Abstract
Two chromatin compartments are present in most mammalian cells; the first contains primarily euchromatic, early replicating chromatin and the second, primarily late-replicating heterochromatin, which is the subject of this review. Heterochromatin is concentrated in three intranuclear regions: the nuclear periphery, the perinucleolar space and in pericentromeric bodies. We review recent evidence demonstrating that the heterochromatic compartment is critically involved in global nuclear organization and the maintenance of genome stability, and discuss models regarding how this compartment is formed and maintained. We also evaluate our understanding of how heterochromatic sequences (herein named heterochromatic associated regions (HADs)) might be tethered within these regions and review experiments that reveal the stochastic nature of individual HAD positioning within the compartment. These investigations suggest a substantial level of functional redundancy within the heterochromatic compartment.
Keywords: heterochromatin, nuclear organization, nucleolus, nuclear periphery, pericentromeric heterochromatin, perinucleolar heterochromatin
INTRODUCTION
A readily evident heterochromatic compartment is present in most differentiated cell nuclei. Chromatin in this compartment is generally more compacted than euchromatin, and although many sequences may be transcribed in heterochromatin, this transcription usually maintains gene silencing, rather than giving rise to protein-coding mRNAs (see [1–3] for reviews). Heterochromatin is commonly described as constitutive (primarily silenced repeat sequences) or facultative (unexpressed developmentally specific genes), but both types of heterochromatin are usually concentrated together in the heterochromatic compartment [2,4]. Spatially, heterochromatin in the compartment is distributed within three intranuclear regions: the nuclear periphery (PH), the perinucleolar region (PNH) and pericentromeric bodies (PCH), (Figure 1A–1B) [2,5]. The size and composition of the compartment often changes dramatically during cell differentiation. For example, the compaction and amount of heterochromatin increases significantly during erythroid differentiation as non-erythroid genes are silenced (Figure 1C–1D)[6]. In some vertebrate classes this culminates in an erythrocyte nucleus in which the entire genome is highly condensed chromatin. In contrast, naïve stem cells and very early embryos have relatively little heterochromatin (Figure 1E–1F)[7–9]. In this review, we explore recent advances, primarily in mammalian cells, that help to define the role of the heterochromatic compartment in nuclear organization and function. We focus on the composition of the heterochromatin associated domains (HADs), the effect of their sequestration on nuclear organization and function, and finally, on the redundancy of the heterochromatic compartment and how this redundancy impacts studies on nuclear organization and function.
Figure 1. A–B. Heterochromatin distribution in mammalian cell.
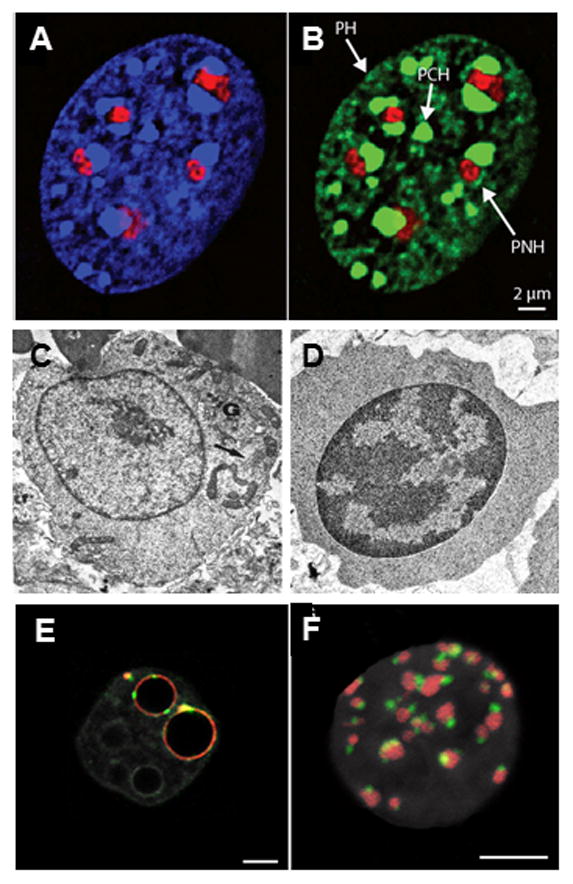
Murine embryonic fibroblast stained with A) DAPI (blue) plus antibodies to fibrillarin (red) to mark nucleoli and B) antibodies to H3K9me3 (green) and fibrillarin (red). PH = peripheral heterochromatin. PNH = perinucleolar heterochromatin. PCH = pericentromeric heterochromatin. From [2]. C–D. Heterochromatin changes during erythropoiesis. Electron micrographs of C) murine proerythroblast and D) late erythroblast showing change in heterochromatin distribution (arrows) during differentiation. From [6] with permission from Nature Publishing Group. E–F. Heterochromatin changes during early development. E) Single confocal section of mouse preimplantation embryo at early 2 cell and F) 16 cell stage showing distribution of pericentromeric (red) and centromeric chromatin (green). DNA is grey, bars = 5 μm. From [9].
Sequestration of HADs into the heterochromatic compartment
The major chromatin marks that define HADs are the constitutive heterochromatin marks, 5-mC, H3K9me and H4K20me (in PCH especially) and the facultative mark, H3K27me3, although there are other marks also involved (see [3,4] for reviews). Condensation and compaction is mediated by the linker histone H1, the H3K9me binding heterochromatin protein 1 (HP1α and β), and the DNA and histone methyl transferases (DNMTs and HMTs). A number of other heterochromatin-binding proteins (HBPs) and histone variants are also involved, but their direct roles in compaction remain less well defined (see [2,3,10] for reviews).
Highly and moderately repetitive HADs, such as centromeric and pericentromeric sequences (satellite DNA), telomeric sequences, endogenous retrotransposons and a portion of the rDNA repeats are commonly constitutively silenced in differentiated cells. Unexpressed developmentally specific HADs are usually subject to facultative silencing via polycomb-mediated H3K27 methylation. If heterochromatic marks are removed experimentally [11,12], or are lost during aging [13,14] or cancer [15,16], inappropriate gene expression (e.g. of retrotransposons [17,18]) and spontaneous recombination between newly exposed repeat sequences (especially rDNA [19,20]) result in genome instability and eventual cell death. It is of no surprise then that most organisms possess redundant silencing pathways to maintain heterochromatin features and thus genome stability [1,2,17,21].
Given the importance of heterochromatin to genome stability, it had been difficult to understand how heterochromatin-poor early embryos and naïve stem cells accommodate active retrotransposons, for example, but recent work suggests that piwi-based RNAi mechanisms [22,23] and/or the deposition of the histone variant, H3.3, and specific histone chaperones [24–26], silences retrotransposons until heterochromatic marks are deposited. Notably, however, some early retrotransposon activity, such as that resulting in the presence of the long non-coding RNA (ncRNA), HERV, in primate cells, is crucial in early embryos and naïve stem cells [27,28]. In aging or cancerous cells, where piRNAs are not prevalent, retrotransposon activity can wreak havoc on chromatin organization [15,18]. This does not preclude the fact, however, that many repeats, such as those in PCH, are transcribed at low levels in differentiated cells and some of these ncRNAs complex with HBPs and serve as adaptors to help target their binding (e.g. [3,29,30]).
Formation of the heterochromatin compartment
An intriguing aspect of the transition from pluripotency involves the rDNA repeats. In naïve mouse embryonic stem cells (mESCs), all or almost all rDNA cassettes are active, while in differentiated cells, approximately half are inactive and condensed into heterochromatin [31]. The silencing is specifically mediated by pRNA, which is transcribed from the rDNA promoter region and binds TIP5 within the nucleolar remodeling complex [32]. Interestingly, this rDNA heterochromatization has recently been shown to be a necessary step for exiting pluripotency. If pRNA is removed, cells remain pluripotent, with very low levels of heterochromatin [33]. Earlier work had shown that knockdown of the pRNA partner, TIP5, disrupts rDNA heterochromatization in fibroblasts, resulting in disorganization and unfolding of PNH, including the non-rDNA sequences localized there [32,34], and that deletion of rDNA cassettes decreases the levels of global heterochromatin [35]. Notably, knockdown of the linker histone H1 has recently been shown to disrupt nucleolar structure as well. Unexpectedly, H1 not only interacted with silencing enzymes in the nucleolus but also with many proteins involved in early ribosomal RNA transcription and processing, suggesting H1 may be involved in maintaining the balance between active and inactive rDNA loci [36]. Taken together, these findings further buttress an attractive model which postulates that silenced rDNA seeds the formation of the heterochromatic compartment [37–39]. Consistent with this model, PCH accumulates around pre-nucleolar bodies at the mid-zygote stage [40].
Mechanism of heterochromatin compartmentalization
Although silenced rDNA may well seed heterochromatin formation, heterochromatin accumulates not only in the PNC, but also the PH and in spatially distinct PCH bodies in some cells. There are two main mechanisms at work in this compartmentalization: first, self-association of repetitive chromatin guided by biophysical principles, and second, tethering of these chromatin domains within the heterochromatic compartment.
Assembly of nuclear compartments within the membrane-free nucleoplasm can be modeled using self-organization principles based on molecular self-association, volume exclusion and phase partitioning [41–44] and in the case of chromatin, polymer physics and the behavior of fractal globules [45–47]. Simply put, early in G1, repetitive sequences, including rDNA, centromeric and telomeric sequences, are present at much higher concentrations than unique sequences and thus encounter one another more often, favoring self-association (the so-called “birds of feather stick together” model [45]. The resulting volume exclusion effects [47–49] drive nuclear organization in what has been termed a dog-on-a-leash mechanism, where strong associations between repetitive sequences cause clustering and adjacent sequences are dragged along on the “leash”[50]. In this way, highly repetitive HADs may act as dominant seeds to probabilistically concentrate any nearby heterochromatin into the PNH, PCH and PH during reorganization after each mitosis. Consistent with a self-assembly model, it has been known for years that the relative compaction of heterochromatin can be reversibly manipulated simply by changes in osmotic strength [51–53].
Crowding and clustering in turn favors concentration of HBPs at their sites of action. Proteins move with fractal kinetics through the compacted chromatin [47], with intermittent corralling within micro-regions (giving rise to anomalous intranuclear diffusion [46]. This increased residence time increases binding efficiency at a particular site since proteins have time to sample most binding sites within the corral. For example, as mentioned above, HP1α and β have been long known to be important global compactors of heterochromatin via their affinity with H3K9me sites and their propensity for oligomerization, which drives heterochromatic spreading [54]. Oligomerization would be favored in corrals with high monomer concentration.
Peripheral tethering of chromatin (so-called lamin-associated domains or LADs [55]) requires the presence of heterochromatic-specific histone marks and is thought to occur through both direct and indirect interactions with the lamina [4,56]. Somewhat surprisingly, the lamins themselves are not required for the peripheral localization of LADs in mESCs [57,58], but their presence is necessary for peripheral heterochromatin localization in differentiated cells [55]. For example, the loss of lamin B1 in aging cells disrupts heterochromatin organization [59,60] and terminally differentiated retinal cells display an “inside-out” heterochromatin arrangement (where heterochromatin occupies the center of the nucleus and euchromatin and nucleoli the periphery) primarily because neither lamin A/C nor Lamin B Receptor (LBR) is expressed in these cells [61]. Both of these latter proteins have been implicated in heterochromatin tethering: the transmembrane LBR interacts with methylated histones and HP1 and lamin A/C is associated with LADs in murine embryonic fibroblasts and pro-B cells [4]. Notably, Kind et al. [62] have shown that lamin A/C (and Barrier to Autointegration Factor (BAF)) associates with both LADs and nucleolar-associated-domains (NADs) in human fibrosarcoma cells. This indicates that lamin A and BAF are not PH specific tethers in human and it will be interesting to determine whether this is true in mouse as well. On the other hand, like LBR, the inner nuclear membrane protein LAP2β has so far only been implicated in PH tethering [63], and it, like BAF, binds specific regions of chromatin during mitosis [64]. Other potentially specific PH tethering candidates include nuclear envelope transmembrane proteins and the proline-rich protein 14 [56]. Nucleoli were not labeled in the latter study, however. Thus, even though much work has been done, only a few proteins that tether heterochromatin specifically to the PH have been identified.
Similarly, few candidates for direct binding of NADs [39] specifically to nucleolar components have been identified. One encouraging finding is that a complex of CTCF and a nucleoplasmin-like protein has been shown to mediate binding of centromeres to the nucleolar protein nucleolin (Modulo) in Drosophila [65]. CTCF also was shown in earlier studies to mediate binding of a transgene to the nucleolus, perhaps through nucleophosmin (aka B23) [66]. One reason few specific tethers have been identified could be that many repetitive sequences are not specifically tethered to one region within the compartment, but rather are able to move among the three regions, as discussed below in “Redundancy of the heterochromatic compartment”. Additionally, most past studies have been focused on identification of protein tethers, and it is only very recently that studies have begun to identify ncRNAs that act as specific adaptors for tethering. A clear example of this is the long ncRNA Firre. This ncRNA is transcribed from a macrosatellite repeat on the murine X chromosome and can organize 3D chromatin topology in trans [67]. Intriguingly, Firre, and also a previously identified CTCF-site-associated long ncRNA, Dxz4, have now been implicated as important mediators in the interaction between CTCF sites on the inactive X with the PNH, maybe in a complex with cohesin and nucleophosmin [68]. The fact that these ncRNAs are transcribed from the inactive X and then employed in tethering it within the heterochromatic compartment (and maintaining its silencing) suggests this may be a more general mechanism for tethering. pRNA, active in the silencing of rDNA discussed under “Formation of the heterochromatin compartment”, is another example of this mechanism, as it is transcribed from the rDNA promotor and is a necessary component of the nucleolar remodeling complex. Much exciting work remains to be done in this area.
Redundancy of heterochromatic compartment
A number of experiments have now shown that many sequences within the heterochromatic compartment can be stochastically shuffled between the PN, PNH and PCH (Figure 2). For example, “mother” LADs become distributed throughout the heterochromatic compartment in daughter cells without affecting cell function (Figure 2A, [69]), a finding supported by earlier photoactivation and photobleaching experiments [70]. The distribution patterns are different from mother to daughter and between daughter nuclei, suggesting that location of a given LAD within a particular heterochromatic region is not necessary. In further support of this interpretation, late-replicating (type B compartment, primarily silent) DNA sequences shown to interact with one another via Hi-C [71] can be found, by using in situ hybridization, in either the PH, PNH or PCH in different lymphoblastoid cells within the same culture (Figure 2B) [72]. In many cases these sequences correspond to LADs, supporting the live cell relocalization experiments. Multiple laboratories have also now shown that many LADs overlap with NAD sequences (e.g. [62,69,73,74]), and in situ hybridization studies have long shown that PCH can overlap with PH or PNH in many tissue types. Taken together, these observations are consistent with a self-assembly model, where inactive sequences may be sequestered into any region of the heterochromatin compartment and this may suffice to induce/maintain silencing (Figure 2C)[37,72].
Figure 2. The redundancy of the heterochromatic compartment.
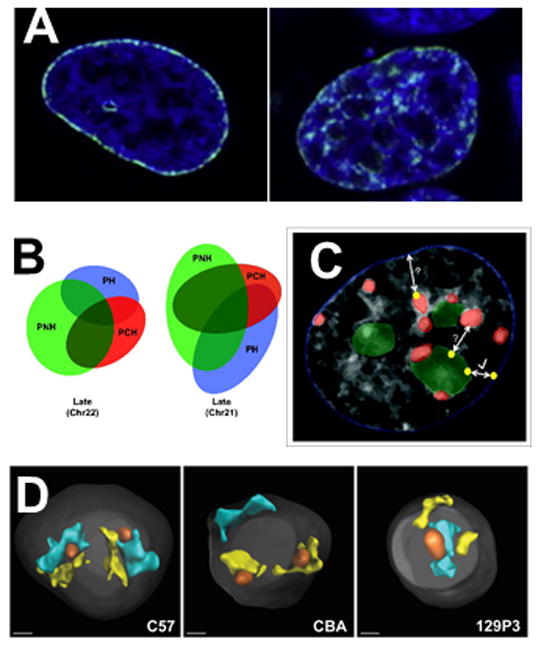
A) Redistribution of mother LADs (left, green) in daughter cell (left, green). DAPI staining of chromatin is blue. The unstained areas in the nucleus on the left are nucleoli. Adapted from the graphical abstract in [65], with permission from Elsevier. B) Venn diagrams depict the overlapping association of two late and one early replicating region with the PH, PCH and PNH in human lymphoblastoid nucleus. The size and overlap of the ellipses is proportional to the percent association of the region with each compartment as determined by DNA FISH. From [68]. C) Cartoon overlaid on DAPI-stained nucleus to represent redundant association sites of late replicating DNA regions (yellow dots) in the PH (blue nuclear outline), PCH (red) and PNH (green). Regions can relocalize from PNH to PH after mitosis or upon the loss of nucleoli (arrow with check mark). Shuttling between the PCH and PH or the PNH and PCH has not been studied directly (bidirectional arrows with question marks). From [68]. D) 3D reconstruction and rendering of chromosome paint images from primary B cells from three different mouse strains showing how the presence or absence of an NOR affects chromosome 12 (cyan) and chromosome 15 (yellow) territory position. When NORs are on both chromosomes (C57), territories tend to associate with the nucleolus (orange), if no NOR is present, the territory associates more often with the periphery (CBA: no NOR on chromosome 12; 129P3: no NOR on chromosome 15). DAPI stain is shown in gray. Scale bar = 1 μm. From [72].
It has not been clear whether this is true for all HADs, however, since, for example, particular centromeric sequences have classically been observed to cluster preferentially in either the PNH or PH, at least in certain cell types [2,5]. Interestingly, a recent paper goes some distance toward shedding light on this preferential localization, at least in human lymphoblastoid cells [75]. These authors showed that PH centromeric sequences in non-proliferating cells became more closely associated with the PNC upon proliferation. The location of the centromeric-bearing chromosome territories did not change substantially, but rather the position of the centromeric sequence within each territory was altered so that it was now closer to nucleoli than the periphery. Thus, at least in this system, it appears centromeric sequences can move between the PH and PNC also, just as LADs and NADs do. It would be interesting to determine whether lamin A/lamin B1 ratios increase when the cells proceed toward proliferation, since Kind et al. (discussed above [62]) have shown that changing these ratios can affect the PNH/PH distribution of HADs.
Another aspect of preferential location to either the PH or PNH is exemplified by experiments performed in B cells from mice of different strains. In the standard laboratory mouse strain, C57BL/6J, chromosomes 12 and 15 possess acrocentric rDNA arrays (aka nucleolar organizing centers or NORs) and thus localize preferentially near nucleoli. However, in other laboratory (and wild) mouse strains, one or both of these chromosomes do not contain an NOR (presumably due to the increased recombination rate between mouse acrocentric sequences) and in these strains, chromosomal territories 12 and 15 associate more frequently with the periphery (Figure 2D) [76]. It is unlikely that lamin A/lamin B1 ratios affect the proximity of an NOR bearing chromosome to a nucleolus, as can be proposed for the centromeric sequences above. One model that explains both these sets of results supposes that association with the PH is a default state, while association with the PNH requires an active tether. The nucleolar tether could be nucleophosmin, nucleolin, lamin A, or BAF, perhaps associated with adaptor RNAs specific to the sequence tethered, or in other cases, an NOR.
A model of the heterochromatic compartment
This gives rise to a model in which HADs are sequestered into the PH, PNH and PCH in a largely stochastic way (Figure 3). The compartmentalization is driven by biophysical forces, and enforced by tethering proteins and ncRNAs adaptors transcribed from the HADs. As exemplified by lamin A, the model supposes that some tethering proteins are present at multiple locations within the heterochromatic compartment, enforcing probabilistic distribution. We also speculate that “promiscuous” transcription from different regions of a particular HAD would yield a constellation of adaptor ncRNAs that could promote binding to different tethering proteins within the PH, PNH or PCH. Affinity for a particular tethering protein could vary, depending on which transcribed sequences were present in higher quantities at the time. This mechanism also would enforce stochasticity.
Figure 3. Model depicting redundant distribution of HADs within heterochromatic compartment.
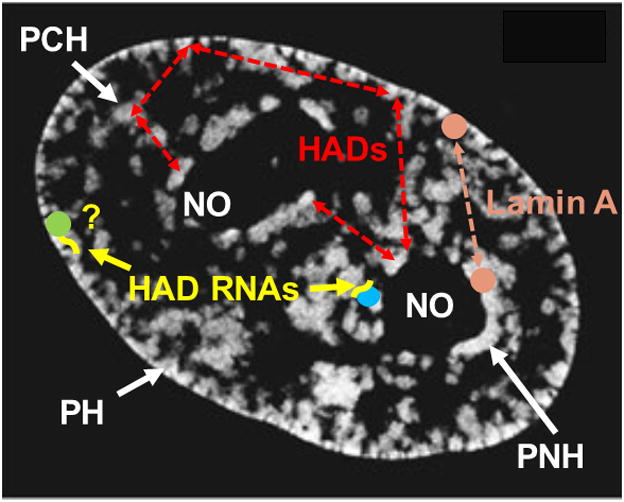
Red dashed arrows indicate that most heterochromatic associated domains (HADs) can be found in any of the three heterochromatic regions in different cells or after mitosis. PH: peripheral heterochromatin. PNH: perinucleolar heterochromatin. PCH: pericentromeric heterochromatin. RNAs: RNA adaptors (yellow) transcribed from HADs. PH RNA adaptors have not yet been identified (yellow “?”). Differently colored balls: tethering proteins, some of which are unique to a region (blue and green) and may tether some HADs specifically, and some of which are present in multiple regions (Lamin A, orange). White regions: DAPI staining. Underlying image is midplane of HeLa nucleus acquired using structured illumination microscopy from [74]. Reproduced with permission from Springer. PCH is not obvious in DAPI-stained HeLa nuclei and is labeled here only for illustrative purposes.
In addition to the probabilistic distribution of HADs, some HADs still associate preferentially with a particular region within the heterochromatic compartment. This finding is more solid for PNH than PH, as in many PH studies, NADs are not characterized. Thus, we only include specific PNH- tethering proteins (blue) and ncRNAs (Firre) in this model. Nevertheless, it is early days in this field, so we may find that ncRNAs serve as specific adaptors at the lamina also (“?” in Figure 3). It should be stressed, however, that even in the cases where nucleolar tethering is observed, localizations are probabilistic, and a sizable fraction of cells still show PH localization.
CONCLUSION
In general, current data indicates that functional cells can contain a given HAD in the PH, PNC or PCH, suggesting that the three regions within the heterochromatic compartment are redundant with respect to function. This redundancy results in a stochastic distribution of HADs throughout a significant fraction of the nuclear volume and helps explain why, although the spatial positioning of chromosome territories in a given cell type is not random, it is probabilistic rather than fixed [70,77]. It will be important to better define the degree and nature of redundancy in future experiments, which should include characterization of HACs in the PH, PNC and PCH. Live cell experiments in primary cell lines maintained in defined medium are the Holy Grail, but further technical advances are necessary before these types of experiments become standard.
Acknowledgments
We thank Thoru Pederson and Tobias Ragoczy for helpful comments on the manuscript. We apologize to authors whose work we could not cite given space considerations in this minireview. The authors were supported by NIH R01 HL65440 and NIH R37 DK44746.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 2.Politz JC, Scalzo D, Groudine M. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu Rev Cell Dev Biol. 2013;29:241–270. doi: 10.1146/annurev-cellbio-101512-122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saksouk N, Simboeck E, Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics & Chromatin. 2015;8:3. doi: 10.1186/1756-8935-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaitre C, Bickmore WA. Chromatin at the nuclear periphery and the regulation of genome functions. Histochem Cell Biol. 2015;144:111–122. doi: 10.1007/s00418-015-1346-y. [DOI] [PubMed] [Google Scholar]
- 5.Padeken J, Heun P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr Opin Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 7.Martin C, Beaujean N, Brochard V, Audouard C, Zink D, Debey P. Genome restructuring in mouse embryos during reprogramming and early development. Dev Biol. 2006;292:317–332. doi: 10.1016/j.ydbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 8.de Wit E, Bouwman BA, Zhu Y, Klous P, Splinter E, Verstegen MJ, Krijger PH, Festuccia N, Nora EP, Welling M, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 9.Aguirre-Lavin T, Adenot P, Bonnet-Garnier A, Lehmann G, Fleurot R, Boulesteix C, Debey P, Beaujean N. 3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development. BMC Dev Biol. 2012;12:30. doi: 10.1186/1471-213X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier K, Brehm A. Chromatin regulation: How complex does it get? Epigenetics. 2014;9:1485–1495. doi: 10.4161/15592294.2014.971580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng JC, Karpen GH. Heterochromatic Genome Stability Requires Regulators of Histone H3 K9 Methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 13.Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Front Genet. 2013;4:274. doi: 10.3389/fgene.2013.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triche T, Capone BS, Merchant MD, Chaudhary P, Ramsingh G. DNA Methylation Changes In Aging Human CD34+ Cells Coincide With Hotspots Of Disordered Methylation In AML At Imprinted and Allelically Methylated Regions. Blood. 2013;122:1193. [Google Scholar]
- 16.Rafique S, Thomas JS, Sproul D, Bickmore WA. Estrogen-induced chromatin decondensation and nuclear re-organization linked to regional epigenetic regulation in breast cancer. Genome Biol. 2015;16:145. doi: 10.1186/s13059-015-0719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci. 2012;37:127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Sullivan JM, Sontam DM, Grierson R, Jones B. Repeated elements coordinate the spatial organization of the yeast genome. Yeast. 2009;26:125–138. doi: 10.1002/yea.1657. [DOI] [PubMed] [Google Scholar]
- 20.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towbin BD, Gonzalez-Sandoval A, Gasser SM. Mechanisms of heterochromatin subnuclear localization. Trends Biochem Sci. 2013;38:356–363. doi: 10.1016/j.tibs.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Gene Dev. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pezic D, Manakov SA, Sachidanandam R, Aravin AA. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Gene Dev. 2014;28:1410–1428. doi: 10.1101/gad.240895.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsasser SJ, Noh K-M, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 26.Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, et al. Systematic Identification of Factors for Provirus Silencing in Embryonic Stem Cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 29.Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell. 2014;54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 30.Alekseyenko AA, Gorchakov AA, Zee BM, Fuchs SM, Kharchenko PV, Kuroda MI. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Gene Dev. 2014;28:1445–1460. doi: 10.1101/gad.241950.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grummt I, Längst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. BBA-Gene Regul Mech. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 2014;15:720–734. doi: 10.1016/j.stem.2014.10.005. This paper demonstrates that heterochromatization of rDNA repeats is necessary for pluripotency exit in mouse ESCs. If pRNA, a required ncRNA component of the nucleolar remodeling complex, is knocked down, preventing rDNA silencing, mESCs remain pluripotent and cannot differentiate. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz K-M, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Gene Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. P Natl Acad Sci USA. 2009;106:17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szerlong HJ, Herman JA, Krause CM, DeLuca JG, Skoultchi A, Winger QA, Prenni JE, Hansen JC. Proteomic characterization of the nucleolar linker histone H1 interaction network. J Mol Biol. 2015;427:2056–2071. doi: 10.1016/j.jmb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feinberg AP. The nucleolus gets the silent treatment. Cell Stem Cell. 2014;15:675–676. doi: 10.1016/j.stem.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Sullivan JM, Pai DA, Cridge AG, Engelke DR, Ganley AR. The nucleolus: a raft adrift in the nuclear sea or the keystone in nuclear structure? Biomol Concepts. 2013;4:277–286. doi: 10.1515/bmc-2012-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matheson TD, Kaufman PD. Grabbing the genome by the NADs. Chromosoma. 2015 doi: 10.1007/s00412-015-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jachowicz JW, Santenard A, Bender A, Muller J, Torres-Padilla ME. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev. 2013;27:2427–2432. doi: 10.1101/gad.224550.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Liu Q, Yan Q, Shi L, Fang Y. Nucleolus-tethering system (NoTS) reveals that assembly of photobodies follows a self-organization model. Mol Biol Cell. 2014;25:1366–1373. doi: 10.1091/mbc.E13-09-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dundr M, Misteli T. Biogenesis of Nuclear Bodies. Cold Spring Harbor Persp Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **43.Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. These authors have extended their 2013 work (showing that Xenopus nucleoli form liquid droplets) to demonstrate that nucleolar formation in C. elegans embryos fits with a phase transition model where the solute/droplet transition is dependent on nucleolar protein concentration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pederson T, King M, Markoc JF. Forces, fluctuations and self-organization in the nucleus. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-06-0357. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibcus JH, Dekker J. The Hierarchy of the 3D Genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsch CC, Langowski J. Anomalous diffusion in the interphase cell nucleus: The effect of spatial correlations of chromatin. J Chem Phys. 2010;133:11. doi: 10.1063/1.3435345. [DOI] [PubMed] [Google Scholar]
- 47.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finan JD, Guilak F. The effects of osmotic stress on the structure and function of the cell nucleus. J Cell Biochem. 2010;109:460–467. doi: 10.1002/jcb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krijger PH, de Laat W. Identical cells with different 3D genomes; cause and consequences? Curr Opin Genet Dev. 2013;23:191–196. doi: 10.1016/j.gde.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Robbins E, Pederson T, Klein P. Comparison of mitotic phenomena and effects induced by hypertonic solutions in HeLa cells. J Cell Biol. 1970;44:400–416. doi: 10.1083/jcb.44.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubner B, Cremer T, Neumann J. Correlative microscopy of individual cells: sequential application of microscopic systems with increasing resolution to study the nuclear landscape. Meth Mol Biol. 2013;1042:299–336. doi: 10.1007/978-1-62703-526-2_21. [DOI] [PubMed] [Google Scholar]
- 53.Walter A, Chapuis C, Huet S, Ellenberg J. Crowded chromatin is not sufficient for heterochromatin formation and not required for its maintenance. J Struct Biol. 2013;184:445–453. doi: 10.1016/j.jsb.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014;24:377–386. doi: 10.1016/j.tcb.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 56.Amendola M, van Steensel B. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr Opin Cell Biol. 2014;28:61–68. doi: 10.1016/j.ceb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan C-M, Gaiano N, Ko MSH, Zheng Y. Mouse B-Type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amendola M, van Steensel B. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 2015;16:610–617. doi: 10.15252/embr.201439789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young AR, Narita M, Perez-Mancera PA, Bennett DC, Chong H, et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Gene Dev. 2013;27:1800–1808. doi: 10.1101/gad.217281.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Gene Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solovei I, Wang Audrey S, Thanisch K, Schmidt Christine S, Krebs S, Zwerger M, Cohen Tatiana V, Devys D, Foisner R, Peichl L, et al. LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- **62.Kind J, van Steensel B. Stochastic genome-nuclear lamina interactions: modulating roles of Lamin A and BAF. Nucleus. 2014;5:124–130. doi: 10.4161/nucl.28825. This paper extends the work of Kind et al., 2013, to show that lamin A associates with heterochromatin at both the periphery and the nucleolus. Reduction of lamin A levels increases the proportion of LADs associated with lamin B1 at the periphery, indicating HADs can be handed off between the PH and PNH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, Gal-Yam EN. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 64.Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, Furukawa K, Ellenberg J, Foisner R. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J Cell Sci. 2004;117:6117–6128. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]
- 65.Padeken J, Mendiburo MJ, Chlamydas S, Schwarz HJ, Kremmer E, Heun P. The Nucleoplasmin Homolog NLP Mediates Centromere Clustering and Anchoring to the Nucleolus. Mol Cell. 2013;50:236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF Tethers an Insulator to Subnuclear Sites, Suggesting Shared Insulator Mechanisms across Species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 67.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **68.Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. This paper implicates Firre ncRNAs, which are transcribed from a cluster of CTCF binding sites on the inactive X, as critical for tethering this chromosome territory to the PNH. Firre is an example of a ncRNA adaptor that mediates HAD tethering to a particular heterochromatic region within the heterochromatic compartment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **69.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. The Bickmore and van Steensel laboratories report that specific LADs can be found in the nucleoplasm after mitosis, preferentially associated with the PNH. This work employs a m6A-Tracer modification of the DamID technique that allows LADs to be tracked in live cells and strongly supports earlier work suggesting that HADs can shuttle between the PH and PNH. [DOI] [PubMed] [Google Scholar]
- 70.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **72.Ragoczy T, Telling A, Scalzo D, Kooperberg C, Groudine M. Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus. 2014;5:626–635. doi: 10.4161/19491034.2014.990863. The presence of previously identified LAD sequences in all three regions (PH, PNH and PCH) of the heterochromatic compartment is documented using in situ hybridization in human lymphoblastoid cell populations. These results directly demonstrate the redundancy within the heterochromatic compartment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ollion J, Loll F, Cochennec J, Boudier T, Escude C. Proliferation-dependent positioning of individual centromeres in the interphase nucleus of human lymphoblastoid cell lines. Mol Biol Cell. 2015;26:2550–2560. doi: 10.1091/mbc.E14-05-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strongin DE, Groudine M, Politz JC. Nucleolar tethering mediates pairing between the IgH and Myc loci. Nucleus. 2014;5:474–481. doi: 10.4161/nucl.36233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **77.Cremer T, Cremer M, Hubner B, Strickfaden H, Smeets D, Popken J, Sterr M, Markaki Y, Rippe K, Cremer C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.05.037. A wonderful summary of the probabilistic nature of chromosome territory distribution and current models of nuclear organization. It encompasses many of the past and present questions involving cell-to-cell vs tissue-type specific variation in nuclear organization, and has an extensive reference list. [DOI] [PubMed] [Google Scholar]
- 78.Hübner B, Cremer T, Neumann J. Correlative Microscopy of Individual Cells: Sequential Application of Microscopic Systems with Increasing Resolution to Study the Nuclear Landscape. In: Shav-Tal Y, editor. Imaging Gene Expression. Humana Press; 2013. pp. 299–336. [DOI] [PubMed] [Google Scholar]; Meth Mol Biol. 1042 [Google Scholar]


