Abstract
Background
Stool DNA testing in inflammatory bowel disease (IBD) patients may detect colorectal cancer and advanced precancers with high sensitivity; less is known about the presence of DNA markers in small IBD lesions, their association with metachronous neoplasia, or contribution to stool test positivity.
Methods
At a single center in two blinded phases, we assayed methylated BMP3 (mBMP3), NDRG4 (mNDRG4), and mutant KRAS in DNA extracted from paraffin-embedded benign lesions and matched control tissues of IBD patients, who were followed for subsequent colorectal dysplasia. Stool samples from independent cases and controls with lesions <1cm or advanced neoplasms were assayed for the same markers.
Results
Among IBD lesions (29 low-grade dysplasia (LGD), 19 serrated epithelial change (SEC), 10 sessile serrated adenoma/polyps), the prevalence of methylation was significantly higher than in mucosae from 44 matched IBD controls (p <0.0001 for mBMP3 or mNDRG4). KRAS mutations were more abundant in SEC than all other groups (p<0.001). Subsequent dysplasia was not associated with DNA marker levels. In stools, the sensitivity of mBMP3 as a single marker was 60% for all lesions <1cm, 63% for LGD ≥1cm and 81% for high-grade dysplasia/CRC, all at 91% specificity (p<0.0001).
Conclusions
Selected DNA markers known to be present in advanced IBD neoplasia can also be detected in both tissues and stools from IBD patients with small adenomas and serrated lesions. Mutant KRAS exfoliated from SEC lesions might raise false-positive rates. These findings have relevance to potential future applications of stool DNA testing for IBD surveillance.
Keywords: Inflammatory bowel diseases, colorectal neoplasms, early detection of cancer, survival analysis, feces/analysis
INTRODUCTION
Patients with inflammatory bowel disease (IBD), including Crohn’s colitis (CD) and chronic ulcerative colitis (CUC), are at increased risk for colorectal cancer (CRC).(1–4) CRC risk increases with IBD duration, extent of colonic involvement, prior neoplasia, family history of CRC, and primary sclerosing cholangitis (PSC).(2, 5–7) Accordingly, patients with colonic IBD and these risks are advised to undergo surveillance every 1–3 years(8) by colonoscopy.(9)
The benefits of this practice are becoming clearer. CRC detected at surveillance may be at earlier stage, and therefore have a better prognosis than when presenting symptomatically(10–12), and evidence suggests that regular colonoscopic surveillance reduces CRC mortality in patients with IBD.(11, 13) Despite these encouraging data, patient compliance with surveillance colonoscopy is poor, even among those at highest risk.(14) Additionally, 17–30% of CRC diagnosed in large IBD surveillance cohorts were interval cancers.(10, 15) CRC missed at surveillance appears to have significantly worse prognosis, with 5-year survival rates of <50%, compared to >80% in those detected per-protocol.(10)
These sobering observations provide a compelling rationale to develop and validate adjunctive tests to surveillance colonoscopy. Genetic and epigenetic markers which accumulate during IBD-associated and sporadic CRC tumorigenesis(16) are attractive candidate targets for non-invasive testing. Mutant KRAS and methylation in the promotors of bone morphogenic protein 3 (BMP3) and N-Myc downstream-regulated gene 4 (NDRG4) have been validated in both tissue and stool for detecting advanced adenomas and CRC in patients without IBD.(17–20) We have also shown that stool DNA testing of methylated markers is sensitive and specific for IBD-associated CRC, high-grade dysplasia (HGD) and low-grade dysplastic (LGD) lesions ≥ 1 cm in diameter.(21)
The spectrum of target lesions most appropriate for surveillance is not yet fully defined. A clinically useful test would exhibit high sensitivity for CRC and HGD. LGD may require further risk stratification to determine which lesions would be at highest risk of progressing to invasive cancer. Current guidelines stratify LGD management by resectability;(22, 23) however, size of the neoplastic lesion also matters. Recent univariate and multivariate analyses on the natural history of IBD patients under surveillance suggest that dysplastic lesions <1 cm may not independently raise the risk of subsequent HGD or CRC.(24) The size of sporadic colorectal neoplasms appears to correlate with the quantity of exfoliated DNA.(18, 25) However, in IBD, neither the full clinical significance of small neoplastic or serrated lesions nor their contributions to measurable DNA in stool are understood at this time. Adding to this uncertainty, early reports on serrated epithelial change (SEC) in IBD patients have reached opposing conclusions with regard to the clinical relevance of this newly described histologic abnormality.(26, 27)
As a first series of steps to address these important knowledge gaps, we performed a cross-sectional analysis in tissues of benign lesions arising in IBD; we hypothesized that these would contain DNA mutations and aberrant methylation markers associated with adenomatous or serrated pathways. We then followed this cohort forward through the medical record to measure the association, if any, between these DNA alterations and the development of subsequent dysplasia. To measure the exfoliation of aberrant DNA from of small neoplasms in IBD, and thus their potential influence on stool DNA test results, we performed a case-control study in stools from independent IBD patients.
ETHICAL CONSIDERATIONS
The study was approved by the Mayo Clinic Institutional Review Board.
MATERIALS AND METHODS
Overall study design
Tissue phase
Archival frozen tissues were studied in case-control fashion to determine optimal cut-off values of markers planned for study in a retrospective cohort of IBD patients with SEC and sessile serrated adenoma/polyp (SSA/P), who were matched to IBD patients with LGD and IBD patients free of neoplasia. DNA extracted from primary lesions or control tissues from patients in the cohort were assayed for mutations in p53, KRAS and BRAF as well as methylation of BMP3 and NDRG4 and then followed forward in the medical record for subsequent dysplasia.
Stool phase
A case-control study was then performed in archival stool samples from independent IBD patients with small (<1cm) lesions, advanced colorectal neoplasms and controls.
Study Population
Tissue phase
Cut-off study
De-identified tissues from primary IBD-CRC tumors and IBD control mucosae were obtained from a frozen tissue archive after matching for age, sex and IBD disease subtype (ulcerative colitis versus the combination of Crohn’s colitis and indeterminate colitis).
Cohort-study
Paraffin-embedded tissue samples were requested from a cohort of IBD patients diagnosed with SEC and SSA/P between 2010 and 2012. IBD controls and cases were matched from an existing cohort identified in a comprehensive list of unique patients with CUC and CD who underwent colonoscopy in 2010, as previously described.(26) To obtain LGD-IBD cases, codes from the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT) were used to search a centralized pathology database at Mayo Clinic (Rochester, MN) between 2000 and 2012. Using the terms “colon”, “active chronic colitis”, “inactive chronic colitis”, and “low-grade dysplasia”, potential cases were identified and electronic records were reviewed by a single examiner to confirm CUC or CD. Patients were excluded if they had synchronous HGD or CRC. All cases underwent pathologic review by an expert GI pathologist (T.C.S.) to ensure proper classification: IBD control, SEC, SSA/P, or LGD arising in chronic colitis. Due to small numbers of samples available, SSA/P were combined with the SEC group for frequency matching against control and LGD groups on age, sex, IBD type, extent, and PSC, prior to DNA extraction.
Stool phase
Stool samples were identified from an archival collection of IBD patients participating in a biospecimen banking protocol. Cases were segregated by findings at surveillance or diagnostic colonoscopy into three groups: those with small (<1 cm diameter) lesions; those with advanced LGD (≥ 1 cm); and those with either HGD or CRC. Controls included IBD patients with no prior colorectal neoplasia and negative for neoplasia after either chromoendoscopy or random surveillance biopsies. Patients with polyposis syndromes, prior solid organ transplant or current chemo- or radio-therapy were excluded. Other than those with prior ileocecal resection for Crohn’s disease, patients with prior colonic surgery were also excluded.
All stools were collected prior to, or at least one week after, colonoscopy bowel preparation and stabilized with EDTA buffer. Samples were returned to the laboratory and normalized with additional buffer to a 1 gm/5 mL stool-to-buffer ratio and homogenized. Aliquots of 42 mL/tube were frozen at −80°C for storage until analysis.
Molecular analysis
Laboratory personnel were blinded to all clinical data.
Tissue phase
DNA extraction
For the cut-off study, DNA was extracted from frozen tissues at the Mayo Clinic Biospecimens Accession and Processing laboratory (Rochester, MN) using the Qiagen micro kit (Qiagen, Valencia, CA) prior to quantification by a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). Upon receipt of tissue-extracted DNA specimens in the research laboratory, quantity was verified by PicoGreen method (Molecular Probes, Eugene, OR). For the cohort study, DNA was extracted from paraffin-embedded tissues by laboratory personnel using the Qiagen micro kit, eluted into 100 µl of buffer and also quantified by the PicoGreen method.
Quantitative allele-specific real time target and signal amplification (QuARTS)
From each sample, 2 µg of DNA was bisulfite treated using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) and eluted in buffer. A triplex QuARTS reaction assayed methylated BMP3, NDRG4 and β-actin as previously described.(28) QuARTS assays also quantified KRAS mutations, in samples with sufficient residual non-bisulfite-treated DNA, which was PCR-amplified with primers flanking KRAS codons 12 and 13 using 20 µl of captured KRAS DNA as a template. The assays then evaluated seven mutations at codons 12 and 13 in two multiplex QuARTS reactions (cassette 1: G12S, G12D, G13D; cassette 2: G12C, G12R, G12V, G12A) and wild type β-actin. Each QuARTS reaction incorporated primers, detection probes, an invasive oligo, FAM (Hologic, Madison WI), Yellow (Hologic), Quasar® 670 (BioSearch Technologies, Novato CA) fluorescence resonance energy transfer reporter cassettes (FRETs), Cleavase® 2.0 (Hologic), GoTaq® DNA polymerase (Promega, Madison WI), MOPS buffer, MgCl2, and deoxyribonucleotide triphosphate (dNTP). Assay plates were run on a LightCycler 480 (Roche, Mannheim Germany) and also contained standards made of engineered plasmids, positive controls, negative controls, and water blanks.
Both methylated candidates and mutant KRAS copy numbers per sample were calculated in reference to standard curves. PCR products were quantified by fluorescence values in relationship to the 1:5 serially-diluted reference standards which reproducibly amplify at 5000, 1000, 200, 40, 8 and 1.6 copies per well, respectively. For values below the analytical threshold, a value of 1 copy was assigned in order to normalize results; the quantitative PCR product (in copies) of methylation markers and mutant KRAS were corrected by copies of bisulfite-treated β-actin and wild type β-actin, respectively. To determine if variability in β-actin levels impacted stool assay results, the analysis was repeated with correction of the PCR product copy counts by input DNA concentration (ng/mL). Samples that did not yield ≥50 strands of β-actin were considered inadequate for further analysis.
DNA sequencing
BRAF V600E mutation and mutations on 5 exons of p53 were determined in the cohort samples by Sanger dye-termination sequencing by the Mayo Clinic Medical Genome Facility after PCR amplification of a 20 ng DNA template. Mutation peaks were called by Mutation Surveyor® software (Softgenetics, State College, PA) and manually verified. Potential mutations in p53 were verified against the dbSNP library (http://www.ncbi.nlm.nih.gov/SNP/).
Stool phase
Aliquots were thawed at room temperature, centrifuged and the supernatant treated with polyvinylpyrrolidone to remove PCR inhibitors and spin filtered to clarify. Sequence-specific DNA biomarker targets were isolated directly from the clarified supernatant using a magnetic bead based oligonucleotide hybrid capture method.(18) QuARTS assays for methylated BMP3, NDRG4, mutant KRAS and β-actin were performed, as above.
Data abstraction
From the electronic medical record, a single examiner (D. H. J.) extracted patients’ sex, vital status, age at index diagnosis, and cigarette smoking history (current, former, or never). IBD characteristics including disease subtype (CUC or CD), duration (in years from diagnosis), and disease extent (relative to the splenic flexure) at historical maximum, and the presence or absence of primary sclerosing cholangitis (PSC) were also abstracted. The pathology record for each patient was reviewed for the development of subsequent colorectal neoplasia (CRN). The date of last colonoscopy or colectomy was used to determine the length of follow-up for time-to-event analysis.
Statistical analysis
Tissue phase
Cut-off values for quantitative assays of methylated BMP3, NDRG4 and mutant KRAS were set at 90% specificity. The sample size was based on the primary study hypothesis that markers would be detected above the cut-off value in at least half of the cases. At least 10 patients in each case subgroup would provide >90% power to detect 0.5 sensitivity against the null hypothesis of 0.1 using a 2-sided test of significance of 5%. Trends in the quantitative differences among groups in the cohort for methylated BMP3, NDRG4 and each KRAS mutation cassette were measured by Wilcoxon rank-sum tests. Potential differences among baseline characteristics of cohort patients were assessed by Wilcoxon rank-sum test for continuous variables; proportional variables were assessed by chi-square or Fisher’s exact test, where appropriate. The cumulative incidence of subsequent CRN was estimated using the Kaplan-Meier method and comparisons between patient subgroups was based on the log-rank test. Univariate proportional hazards models were constructed to examine the association of metachronous CRN with index lesion type (control, LGD, SEC and SSA/P), lesion size and levels of each DNA marker. Neoplastic events occurring within 6 months of the index diagnosis were excluded to prevent inclusion of lesions which may have been present but missed at the index colonoscopy.
Stool phase
Cut-off values for each DNA marker were also set to 90% specificity as above.
RESULTS
Study flows for tissue and stool phases are depicted in Figure 1.
Tissue cut-off study
Baseline characteristics
Seventeen frozen IBD-CRC and 12 IBD-controls were provided for analysis. Median age was 54 years (interquartile range [IQR], 48 – 62) and 53 years (IQR, 44 – 58) for IBD-CRC and control patients, respectively. Ten of 17 patients (59%) with IBD-CRC and 7/12 IBD controls (58%) were men. Median IBD disease duration was significantly greater among IBD-CRC at 20 years (IQR, 9 – 27) compared to 5 years (IQR, 1 – 14) among IBD-controls (p = 0.007).
DNA methylation and mutations
All frozen control tissues had sufficient DNA for methylation assays. Each of the DNA markers studied was significantly greater among cases than controls. The median copy number of mBMP3 was 14.7 (IQR, 1 – 58) in IBD-CRC and 0.4 (IQR, 0.17 – 0.7) in controls (p < 0.0001). Median mNDRG4 copies were 35 (IQR, 8 – 58) in IBD-CRC and 0.5 (IQR, 0.26 – 0.85) in controls (p <0.0001). One control did not have sufficient amplification of wild-type β-actin. The median number of KRAS cassette 1 (KRAS1) (representing mutations of G12S, G12D, or G13D) copies was 0.1 (IQR, 0.006 – 11) among IBD-CRC and 0.03 (IQR, 0 – 0.6) among IBD controls (p = 0.07). Additionally, the median copy number of KRAS2 (representing mutations of G12C, G12R, G12V, or G12A) was 0.07 (IQR, 0 – 12) among IBD-CRC and 0 (IQR, 0 – 0) among IBD controls (p = 0.01).
The 90th percentile value for mBMP3 copies among IBD controls was 1.08. Among IBD-CRC, 13/17 had mBMP3 copies above this threshold for a sensitivity of 76% (95% CI, 50 – 92) at 92% specificity (95% CI, 62 – 96). Fifteen IBD-CRC were above the mNRGR4 cut–off of 2.56 copies for an estimated sensitivity of 88% (95% CI, 62 – 98) also at 92% specificity. The combination of mBMP3 and mNDRG4 was positive in all IBD-CRC, but also in 1 additional control (2/12), with a slightly decreased specificity of 83% (95% CI, 51 – 97).
Cut-off values for KRAS1 and KRAS2 were 1.67 copies and 0.39 copies, respectively. KRAS mutations were less sensitive and specific than the DNA methylation markers. The combination of all measured KRAS mutations showed 47% sensitivity (95% CI, 23 – 71) at specificity of 81% (95% CI, 48 – 97).
Tissue cohort study
Baseline characteristics
After matching, pathologic review and DNA quantification, a total of 102 patients (44 IBD control, 29 LGD-IBD, 19 SEC, 10 SSA/P) with paraffin-embedded tissues were available for molecular analysis. Median age at index and proportions of patients with PSC, extensive disease, and smoking history were not significantly different between groups (Table 1). Median disease duration was lower in the SSA/P and LGD-IBD groups (p = 0.03). After exclusion of patients with insufficient DNA for marker analysis, there was a significantly higher proportion of males in the SEC group (79%) as compared to other groups (p = 0.03). A higher proportion of SEC lesions were sampled by random biopsy, resulting in a smaller size estimate (p=0.01) (Table 1).
Table1.
Clinical and molecular characteristics of cohort study lesions
| IBD control |
LGD-IBD | SEC | SSA/P | P-value* | |
|---|---|---|---|---|---|
| N | 44 | 29 | 19 | 10 | |
| Median age, years (IQR) | 56 (52–60) | 58 (51–61) | 60 (53–64) | 48 (42–57) | 0.16 |
| Males, n (%) | 18 (41%) | 15 (52%) | 15 (79%) | 7 (70%) | 0.03 |
| CUC, n (%) | 34 (77%) | 20 (69%) | 14 (74%) | 4 (40%) | 0.13 |
| Median IBD duration at index, years (IQR) | 21 (13–30) | 12 (6–21) | 23 (11–31) | 10 (2–22) | 0.03 |
| Extensive disease, n (%) | 37 (84%) | 26 (90%) | 14 (74%) | 7 (70%) | 0.36 |
| PSC, n (%) | 10 (23%) | 6 (21%) | 7 (37%) | 0 (0%) | 0.16 |
| Median lesion size, mm (IQR) | - | 3 (1–8.5) | 1 (1–3) | 4.5 (4–6.25) | 0.01 |
| mBMP3+, n (%) | 5 (11%) | 4 (14%) | 9 (47%) | 8 (80%) | <0.0001 |
| mNDRG4+, n (%) | 4 (9%) | 12 (41%) | 12 (63%) | 8 (80%) | <0.0001 |
| KRAS+, n (%)** | 6/42 (14%) | 6/28 (21%) | 6/12 (50%) | 1/8 (13%) | 0.06 |
| Any marker+, n (%) | 10 (23%) | 16 (55%) | 15 (79%) | 9 (90%) | <0.0001 |
Trend across all groups
KRAS mutations could not be assayed in all samples
CRN, colorectal neoplasia; CUC, chronic ulcerative colitis; IBD, inflammatory bowel disease; IQR, inter-quartile range; LGD, low-grade dysplasia; PSC, primary sclerosing cholangitis; SEC, serrated epithelial change, SSA/P, sessile serrated adenoma/polyp
DNA methylation and mutations
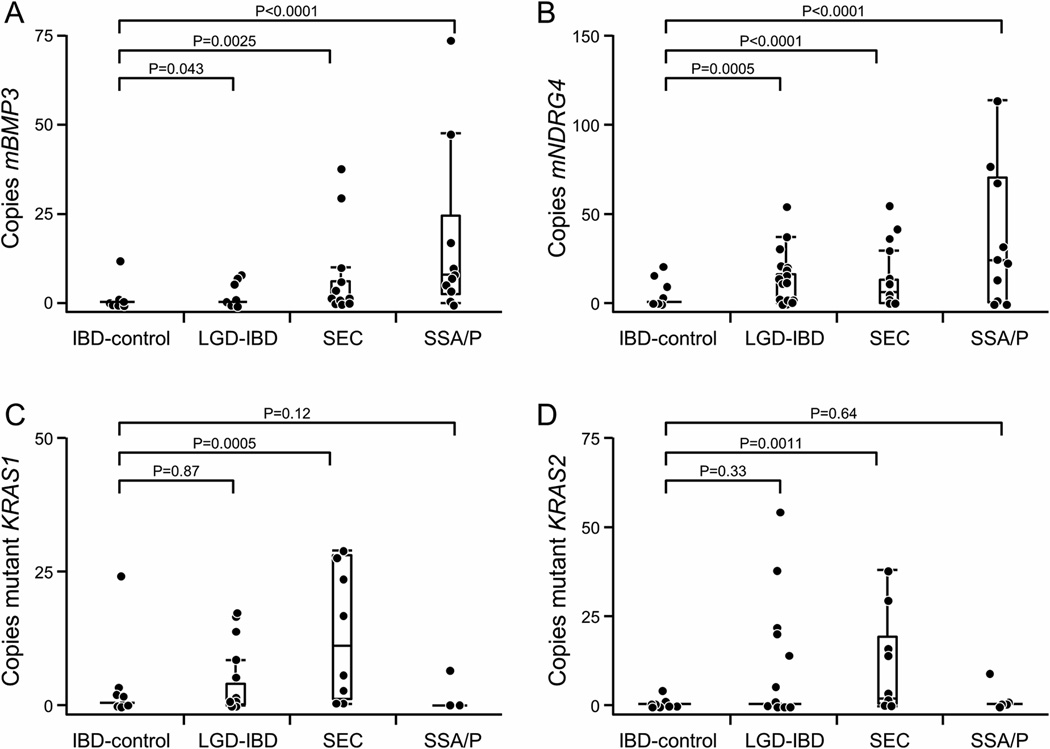
For the 102 IBD patients in the cohort study, mBMP3 and mNDRG4 levels were significantly higher for each lesion category compared to controls (Figure 2, A and B). The median levels of mutant KRAS1 and KRAS2 appeared elevated in SEC but not for controls, LGD or SSA/P (Figure 2, C and D). Applying cut-offs from the frozen tissue analysis to the cohort of FFPE samples, 5/44 IBD-controls (11%), 4/29 LGD-IBD (14%), 9/19 SEC (47%), and 8/10 SSA/P patients (80%) were positive for mBMP3 (p <0.0001). For mNDRG4, 4/44 controls (9%), 12/29 LGD (41%), 12/19 SEC (63%), and 9/10 SSA/P patients (90%) were positive (p<0.0001). Mutant KRAS was found in tissues of 6/42 IBD-controls (14%), 6/28 LGD-IBD (21%), 6/12 SEC (50%), and 1/8 SSA/P patients (13%) (p = 0.06). Any marker in the panel was positive in tissue from 10/44 IBD-controls (23%), 16/29 LGD-IBD (55%), 15/19 SEC (79%), and 9/10 SSA/P lesions (90%) (p <0.0001) (Table 1).
Figure 2.
Mutant BRAF was assayed from 35 SEC samples and all 10 SSA/P samples. A verified V600E mutation was found in 3/13 SEC tissues (9%) and 4/10 SSA/P (40%), and a V600M mutation found in 1/35 SEC samples (3%). Sufficient DNA was available for p53 sequencing on only 10 SEC samples and all 10 SSA/P. Only 1 SEC sample contained a verified p53 mutation. Due to low abundance among the SEC samples, BRAF and p53 mutations were not considered for further study.
Time to subsequent dysplasia
Further chart review excluded 13 patients from analysis for development of subsequent CRN due to colectomy or subsequent CRN in less than six months, or lack of subsequent colonoscopy, leaving 89 patients for analysis (Figure 1). There were significantly more men in the SEC group (p = 0.03) and their duration of disease was longer than other groups (p = 0.05). Otherwise, there was no significant difference in median age, IBD subtype, smoking status, disease extent, or presence of PSC between the cohort groups. Median follow-up to last colonoscopy or colectomy was 43 months (IQR, 27 – 53); the SSA/P group had significantly shorter follow-up time compared to other groups (p = 0.0007) (Supplemental Table 1).
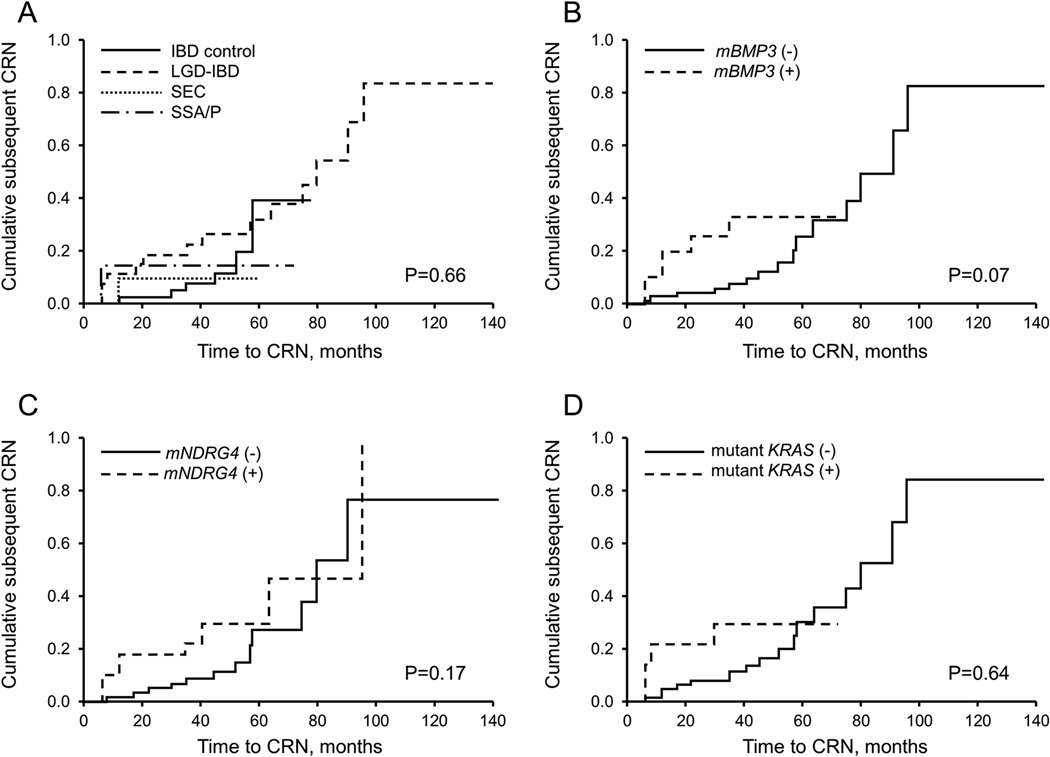
A total of 23 patients went on to develop subsequent CRN; these included 6/44 IBD controls (14%), 14/27 LGD-IBD (52%), 1/11 SEC (9%), and 2/7 SSA/P (29%). The cumulative incidence of subsequent dysplasia was not significantly different between groups (p = 0.66, likelihood ratio) (Figure 3, A). There was only one case of subsequent CRC, which occurred in the LGD-IBD group (Supplemental Table 2). Proportional hazards models did not show significant association between marker levels and subsequent CRN (Figure 3, B–D). There were no significant differences in cumulative subsequent CRN by sex, disease duration, or lesion size.
Figure 3.
Stool case-control study
Among 99 patients participating in a stool registry study between February 2008 and September 2013, 80 eligible patients were identified; these included 46 controls, 10 with lesions <1 cm, and 24 with advanced dysplasia or CRC. Among patients with small lesions, 4 were SEC (single random biopsy), 1 SSA/P (4 mm), 4 polypoid LGD (2, 2, 5 and 9 mm, respectively) and 1 focal flat LGD (random biopsy only). Among those with advanced neoplasia, there were 8 with LGD ≥ 1 cm, 8 with HGD, and 8 with CRC. Baseline characteristics were similar with the exception of IBD subtype and disease duration. Crohn’s colitis was more common among controls (23/46, 51%) than those with small (1/10, 10%) or advanced lesions (3/24, 13%) (p = 0.001), but the case groups were not significantly different from each other (p = 0.67). Median IBD disease duration was significantly lower in controls, but not significantly longer in those with small compared to advanced lesions (p = 0.42) (Supplemental Table 3).
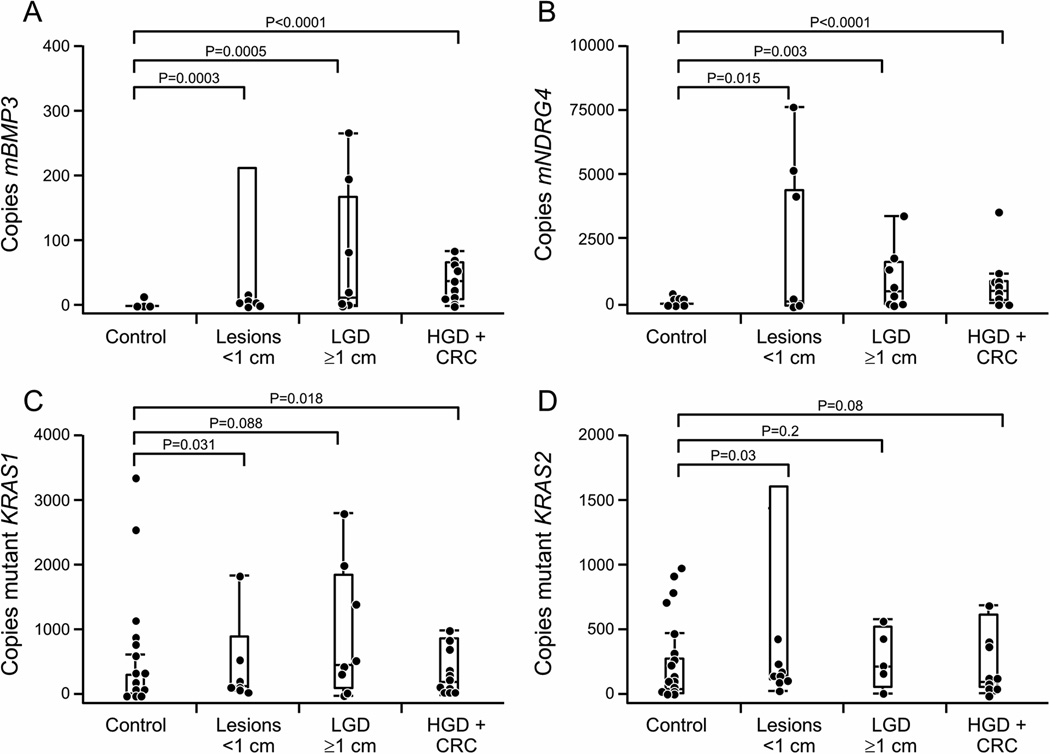
All marker levels were significantly greater in stools from patients with neoplasia than controls. Stool β-actin levels correlated with neoplasia grade in cases (p=0.0017) and histologic severity of inflammation (p=0.01) among controls. Therefore, cut-off values were selected at the 90th percentile values for each individual marker in control patient stool samples using copy numbers corrected by input DNA concentration in ng/mL rather than by copies of β-actin QuARTS product (Figure 4, A–D). Methylated BMP3 alone performed best; at 91% specificity, sensitivity was 60% for diminutive lesions (6/10), 63% for advanced LGD (5/8) and 81% for the combination of HGD and CRC (13/16). At the other extreme, mutant KRAS alone was positive in 2/10 small lesions (20%), 2/8 LGD ≥1cm (25%), and only 3/19 HGD/CRC (19%); this rate was not significantly greater than among controls, 4/46 (9%) (p=0.47). Combining KRAS with methylation markers identified a single additional LGD ≥ 1cm, but at a cost of 4 more false positives, thereby reducing the specificity to 80% (Table 2).
Figure 4.
Table 2.
Stool samples positive for concentration-corrected DNA markers
| IBD-controls (n=46) |
Lesions <1cm (n=10) |
LGD (≥1 cm)(n=8) |
HGD/CRC (n=16) |
P-value | |
|---|---|---|---|---|---|
| mBMP3+, n (%) | 4 (9%) | 6 (60%) | 5 (63%) | 13 (81%) | <0.0001 |
| mNDRG4+, n (%) | 4 (9%) | 4 (40%) | 5 (63%) | 12 (75%) | <0.0001 |
| mBMP3 or mNDRG4+, n (%) | 5 (11%) | 6 (60%) | 5 (63%) | 13 (81%) | <0.0001 |
| KRAS+, n (%) | 4 (9%) | 2 (20%) | 2 (25%) | 3 (19%) | 0.47 |
| Any marker+, n (%) | 9 (20%) | 6 (60%) | 6 (75%) | 13 (81%) | <0.0001 |
CRC, colorectal cancer; HGD, high-grade dysplasia; IBD, inflammatory bowel disease; LGD, low-grade dysplasia.
Marker copy numbers were not significantly associated with IBD extent, PSC, IBD subtype or histologic disease severity after stratification by case and control status.
DISCUSSION
In this exploratory study in patients with IBD, we observed that selected DNA tumor markers were commonly present in small adenomatous and serrated lesions, and that such markers can be detected in stool. Specifically, small adenomas occurring in IBD frequently contained aberrant methylation of BMP3 and NDRG4. SSA/Ps harbored molecular features consistent with the serrated pathway, specifically aberrant DNA methylation and BRAF mutations.
While SEC lesions appear to frequently contain mutant KRAS, this marker was not a significant predictor of subsequent dysplasia, and showed low specificity in both tissue and stool comparisons. From these findings, we hypothesize that SEC is likely a hyperplastic phenomenon. This is supported by reports in which the risk of subsequent dysplasia in IBD patients with isolated SEC appears low,(26, 29) and concordant with the favorable outcomes reported in patients with isolated hyperplastic polyps in IBD.(30) This finding will require larger studies for corroboration.
DNA methylation and KRAS mutations in the stools of IBD patients may be attributable to small and diminutive lesions; this observation informs the interpretation of non-invasive tests in development as potential adjuncts to surveillance colonoscopy. Small lesions have been excluded as an endpoint in prior case-control assessments of stool DNA for detection of both IBD-associated (21) and sporadic CRN,(25, 28) but were shown to account for at about 30% of positive stool DNA tests in a large average-risk cohort study.(18) The present data suggest that in IBD patients, both serrated and adenomatous lesions may be detectable by stool assay, and that lesion size correlates with discrimination, as has been observed in sporadic CRN.(18, 28)
The interpretation of stool DNA markers is also nuanced by the observation that stool β-actin levels appear to be increased by both inflammatory severity and the presence of neoplasia. Elevated human DNA levels with inflammation or neoplasia could be respectively attributable to exudation of inflammatory cells with colitis and to increased shedding of colonic epithelial cells with neoplasm-related hyper-proliferation. Therefore, the use of β-actin (a measure of total human DNA) as a denominator to correct tumor marker levels has potential to diminish sensitivity due to a possible confounding effect. These findings indicate that better approaches to tumor marker normalization are needed for stool testing in setting of IBD.
We acknowledge several study limitations. The overall sample size was reduced by the available tissue volume in the SEC group, as the majority of these lesions were identified on random biopsy only. Additionally, very few SSA/P cases were identified after a rigorous search, underscoring the rarity of this lesion in IBD.(26) However, we were able to stringently match serrated lesions as a group to IBD controls and IBD patients with LGD, permitting control for other known prognostic variables, including PSC, disease duration, disease extent, and age. While all index lesions in the time to subsequent dysplasia analysis were reviewed by a senior GI pathologist, the endpoint diagnoses did not undergo research review, and this could potentially have resulted in misclassification. However, only 3 of the index lesions in the cohort were re-classified in the initial pathology review, showing 96% agreement with the initial clinical diagnosis. We were also conservative to avoid potentially missed subsequent CRN at the time of index by excluding subsequent neoplastic lesions diagnosed within six months of study index. Sample sizes available for the stool study did not permit us to perform multivariate assessments for potential confounding.
Molecular testing of small neoplasms in IBD patients demonstrated adenomatous and serrated pathway profiles, and suggested a hyperplastic pathway hypothesis for SEC. IBD patients with these lesions appear to exfoliate DNA markers at increased levels, relative to IBD control patients. These findings are relevant to the design and interpretation of potential future IBD-specific stool surveillance tests, as small or diminutive lesions may contribute to positive test results.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Maxine and Jack Zarrow Family Foundation of Tulsa Oklahoma, the Paul Calabresi Program in Clinical-Translational Research (NCI CA90628), and a Pilot & Feasibility award from the Mayo Clinic Center for Cell Signalling in Gastroenterology (P30DK084567). Biospecimens were partially provided by the Clinical Core of the Mayo Clinic Center for Cell Signalling in Gastroenterology (P30DK084567). Additional partial support was provided by the Carol M. Gatton endowment for Digestive Diseases Research. Reagents for tissue QuARTS assays were provided by Exact Sciences (Madison, WI). Stool QuARTS assays were performed by Exact Sciences.
Footnotes
Role of Authors:
Johnson: concept and design, data collection, data analysis, manuscript draft
Taylor: data analysis, critical revision
Aboelsoud: data collection, data analysis
Foote: data collection, data analysis.
Yab: data collection, data analysis
Cao: data collection, data analysis
Smyrk: concept and design, data collection, pathologic review, critical revision
Loftus: concept and design, critical revision
Mahoney: data analysis/interpretation, critical revision
Ahlquist: concept and design, data analysis/interpretation, critical revision, obtained funding
Kisiel: concept and design, data analysis, critical revision, obtained funding, study supervision
Presented in part at the 115th Annual Meeting of the American Gastroenterological Association, May 4, 2014, Chicago, IL
REFERENCES
- 1.Herrinton LJ, Liu L, Levin TR, et al. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology. 2012;143:382–389. doi: 10.1053/j.gastro.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 2.Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10:639–645. doi: 10.1016/j.cgh.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Munkholm P. Review article: the incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther. 2003;18(Suppl 2):1–5. doi: 10.1046/j.1365-2036.18.s2.2.x. [DOI] [PubMed] [Google Scholar]
- 4.Iacucci M, Uraoka T, Fort Gasia M, et al. Novel diagnostic and therapeutic techniques for surveillance of dysplasia in patients with inflammatory bowel disease. Can J Gastroenterol Hepatol. 2014;28:361–370. doi: 10.1155/2014/825947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns SR, Scholefield JH, Steele RJ, et al. Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups (update from 2002) Gut. 2010;59:666–689. doi: 10.1136/gut.2009.179804. [DOI] [PubMed] [Google Scholar]
- 6.Nuako KW, Ahlquist DA, Mahoney DW, et al. Familial predisposition for colorectal cancer in chronic ulcerative colitis: a case-control study. Gastroenterology. 1998;115:1079–1083. doi: 10.1016/s0016-5085(98)70077-0. [DOI] [PubMed] [Google Scholar]
- 7.Colonoscopic Surveillance for Prevention of Colorectal Cancer in People with Ulcerative Colitis, Crohn's Disease or Adenomas. London: National Institute for Health and Clinical Excellence; 2011. [PubMed] [Google Scholar]
- 8.Farraye FA, Odze RD, Eaden J, et al. AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology. 2010;138:746–774. 774 e741–774 e 744. doi: 10.1053/j.gastro.2009.12.035. quiz e712–743. [DOI] [PubMed] [Google Scholar]
- 9.Laine L, Kaltenbach T, Barkun A, et al. SCENIC international consensus statement on surveillance and management of dysplasia in inflammatory bowel disease. Gastroenterology. 2015;148:639–651. e628. doi: 10.1053/j.gastro.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Choi CH, Rutter MD, Askari A, et al. Forty-Year Analysis of Colonoscopic Surveillance Program for Neoplasia in Ulcerative Colitis: An Updated Overview. Am J Gastroenterol. 2015;110:1022–1034. doi: 10.1038/ajg.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi PM, Nugent FW, Schoetz DJ, Jr, et al. Colonoscopic surveillance reduces mortality from colorectal cancer in ulcerative colitis. Gastroenterology. 1993;105:418–424. doi: 10.1016/0016-5085(93)90715-o. [DOI] [PubMed] [Google Scholar]
- 12.Collins PD, Mpofu C, Watson AJ, et al. Strategies for detecting colon cancer and/or dysplasia in patients with inflammatory bowel disease. Cochrane Database Syst Rev. 2006:CD000279. doi: 10.1002/14651858.CD000279.pub3. [DOI] [PubMed] [Google Scholar]
- 13.Ananthakrishnan AN, Cagan A, Cai T, et al. Colonoscopy is associated with a reduced risk for colon cancer and mortality in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2015;13:322–329. e321. doi: 10.1016/j.cgh.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velayos FS, Liu L, Lewis JD, et al. Prevalence of colorectal cancer surveillance for ulcerative colitis in an integrated health care delivery system. Gastroenterology. 2010;139:1511–1518. doi: 10.1053/j.gastro.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Mooiweer E, van der Meulen-de Jong AE, Ponsioen CY, et al. Incidence of Interval Colorectal Cancer Among Inflammatory Bowel Disease Patients Undergoing Regular Colonoscopic Surveillance. Clin Gastroenterol Hepatol. 2015;13:1656–1661. doi: 10.1016/j.cgh.2015.04.183. [DOI] [PubMed] [Google Scholar]
- 16.Itzkowitz SH, Harpaz N. Diagnosis and management of dysplasia in patients with inflammatory bowel diseases. Gastroenterology. 2004;126:1634–1648. doi: 10.1053/j.gastro.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Heigh RI, Yab TC, Taylor WR, et al. Detection of colorectal serrated polyps by stool DNA testing: comparison with fecal immunochemical testing for occult blood (FIT) PLoS One. 2014;9:e85659. doi: 10.1371/journal.pone.0085659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 19.Melotte V, Lentjes MH, van den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- 20.Loh K, Chia JA, Greco S, et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer. 2008;47:449–460. doi: 10.1002/gcc.20552. [DOI] [PubMed] [Google Scholar]
- 21.Kisiel JB, Yab TC, Nazer Hussain FT, et al. Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2013;37:546–554. doi: 10.1111/apt.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farraye FA, Odze RD, Eaden J, et al. AGA medical position statement on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology. 2010;138:738–745. doi: 10.1053/j.gastro.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 23.Itzkowitz SH, Present DH. Consensus conference: Colorectal cancer screening and surveillance in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:314–321. doi: 10.1097/01.mib.0000160811.76729.d5. [DOI] [PubMed] [Google Scholar]
- 24.Choi CH, Ignjatovic-Wilson A, Askari A, et al. Low-Grade Dysplasia in Ulcerative Colitis: Risk Factors for Developing High-Grade Dysplasia or Colorectal Cancer. Am J Gastroenterol. 2015;110:1461–1471. doi: 10.1038/ajg.2015.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lidgard GP, Domanico MJ, Bruinsma JJ, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol. 2013;11:1313–1318. doi: 10.1016/j.cgh.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Johnson DH, Khanna S, Smyrk TC, et al. Detection rate and outcome of colonic serrated epithelial changes in patients with ulcerative colitis or Crohn's colitis. Aliment Pharmacol Ther. 2014;39:1408–1417. doi: 10.1111/apt.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parian AM, Koh JM, Badamas J, et al. Serrated Epithelial Changes Are Associated With Colorectal Dysplasia in Inflammatory Bowel Disease. Gastroenterology. 2013;144:S-11. [Google Scholar]
- 28.Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–256. doi: 10.1053/j.gastro.2011.10.031. quiz e225–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atwaibi M, Batts KP, Weinberg DI, et al. Flat Serrated Change: Does it Predict the Development of Colonic Mucosal Dysplasia in Inflammatory Bowel Disease? Gastroenterology. 2012;142:S-665. [Google Scholar]
- 30.Shen J, Gibson JA, Schulte S, et al. Clinical, Pathologic, and Outcome Study of Hyperplastic and Sessile Serrated Polyps in Inflammatory Bowel Disease. Hum Pathol. 2015 doi: 10.1016/j.humpath.2015.06.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.