Abstract
The differences between countries in national income, growth, human development and many other factors are used to classify countries into developed and developing countries. There are several classification systems that use different sets of measures and criteria. The most common classifications are the United Nations (UN) and the World Bank (WB) systems. The UN classification system uses the UN Human Development Index (HDI), an indicator that uses statistic of life expectancy, education, and income per capita for countries' classification. While the WB system uses gross national income (GNI) per capita that is calculated using the World Bank Atlas method. According to the UN and WB classification systems, there are 151 and 134 developing countries, respectively, with 89% overlap between the two systems. Developing countries have limited human development, and limited expenditure in education and research, among several other limitations. The biggest challenge facing genomic researchers and clinicians is limited resources. As a result, genomic tools, specifically genome sequencing technologies, which are rapidly becoming indispensable, are not widely available. In this report, we explore the current status of sequencing technologies in developing countries, describe the associated challenges and emphasize potential solutions.
1. Introduction
Countries are generally classified into developed and developing countries using several classifying systems. There are many factors used to rank/classify countries such as the national income, human development and growth rate. For instance, the United Nations (UN) uses the UN Human Development Index (HDI) that is a summary measure of the average achievements in key aspects of human development, and the World Bank (WB) uses gross national income (GNI) per capita for this form of classification (United Nations, 2014, World Bank, 2016). These classifications are used to assess how a country is progressing, as well as what needs remain unmet, such as funding or long term strategies. Health, education, and scientific research are among the fields that need attention in the developing countries since they are tightly connected with the human development and the country's national income. Genomics and other ‘omics’ disciplines have proven to be valuable tools to be effectively applied to many fields including food, drug, and health development. The ability to become proficient and deploy ‘omics’ offers opportunities for developing nations to advance knowledge as well as improve health risk identification, diagnoses, treatment and prevention.
Genome sequencing represents a landmark in the way scientists study the genetic information of living organisms. In the 1970s, the first generation of genome sequencing methods was introduced and was industrialized and widely distributed in the following years (Sanger et al., 1977). This generation of genome sequencing methods had high accuracy and a relatively long read-length. However, it was also expensive and slow, which limited the utilization of the genome sequencing technologies to only institutions that could afford the high costs of establishing and running genome sequencing facilities. For instance, the genome sequencing component of the Human Genome Project (HGP) cost 100 million U.S. dollars and required about 13 years to complete (El-Metwally et al., 2014a). Obviously, only research institutions in developed countries were able to adopt this technology.
Subsequently, several next-generation sequencing (NGS) technologies were introduced, providing a revolutionary means for extracting the genetic information in massive amounts and within reasonable costs. NGS enabled scientists to extend the genome sequencing applications to new dimensions and invent novel applications in several fields (El-Metwally et al., 2014b). NGS is characterized by its high-throughput nature that resulted in millions of short-read sequences that were assembled to build the sequences of whole molecules using sophisticated programs that required powerful computers (El-Metwally et al., 2013). In spite of the remarkable increase in speed and decrease in costs, establishing a genome sequencing facility with NGS technology remains challenging, to the developed world, in part because the estimated cost of establishing a facility ranges between $100 K to $700 K U.S. dollars (El-Metwally et al., 2014a). Further, the costs are greater when establishing NGS facility in developing world due to the costs of shipments, customs and profit margin for local companies. This level of expense far exceeds the available funds for scientists in most developing countries.
In this report, we employed the WB classification and statistics, and the Genome OnLine Database (GOLD) genome sequencing projects' data to evaluate the utilization of genome sequencing technology in developing countries, and point out the challenges that confront the field, and proposed possible solutions.
2. Current state of sequencing technologies in developing countries
To evaluate the current status of the utilization of sequencing technologies in developing countries, we analyzed the genome sequencing projects' data available through the Genome OnLine Database (GOLD) (Reddy et al., 2015), as well as the WB classification of countries and statistics concerning worldwide expenditures for research and development (R&D) (World Bank, 2015). The GOLD database is the world's most comprehensive resource of information about the genome sequencing projects worldwide. As of November 2015, GOLD contained information for about 63,325 genome projects, including the sequencing center(s) involved in each project, which allowed us to map the projects to the countries. We excluded projects with missing information (4935) or projects involve centers from more than one country (despite the classification of the countries) (407). For the remaining projects (57,983), we mapped the sequencing centers to the related countries and obtained the number of centers and genome projects per country. Subsequently, we associated this information with the country's R&D expenditures.
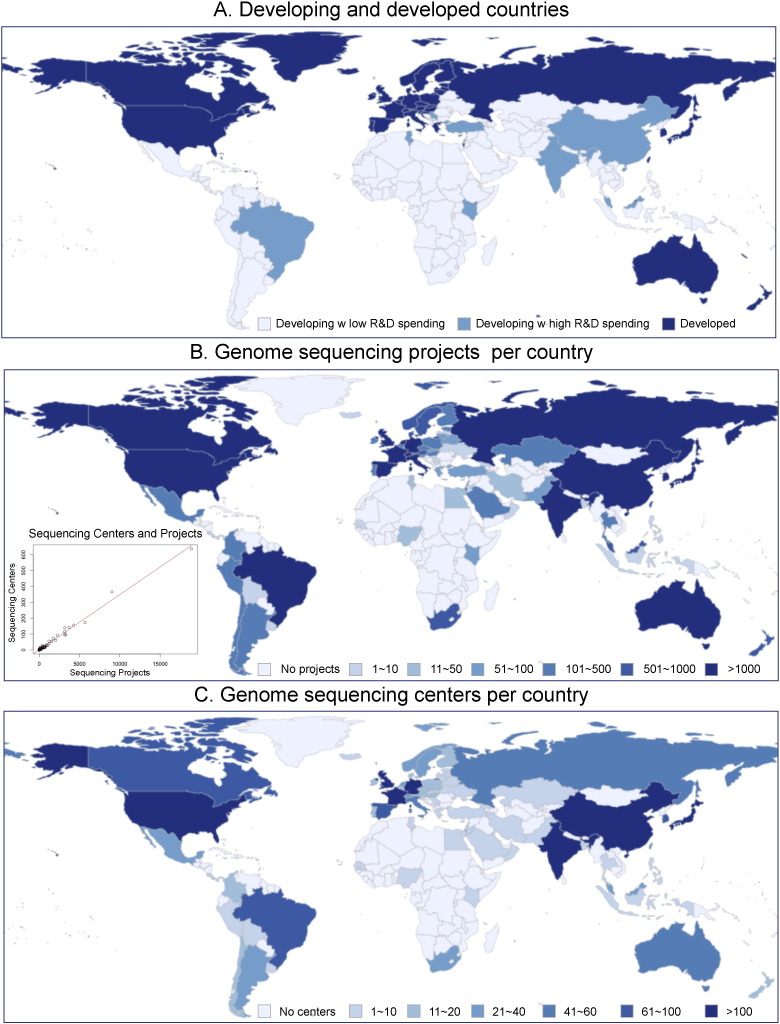
Using this data, we calculated the average R&D expenditures of the developing countries and classified them into two classes based on the R&D spending. Developing countries with R&D expenditures greater than twice the average (eight countries — see Fig. 1A) were put into a separate class (Fig. 1A). The correlation between the genome sequencing projects and sequencing centers worldwide is illustrated in Fig. 1B–C. Developed countries have more sequencing centers and therefore, are able to sequence more genomes. Developing countries with higher R&D spending, such as China, India and Brazil, are also important contributors to the field (Fig. 1B–C). This analysis suggests that genome sequencing technologies are underutilized in developing countries and that limited R&D funding is a major obstacle.
Fig. 1.
Current state of genome sequencing technologies worldwide. A) The distribution of developed and developing countries. The developing countries of high R&D spending are Brazil, China, India, Kenya, Malaysia, Serbia, Tunisia and Turkey. B) Genome sequencing projects per country. C) Genome sequencing centers per country. The insert in B shows the correlation between numbers of genome sequencing centers and the numbers of sequenced genomes per country. The analysis was performed based on data from GOLD and World Bank (see text).
3. Challenges facing sequencing technologies in developing countries
Advancements in genome sequencing technologies resulted in remarkable increases in accuracy and read-length, as well as decreases in overall cost, which has spurred a significant increase in the utilization of these technologies and expanded the applications of these technologies beyond sequencing genomes to include genome resequencing, RNA sequencing, exome sequencing and targeted sequencing in the developed world (Sims et al., 2014). However, these technologies are underutilized in the majority of research laboratories and educational institutions in the developing world. This is mainly due to the following factors.
3.1. The high cost of establishing and maintaining a sequencing facility
The significant decrease in sequencing costs over the last decade does not reflect in the prices of most of the sequencing instruments. It was estimated that the establishment of a NGS facility costs $100 K to $700 K U.S. dollars for the sequencing instruments alone (Glenn, 2011). As indicated above, in the developing countries, this cost can increase more due to the addition of other costs such as shipment and customs. In addition, infrastructure and computational facilities are required. Furthermore, the limited funding for the operational costs of such facility (cost of reagents and instrument maintenance) is one of the main factors that limit genome sequencing facilities in developing countries. This is not affordable for most educational, research and clinical laboratories in developing countries.
3.2. Lack of skilled personnel
Genome sequencing is an interdisciplinary field that requires knowledge in biology, chemistry and bioinformatics. The personnel working in this field require adequate training. Developing countries, however, have limited capacities in education and human development (United Nations, 2014). Insufficient training, therefore, is a formidable obstacle that limits the use of such facilities.
3.3. Limited access to tools for genomic data manipulation and analysis
The genomic data manipulation and analysis tools are an indispensable component of genomic research. Although several tools are distributed under different open source licenses, many advanced tools are commercialized by companies or require complicated licensing procedures to be used (El-Metwally et al., 2013). Such licensure fees are frequently beyond the abilities of institutions in the developing world.
3.4. Weak or absence of a regulatory framework
The genome sequencing research and clinical applications can involve sensitive information such as personal data (name, gender, date of birth, race), medical history, and family history with diseases. Such information should be handled carefully with restricted regulations to protect the privacy and maintain the anonymity of the source of the sample, as is the case for the most part in the developed world. However, the developing world genome sequencing research is rare or absent, in addition to an overall weak infrastructure, weak or non-existent regulatory frameworks (established legal and ethical conventions that stipulate quality and proficiency standards, specimen shipment requirements and the like) can be viewed as limiting a country's capacity. The scrutiny and rules set by the international research ethics communities are not imposed and resources for enforcement are for the most part non-existent (Conley et al., 2010).
3.5. Internet access limitations and limited access to up-to-date scientific literature
Having access to up-to-date literature is crucial for working in rapidly-evolving research fields. The recent growth of open access publications has greatly assisted in making contemporary literature available to scientists from developing countries. However, highly respected journals that require subscription fees are unaffordable for developing countries which means that important new knowledge is inaccessible (Djikeng et al., 2012). In addition, genomic data download and manipulation require fast and stable Internet connections that are not always available in developing countries, especially in Africa (Karikari et al., 2015, Slippers et al., 2011).
3.6. Low quality of outsourcing services
The lack of access to genome sequencing facilities in developing countries makes outsourcing the only available option to utilize these technologies. However, outsourcing companies provide low-quality services in many developing countries. Typically, the companies do not provide the sequencing service. Instead, they act as local representatives of companies in other countries. They send the samples to companies abroad to be sequenced and the results are sent back once completed. The improper handling of the samples, transport, and long service timing, all together are usually producing a low quality service. Our personal experience and observations show that this is an expensive and time-consuming process that may take up to several months. Furthermore, the entire outsourcing process can fail at any stage, which wastes more time and cost (Awad et al., 2015).
4. Potential solutions
Governments as well as academic and research institutes can take several actions to overcome the challenges confronting the development and utilization of this indispensable technology. We suggest potential solutions that can be part of a larger plan for the sustainable development of genome sequencing technologies in developing countries.
4.1. Increase research funds for genomic research
By realizing the importance of genomic research and its applications on health, drugs and food security, governmental policies should prioritize research funding for genomics. Our analysis of WB and GOLD data demonstrates that the utilization of these technologies in developing countries with sufficient R&D spending is comparable to developed countries (Fig. 1B–C). One clear example is the governmental fund by the National Research Foundation in South Africa which has been allocated for genomic research, hence boosting national development and sustaining a leading position for South Africa in genomic research in the continent (Karikari, 2015, Tastan Bishop et al., 2015).
4.2. Establishment of centralized sequencing facilities (centers of excellence)
Centers of excellence could be a very promising solution for providing high quality sequencing services for several laboratories and research groups. These centers can be established at the national or regional level. For example, the African Center of Excellence for Genomics of Infectious Diseases (ACEGID) at Redeemer's University (Nigeria) has been established by the support of the WB and NIH to serve several institutions in the surrounding region, including Senegal, Nigeria, and Sierra Leone (Folarin et al., 2014).
4.3. Fostering international collaborations
Collaboration between developed and developing countries in genomic research will undoubtedly result in a substantial increase in scientific capacity. Several initiatives have been set up to support such collaborations (Kumar, 2012). A recent example is the public health collaboration by The Genome Science Program at the LA National Laboratory, USA and research institutions in several developing countries including Jordan, Uganda, and Gabon (Cui et al., 2015). The developed countries can also provide support for establishing the centers of excellence (see above) by providing funding, equipment and training.
4.4. Create effective training programs
Developing the researchers' skills is one of the most important aspects in advancing genome research in developing countries. This development should be in both experimental and computational skills. Education and training in the basics of genomics and bioinformatics may begin at the level of secondary schools. More advanced training can be at the undergraduate and graduate levels (Karikari et al., 2015, Tastan Bishop et al., 2015).
4.5. Provide means of accessing up-to-date genomic data and publications
Several scientific publishers have adopted the open access publication model for all their journals including PLOS, Frontiers, and BMC, while other high-profile publishers provide open access in some of their journals such as Nature Communications and Science Advances. The World Health Organization (WHO) and the Food and Agriculture Organization (FAO) have also provided two initiatives for allowing access to scientific literature in developing countries: HINARI (http://www.who.int/hinari/en/) and AGORA (http://www.aginternetwork.org/en/) (Djikeng et al., 2012). Despite these efforts, additional strategies are required to effectively tackle the gap in access to new knowledge. Creating central academic libraries that have access to non-open access journals, and establishing agreements between the universities and research centers in the developing countries and scientific publishers to grant access to scientific articles for free would fill this gap.
4.6. Utilize modern sequencing technologies that minimize the infrastructure requirements
Modern genome sequencing technologies, such as MinION by Oxford Nanopore Technologies, have minimized the requirements of adopting this technology to an affordable device, preparation kit, stable Internet connection and standard PC. Although the technology is still new and developing, it nonetheless represents a promising solution for utilizing a wide range of genome sequencing applications and with minimal laboratory and computational skills.
4.7. Developing alternative experimental methods
Occasionally, alternative experimental methods could be the best choice in some cases where sequencing facilities are not available. In our recent work, we have proposed an alternative restriction enzyme-based method for bacterial identification using standard microbiology techniques (Awad et al., 2015). Developing such alternative experimental methods is more fruitful when combined with other computational methods.
5. Conclusion
Several challenges confronting researchers and scientists in developing countries have delayed their ability to participate in the genomic revolution. While, many developing countries including India, South Africa, Mexico, and Brazil were able to make significant strides in utilizing genomic technologies through the availability of sufficient funds, establishing institutions for genomics, and the training of personnel, the situation remains unaltered in several regions of the world, especially Africa. We recommend increasing research funding, establishing centers of excellences, encouraging international collaborations and organizing specialized training programs as possible potential solutions for the sustainable future improvement of genomic research in developing countries.
Acknowledgments
The authors would like to thank the Editor for the thorough insight and valuable comments and suggestions, which were helpful in improving the paper. KAM would like to thank the University of Sharjah, UAE for administrative support.
References
- Awad M., Ouda O., El-Refy A., El-Feky F.A., Mosa K.A., Helmy M. FN-identify: novel restriction enzymes-based method for bacterial identification in absence of genome sequencing. Adv. Bioinformatics. 2015;14 doi: 10.1155/2015/303605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley J.M., Doerr A.K., Vorhaus D.B. Enabling responsible public genomics. Health Matrix Clevel. 2010;20:325–385. [PubMed] [Google Scholar]
- Cui H.H., Erkkila T., Chain P.S.G., Vuyisich M. Building international genomics collaboration for global health security. Front. Public Health. 2015;3:264. doi: 10.3389/fpubh.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Ommeh S., Sangura S., Njaci Isaac, Ngara M. Genomics and potential downstream applications in the developing world. In: Nelson K.E., Jones-Nelson B., editors. Genomics Applications 335 for the Developing World. NY, USA. 2012. pp. 335–356. [Google Scholar]
- El-Metwally S., Hamza T., Zakaria M., Helmy M. Next-generation sequence assembly: four stages of data processing and computational challenges. PLoS Comput. Biol. 2013;9:e1003345. doi: 10.1371/journal.pcbi.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Metwally S., Ouda O.M., Helmy M. Next Generation Sequencing Technologies and Challenges in Sequence Assembly. Springer; New York, New York, NY: 2014. Challenges in the next-generation sequencing field. [Google Scholar]
- El-Metwally S., Ouda O.M., Helmy M. Next Generation Sequencing Technologies and Challenges in Sequence Assembly. Springer; New York, New York, NY: 2014. Novel next-generation sequencing applications. [Google Scholar]
- Folarin O.A., Happi A.N., Happi C.T. Empowering African genomics for infectious disease control. Genome Biol. 2014;15:515. doi: 10.1186/s13059-014-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- Karikari T.K. Bioinformatics in Africa: the rise of Ghana? PLoS Comput. Biol. 2015;11:e1004308. doi: 10.1371/journal.pcbi.1004308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikari T.K., Quansah E., Mohamed W.M.Y. Developing expertise in bioinformatics for biomedical research in Africa. Appl. Transl. Genomics. 2015;6:31–34. doi: 10.1016/j.atg.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., editor. Genomics and Health in the Developing World. first ed. Oxford University Press; Oxford, UK: 2012. [Google Scholar]
- Reddy T.B.K., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–D1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- Slippers B., Majozi T., Nelwamondo F.V., Steenkamp C.M., Van Heerden E., Wright C.Y. Internet access constrains science development and training at South African universities. S. Afr. J. Sci. 2011;107 (1 Pages) [Google Scholar]
- Tastan Bishop Ö., Adebiyi E.F., Alzohairy A.M., Everett D., Ghedira K., Ghouila A., Kumuthini J., Mulder N.J., Panji S., Patterton H.-G. Bioinformatics education—perspectives and challenges out of Africa. Brief. Bioinform. 2015;16:355–364. doi: 10.1093/bib/bbu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations . 2014. Human Development Report 2014. [Google Scholar]
- World Bank World Bank: research and development expenditure data [www document] 2015. http://data.worldbank.org/indicator/GB.XPD.RSDV.GD.ZS/countries URL. (accessed 12.29.15)
- World Bank World Bank: country and lending groups data [www document] 2016. http://data.worldbank.org/about/country-and-lending-groups URL. (accessed 12.29.15)