Abstract
Purpose
Inflammatory marker expression in stage III melanoma tumors was evaluated for association with outcome, using two independent cohorts of stage III melanoma patients’ tumor tissues.
Experimental Design
Fifteen markers of interest were selected for analysis, and their expression in melanoma tissues was determined by immunohistochemistry. Proteins associating with either overall survival (OS) or relapse-free survival (RFS) in the retrospective discovery tissue microarray (TMA) (n = 158) were subsequently evaluated in an independent validation TMA (n = 114). Cox proportional hazards regression models were used to assess the association between survival parameters and covariates, the Kaplan-Meier method to estimate the distribution of survival, and the log-rank test to compare distributions.
Results
Expression of CD74 on melanoma cells was unique, and in the discovery TMA it associated with favorable patient outcome (OS: HR, 0.53, P = 0.01 and RFS: HR, 0.56; P = 0.01). The validation dataset confirmed the CD74 prognostic significance and revealed that the absence of MIF and iNOS was also associated with poor survival parameters. Consistent with the protein observation, tumor CD74 mRNA expression also correlated positively (p = 0.003) with OS in the melanoma TCGA dataset.
Conclusions
Our data validate CD74 as a useful prognostic tumor cell protein marker associated with favorable RFS and OS in stage III melanoma. Low or negative expression of MIF in both TMAs, and of iNOS in the validation set also provided useful prognostic data. A disease-specific investigation of CD74’s functional significance is warranted, and other markers appear intriguing to pursue.
INTRODUCTION
At this time, the validated prognostic factors in patients with stage III melanoma are clinical and pathological features: (1) specifically, the pattern of regional metastases (lymph node and/or in-transit), the size and number of regional metastases, and the ulceration status of the primary tumor (2). As outcomes remain widely variable for stage III melanoma patients, it is critical to identify clinically useful molecular markers that may improve our risk models, thereby increasing our ability to provide better clinical care.
Previous studies suggested that intrinsic tumor cell inflammatory mediators are associated with poor prognosis, yet no molecular tumor markers have been proven to be prognostic. To date, one tumor infiltrating lymphocyte (TIL) molecular marker, (CD2; T and NK cells identified) was identified as associating with favorable outcomes, heralding an era of molecular marker usage (3–5) in melanoma. In our own studies, we have previously observed impaired outcomes in patients with constitutive expression of inducible nitric oxide synthase (iNOS) and associated production of nitric oxide (NO) in melanoma cells (6,7). From the work of us and many others, a cohesive model is emerging in which poor clinical outcome is associated with varying concentrations of cellular NO in many cancers (8–14) including stage III melanoma (12,15–16). Currently, immunohistochemical evaluation of iNOS is routinely performed at the Clinical Laboratory Improvement Amendments (CLIA)-approved laboratory in our institution (MD Anderson Cancer Center) to inform clinical trials; however, determining the power of this marker compared with that of other markers, is a priority at this time. Therefore in this study, we applied the REporting recommendations for tumor MARKer prognostic studies (REMARK) criteria (17) to test the prognostic value of a set of inflammatory markers, using separate discovery and validation tissue microarrays (TMA) associated with known long-term survival data. Our analyses have determined that iNOS and MIF expression in patient melanoma cells had varying statistical association with poor outcome; however a strong favorable patient outcome was indicated by the expression of CD74, which we now report as a new prognostic marker for patients with stage III melanoma.
MATERIALS AND METHODS
Patient Samples and Data Sets
Discovery and Validation Data Sets
Two independent TMAs of stage III melanoma (regional metastases) were employed. Under the IRB-approved protocols, patients were identified whom archival material was available, and that had clinical follow-up of at least 5 years for surviving patients. Patient data and primary tumor characteristics were collected for all patients. Patient records were reviewed for clinical events including development of local recurrence, development and pattern(s) of distant metastasis, and overall survival. Patient demographics and survival status are detailed in Table 1. In accordance with REMARK criteria, the TMA from the John Wayne Cancer Institute (JWCI) served as the discovery set in which candidate biomarkers were identified. This TMA was prepared from formalin-fixed paraffin-embedded (FFPE) lymph node tumors, and contained two core samples from each specimen; tumors from 158 patients with defined clinical outcomes and follow-up data (15). The associations between markers and survival outcomes were then evaluated in an independent and more contemporary TMA of sequential stage III patients’ tumors (n = 114) from MD Anderson Cancer Center (MDACC). The MDACC TMA served as the validation (17) and was prepared as triplicate core samples from patients who had undergone standard-of-care lymphadenectomy for stage IIIB/C melanoma and for whom clinical data were available. While MD Anderson Cancer Center patients often later received several adjuvant therapies, including Ipilimumab, BRAF inhibitors, vaccine, biochemotherapy; John Wayne Cancer Institute patients predominantly received additional surgeries or an experimental vaccine.
Table 1.
Patient Demographics and Survival Status
| Discovery Set (JWCI) Measure | All Patients (N=158) |
|---|---|
|
| |
| Gender, n (%) | |
| Male | 90 (57.0) |
| Female | 68 (43.0) |
|
| |
| Age at Surgery (years), median (range) | 51.9 (11.5–94.7) |
|
| |
| Positive Lymph Node, n (%) | |
| 1 | 53 (33.5) |
| 2–3 | 34 (21.5) |
| ≥4 | 71 (45.0) |
|
| |
| Ulceration, n (%) | |
| Yes | 31 (29.5) |
| No | 74 (70.5) |
| Missing | 53 |
|
| |
| Breslow thickness (mm) | |
| N | 115 |
| Median (range) | 2.0 (0.35–15.00) |
|
| |
| Overall Survival, n (%) | |
| Alive | 52 (32.9) |
| Dead | 106 (67.1) |
|
| |
| Recurrence-Free Survival, n (%) | |
| Alive with no disease | 22 (13.9) |
| Dead and/or disease progressed | 136 (86.1) |
| Validation Set (MDACC) Measure | All Patients (N=114) |
|---|---|
|
| |
| Gender, n (%) | |
| Male | 79 (69.3) |
| Female | 35 (30.7) |
|
| |
| Age at Surgery (years), median (range) | 55.4 (25.0–85.4) |
|
| |
| Positive Lymph Nodes, n (%) | |
| 0–1 | 23 (20.2) |
| 2–3 | 34 (29.8) |
| ≥4 | 57 (50.0) |
|
| |
| Ulceration, n (%) | |
| Yes | 64 (79.0) |
| No | 17 (21.0) |
| Missing | 33 |
|
| |
| Breslow thickness (mm) | |
| N | 95 |
| Median (range) | 3.0 (0.25–33.00) |
|
| |
| Overall Survival, n (%) | |
| Alive | 30 (26.3) |
| Dead | 84 (73.7) |
|
| |
| Recurrence-Free Survival, n (%) | |
| Alive with no disease | 12 (10.5) |
| Dead and/or disease progressed | 102 (89.5) |
Biomarker Assessments
A set of 15 inflammatory protein markers were selected in part based on their involvement with cancer inflammation by us or others (13,18–19). The final list was also dependent on availability of antibodies optimized for use in IHC analysis in this study, having been previously tested and optimized for specificity and validated with use of tumor tissue and controls (see, Supplementary Material S1). Therefore, this is not an inclusive nor in any means a complete list of such markers, and we continue to test and validate additional antibodies for use in IHC analysis to expand similar testing.
The protein markers assessed for this report were arginase, IL-1α, IL-1β, IL-24, COX-2, CD70, CD74, iNOS, macrophage migration inhibitory factor (MIF), CXCL9, CXCL10, CXCR3, and Ki-67. NO-driven post-translational modifications (PMTs) were also tested since antibodies to these have recently become available and are of much interest to our work on iNOS; these were Nitrotyrosine (NT) and S-nitroso-cysteine (SNO) and therefore serve as surrogate markers of NO production and ONOO- availability.
Immunolabeling was scored separately for two variables: [1] percentage of positively stained cells; and [2] intensity of staining of the positive cells as considered standard by many pathologists and used by us previously (20). The complete absence of protein markers was defined as <5% of tumor cells (or infiltrating lymphocytes) expression when performed in parallel with positive expression in controls, such as endothelial cells in the same tissues. Thus, expression of a given protein marker was considered positive if the average positivity was at least 5% and evident intensity of the staining of which quantifications detailed below. For each patient sample, two (JWCI) or three (MDACC) independent cores were read on the respective TMA, and for each of these cores two independent readings were performed, and results averaged. Percentage scoring was considered positive if the average positivity was at least 5%. Intensity scoring staining was defined as follows: “0,” none; “1,” light; “2,” moderate; and “3,” intense, and reported as positive if an average score of at least 1 was obtained.
All IHC analyses were independently interpreted by two readers without knowledge of the clinical data. Discrepancies in scoring were reconciled by a consensus reading between the readers.
Public TCGA data repositories (https://tcga-data.nci.nih.gov and http://gdac.broadinstitute.org/) were used to obtain Skin Cutaneous Melanoma (SKCM) sample data. Expression of mRNA for the selected markers and the associated patient clinical data were analyzed (21). Normal tissue gene expressions (2921 samples) were downloaded from the GTEx Portal (http://www.gtexportal.org). We converted the expression values to transcripts per million (TPM), which was found to be the most comparable metric across samples and experiments (22).
Statistical Analysis
Two time-to-event outcomes were considered: overall survival (OS), and relapse free survival (RFS) which was defined as local/regional or distant relapse. Both endpoints were calculated from the date of lymph node surgery. For OS, patients who remained alive were censored at the date of their last follow-up. For RFS, disease-free patients were also censored at the date of last follow-up.
Univariate Cox proportional hazards regression models were fit to determine the association between each marker and each survival outcome. Multivariable Cox models were fit for each marker that adjusted for disease and demographic covariates. The Kaplan-Meier method was used to estimate the distribution of survival times, and the log-rank test was used to compare distributions. Statistical analyses were performed with use of R v. 3.1.1 and SAS version 9.3 for Windows. All statistical tests used a significance level of 5%. No adjustments for multiple testing were made.
RESULTS
Patient Characteristics
Table 1 details the characteristics of the two patient cohorts used to prepare the discovery and validation TMAs from JWCI and MDACC respectively. Response Evaluation Criteria in Solid Tumors (RECIST) criteria (23) were used to define progression after therapy for both cohorts. The cohorts were similar; in the MDACC set, 69% of the patients were male and the median age at surgery was 55 years, in the JWCI set, 57% were male with a median age of 52 years. Ulceration status of the primary tumor was lacking in approximately one-third of both sets, and mutated BRAF status was available only from the MDACC cohort. Fortunately both cohorts had significant follow-up periods of more than 10 years, which provided a rich data set, with 86% of JWCI patients progressing or dead as were 90% of the patients in the MDACC cohort. Although both TMAs were prepared from tumor biopsy specimens from stage III patient, the MDACC TMA consisted primarily of stage IIIB/C patients for which larger tumor samples were available. As a result, the MDACC cohort contained more patients with advanced disease, overall.
Univariate Analysis
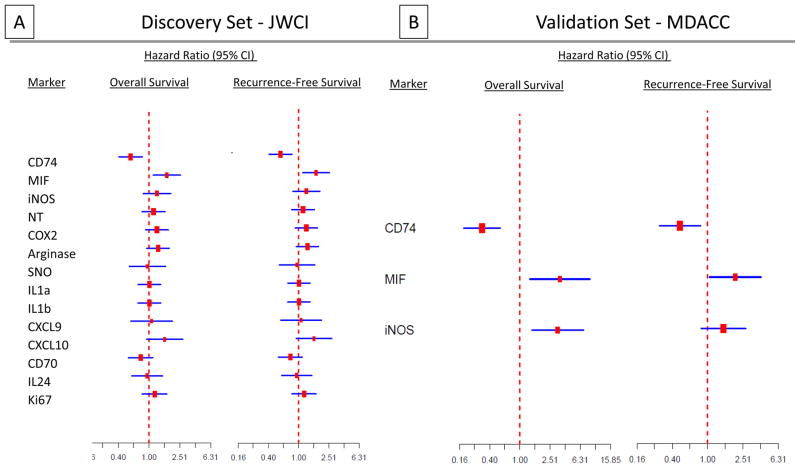
The strategy and the flow of samples for analyses are presented in Supplementary Material S2. Results of Cox proportional hazards regression models for the associations of the 15 markers with OS and RFS in the JWCI discovery set are shown in Figure 1A (intensity) and Supplementary Figure S1A (percentage). Tumor CD74 staining intensity (Hazard Ratio [HR] 0.45; P < 0.001) and staining percentage (HR 0.46, P =< 0.001), both correlated significantly with a lower risk of death and disease recurrence or death (HR 0.58, P = 0.002 for intensity; HR 0.51, P = 0.002 for percentage). Tumor cell MIF expression intensity ((HR 1.57, P = 0.06) and percentage (HR 1.63; P = 0.05) were marginally associated with poorer OS, and significantly associated with worse RFS (HR 1.70, P = 0.01 for intensity; HR 1.60; P = 0.026 for percentage) (Figure 1A; Supplementary Figure S1A).
Figure 1.
Univariate analyses of survival models with individual markers in discovery and validation datasets. Forest plots of OS and RFS hazard ratios and associated 95% confidence intervals for specific protein expression from univariate Cox proportional hazards regression models presented as intensity of staining in discovery (A) and validation (B) datasets
The expression of CD74 and of MIF in tumor was then assessed for association with OS and RFS in the validation TMA (Figure 1B; Supplementary Figure S1B). As in the discovery set, CD74 expression in tumor cells correlated with both improved OS (HR 0.32; P < 0.001 for intensity; HR 0.33, P < 0.001 for percentage) and RFS (HR 0.49, P = 0.008 for both). MIF expression in tumor cells was also significantly associated with poorer OS (HR 3.39, P = 0.009 for intensity; HR 4.86; P < 0.001 for percentage) and RFS (HR 2.05, P = 0.035 for intensity; HR 1.92; P = 0.038 for percentage). Staining percentage and intensity of CD74 and MIF in all samples is summarized in Supplementary Table S1.
As we previously observed significant associations of tumor iNOS expression and disease progression in an earlier cohort of MDACC melanoma patients (12,13), it was noted that such an association was observed in the MDACC TMA (data not shown); iNOS percentage and intensity in tumor cells was again significantly associated with OS (HR 3.18, P = 0.004 for intensity; HR=3.74, P = 0.002 for percentage) but not with RFS (HR 1.51, P = 0.165 for intensity; HR 1.64, P = 0.103 for percentage. However, iNOS percentage and intensity in tumor cells was not significant in the JWCI discovery TMA. We concluded that iNOS remains of interest, but clearly not as a powerful prognostic marker of either CD74 positivity or MIF negativity.
Multivariable Analysis
Multivariable Cox models were fit to assess the association of OS and RFS for all patients in the JWCI cohort, with variables of gender, age, number of positive lymph nodes, primary tumor Breslow thickness (BT), and CD74 and MIF intensity (Table 2). CD74 intensity in tumor remained a significant predictor of OS (HR 0.53, P = 0.01) and RFS (HR 0.56; P = 0.01). MIF intensity in tumor was not significantly associated with OS (P = 0.08), but it was significantly associated with RFS (HR 1.85, P = 0.02). Among the other characteristics, only having ≥4 lymph nodes involved was significantly associated with OS (HR 2.04, P = 0.01)
Table 2.
Multivariable Cox Analyses on Survival Models in Discovery Set (N=103)
| Multivariable Cox Regression Model | Overall Survival | Recurrence-Free Survival | |||||
|---|---|---|---|---|---|---|---|
| Parameter | Level | HR | 95% CI | P-value | HR | 95% CI | P-value |
| CD74 Tumor Intensity | Positive | 0.53 | 0.32, 0.88 | 0.01 | 0.56 | 0.35, 0.89 | 0.01 |
| MIF Tumor Intensity | Positive | 1.67 | 0.94, 2.96 | 0.08 | 1.85 | 1.10, 3.10 | 0.02 |
| Breslow Thickness | 0–1 | ---- | ---- | ---- | ---- | ---- | ---- |
| >1–2 | 0.68 | 0.35, 1.34 | 0.27 | 0.62 | 0.34, 1.13 | 0.12 | |
| >2–4 | 1.44 | 0.74, 2.79 | 0.29 | 0.82 | 0.45, 1.50 | 0.51 | |
| >4 | 0.90 | 0.46, 1.75 | 0.75 | 0.83 | 0.46, 1.52 | 0.55 | |
| Positive Lymph Nodes | 1 | ---- | ---- | ---- | ---- | ---- | ---- |
| 2–3 | 1.29 | 0.62, 2.70 | 0.49 | 0.98 | 0.50, 1.92 | 0.95 | |
| >=4 | 2.04 | 1.19, 3.50 | 0.01 | 1.71 | 1.05, 2.79 | 0.03 | |
| Age at Surgery | Continuous, Per Year | 1.02 | 1.001, 1.03 | 0.03 | 1.01 | 0.99, 1.02 | 0.35 |
| Gender | Male | 1.15 | 0.69, 1.94 | 0.59 | 1.16 | 0.73, 1.84 | 0.54 |
HR: Hazard Ratio; CI: Confidence Interval
Multivariable Cox proportional hazards regression analysis was also performed for these same markers in the MDACC cohort (Table 3). Positive CD74 expression in tumor cells was again significantly associated with OS (HR 0.27, P < 0.001) and RFS (HR 0.41, P = 0.005). Tumor cell MIF intensity was also significantly associated with both inferior OS (HR 3.02, P = 0.023) and RFS (HR 3.60, P = 0.003). Furthermore, iNOS intensity in tumor was now significantly associated with OS (data not shown; HR 2.82, P = 0.024). No other clinical or tumor factors were significantly associated with OS or RFS in this cohort. BRAF mutation data were available only for the MDACC validation cohort, so in an exploratory analysis, we also reported that BRAF mutation status was not significantly associated with either OS or RFS (Supplementary Table S2).
Table 3.
Multivariable Cox Analyses on Survival Models in Validation Set (N=79)
| Multivariable Cox Regression Model | Overall Survival | Recurrence-Free Survival | |||||
|---|---|---|---|---|---|---|---|
| Parameter | Level | HR | 95% CI | P-value | HR | 95% CI | P-value |
| CD74 Tumor Intensity | Positive | 0.27 | 0.14, 0.51 | <0.001 | 0.41 | 0.22, 0.76 | 0.005 |
| MIF Tumor Intensity | Positive | 3.02 | 1.16, 7.83 | 0.02 | 3.60 | 1.57, 8.26 | 0.003 |
| Breslow Thickness | 0–1 | ---- | ---- | ---- | ---- | ---- | ---- |
| >1–2 | 0.81 | 0.31, 2.11 | 0.66 | 0.73 | 0.29, 1.84 | 0.51 | |
| >2–4 | 0.99 | 0.44, 2.25 | 0.99 | 0.61 | 0.27, 1.39 | 0.24 | |
| >4 | 1.12 | 0.48, 2.57 | 0.80 | 1.02 | 0.47, 2.19 | 0.96 | |
| Positive Lymph Nodes | 0–1 | ---- | ---- | ---- | ---- | ---- | ---- |
| 2–3 | 1.24 | 0.49, 3.09 | 0.65 | 0.74 | 0.31, 1.77 | 0.50 | |
| ≥4 | 1.46 | 0.68, 3.14 | 0.37 | 1.26 | 0.62, 2.55 | 0.52 | |
| Age at Surgery | Continuous, Per Year | 1.01 | 0.99, 1.03 | 0.47 | 1.00 | 0.99, 1.02 | 0.81 |
| Gender | Male | 1.24 | 0.68, 2.29 | 0.48 | 1.49 | 0.79, 2.80 | 0.22 |
HR: Hazard Ratio; CI: Confidence Interval
While MDACC patients received several different adjuvant therapies, such as, Ipilimumab, BRAF inhibitors, vaccine, biochemotherapy (Supplementary Table S3), JWCI TMA patients were predominantly received additional surgery or an experimental vaccine. Even when adjuvant therapy was included in our multivariable analyses, CD74 and MIF intensity persisted as significant. We choose not to present this data due to the variation of the treatment selections between two TMAs, and smaller numbers of groups receiving each treatment. The association of these markers with response and prognosis after to various immunotherapies and targeted therapies, or their combinations, remains of great interest and will be pursued.
Detailed Analysis of CD74 Expression
As described above, CD74 expression in melanoma cells correlated with improved OS and RFS in both cohorts, in univariate and multivariate analysis. Analysis of both the quartiles of percentage and intensity staining demonstrated a near dose-dependent difference in CD74 expression with OS and RFS (Supplementary Figure S2). Patients with >75% of their tumor cells expressing CD74, as well as those with the higher intensity of staining generally had the best outcomes, suggesting a quantitative relationship. The difference between OS or RFS and CD74 tumor cell intensity categories (P < 0.001 and P = 0.013, respectively) and tumor cell percentage categories in the validation dataset (P = 0.011 for both) were statistically significant.
As an additional finding, we further noted that CD74 was positive in the TIL. Analysis of this TIL data indicated that CD74 expression on TIL was also associated with improved OS (HR 0.54, P < 0.0001 for percentage; HR 0.61, P = 0.004 for intensity), and RFS (HR of 0.65, P = 0.002 for percentage; HR 0.68, P = 0.001 for staining intensity). Although our studies did not measure CD2, we assume all TIL are CD2 positive, which also will be tested in further studies. Therefore CD74 protein expression in both tumor cells and TIL predicts better OS and RFS, indicating that most patients with higher CD74 percentage or intensity of protein expression had longer survival.
Survival Analysis of the Markers of Interest in Stage III Melanoma Patients
Figure 2 presents Kaplan-Meier plots of OS and RFS by a classifier that uses both CD74 and MIF intensity, depicting the major outcome of this study. A statistically significant relationship between OS and combined markers intensity (P < 0.001) as well as RFS and combined markers intensity (P < 0.001) was determined in our discovery data set. Similarly, there was a statistically significant relationship between OS or RFS and CD74+/MIF− intensity in tumor cells in the MDACC validation data set. Patients with CD74+/MIF− intensity in melanoma cells had the longest OS (P < 0.001) and RFS (P = 0.003) among patients with stage III melanoma (P = 0.003). We also observed similar patterns of significance for OS and RFS by combined CD74 and MIF as the percentage of expression in tumors (Supplementary Figure S3).
Figure 2.
Clinical outcomes by combined markers of CD74 and MIF in tumor cells in stage III melanoma patients in discovery (A) and validation (B) datasets. Kaplan-Meier analyses are shown for OS and RFS for their expression by intensity of staining.
TCGA Results Confirm CD74 Association with Good Survival
Recently the cutaneous melanoma TCGA dataset has become available (21), and CD74 expression was interrogated. Our analysis was restricted to patients for whom the TCGA tumor biopsy was collected at the first diagnosis of stage III disease (n = 98). Publicly available RNAseq data were used to categorize tumors as having low (lowest quartile) versus high (top three quartiles) CD74 mRNA expression. Including all stage III patients, longer OS was observed in patients with high CD74 (n = 73; median, 73 months) compared with patients with low CD74 expression (n = 25; median, 27.6 months, P = 0.03) (Figure 3A). Considering that this difference might have been due to cumulative CD74 expression in tumor plus TIL, the same analysis was also performed using data only from patients with low lymphocyte scores (21). This analysis again identified longer OS in patients with high CD74 expression (n = 37; median, 69 months) compared to patients with low CD74 (n = 12; median, 16.8 months, P = 0.003) (Figure 3B). These data suggest that tumor cell CD74 expression correlates with OS in the TCGA cohort. Of note, in comparing tumor CD74 mRNA expression in cutaneous melanoma samples (n = 470) with normal tissue expression (n = 2921), only the spleen and lymphocytes had a higher median expression than melanoma, yet the highest individual expressing samples were in melanoma (Figure 3C).
Figure 3.
Expression characteristics and survival outcomes of CD74 mRNA expression in TCGA database. Survival analysis of all stage III samples (A) and a group of patient with stage III disease and low lymphocyte infiltration in their tumors (B), as well as tumor CD74 mRNA expression in normal tissues (C) in comparison with melanoma samples.
DISCUSSION
Improved risk models of patients with stage III melanoma will improve the treatment of patients with this disease, however molecular markers are lacking. Our analysis of independent cohorts (two for protein in TMA, and one for mRNA in TCGA) of patients’ tumors strongly implicates increased CD74 expression as a new marker of good prognosis in stage III melanoma. Previously we had considered inflammatory markers to be associated with poor prognosis, which was the case for MIF in both cohorts, and for iNOS in the MDACC cohort. The surprising finding of higher expression of CD74 having a strong association with good prognosis is most intriguing and elucidating the mechanism involved are likely to open new avenues for melanoma research.
The functional role(s) of CD74 is not well understood. Historically, CD74 was known primarily as the MHC Class II invariant chain, and functions in the molecular processing of MHC II through the Golgi (24). It has a potential role in the anti-tumor immune response (25), and also been shown to be expressed in inflammatory disorders and other inflammation-associated cancers (26). Our unexpected finding of high expression of CD74 in a subset of samples in both the discovery and validation TMAs of melanoma patients from two different institutions, along with highly significant associations with improved survival in both univariate and multivariable analysis of both TMAs, strongly suggests a previously unappreciated biologic role for this curious molecule. Irrespective of the mechanism(s) involved, CD74 now meets the criteria as a validated marker of improved prognosis in stage III melanoma patients. Similar findings that high expression of CD74 is an independent prognostic factor for OS were recently reported in mesothelioma (27).
Numerous inflammation markers have been identified in melanoma cell lines and patient samples with use of gene expression array methods; these markers include iNOS, arginase, VEGFA, CXCL-10, IL-8, IL-1α, IL-1β, and TNFSF9 (19, 28).. More recent results from our laboratory have continued to support association of iNOS protein with nitrotyrosine (NT), COX-2, pSTAT3, and arginase, consistent with the report of Johansson and colleagues (18). Because iNOS expression and NO production lead to tumor progression, in this study we asked whether we could identify other markers or even a signature of common inflammatory markers. Although many markers have been noted extensively in the literature, in melanoma we have identified inflammatory cytokines IL-1α IL-1β and IL-6 (29) but none of these proved prognostic in our controlled and parallel studies shown here. In a previous functional study we observed that an MIF-CD74 autocrine interaction upregulated by exogenous IFN-γ, promoted the phosphorylation of AKT Ser473 and drove expression of such inflammatory cytokines (30), presenting a potentially protumor circuitous or alternative function when MIF is present, which may be likely in a subset of tumors after exposure to IFN-γ. These findings could be specifically important as we observed this effect regardless of the BRAF and NRAS mutation status. This implies that the augmentation of CD74-MIF signaling by IFN-γ is a universal phenomenon in melanoma. In that functional study, we were able to show all the melanoma lines expressed MIF and the inhibition of its binding to CD74 by using ISO-1 suppressed the phosphorylation of AKT Ser 473, as well as the expression of BCL-2, IL-6 and IL-8. Another important finding in that study was the expression characteristics of CD74 in a melanoma progression array. We reported that CD74 expression in primary and metastatic lesions was significantly higher than that in nevi (p<0.0001). These results suggested that melanoma cell lines express CD74 and that expression is associated with the progression from benign nevi to clinically evident melanoma. However, the TMA we used in that study was not designed to evaluate CD74’s role in melanoma prognosis. Therefore, we concluded that CD74 expression was associated with melanoma progression, however from our new data it represents a good prognostic marker if expressed in the absence of MIF in the microenvironment. Another important feature is that its role in MHC processing potentiates antigen expression, and its association with immunotherapy responses is now a high priority to resolve.
Low MIF was validated as a good prognostic finding in this report. In an attempt to understand possible interactions, it is known that MIF is a pleiotropic proinflammatory protein for which the effects include the promotion of cytokine expression and inhibition of apoptosis (31). It was recently shown that MIF could also play a role as a dominant chemotactic cytokine by physical interaction with its many receptors (CXCR2, CXCR4, and CD74) in human mesenchymal stem cell recruitment to tumors (32). CD74 has been reported to bind MIF and to be required for MIF-dependent cellular responses such as PGE2 production and proliferation (33). Although the role of surface expressed CD74 in chemokine-mediated signaling has not been well studied, we originally hypothesized that positive expression of both MIF and CD74 expression in tumor cells would predict poor survival of melanoma patients via activation of tumor survival pathways. Our results of CD74 in the absence of MIF providing value as a good prognostic marker was most significant, so that low MIF and high CD74 provides another intriguing model for functional analysis. CD74 appears to have at least two functions: [1] originally recognized as the MHCII invariant chain, loading antigen into MHC for cell surface expression; and [2] as a cell surface receptor for MIF in the presence of IFNγ(19). The intracellular portion of CD74 lacks signaling domains, but has been predicted to interact with downstream signaling molecules (33). Another consideration is that CD74 is a homotrimer, and MIF engagement of CD74 may affect the oligomerization of the intracellular domain that is necessary for downstream signaling; alternatively there may be unknown variants of CD74 unique to melanoma. Regardless of the mechanisms yet to be revealed, it is clear from our data that CD74 expression in both the melanoma cell and infiltrating lymphocytes indicates a survival advantage for the patient. Studies are underway to assess whether the CD74 expression alone would predict a favorable response to immunotherapies; conversely if the IFNγ–driven product of MIF is present in the tumor site, perhaps other means to inhibit them or targeted therapy approaches might prioritized. While this scenario is hypothetical, we currently envision that these or other inflammatory markers are likely to be useful beyond their prognostic value and may help make predictive treatment decisions.
In conclusion, we have identified and validated the protein expression of CD74 and of MIF as providing survival information, and propose that CD74+ and MIF− together be considered to form a “signature” for stage III melanoma prognosis. Our data provides clear justification for including these markers for testing in larger stage III data cohorts to address their value in refining the current substaging data for stage III melanomas, and for testing these markers in prospective clinical trials to determine whether this signature adds value to either targeted or immunotherapy treatments. It is our hope that these two-protein markers (and others to be discovered), either together as a “signature” or individually, will help clinicians make better personalized treatment suggestions and help patients make better treatment decisions.
Supplementary Material
Translational Relevance.
Although gene mutations are now providing predictive information for targeted therapy, molecular markers to inform prognosis remain elusive. It is also known that chronic inflammation associates with melanoma patient disease progression, and resolution of inflammation in experimental models leads to development of adaptive immunity as well as improved outcomes to chemotherapy. Therefore, we tested 15 selective molecular inflammation-related markers in melanoma stage III patient tumor cells for association with outcomes, and identified three markers of importance. Protein expression of two (iNOS and MIF) had predicted statistical association with poor outcome; surprisingly, CD74 associated strongly with a favorable patient outcome; this is intriguing as CD74 has been hypothesized to be linked to development of specific adaptive immune response as it is known as the MHC Class II invariant chain. The presence and positive role of CD74 is novel, and we believe our findings will lead to approaches for improved personalized treatment.
Acknowledgments
Financial Support
This work was supported by the MD Anderson Cancer Center SPORE in Melanoma P50 CA093459 (SE, MAD, RB, VGP, JEG, and EAG) funded from the National Institutes of Health, the MD Anderson’s Cancer Center Support Grant P30- CA016672 (EAG) funded from the National Institutes of Health, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (KT, DSH, DLM, and EAG), The University of Texas, M.D. Anderson Cancer Center Melanoma Moon Shots Program (MAD, JEG, EAG), the Miriam and Jim Mulva Foundation, and AIM at Melanoma Foundations.
The authors thank to the members of our Melanoma Informatics, Tissue Resources and Pathology Core, who provided the deidentified tissues for the validation TMA used in this project as well as maintain accurate patient information available for our studies. We also express our appreciation to Ms. Sandra A. Kinney for her excellent technical assistance in our immunohistochemistry studies and Ms. Tamara K. Locke for her editorial assistance.
Footnotes
Conflict of Interest: There is no potential conflict of interest to disclose.
References
- 1.Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch CM, Gershenwald JE, Soong SJ, et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J Clin Oncol. 2010;28:2452–9. doi: 10.1200/JCO.2009.27.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harcharik S, Bernardo S, Moskalenko M, et al. Defining the role of CD2 in disease progression and overall survival among patients with completely resected stage-II to -III cutaneous melanoma. J Am Acad Dermatol. 2014;70:1036–44. doi: 10.1016/j.jaad.2014.01.914. [DOI] [PubMed] [Google Scholar]
- 4.Sivendran S, Chang R, Pham L, et al. Dissection of immune gene networks in primary melanoma tumors critical for antitumor surveillance of patients with stage II-III resectable disease. J Invest Dermatol. 2014;134:2202–11. doi: 10.1038/jid.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogunovic D, O’Neill DW, Belitskaya-Levy I, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;106:20429–34. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimm EA, Sikora AG, Ekmekcioglu S. Molecular pathways: inflammation-associated nitric-oxide production as a cancer-supporting redox mechanism and a potential therapeutic target. Clin Cancer Res. 2013;19:5557–63. doi: 10.1158/1078-0432.CCR-12-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomsen LL, Lawton FG, Knowles RG, et al. Nitric oxide synthase activity in human gynecological cancer. Cancer Res. 1994;54:1352–1354. [PubMed] [Google Scholar]
- 9.Cobbs CS, Brenman JE, Aldape KD, et al. Expression of nitric oxide synthase in human central nervous system tumors. Cancer Res. 1995;55:727–730. [PubMed] [Google Scholar]
- 10.Gallo O, Masini E, Morbidelli L, et al. Role of nitric oxide in angiogenesis and tumor progression in head and neck cancer. J Natl Cancer Inst. 1998;90:587–96. doi: 10.1093/jnci/90.8.587. [DOI] [PubMed] [Google Scholar]
- 11.Reveneau S, Arnould L, Jolimoy G, et al. Nitric oxide synthase in human breast cancer is associated with tumor grade, proliferation rate, and expression of progesterone receptors. Lab Invest. 1999;79:1215–25. [PubMed] [Google Scholar]
- 12.Ekmekcioglu S, Ellerhorst J, Smid CM, et al. Inducible nitric oxide synthase and nitrotyrosine in human metastatic melanoma tumors correlate with poor survival. Clin Cancer Res. 2000;6:4768–4775. [PubMed] [Google Scholar]
- 13.Ekmekcioglu S, Ellerhorst JA, Prieto VG, et al. Tumor iNOS predicts poor survival for stage III melanoma patients. Int J Cancer. 2006;119:861–866. doi: 10.1002/ijc.21767. [DOI] [PubMed] [Google Scholar]
- 14.Borghese F, Clanchy FI. CD74. an emerging opportunity as a therapeutic target in cancer and autoimmune disease. Expert Opin Ther Targets. 2011;15:237–51. doi: 10.1517/14728222.2011.550879. [DOI] [PubMed] [Google Scholar]
- 15.Haq R, Yokoyama S, Hawryluk EB, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci U S A. 2013;110:4321–6. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granados-Principal S, Liu Y, Guevara ML, et al. Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 2015;17:25. doi: 10.1186/s13058-015-0527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 18.Johansson CC, Egyhazi S, Masucci G, et al. Prognostic significance of tumor iNOS and COX-2 in stage III malignant cutaneous melanoma. Cancer Immunol Immunother. 2009;58:1085–1094. doi: 10.1007/s00262-008-0631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanese K, Grimm EA, Ekmekcioglu S. The role of melanoma tumor-derived nitric oxide in the tumor inflammatory microenvironment: its impact on the chemokine expression profile, including suppression of CXCL10. Int J Cancer. 2012;131:891–901. doi: 10.1002/ijc.26451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieto VG, Mourad-Zeidan AA, Melnikova V, et al. Galectin-3 expression is associated with tumor progression and pattern of sun exposure in melanoma. Clin Cancer Res. 2006;12:6709–15. doi: 10.1158/1078-0432.CCR-06-0758. [DOI] [PubMed] [Google Scholar]
- 21.Network TCGA. Genomic Classification of Cutaneous Melanoma. Cell. doi: 10.1016/j.cell.2015.05.044. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 23.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Karakikes I, Morrison IE, O’Toole P, et al. Interaction of HLA-DR and CD74 at the cell surface of antigen-presenting cells by single particle image analysis. FASEB J. 2012;26:4886–96. doi: 10.1096/fj.12-211466. [DOI] [PubMed] [Google Scholar]
- 25.Basha G, Omilusik K, Chavez-Steenbock A, et al. A CD74-dependent MHC class I endolysosomal cross-presentation pathway. Nat Immunol. 2012;13:237–45. doi: 10.1038/ni.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beswick EJ, Reyes VE. CD74 in antigen presentation, inflammation, and cancers of the gastrointestinal tract. World J Gastroenterol. 2009;15:2855–61. doi: 10.3748/wjg.15.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otterstrom C, Soltermann A, Opitz I, et al. CD74: a new prognostic factor for patients with malignant pleural mesothelioma. Br J Cancer. 2014;110:2040–6. doi: 10.1038/bjc.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin Y, Deng W, Ekmekcioglu S, et al. Identification of unique sensitizing targets for anti-inflammatory CDDO-Me in metastatic melanoma by a large-scale synthetic lethal RNAi screening. Pigment Cell Melanoma Res. 2013;26:97–112. doi: 10.1111/pcmr.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Y, Ekmekcioglu S, Liu P, et al. Constitutive aberrant endogenous interleukin-1 facilitates inflammation and growth in human melanoma. Mol Cancer Res. 2011;9:1537–50. doi: 10.1158/1541-7786.MCR-11-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanese K, Hashimoto Y, Berkova Z, et al. Cell Surface CD74-MIF Interactions Drive Melanoma Survival in Response to Interferon-gamma. J Invest Dermatol. 2015 doi: 10.1038/jid.2015.259. [DOI] [PubMed] [Google Scholar]
- 31.Fan H, Hall P, Santos LL, et al. Macrophage migration inhibitory factor and CD74 regulate macrophage chemotactic responses via MAPK and Rho GTPase. J Immunol. 2011;186:4915–24. doi: 10.4049/jimmunol.1003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lourenco S, Teixeira VH, Kalber T, et al. Macrophage migration inhibitory factor-CXCR4 is the dominant chemotactic axis in human mesenchymal stem cell recruitment to tumors. J Immunol. 2015;194:3463–74. doi: 10.4049/jimmunol.1402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leng L, Metz CN, Fang Y, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–76. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.