Summary
Defining the molecular regulators of hematopoietic stem and progenitor cells (HSPCs) requires in vivo functional analyses. Competitive bone marrow transplants (BMTs) compare control and test HSPCs to demonstrate the functional role of a genetic change or chemical perturbation. Competitive BMT is enabled by antibodies that specifically recognize hematopoietic cells from congenic mouse strains due to variants of the cell surface protein CD45, designated CD45.1 and CD45.2. The current congenic competitor strain, B6.SJL-Ptprca Pepcb/BoyJ (CD45.1), has a substantial inherent disadvantage in competition against the C57BL/6 (CD45.2) strain, confounding experimental interpretation. Despite backcrossing, the congenic interval over which the B6.SJL-Ptprca Pepcb/BoyJ strain differs is almost 40 Mb encoding ∼300 genes. Here, we demonstrate that a single amino acid change determines the CD45.1 epitope. Further, we report on the single targeted exon mutant (STEM) mouse strain, CD45.1STEM, which is functionally equivalent to CD45.2 cells in competitive BMT. This strain will permit the precise definition of functional roles for candidate genes using in vivo HSPC assays.
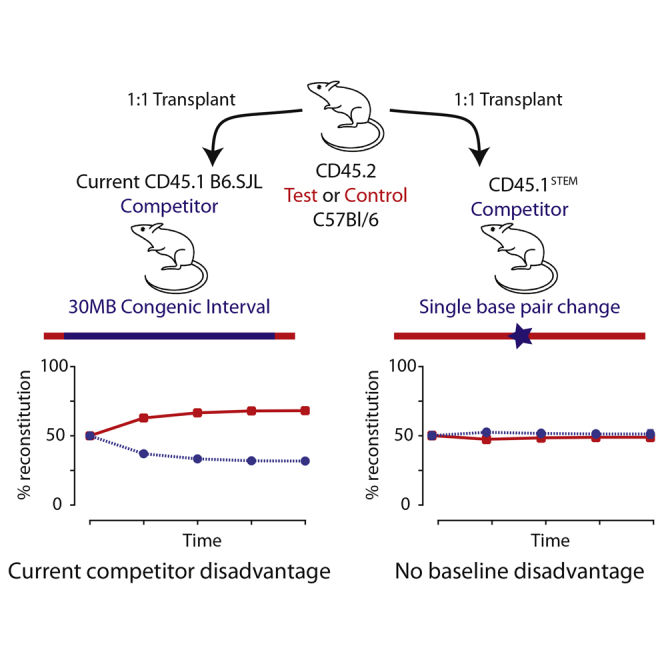
Graphical Abstract
Highlights
-
•
Competitive transplantation is a fundamental tool for examining HSPC biology
-
•
The congenic interval of the B6.SJL-Ptprca Pepcb/BoyJ mouse affects HSPC function
-
•
CD45.1 and CD45.2 epitopes differ by one amino acid
-
•
A single amino acid change in the C57BL/6N strain creates the CD45.1STEM competitor
Competitive bone marrow transplantation experiments are used in the study of hematopoietic stem cell function. Scadden and colleagues describe the generation of a single targeted exon mutant mouse strain, CD45.1STEM, that avoids inherent competitive imbalance seen with current models and acts as an internal control when experiments are performed in a pure C57BL/6 background, reducing the number of animals required.
Introduction
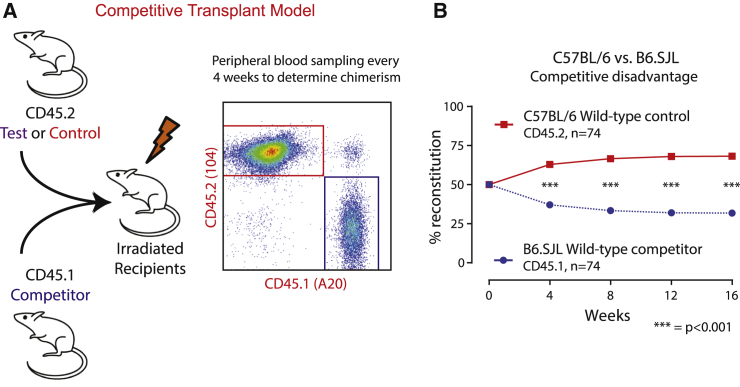
The development of transgenic and knockout mouse models has permitted an examination of how the gain of or loss of a particular gene affects the fitness of hematopoietic stem and progenitor cells (HSPCs). One commonly used approach is to transplant recipient mice with an equal combination of normal and genetically modified HSPCs. By following the progeny of the transplanted cells in the recipient mice over the course of ≥16 weeks, one can identify genetic modifications that give the HSPC a functional advantage or disadvantage compared with wild-type (WT) cells. This competitive transplant approach is a critical tool for assessing the in vivo functional impact of genetic (knockout, transgenic, knockin) or chemical modifications, and has been extremely useful in advancing HSPC biology (Figure 1A).
Figure 1.
The Current B6.SJL Strain Shows an Inherent Competitive Disadvantage
(A) A model of a typical competitive bone marrow transplantation experiment. In the experiment, bone marrow cells from a TEST (CD45.2) mouse are combined with an equal number of bone marrow cells from a COMPETITOR (CD45.1) mouse and transplanted into irradiated recipient mice. The peripheral blood chimerism is followed for 16–20 weeks as a functional assay of hematopoietic stem cell fitness.
(B) HSPCs derived from the current B6.SJL (CD45.1) competitor strain show an inherent competitive disadvantage when compared with HSPCs from wild-type C57BL/6 mice. In these experiments, the bone marrow from three littermate C57BL/6 mice or three littermate B6.SJL mice were pooled. 500,000 nucleated bone marrow cells from each donor strain were combined (1 million cells total) and transplanted by intravenous injection in lethally irradiated recipients. Four experiments were performed, two in competition with WT C57BL/6J donors and two in competition with WT C57BL/6NJ donors. Results represent the mean ± SEM. Given the large number of replicates (74 recipient mice in each arm), the error bars are not visible as they are smaller than the squares. ∗∗∗p < 0.001.
In the competitive transplantation model, a means of distinguishing normal and genetically modified stem cells is essential. Fluorescent protein tagging is theoretically attractive, but the efficiency of labeling, effects of the fluorophore expression on cell function, and immunogenicity of the non-native protein are limitations that compromise its utility. The most commonly used approach of in vivo tracking takes advantage of polymorphisms in the extracellular domain of the transmembrane receptor tyrosine phosphatase protein CD45 (Ly5, Ptprc, B220), a 220-kDa protein expressed on all subsets of leukocytes.
The CD45.1 and CD45.2 alleles differ by only five amino acids within the extracellular domain (Zebedee et al., 1991), resulting in epitope changes that permit specific recognition by monoclonal antibodies (Shen, 1981). The majority of the commonly used mouse strains express the CD45.2 allele. Backcrossing of mice expressing the CD45.1 allele (SJL) into the C57BL/6 background (CD45.2) has resulted in the development of the mouse strain B6.SJL-PtprcaPepcb/Boy (B6.SJL). As the mice have been backcrossed over many generations, they have been termed congenic, with the presumption that they differ only at the CD45 locus. Table 1 contains a description of the nomenclature for the mouse strains described in this article.
Table 1.
A Description of the Mouse Strains Used in the Article
| Formal Name | Commonly Referred to as | CD45 Isoform | Strain Notes |
|---|---|---|---|
| C57BL/6J | Black 6 Bl6 C57 |
CD45.2 | The C57BL/6 syngeneic mouse has been bred for many generations at Jackson Laboratory (J) and at the NIH (N), leading to subtle strain differences. The NIH N substrain is also bred at Jackson Laboratory resulting in the NJ designation. The N substrain is used in the International Mouse Knockout Program to generate new mutations in an effort to standardize reagents |
| C57BL/6N | |||
| C57BL/6NJ | |||
| B6.SJL-PtprcaPepcb/Boy | B6.SJL Pep-Boy |
CD45.1 | This is the currently used congenic strain. Despite backcrossing, the SJL mouse differs from the C57BL/6 mouse by ∼40 Mb and ∼300 genes |
| C57BL/6N-CD45.1STEM | 45.1STEM | CD45.1 | A single base/single amino acid change in the background of the C57BL/6N strain results in the CD45.1STEM strain |
A competitive transplant is typically performed by transplanting an equal number of CD45.1 and CD45.2 donor cells into the background of CD45.1, CD45.2, or heterozygous (CD45.1/2) mice. As the recipient mice are given a myeloablative dose of γ-irradiation prior to the transplant, there are few (usually less than 5%, mostly T lymphocytes) residual host cells, and thus the more-readily available and less expensive CD45.2 mice are generally used as transplant recipients (Figure 1A). The advantage of using F1 CD45.1/2 heterozygote recipient mice is that the host residual cells are doubly stained by both CD45.1 and CD45.2 monoclonal antibodies, and thus can be distinguished from the transplanted cells.
Hundreds of competitive transplants have been reported in the literature. Most of these are performed to test the impact of a genetically modified test gene. Unfortunately, the WT congenic B6.SJL (CD45.1) HSPCs have an inherent disadvantage over WT C57BL/6 (CD45.2) HSPCs, with reported defects in homing efficiency, reduction in transplantable long-term hematopoietic stem cells, and a cell-intrinsic engraftment defect as demonstrated by Waterstrat et al. (2010) and us (Figure 1B). This advantage is particularly marked if C57BL/6J mice are used as recipients and is mildly attenuated when B6.SJL mice are used as recipients, suggesting that B6.SJL cells may be impaired in their ability to interact with C57BL/6J stromal cells (Waterstrat et al., 2010). This lack of competitive balance between the two putatively congenic strains has called into question the interpretation of prior results concerning the impact of modified genes and has created a conundrum for the field.
In order to verify that changes in HSPC function are due to the modified gene and not to the inherent imbalance of the strains currently used in competitive transplantation, an additional set of control experiments is required. To compensate for the baseline engraftment defect of the B6.SJL, which varies according to the experimental protocol (e.g., number of cells transplanted, conditioning regimen, and strain of recipient), a competitive transplant using WT mice must be performed in parallel. This requires essentially double the number of mice and is not consistently performed, resulting in ambiguity as to whether it is the modified gene or the mouse strain that accounts for differential engraftment.
The molecular basis for the inherent advantage of C57BL/6J (CD45.2) HSPCs over B6.SJL (CD45.1) is not known. The CD45 gene is inherited within a locus that comprises almost 40 megabases and more than 300 predicted genes (Waterstrat et al., 2010). The basis for the discordant HSPC function of the two genotypes is presumably within this 40 Mb region and may include immunologically or functionally active gene products.
Several international consortia, including KOMP (Knock-Out Mouse Program) and EUCOMM (European Conditional Mouse Mutagenesis Program), have ongoing programs to generate new strains in the uniform C57BL/6N background (Sacca et al., 2013). Thus, the ideal control for HSPC characterization of these new strains is a functionally equivalent C57BL/6N mouse with the minimal change in the CD45 gene to render it detectable by a CD45.1 antibody. Here, we define the specific epitope difference between the CD45.1 and CD45.2 alleles and describe the creation and testing of a CD45.1 knockin point mutation in the background of the most commonly used C57BL/6N mouse strain. Our data demonstrate that, contrary to the B6.SJL strain, HSPC derived from this mouse are functionally equivalent. We therefore propose that this mouse represents the ideal competitor for competitive HSPC transplant, reducing the number of controls required when working in the C57BL/6N background; we have named it CD45.1STEM (single targeted exon mutation).
Results
A Single Amino Acid Change Confers the Differential Recognition of CD45.1 and C45.2 Antibodies
The CD45 open reading frame (Gene 19,264, NM_001111316) is 3,882 base pairs in length and encodes a 1,293 amino acid protein (NP_001104786). This transcript comprises at least 33 exons (possibly up to 35 exons with a number of variant transcripts) and the regulatory importance of the corresponding introns is not known (Holmes, 2006). The extracellular domain itself is made up of approximately 15 exons and 544 amino acids. The crystal structure of the CD45 intracellular domain, the enzymatic protein phosphatase, has been solved (Nam et al., 2005). However, the structure of the extracellular domain is not yet known.
High-specificity commercially available antibodies exist for distinguishing the hematopoietic cells from mice of a CD45.1 or CD45.2 background; clone A20 specifically recognizes CD45.1, while clone 104 specifically recognizes CD45.2. The CD45 epitope that forms the basis of this specificity has not previously been defined.
An examination of the sequence differences between CD45.1 and CD45.2 reveals numerous base changes throughout the transcript (Zebedee et al., 1991). However, despite these changes, the CD45.1 and CD45.2 proteins differ only by five amino acids in the extracellular domain (Zebedee et al., 1991). Thus, we felt that one of these amino acids, or a combination thereof, most likely represents the epitope of the two monoclonal antibodies that differentially recognize the CD45.1 and CD45.2 isoforms.
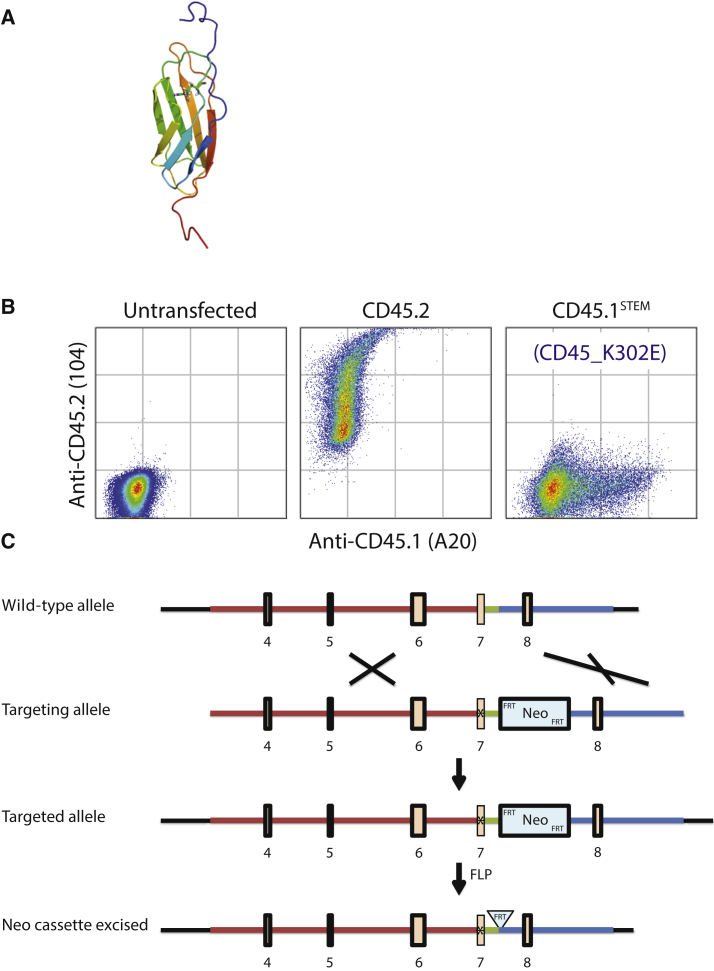
Using site-directed mutagenesis, we found that a single A to G point mutation (base pair 904, AAA → GAA) resulting in a lysine to glutamic acid (K302 → E302) amino acid change, formed the entire basis of the CD45 antibody specificity. The glutamic acid alteration occurs in an exposed loop of the first of three fibronectin type 3 domains found in the extracellular domain of CD45. The homologous arginine residue in the X-ray structure of the human fibronectin type 3 protein is highlighted in the ribbon model shown in Figure 2A.
Figure 2.
A Single Amino Acid Change Determines the Specificity of CD45.1 and CD45.2 Antibodies
(A) A ribbon model of the extracellular fibronectin type III domain. The K302E mutation is highlighted.
(B) The single K302E amino acid change determines CD45.1 versus CD45.2 antibody specificity of the Ptprc (CD45) gene. 293T human embryonic kidney cells were transiently transfected with a plasmid containing the original CD45.2 open reading frame, or the open reading frame with the K302E mutation, and then stained with anti-CD45.1 (clone A20) or anti-CD45.2 (clone 104) antibodies.
(C) Design of the CD45.1STEM mouse bearing a single point mutation targeting the Ptprc gene in the background of the C57BL/6N strain. The long homology (red), middle homology (green), and short homology (blue) arms are indicated. See also Figure S1.
The antibody specificity that results from this point mutation was validated by transient transfection of these variant CD45 open reading frames into 293T human embryonic kidney (HEK) cells. Flow cytometry using the commercially available anti-CD45.1 (A20) and anti-CD45.2 (104) antibodies confirmed the switch in antibody recognition that resulted from the K302E amino acid change (Figure 2B).
The C57BL/6N-CD45.1STEM Knockin Mouse Was Generated with a Single Amino Acid Change
We designed and generated a mouse bearing the single CD45 A904 →G904 point mutation in the background of a C57BL/6N embryonic stem cell line (Figures 2C and S1) using the contracted services of inGenius Targeting Laboratories (iTL). The mice were crossed with an Flp recombinase mouse to remove the FRT/neomycin resistance cassette (NeoR) and then bred to homozygosity (Figure S2).
The CD45.1STEM Hematopoietic Stem Cells Demonstrate Equivalence in Competitive Transplantation Assays
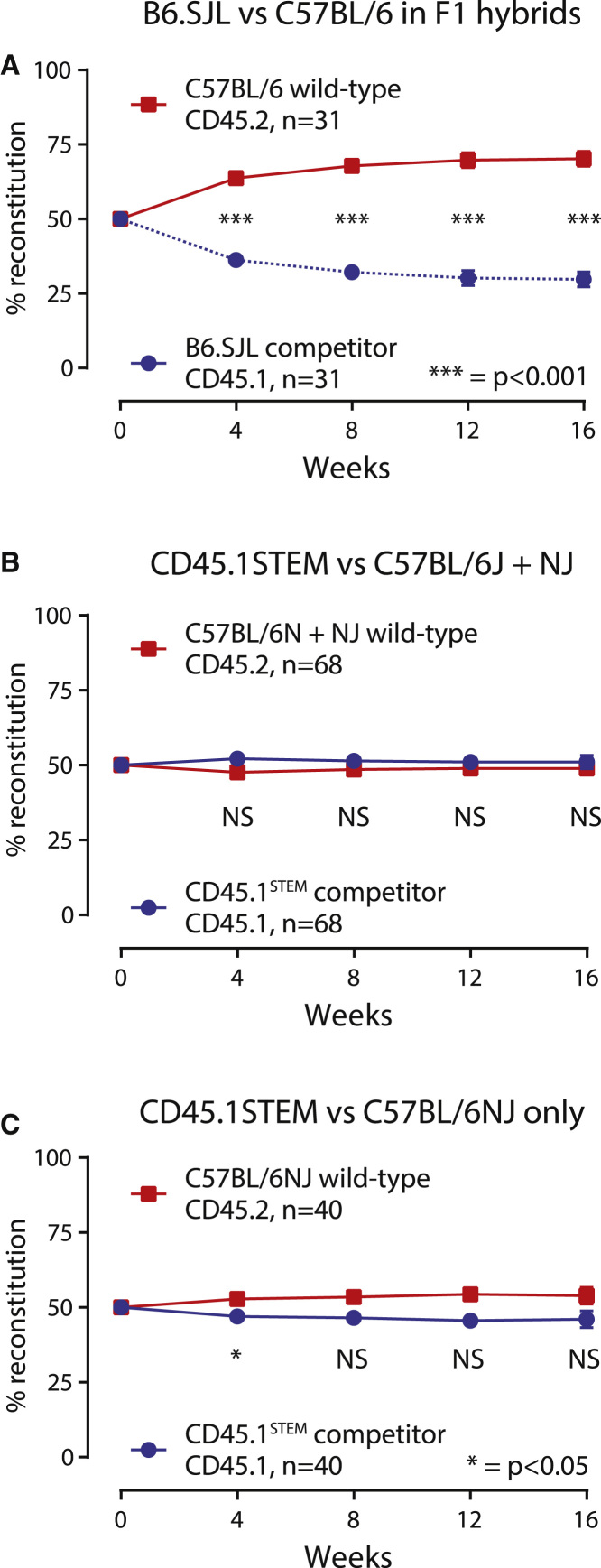
Competitive bone marrow transplantation assays were performed to compare the fitness of the conventional CD45.1-expressing strain B6.SJL and our CD45.1STEM strain against CD45.2 C57BL/6J or C57BL/6NJ (Table 1 and Figure 3). The assays were performed using both WT C57BL/6NJ and well as B6.SJL-PtprcaPepcb/Boy∗C57BL/6NJ F1 hybrid recipients.
Figure 3.
The CD45.1STEM Mouse Is Functionally Equivalent to C57BL/6 in Competitive Transplant Assays
For all competitive transplantation experiments, 500,000 nucleated bone marrow cells from each pool of CD45.2 and CD45.1 donors were co-transplanted into lethally irradiated recipients. The percent reconstitution was tracked in the peripheral blood over a period of 16 weeks.
(A) HSPCs derived from the current B6.SJL (CD45.1) competitor strain have an inherent competitive disadvantage when compared with HSPCs from wild-type C57BL/6 (CD45.2) mice even when transplanted into the background of F1 B6.SJL∗C57BL/6J hybrid recipients. Three experiments, two in competition with wild-type C57BL/6J donors and one in competition with wild-type C57BL/6NJ donors, were performed (31 recipient mice in total).
(B) HSPCs derived from the CD45.1STEM competitor strain show functional equivalence when compared with HSPCs from wild-type C57BL/6 mice. Four experiments, two in competition with wild-type C57BL/6J donors and two in competition with wild-type C57BL/6NJ donors, were performed. The recipients in these four experiments were a mixture of C57BL/6J, C57BL/6NJ, and F1 B6.SJL∗C57BL/6J hybrids (68 recipient mice in total).
(C) HSPCs derived from the CD45.1STEM competitor strain show functional equivalence when compared specifically with HSPCs from the wild-type C57BL/6NJ substrain of mice. Two experiments were performed (40 recipient mice in total). The results represent the mean ± SEM Given the large number of replicates, the error bars are not visible as they are smaller than the squares.
∗p < 0.05, ∗∗∗p < 0.001. See also Figure S3.
Donor mice were matched for age and gender. Donor cells were pooled from three mice to minimize individual variation, purified over a Ficoll density gradient, and pooled at a 1:1 ratio. A total of 1 million cells (500,000 cells from each genotype) were transplanted by tail vein injection into lethally irradiated mice, and the ratio of transplanted cells was confirmed by flow cytometry. Recipient mice were conditioned with two doses of 600 cGy radiation (separated by 8–12 hr, last dose given 18–24 hr prior to transplantation). This resulted in less than 5% residual recipient marrow during the 4 month post-transplantation analysis.
WT B6.SJL HSPC showed a consistent and inherent competitive disadvantage over WT HSPCs from C57BL/6 mice (Figure 1B; n = 74), consistent with previous results (Waterstrat et al., 2010). This effect remained apparent when transplanting into F1 B6.SJL∗C57BL/6J hybrids (Figure 3A; n = 31), arguing against an immunological cause of rejection.
In comparison, HSPCs derived from CD45.1STEM HSPCs were functionally equivalent to C57BL/6 HSPCs in hematopoietic reconstitution at 16 weeks (Figure 3B; n = 68). They were also functionally equivalent when compared specifically with HSPCs from the C57BL/6NJ strain (Figure 3C; n = 40), arguing against any subtle substrain variation.
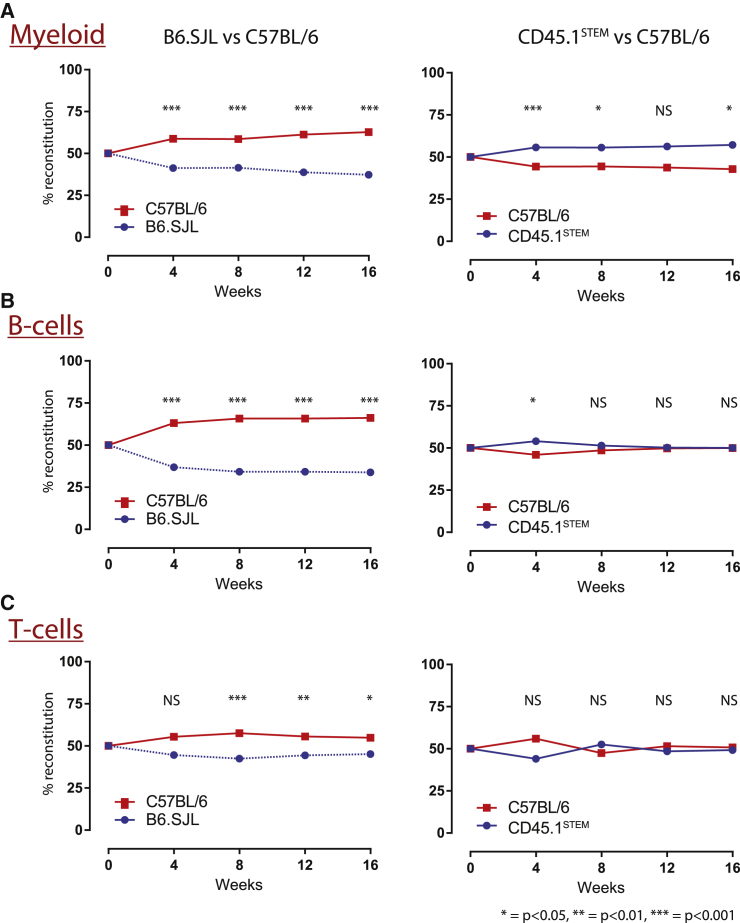
The disadvantage of the B6.SJL HSPC was most pronounced in the myeloid (Figure 4A) and B-lymphoid (Figure 4B) lineages and less pronounced in the T-lymphoid lineage (Figure 4C). Of note, there was a subtle but statistically significant advantage of the CD45.1STEM HSPCs in reconstitution of the myeloid lineage at 16 weeks (Figure 4A, left panel, p = 0.018). However, since reconstitution of the other lineages is equally balanced, and since there was statistical equivalence in aggregate, we do not regard this as reflecting a biologically meaningful difference, but investigators should be aware of this possibility. Rather, we think the result is due to our experimental design. By design, we transplanted a small number of donor cells (only 500,000 cells per strain) to stress the stem cell compartment, and this resulted in increased mouse to mouse variability (Figure S3A).
Figure 4.
The CD45.1STEM Strain Shows Improved Reconstitution across Hematopoietic Lineages
(A–C) Assessment of hematopoietic reconstitution within myeloid (A), B-lymphoid (B), or T-lymphoid (C) lineages for mice transplanted with B6.SJL (left panels) or CD45STEM (right panels) donor cells in competition with wild-type C57BL/6 HSPCs. Four experiments, two in competition with wild-type C57BL/6J donors and two in competition with wild-type C57BL/6NJ donors, were performed. The results represent the mean ± SEM. Given the large number of replicates (left panels, n = 74; right panels, n = 68), the error bars are not visible as they are smaller than the squares. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
The functional equivalence of the C57BL/6N-CD45.1STEM HSPCs with C57BL/6 strains was consistent with our hypothesis that the single point mutant knockin mouse would act as an improved competitor strain.
Discussion
Our data demonstrate that, by introducing a single point mutation into the genetic background of a C57BL/6N mouse, we could generate a knockin strain that is identifiable using the commercially available CD45.1 antibody (clone A20). HSPCs from this knockin strain, named C57BL/6N-CD45.1STEM (single targeted exon mutation) are equivalent in competitive transplant assays when compared with HSPCs from either C57BL/6N or C57BL/6J mice.
This has important implications for competitive transplantation experiments. Because they have the same reconstitution potential, modified HSPCs from any C57BL/6N or C57BL/6J strain can be directed compared with our CD45.1STEM strain without an additional control group. This contrasts with the situation when using the current B6.SJL competitor strain. Given the inherent disadvantage of WT B6.SJL HSPCs, one requires both a test competitive transplant (CD45.2 test versus B6.SJL) as well as a control competitive transplant (CD45.2 WT versus B6.SJL) in order to correctly determine HSPC fitness. As such, the C57BL/6N-CD45.1STEM strain provides a better competitor and offers the possibility of reducing the number of mice and cost associated with competitive HSPC transplantation assays.
We identified the CD45 epitope that is recognized by the anti-CD45.1 and anti-CD45.2 antibodies. Then, by engineering a mouse strain with a single point mutation, resulting in this single amino acid change in a C57BL/6N background, we avoided alterations in introns or other regulatory domains and created a strain that allows for a truly equivalent competitive bone marrow transplant.
We would like to highlight a few important methodological variables in performing competitive HSPC transplantation assays. (1) We stress the importance of pooling bone marrow cells from a minimum of three donor mice. Despite the fact that these mice may be genetically identical WT littermates, there exist subtle variations in reconstitution ability between individuals that can be minimized by pooling multiple donors. (2) Slight differences in reconstitution ability are exacerbated by transplanting low numbers of HSPCs (Lacombe et al., 2010). As we transplanted only 500,000 bone marrow cells from each strain, this led to impressive mouse to mouse variability that was overcome by performing multiple biological replicates (Figure S3A). (3) The high fractionated dose of radiation (2 × 600 cGy) given as pre-transplant conditioning ensures that there are few residual endogenous recipient cells, although in all cases we would advocate the use of F1 hybrid CD45.1∗CD45.2 recipients to permit distinguishing the doubly labeled residual host cells (Figure S3B).
In summary, the C57BL/6N-CD45.1STEM mouse represents a technical advance for investigators who wish to accurately quantify the fitness of genetically, or otherwise modified, HSPCs derived in the common background of the C57BL/6N or C57BL/6J mouse strains. Unlike the previous competitor strain, HSPCs from the CD45.1STEM mouse have comparable reconstitution potential, eliminating the need for a separate control group. For these reasons, we propose that the CD57Bl/6N-CD45.1STEM mouse should become the standard competitor in competitive bone marrow transplantation assays.
Experimental Procedures
Site-Directed Mutagenesis
The mouse Ptprc cDNA was purchased from Genecopeia in a Gateway-technology-compatible vector (clone MOC23994). Using the QuickChange II kit (Agilent Technologies), PCR reactions were designed to introduce the A904G mutation into the plasmid. Following bacterial transformation, individual colonies were screened by Sanger sequencing. The desired mutants were recombined using LR clonase II (Thermo Fisher) into the pLEX_307 lentiviral expression vector, where expression is driven by the eIF1a promoter (a kind gift from the Broad Institute, Cambridge, MA).
Screening of Mutations for Antigenicity
HEK293T cells were transfected (FuGENE, Promega) with pLEX_307 plasmids driving expression of the native or mutant mouse Ptprc protein described above. Four days following transfection, cells were trypsinized and the single-cell suspension was incubated with fluorochrome-conjugated antibodies CD45.1-FITC (A20) and CD45.2-APC (104) (eBioscience). The cells were analyzed on an LSR II instrument (BD Biosciences).
Design of the Targeting Vector
An ∼8.70 kb region used to construct the targeting vector was first subcloned from a positively identified C57BL/6 bacterial artificial clone (BAC; RP23:131J7). The region was designed such that the long homology arm (LA) extends ∼6.19 kb 5′ to the site of the point mutation (A→G) in exon 7 and the FRT flanked NeoR cassette was inserted 400 bp 3′ to the point mutation. The short homology arm (SA) extends 2.11 kb 3′ to the FRT flanked NeoR cassette. The targeting vector was constructed using Red/ET recombineering technology.
The BAC was subcloned into a ∼2.4 kb backbone vector (pSP72; Promega) containing an ampicillin selection cassette for retransformation of the construct prior to electroporation. A pGK-gb2 FRT NeoR cassette was inserted into the gene as described in the project schematic. The total size of the targeting construct (including the vector backbone and NeoR cassette) was 14.25 kb.
The mutation was engineered by overlap extension PCR. Two primary PCR fragments that overlap 12 bp just 5′ to the point mutation were generated using primers PT1/PT2 and PT3/PT4. The A → G mutation was engineered into primer PT3. The two primary products were then mixed and used as a template in a secondary PCR reaction in which the PT1/PT4 primer pair amplifies the entire sequence containing the point mutation. A 5′ endogenous restriction enzyme site, PmlI, was used with the 3′ endogenous restriction enzyme site, PacI, for insertion of the mutant PCR fragment into the subcloned vector (Figure S1).
The targeting vector was confirmed by restriction analysis after each modification step and by sequencing using primers designed to read from the selection cassette into the SA (N1) and the genomic sequence containing the point mutations (N2). P6 and T73 primers anneal to the BAC subclone sequence and read into the 5′ and 3′ ends of the subcloned vector. Sequencing with primers PROC PT2 confirmed that no errors were introduced into the PCR modified region.
Generation of Knockin Mice
The linearized targeting plasmid DNA was electroporated into embryonic stem cells. After selecting with G418, resistant colonies were screened for homologous recombination of the Ptprc targeted allele by PCR and Southern blot analysis. The clones with homologous recombination were identified and isolated. These embryonic stem cells were injected into blastocysts from BALB/c mice. The blastocysts were transferred to pseudopregnant ICR foster mothers, and chimeric males were obtained. Subsequently, chimeric mice were mated to mice constitutively expressing the Flp recombinase to produce F1 heterozygous, targeted, NeoR cassette-deleted mice. The F1 mice were crossed to produce F2 homozygous mutants (CD45.1STEM) and WT littermate controls.
To screen for deletion of the NeoR cassette, primer set NDEL1 and NDEL2 were used. The PCR reaction consisted of 30 cycles (denature at 94° for 30 s, anneal at 60° for 30 s, extension at 72° for 60 s). The PCR product of the WT reaction is 295 bp. After neomycin deletion, one set of FRT sites remain (70 bp). A second band with a size of 365 confirmed deletion of the NeoR cassette. The intact NeoR cassette is not amplified by this PCR reaction because of its large size (Figure S2A).
Genotyping Mice for the CD45.1STEM Point Mutation
Genotype confirmation to identify the A904G point mutation was done by sequencing the PCR product amplified by primers SC1 (upstream) and REVG1 (downstream). The PCR reaction consisted of 30 cycles (denature at 94° for 30 s, anneal at 55° for 30 s, extension at 72° for 60 s). The amplified product of the SC1/REVG1 reaction is 678 bp, and the SC1 primer can be used in the subsequent sequencing reaction (Figure S2B).
Competitive Transplantation Assays
All experiments were performed in accordance with the institutional animal care and use committee guidelines at Massachusetts General Hospital.
Transplantation experiments were performed using 8- to 12-week-old, age-matched female mice. Total bone marrow nucleated cells were extracted from the long bones of C57BL/6NJ mice (Jackson Laboratory), B6.SJL-PtprcaPepcb/BoyJ (Jackson Laboratory), and CD45.1STEM, then purified using Ficoll-Paque Plus (GE Healthcare). For each experiment, the bone marrow was extracted from three donor mice per group and then pooled prior to transplantation.
Twenty-four hours prior to transplantation, recipient mice were lethally irradiated with two 600 cGy doses separated by 6 hr. For each recipient, a total of one million nucleated bone marrow cells were injected in the tail vein: 500,000 cells from a C57BL/6 donor group mixed with 500,000 cells of either B6.SJL-PtprcaPepcb/BoyJ or CD45.1STEM donor group.
Peripheral blood was sampled every 4 weeks, and the red blood cells were lysed with ACK buffer (Lonza). The leukocytes were resuspended in PBS supplemented with 2% fetal bovine serum and EDTA (1 mM). The cells were labeled (30 min, 4°) with the following antibodies: CD3-BUV395, CD45.1-FITC, B220-PE, CD11b-PE-Cy7, CD45.2-APC. The cells were analyzed on an LSRII instrument (BD Biosciences).
Statistical Analysis of the Competitive Transplants
Following transplantation of the 1:1 mixture of CD45.1+ and CD45.2+ cells, the ratio of CD45.1 cells to CD45.2 cells was assessed by flow cytometry to account for slight imbalances in mixing. This “time 0 real ratio” was used to adjust the subsequent ratios of CD45.1 and CD45.2 cells in the peripheral blood at weeks 4, 8, 12, and 16. In this manner, the data can be presented to most clearly demonstrate the deviation, or lack of deviation, from the starting point of 50%.
At each time point, we analyzed the difference between CD45.1 reconstitution and CD45.2 reconstitution. This was done via the Wilcoxon signed-rank test which was used to compare CD45.1 and CD45.2 reconstitution and obtain raw p values; then, in order to adjust for multiple testing, the Benjamini-Hochberg procedure was used to get adjusted p values.
Author Contributions
F.E.M. and D.B.S. designed the experiments. F.E.M. performed the mutagenesis experiment. F.E.M. and D.B.S. performed the competitive transplantation assays. D.B.S. performed flow cytometric analysis. D.T.S. supervised the project and edited the manuscript.
Acknowledgments
We would like to thank Youmna Kfoury, Jonathan Hoggatt, Jacqueline Bachand, Cam Dung Le, Katrina Maxcy, and Alexa Carver. We would like to thank the staff of ingenious Targeting Laboratories, especially AnnMarie Degruccio and Anne Schirmer. David Tan provided the mouse graphic in Figure 1. Brian D. Sykes created the ribbon structure in Figure 2A. Jiantao Shi helped with the statistical analysis. We would also like to thank the Harvard Stem Cell Institute (HSCI) for the Seed Grant that provided funds for the generation of this mouse strain as well as members of the CRM/HSCI Flow Cytometry Core. F.E.M. was supported by a Clinician-Scientist training award from the Canadian Institutes of Health Research. D.B.S. was supported by the American Society of Hematology, the Leukemia and Lymphoma Foundation, and Alex's Lemonade Stand Foundation. D.T.S. was supported by the Gerald and Darlene Jordan Chair in Medicine at Harvard University.
Published: May 12, 2016
Footnotes
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.04.010.
Contributor Information
Francois E. Mercier, Email: fmercier@mgh.harvard.edu.
David T. Scadden, Email: dscadden@mgh.harvard.edu.
Supplemental Information
References
- Holmes N. CD45: all is not yet crystal clear. Immunology. 2006;117:145–155. doi: 10.1111/j.1365-2567.2005.02265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J., Herblot S., Rojas-Sutterlin S., Haman A., Barakat S., Iscove N.N., Sauvageau G., Hoang T. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. 2010;115:792–803. doi: 10.1182/blood-2009-01-201384. [DOI] [PubMed] [Google Scholar]
- Nam H.-J., Poy F., Saito H., Frederick C.A. Structural basis for the function and regulation of the receptor protein tyrosine phosphatase CD45. J. Exp. Med. 2005;201:441–452. doi: 10.1084/jem.20041890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacca R., Elder B., Wasson K. 2013. The C57BL/6 Mouse: the Role of the C57BL/6N Mouse in the Creation of Future Genetically Engineered Models.http://www.criver.com/files/pdfs/rms/c57bl6/rm_rm_r_c57bl6_white_paper.aspx accessed 19 January 2016. [Google Scholar]
- Shen F. Monoclonal antibodies to mouse lymphocyte differentiation alloantigens, in: monoclonal antibodies and T-cell hydridomas. In: Hammerling G.J., Hammerling U., Kearney J.F., editors. Perspectives and Technical Advances. Elsevier/North-Holland; 1981. pp. 25–31. [Google Scholar]
- Waterstrat A., Liang Y., Swiderski C.F., Shelton B.J., Van Zant G. Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood. 2010;115:408–417. doi: 10.1182/blood-2008-03-143370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee S.L., Barritt D.S., Raschke W.C. Comparison of mouse Ly5a and Ly5b leucocyte common antigen alleles. Dev. Immunol. 1991;1:243–254. doi: 10.1155/1991/52686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.