCF10A cells resuspended in 300 μl assay medium which contained a reduced 2% serum amount and no EGF, as described in (22), were seeded in the invasion chambers and incubated at 37°C. After 24 hours non-invading cells on top of the invasion inserts were removed with a cotton swab. The invaded cells on the bottom of the inserts were fixed with methanol at −20°C for 30 min. and stained with 0.1% crystal violet in 200 mM MES at room temperature for 1 hour. The invasion chambers were photographed using Olympus IX51 microscope equipped with a PlanN 2X NA 0.06 objective, and the total invaded cells per chamber were counted. The presented data represents triplicates ± SEM for each experimental condition.
Flow cytometry analysis and FACS
The cells were trypsinized, counted and washed three times with sterile 2% BSA in PBS. For antibody incubation 1 × 106 cells (scaled up for FACS) was resuspended in 100 μl ‘wash’ buffer and incubated with indicated antibodies for 30 min. at 4°C. Following antibodies were used: APC-CD49f (BioLegend, #313615) 5 μl/reaction, FITC-EpCAM (BD Biosciences, #347197) 20 μl/reaction, FITC-CD49f (BioLegend, #313606) 5 μl/reaction, APC-EpCAM (BD Biosciences, #347200) 5 μl/reaction. The cells were then washed three times and sieved through a cell strainer. Flow cytometry analysis and FACS were performed using FACSAriaII flow cytometer (BD Biosciences).
Orthotopic xenograft model of breast cancer metastasis
All animal procedures were performed under the guidance of an approved Mayo Clinic Institutional Animal Care and Use Committee protocol (protocol no. A330–13). 8–10 week old female NOD/SCID mice were anesthetized with ketamine 100 mg/kg and xylazine 10 mg/kg by intraperitoneal injection. The mice also received buprenorphine for pain relief at 0.1 mg/kg. The abdomen was shaved and sterilized using providone iodine swabsticks (PDI) and ethanol. The left number four mammary fat pad was exposed by performing a small Y-shaped incision in the skin. Suspension of MDA-MB-231 cells was prepared in a mix of 50% DMEM (Gibco) and 50% phenol red-free Matrigel (BD Biosciences). 1 × 105 MDA-MB-231 cells in 50 μl were injected into the number four fat pad using a 28-gauge insulin syringe (BD Biosciences). The incision was closed with 7 mm surgical clips (Cell Point Scientific). Mice were subcutaneously injected with 0.9% sodium chloride (Baxter) and placed on a warming bed during recovery. Mice were sacrificed at 6 weeks post-surgery. Tissues (primary tumors and lungs) were formalin fixed and paraffin embedded for further analysis.
Biophotonic imaging of mice
Mice were injected intraperitoneally with 200 mg/kg body weight of D-luciferin (Gold Biotechnology) in PBS, anesthesized with 1.5–2% isoflurane (Piramal Healthcare) using XGI-8 anesthesia system (Xenogen), and imaged in IVIS 200 Imaging Spectrum System biophotonic imager (Xenogen). Imaging was performed within 5–10 min after injection. A region of interest (ROI) was drawn around each tumor and quantified. At the time of experimental end points animals were injected with D-luciferin prior to sacrifice, and primary tumor and organs were removed and imaged. Analysis was performed using Living Image software (Xenogen). Luminescence is shown as photons/sec/ROI.
Immunohistochemistry
Formalin-fixed, paraffin-embedded primary tumors and lungs were sectioned and stained, either with hematoxylin-eosin or specific antibodies. Briefly, the tissue sections mounted on glass slides were de-paraffinized and rehydrated with distilled water, followed by a 25-minute incubation in Target Retrieval Solution pH 6 (Dako) heated to 100°C. After a 25-minute incubation at room temperature, the sections were blocked in 3% H202 for 5 min, then in serum-free protein block (Dako) for another 5 min. Primary antibody incubations were performed at room temperature for 1 hour. Detection of exogenous TGFBRI used anti-HA tag rat monoclonal (Roche, #11867423001) 1:100. EnVision+ System HRP DAB+ (Dako, #K4007) anti-mouse and Rat-on-Mouse secondary antibody kit (Biocare, #RT517) were used for secondary antibody incubations, performed for 30 min., followed by a 5-minute incubation with the DAB+ chromogen, and counter-stain with hematoxylin, all at room temperature. Stained sections were scanned and images were captured using the T2 ScanScope console and Aperio ImageScope software (Aperio Technologies).
Statistics
Data analysis was performed using Microsoft Excel and GraphPad Prism. Bar graphs represent means ± SEM, as indicated. Student’s t-test or Mann-Whitney test was performed, as indicated, to assess statistical significance.
RESULTS
TGFβ-induced downregulation of MYC expression leads to increased metastasis
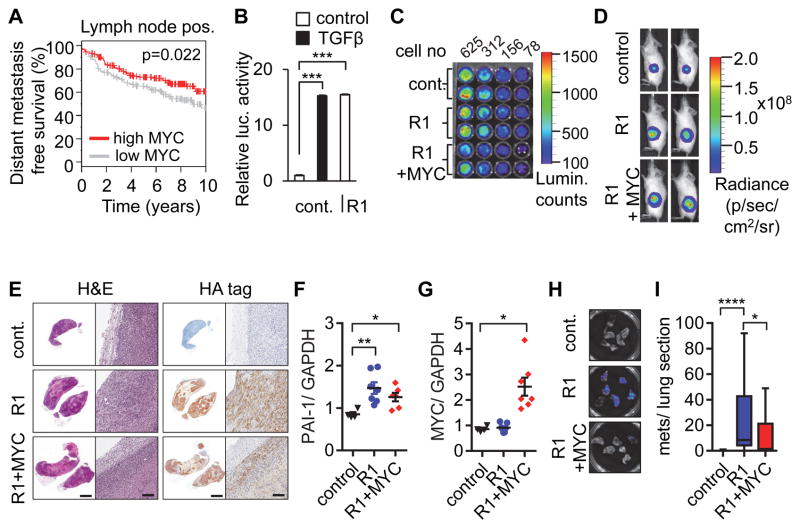
In a recent study we found that MYC inhibits breast cancer metastasis in experimental models(11). Corroborating the clinical relevance of this tumor suppressor activity of MYC, we performed a meta-analysis of published datasets using the Gene expression-based Outcome for Breast cancer Online (GOBO) (25), and found that MYC expression in node-positive breast cancers is associated with improved distant metastasis-free survival (Fig. 1A). Because TGFβ is a key driver of breast cancer metastasis, we hypothesized that MYC downregulation is required for TGFβ-induced breast cancer metastasis, and thus that forced overexpression of MYC would inhibit TGFβ-induced metastasis. We tested this hypothesis using metastatic MBA-MB-231 breast cancer cells in an orthotopic xenograft model. As a surrogate for exogenous TGFβ in the in vivo experiments, we transduced cells with the HA-tagged constitutively active TGFβ Receptor I (CA TGFBR1) (R1) (26), which effectively activated a SMAD-responsive luciferase reporter construct (Fig. 1B). MDA-MB-231 cells were transduced with a retrovirus conferring constitutive expression of MYC or retroviral control, then transduced with CA TGFBR1 lentivirus or control, and then injected into the mammary gland of immunocompromised mice. All cells were additionally transduced with lentiviral firefly luciferase to enable tumor monitoring (Fig. 1C–D). Primary tumors from mice sacrificed after 6 weeks showed sustained tumor expression of HA-tagged constitutively active TGFβ receptor 1 (CA TGFBR1) (Fig. 1E) and increased expression of TGFβ-responsive plasminogen activator inhibitor-1 (PAI-1) transcript levels (Fig. 1F) in tumors transduced with the CA TGFBR1 construct, as well as sustained MYC overexpression in the MYC-transduced cells (Fig. 1G). Consistent with our hypothesis, MYC expression significantly decreased the number of lung metastases induced by CA TGFBR1 (Fig. 1H–I), showing that TGFβ-induced metastasis can be inhibited by expression of MYC.
Figure 1. MYC overexpression inhibits TGFβ-induced metastasis.
(A) High MYC expression is associated with longer metastasis free survival. Kaplan-Meier analysis for n=346 lymph node positive breast cancer patients stratified into two equally sized groups by MYC expression levels. (B) Constitutively active TGFBR1 (R1) acts as a surrogate for exogenous TGFβ in activating TGFβ-responsive luciferase reporter construct. (C) Bioluminescence analysis of decreasing cell numbers of MDA-MB-231 cells transduced with a luciferase construct, with either control or MYC, and control or R1. (D) Growth of orthotopic xenografts was monitored by bioluminescent imaging of the animals (n=7 per group). Representative images of mice after 6 weeks. (E) IHC staining of representative tumors with H&E or an anti-HA tag antibody to detect R1 expression (left image of pair, scale bar 2 mm; right image of pair, scale bar 100 mm). (F–G) Transcript levels of (F) PAI-1 (as an indicator of TGFβ signaling) and (G) MYC expression in primary tumors, presented as relative quantification, normalized to GAPDH. (H) Sample bioluminescence images of lungs. (I) MYC overexpression decreases number of lung metastases induced by constitutively active TGFBR1. Results are presented as number of metastases per section in each group. Error bars, SEM; * p<0.5; ** p<0.01; *** p<0.001; **** p<0.0001.
TGFβ-induced downregulation of MYC is necessary for induction of integrin αvβ3
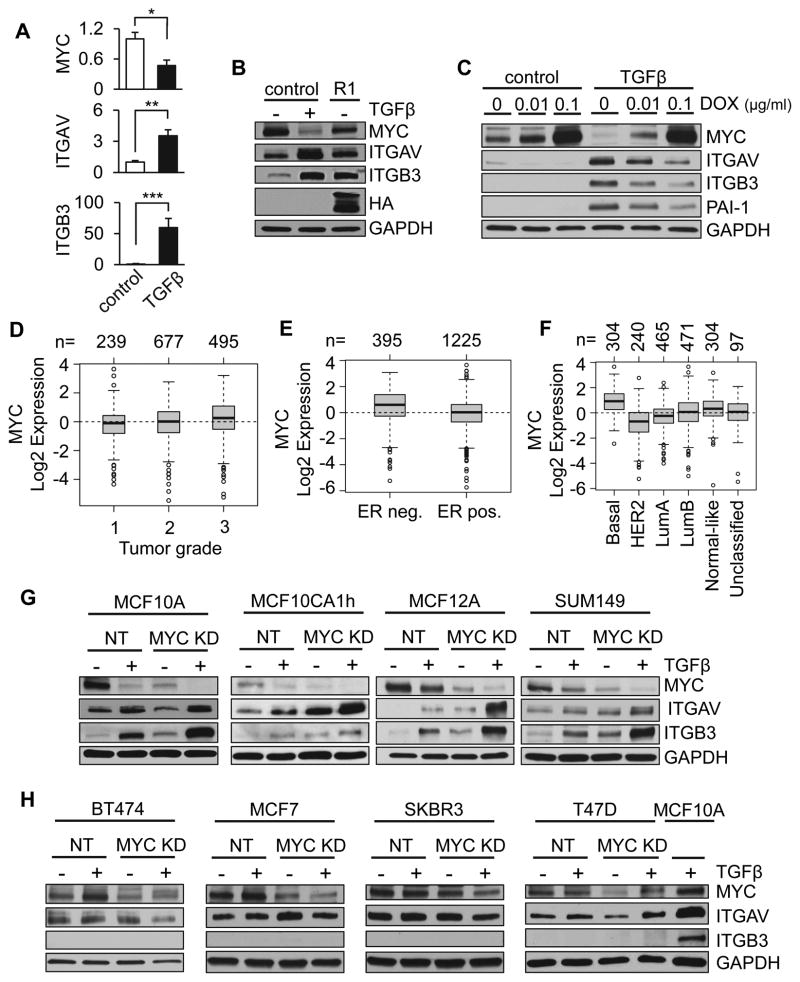
TGFβ is known to be able to downregulate MYC (19, 20), and MYC has been recently shown to decrease expression of integrin αvβ3 in breast cancer models (11). We found that these effects are linked: in MCF10A breast epithelial cells, an established model for TGFβ-dependent regulation of MYC (27), TGFβ treatment led to downregulation of MYC transcription and protein expression, while upregulating transcript and protein levels of both integrin αv and β3 (Fig. 2A). Similar results were obtained by exogenous expression of CA TFBR1 (26), although expression of this transgene was less effective at regulating MYC, ITGAV, and ITGB3 than treatment with TGFβ (Fig. 2B). Of note, exogenous expression of MYC inhibited TGFβ-induced upregulation of integrin subunits αv and β3 in a dose-dependent manner (Fig. 2C), demonstrating that TGFβ-induced upregulation of integrin subunits αv and β3 is dependent upon downregulation of MYC.
Figure 2. TGFβ modulates integrin αvβ3 levels through regulation of MYC in basal subtype breast cancers.
(A) MCF10A cells treated with TGFβ decreased transcription of MYC and increased transcription of αv and β3 integrins. Results are presented as relative quantification, normalized to GAPDH. Error bars, SEM. (B) Constitutively active TGFBR1 (R1) acts as a surrogate for exogenous TGFβ and decreases MYC while increasing integrin αv and β3. (C) MYC overexpression under control of a dox-inducible promoter inhibits TGFβ-induced upregulation of integrin subunits αv and β3 in a dose-dependent manner. (D–F) Meta-analysis of MYC expression using Gene expression-based Outcome for Breast cancer Online (GOBO); number of patients indicated above plots. MYC expression by breast cancer stage (D; p=1e-5), by estrogen receptor (ER) status (E; p<1e-5), and intrinsic subtype classified by PAM50 (F; p<1e-5). (G–H) Expression of MYC, integrin αV (ITGAV), and integrin β3 (ITGB3) in response to MYC knockdown (KD) and TGFβ treatment in basal cell lines MCF10A, MCF10CA1h, MCF12A, and SUM149 (G) and luminal cell lines BT474, MCF7, SKBR3, and T47D (H). Error bars, SEM; * p<0.5; ** p<0.01; *** p<0.001.
TGFβ-dependent regulation of integrin αvβ3 is specific to basal breast cancer cells
To begin to evaluate the clinical impact of TGFβ-dependent downregulation of MYC on breast cancer metastasis, we performed a meta-analysis of MYC expression using the GOBO server, and found that MYC expression was significantly increased with cancer grade (Fig. 2D), and was elevated in ER-negative cancers vs ER-positive cancers (Fig. 2E). Strikingly, MYC expression was significantly elevated in basal breast cancers compared to other subtypes in these datasets (Fig. 2F), as has been noted previously (28, 29). To evaluate the impact of subtype-dependent differences on TGFβ- and MYC-dependent regulation of integrin αvβ3, we tested the effect of MYC knockdown and TGFβ treatment in a selection of breast cancer cell lines. We found that knockdown of MYC in basal subtype MCF10A and breast cancer cell lines led to increased levels of integrin subunits αv and β3 (Fig. 2G), while similar effects were not observed in luminal subtype cell lines (Fig. 2H). Responsiveness of cell lines to TGFβ was verified by checking transcriptional levels of PAI-1 (Supplemental Fig. S1).
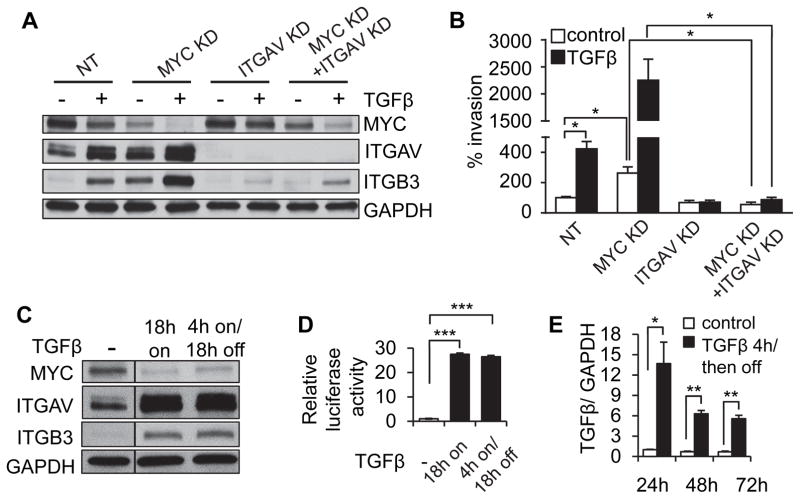
Simultaneous TGFβ treatment and MYC knockdown potentiates cell invasion through highly amplified levels of integrin αvβ3
In evaluating TGFβ treatment and MYC knockdown in basal subtype breast cancer cell lines, we made the unexpected discovery that MYC knockdown can potentiate the TGFβ-induced increase in integrin αv and β3 levels (Fig. 2G, 3A). The striking increase in αv and β3 levels induced by combined TGFβ treatment and MYC knockdown in MCF10A cells was reflected in equally striking potentiation of cellular invasiveness in transwell assays (Fig. 3B; similar results with SUM149 cells, Supplemental Fig. 3). We further found that potentiation of invasion in MCF10A cells was specifically dependent on expression of integrin αvβ3, since the effect could be blocked by knocking down integrin αv (Fig. 3A–B), and we verified that the observed differences in cell invasion were independent of MYC-driven effects on cell proliferation (Fig. 3B; Supplemental Fig. 2A–B), which are not impacted by integrin αv expression level.
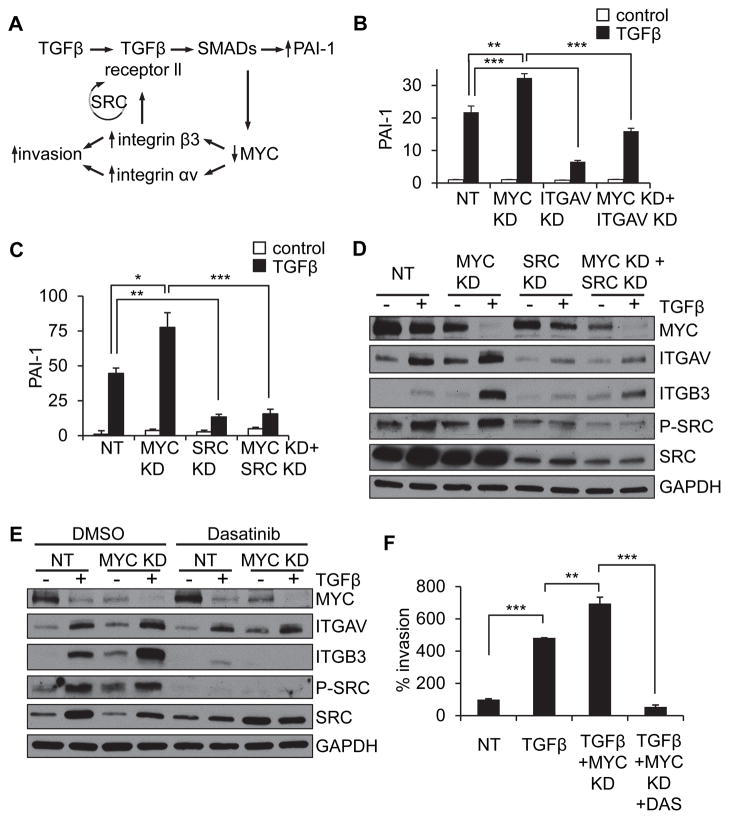
Figure 3. MYC reduction potentiates TGFβ-induced upregulation of integrin αvβ3 and an invasive cellular phenotype.
(A–B) TGFβ treatment and MYC knockdown of MCF10A cells stimulate integrin αvβ3 through a potentiation effect that is dependent upon expression of integrin αvβ3, assessed by protein expression (A) and cell invasion (B). (C–E) MCF10A cells continuously exposed to TGFβ or exposed for a short period of time and then cultured in the absence of exogenous TGFβ showed persistent expression of MYC, integrin αv and integrin β3 (C) and TGFβ-responsive reporter construct (D) through consistent activation of TGFβ expression (E). Transcriptional results are presented as relative quantification, normalized to GAPDH. Error bars, SEM; * p<0.5; ** p<0.01; *** p<0.001.
The invasion assays of TGFβ pre-treated cells were performed in the absence of exogenous TGFβ; thus, the observed TGFβ-stimulated invasion suggested a persistent mechanism of signaling after its removal from the media. We found that the effects of TGFβ on downregulation of MYC, upregulation of integrin αvβ3, and SMAD-dependent signaling in MCF10A cells persisted after removing exogenous TGFβ from the cell culture medium (Fig. 3C–D), and were associated with endogenous TGFβ transcription that persisted once induced by exogenous TGFβ (Fig. 3E). These results suggest that the effects of TGFβ on MYC-dependent inhibition of proliferation and increased invasion, once induced, will require active targeting of susceptible pathways to reverse.
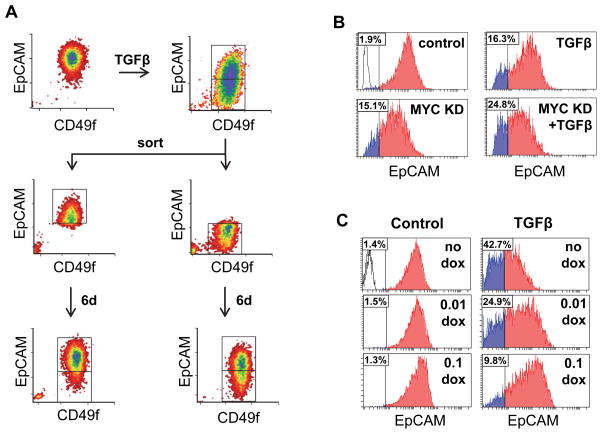
Treatment of mammary epithelial cells with TGFβ has been shown to induce features of a claudin-low phenotype (30, 31), including highly increased invasiveness, acquisition of progenitor-like characteristics, and altered expression of many transcripts, including downmodulation of the epithelial marker EpCAM (32). Flow-analysis of MCF10A cells treated 3d with TGFβ revealed the induction of an EpCAM-low population; when the TGFβ–treated cells were sorted by EpCAM expression and cultured in the absence of TGFβ for six additional days, we found that the EpCAM-low-population differentiated into EpCAM-high parental-like cells, while the EpCAM-high cells maintained their differentiated status (Fig. 4A). We further found that MYC KD in MCF10A cells induced a similar induction of the EpCAM-low population and that this was potentiated by TGFβ (Fig. 4B), and that the TGFβ-induced effect was blocked by dox-dependent exogenous expression of MYC (Fig. 4C). These results suggest that modulation of MYC levels is fundamental to the phenotypic alterations induced by TGFβ
Figure 4. MYC mediates TGFβ-induced generation of EpCAM-high and Ep-CAM-low cell populations.
(A) MCF10A cells exposed to TGFβ for 3d were sorted into EpCAM-high and EpCAM-low populations, cultured for six additional days in the absence of TGFβ, and then reanalyzed for EpCAM expression. (B) TGFβ treatment and MYC knockdown for 3d stimulate development of the EpCAM-low population of MCF10A cells. (C) MYC overexpression under control of a dox-inducible promoter inhibits TGFβ-induced development of the EpCAM-low population in a dose-dependent manner after 4d treatment of MCF10A cells. Insets in histograms indicate percent of population below threshold.
MYC knockdown potentiates TGFβ-induced, integrin αvβ3-mediated invasion in a SRC-dependent mechanism
It has been previously shown that integrin αvβ3 can lead to SRC activation and that integrin β3-dependent SRC activation can lead to activation of TGFβ signaling (33–38). Based on these previous findings, we hypothesized that decreased MYC and consequent increased expression of integrin β3 could activate a SRC-dependent positive feedback loop (Fig. 5A). Consistent with this hypothesis, we found that MYC knockdown led to potentiation of TGFβ-induced increase of the SMAD-dependent PAI-1 gene expression levels in MCF10A cells, and that this effect could be inhibited by reduction of integrin αvβ3 (Fig. 5B) or SRC (Fig 5C). We also found that the potentiating effect of MYC knockdown on TGFβ-induced integrin αv and β3 expression could be inhibited either by SRC knockdown (Fig. 5D), or by pharmacological inhibition of SRC using Dasatinib (Fig. 5E, Supplemental Fig. 4A), or by the alternative SRC inhibitor PP2 (Supplemental Fig. 4B). Furthermore, cellular invasion induced by MYC knockdown and TGFβ treatment could also be inhibited by Dasatinib (Fig. 5F).
Figure 5. MYC knockdown potentiates TGFβ-induced upregulation of integrin αvβ3 through a SRC-dependent mechanism.
(A) A schematic of the proposed model of signaling. (B–C) TGFβ/MYC KD potentiated PAI-1 expression is blocked by ITGAV KD (B) or SRC KD (C) in MCF10A cells. Transcriptional results are presented as relative quantification, normalized to GAPDH (C,D) SRC knockdown (D) or treatment with SRC kinase inhibitor Dasatinib (E), inhibit MYC knockdown-induced potentiation of TGFβ-induced increase in integrin αvβ3 in MCF10A cells. (F) Dasatinib inhibits MYC knockdown-induced potentiation of TGFβ-induced invasion in MCF10A cells. Error bars, SEM; * p<0.5; ** p<0.01; *** p<0.001.
Pharmacological MYC inhibition upregulates integrin αvβ3 and increases cell invasion in a SRC-dependent manner
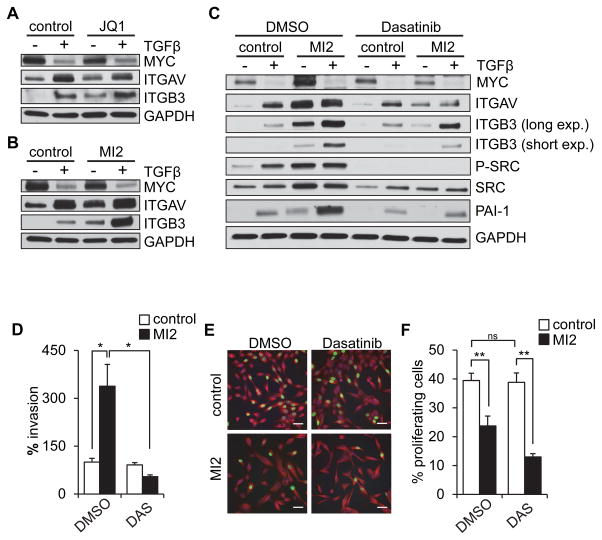
As inhibition of MYC represents a potential therapeutic strategy for breast cancer, we next considered whether pharmacological inhibitors of MYC might also potentiate αvβ3-driven cellular invasion. We evaluated the effects of two different MYC inhibitors on MCF10A cells. Similar to the effects with MYC knockdown, we found that both JQ1 (Fig. 6A) and MI2 (Fig. 6B) led to dose-dependent increases in αv and β3 integrin subunits (Supplemental Fig. 5A–C), with comparable results found with another basal subtype cell line MCF12A (Supplemental Fig. 5D). Also similar to the effects of MYC knockdown, pharmacological inhibition of MYC with Dasatinib potentiated TGFβ-induced upregulation of αvβ3 in a SRC-dependent fashion in MCF10A cells (Fig. 6C). Furthermore, MYC inhibition with MI2 stimulated cell invasion, and this effect could also be blocked by Dasatinib (Fig. 6D) which also enhanced the antiproliferative effects of MYC inhibition (Fig 6E–F). These results suggest that inhibition of MYC in basal breast cancer could have optimal therapeutic benefit when combined with therapeutic inhibition of SRC.
Figure 6. Pharmacological MYC inhibition upregulates integrin αvβ3 and increases cell invasion in a SRC-dependent fashion.
(A–B) MYC inhibition by JQ1 (A) and MI2 (B) increase αv and β3 protein levels and potentiate the TGFβ-induced effect in MCF10A cells. (C) Potentiation of TGFβ-induced upregulation of integrin αv and β3 by MI2-inhibited MYC is blocked by Dasatinib in MCF10A cells. (D) Dasatinib (DAS) blocks MI2-dependent activation of invasion. (E–F) Dasatinib further activates MI2-dependent inhibition of proliferation in MCF10A cells, assessed by EdU (images, E; quantification, F). Scale bars, 50 μm. Error bars, SEM; * p<0.05; ** p<0.01.
DISCUSSION
Basal subtypes of breast cancer have poor prognosis, with high breast cancer mortality within the first few years after diagnosis and the highest risk of recurrence after therapy (39, 40). The poor outcomes for basal breast cancer patients are related to the lack of available targeted therapies, by contrast with the more common luminal subtypes for which standard therapies have been developed, as also to the intrinsically aggressive nature of the basal breast cancer disease (41). As a consequence, considerable effort has been devoted to identifying signaling pathways in basal subtype tumors that can be therapeutically targeted with sufficient specificity so as to minimize adverse effects in normal tissue, towards development of personalized therapy for women with basal breast cancer. Here, we have identified a signaling axis connecting TGFβ, MYC, and integrin αvβ3 that controls both cellular proliferation and invasiveness, the key features of malignancy that underlie mortality, and we show that this pathway is specific for basal breast cancer cells. We found that TGFβ-induced suppression of MYC transcript and protein levels specifically increased abundance of integrin αvβ3, thus promoting invasion and metastasis (Fig. 1, 2). We demonstrated that knockdown or pharmacological inhibition of MYC effectively activates the feedback loop driving invasive and metastatic properties in basal breast cancer cells (Fig. 2–6), that this feedback loop can be interrupted by SRC inhibition (Fig. 5), and that pharmacological inhibition of SRC is sufficient to inhibit the pro-invasive effects while maintaining the antiproliferative characteristics of MYC inhibitors (Fig. 6). Our findings have important implications for using MYC as a therapeutic target and suggest that in certain cases MYC inhibition could potentially result in deleterious clinical responses, by promoting increased tumor metastasis. Based on our findings we propose a novel therapeutic strategy for patients who have the basal subtype of breast cancer through simultaneous targeted inhibition of MYC and SRC. We have demonstrated that this strategy has the potential to block the proliferative capacity of MYC while abrogating the activation of invasion and metastasis mediated through SRC signaling.
We found that the TGFβ/MYC-dependent regulation of integrin αvβ3, including the potentiating effects of MYC knockdown and active TGFβ signaling, were specific to basal subtype breast cancer cells lines (Fig. 2G,H). In luminal cell lines not only did TGFβ fail to downregulate MYC and increase levels of integrin αv or β3, but MYC knockdown itself had no effect on integrin αvβ3 (Fig. 2H). One possible contributor to the observed specificity of this signaling axis is the increased expression of integrin β3 in basal breast cancer cell lines as compared to luminal-derived cell lines; of note, we have previously reported that integrin β3 is required for MYC-knockdown-induced cellular invasiveness (11).
One limitation of the study is the focus on the basal subtype of breast cancer, while at present clinical decisions are made on the basis of histological analyses of marker expression. Breast cancers that lack expression of ER, PR, and Her2, designated as triple-negative cancers, are frequently also basal subtype, while the majority of basal subtype cancers are also histologically triple negative (41). At present, the biology and treatment of basal subtype cancers that express hormone receptors or Her2 has not been clearly defined, although preliminary data suggesting that basal subtype tumors that are not triple negative behave similarly to the larger population of triple negative basal subtype tumors (42). Another limitation is that it is not clear whether the TGFβ-MYC-Integrin αvβ3 prometastatic pathway is limited to breast cancers, or whether it may also be found in tumors developing in other organs. Systematic analyses directly comparing multiple cancer types have confirmed that basal subtype breast cancers are substantially different from luminal subtype breast cancers, reinforcing concepts that these represent very different diseases with distinct etiologies (43). These studies suggest that basal breast cancers share many molecular similarities with serous ovarian cancers, including a central role for MYC signaling; the potential that the TGFβ-MYC-Integrin αvβ3 signaling axis could exist in serous ovarian cancer cells offers an intriguing direction for future study.
Our results have particular relevance for clinical studies which therapeutically target MYC, particularly for basal breast cancer patients. The majority of current clinical trials as listed on www.clinicaltrials.gov that target MYC employ agents that inhibit bromodomain and extra-terminal (BET) family members. Selective inhibitors of BET bromodomains, of which the agent JQ1 used in our investigation is one of the best studied (44), constitute a family of agents that regulate a broad range of transcriptional events, although inhibition of MYC transcription is believed to underlie their therapeutic effect (45, 46). Several other current clinical trials are testing the utility of DCR-MYC, a small interfering RNA targeting MYC (47), and these have shown promising initial results for patient tolerance (10). Our findings predict that while these agents may effectively inhibit basal breast cancer cell proliferation, there is likely to be increased incidence of relapse with metastasis. Importantly, our results point also to a solution to this problem, in that inhibitors of MYC expression can be combined with agents that target SRC, such as dasatinib. While dasatinib has shown limited efficacy as a single agent for triple negative (48) and hormone receptor positive breast cancer patients (49), our results suggest that combination therapy of a MYC inhibitor with dasatinib could confer both antiproliferative and antimetastatic effects. Clinical trials evaluating such a combination therapy in basal breast cancer patients will be required to test this strategy.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the NCI (CA187692) and the Mayo Clinic Breast Cancer SPORE (CA116201).
Footnotes
Author Contributions
D.C.R. supervised the study. M.A.C. and D.C.R. conceived and designed experiments and developed methodology. M.A.C.., M.E.M., T.A.S., E.M., C.M., S.P.E., J.A.C., E.S.R., and D.C.R. analyzed and interpreted data, and wrote the manuscript.
Conflict of Interest: No potential conflicts of interest to disclose.
References
- 1.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Prat A, Karginova O, Parker JS, Fan C, He X, Bixby L, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat. 2013;142:237–55. doi: 10.1007/s10549-013-2743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–67. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 6.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer. 2010;1:629–40. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015 doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 10.Tolcher AW, Papadopoulos KP, Patnaik A, Rasco DW, Martinez D, Wood DL, et al. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J Clin Oncol (Meeting Abstracts) 2015;33:11006. [Google Scholar]
- 11.Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of alphav and beta3 integrin subunits. Nat Cell Biol. 2012;14:567–74. doi: 10.1038/ncb2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013;2:47–63. doi: 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- 15.Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, et al. TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest. 2003;112:1116–24. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–99. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor MA, Lee YH, Schiemann WP. Role of TGF-beta and the tumor microenvironment during mammary tumorigenesis. Gene Expr. 2011;15:117–32. doi: 10.3727/105221611x13176664479322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietenpol JA, Holt JT, Stein RW, Moses HL. Transforming growth factor beta 1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc Natl Acad Sci U S A. 1990;87:3758–62. doi: 10.1073/pnas.87.10.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagi K, Furuhashi M, Aoki H, Goto D, Kuwano H, Sugamura K, et al. c-myc is a downstream target of the Smad pathway. J Biol Chem. 2002;277:854–61. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- 21.Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol Cell Biol. 2004;24:2546–59. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 23.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–71. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wieser R, Wrana JL, Massague J. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. EMBO J. 1995;14:2199–208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CR, Kang Y, Massague J. Defective repression of c-myc in breast cancer cells: A loss at the core of the transforming growth factor beta growth arrest program. Proc Natl Acad Sci U S A. 2001;98:992–9. doi: 10.1073/pnas.98.3.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandriani S, Frengen E, Cowling VH, Pendergrass SA, Perou CM, Whitfield ML, et al. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS One. 2009;4:e6693. doi: 10.1371/journal.pone.0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruna A, Greenwood W, Le Quesne J, Teschendorff A, Miranda-Saavedra D, Rueda OM, et al. TGFbeta induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun. 2012;3:1055. doi: 10.1038/ncomms2039. [DOI] [PubMed] [Google Scholar]
- 31.Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8:e1002723. doi: 10.1371/journal.pgen.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desgrosellier JS, Barnes LA, Shields DJ, Huang M, Lau SK, Prevost N, et al. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15:1163–9. doi: 10.1038/nm.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galliher AJ, Schiemann WP. Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 2006;8:R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galliher AJ, Schiemann WP. Src phosphorylates Tyr284 in TGF-beta type II receptor and regulates TGF-beta stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res. 2007;67:3752–8. doi: 10.1158/0008-5472.CAN-06-3851. [DOI] [PubMed] [Google Scholar]
- 36.Parvani JG, Galliher-Beckley AJ, Schiemann BJ, Schiemann WP. Targeted inactivation of beta1 integrin induces beta3 integrin switching, which drives breast cancer metastasis by TGF-beta. Mol Biol Cell. 2013;24:3449–59. doi: 10.1091/mbc.E12-10-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parvani JG, Taylor MA, Schiemann WP. Noncanonical TGF-beta signaling during mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2011;16:127–46. doi: 10.1007/s10911-011-9207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor MA, Parvani JG, Schiemann WP. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-beta in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 2010;15:169–90. doi: 10.1007/s10911-010-9181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engebraaten O, Vollan HK, Borresen-Dale AL. Triple-negative breast cancer and the need for new therapeutic targets. Am J Pathol. 2013;183:1064–74. doi: 10.1016/j.ajpath.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 40.Hattangadi-Gluth JA, Wo JY, Nguyen PL, Abi Raad RF, Sreedhara M, Niemierko A, et al. Basal subtype of invasive breast cancer is associated with a higher risk of true recurrence after conventional breast-conserving therapy. Int J Radiat Oncol Biol Phys. 2012;82:1185–91. doi: 10.1016/j.ijrobp.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross SJ, Critchlow SE. Emerging approaches to target tumor metabolism. Curr Opin Pharmacol. 2014;17:22–9. doi: 10.1016/j.coph.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Finn RS, Bengala C, Ibrahim N, Roche H, Sparano J, Strauss LC, et al. Dasatinib as a single agent in triple-negative breast cancer: results of an open-label phase 2 study. Clin Cancer Res. 2011;17:6905–13. doi: 10.1158/1078-0432.CCR-11-0288. [DOI] [PubMed] [Google Scholar]
- 49.Mayer EL, Baurain JF, Sparano J, Strauss L, Campone M, Fumoleau P, et al. A phase 2 trial of dasatinib in patients with advanced HER2-positive and/or hormone receptor-positive breast cancer. Clin Cancer Res. 2011;17:6897–904. doi: 10.1158/1078-0432.CCR-11-0070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.