Abstract
Effective use of agricultural residual biomass may be beneficial for both local and global ecosystems. Recently, biochar has received attention as a soil enhancer, and its effects on plant growth and soil microbiota have been investigated. However, there is little information on how the physical, chemical, and biological properties of soil amended with biochar are affected. In this study, we evaluated the effects of the incorporation of torrefied plant biomass on physical and structural properties, elemental profiles, initial plant growth, and metabolic and microbial dynamics in aridisol from Botswana. Hemicellulose in the biomass was degraded while cellulose and lignin were not, owing to the relatively low-temperature treatment in the torrefaction preparation. Water retentivity and mineral availability for plants were improved in soils with torrefied biomass. Furthermore, fertilization with 3% and 5% of torrefied biomass enhanced initial plant growth and elemental uptake. Although the metabolic and microbial dynamics of the control soil were dominantly associated with a C1 metabolism, those of the 3% and 5% torrefied biomass soils were dominantly associated with an organic acid metabolism. Torrefied biomass was shown to be an effective soil amendment by enhancing water retentivity, structural stability, and plant growth and controlling soil metabolites and microbiota.
In African dryland landscapes, improving nutrient-poor soils is important for increasing agricultural productivity, particularly because a significant population growth is expected in this region over the next 100 years. In the Republic of Botswana in southern Africa, Jatropha curcas L. has received attention as a biomass resource1,2 although has exhibited unsatisfactory growth due to the arid climate, chilling injury, and oligotrophic soil conditions (aridisols)3,4. Therefore, methods of soil amendment are expected to promote its agricultural production in nonfarming lands.
In dryland ecosystems, such as arid African landscapes, termites, which build termite mounds, play a key role in soil amelioration5. Their effects may be artificially achieved through soil amendment using charcoal-like soil enhancers6,7. Charcoal has a porous structure and harbors soil microbes8 that play roles in soil enrichment. Activated charcoal has been reported to increase nutrients, reduce nutrient leaching, enhance nutrient uptake, and increase crop production9,10. Recently, biochar, which is made from post-harvest biomass residues, has been studied for its use to amend soils in various African countries11,12.
Torrefied biomass, which is a kind of biochar made at low temperature under anaerobic conditions, is made by the torrefaction of plant biomass derived from grasses and/or woods. Torrefied biomass revealed isothermal pyrolyzed biomass at relatively low temperature ranges of 200 °C–300 °C13,14. The treatment evaporates the internal water from the biomass with an economic use of energy15; therefore, this type of biochar exploits this resource of carbon-rich material. Watanabe et al. characterized torrefied Jatropha biomass components, and they suggest that detoxification of phorbol ester by thermal degradation renders it suitable as a soil amendment16.
The beneficial effects of biochar on plant growth and soil microbiota have also been investigated12,13,17,18. For example, Anders et al.19 reported that biochar enhanced the positive correlation between nutrients and microbiota more in nutrient-poor soils than in nutrient-rich soils. Fox et al.20 also reported that soil amended with biochar enhanced the microbially mediated nutrient mobilization of S and P resulting in an improvement in plant growth. However, the relationship between the metabolites and microbiota has not been evaluated in soil amended with biochar.
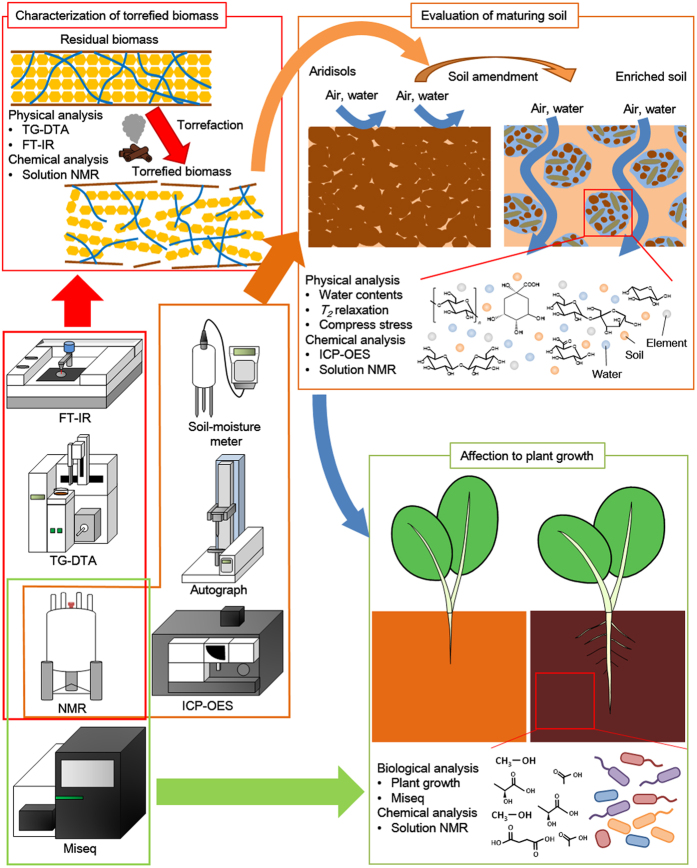
A soil study focused on the humic substance or microbial properties in forest soils using nuclear magnetic resonance (NMR)21,22. However, a comprehensive approach based on physical, chemical, and biological viewpoints was greatly anticipated to evaluate the effects of biochar on soil amendment. We had evaluated various soil properties, such as plant degrading abilities, using some analytical strategies and compared NMR and other meta-analytical methods23,24. In this study, we focused on torrefied biomass and evaluated the soil amendment effects of this biomass in an aridisol from Botswana. We evaluated water retentivity, chemical components, effect on plant growth, and metabolic and microbial variations in the soil (Fig. 1).
Figure 1. Schematic representation of this study.
Effects of soil amendment with torrefied biomass were evaluated using three steps: (1) characterization of Jatropha curcas pyrolysis profiles, (2) evaluation of soil physical and chemical properties, and (3) evaluation of plant growth ability using J. curcas seedlings.
Results and Discussion
Torrefication profile of J. curcas
To prepare the torrefied biomass, the thermal degradation profile of J. curcas was characterized by thermogravimetric (TG)-differential thermal analysis (DTA), attenuated total reflectance (ATR)-Fourier transform infrared (FTIR), grain size distribution, and 1H-13C heteronuclear single quantum coherence (HSQC) NMR spectra (Figs S1–S4). Dehydration occurred at approximately 70 °C and degradation of the hemicellulose components at approximately 160 °C–280 °C (Fig. S1). The torrefied biomass bonds, revealed by the vibration of the benzene C–H bond, the syringyl C–O vibration, and cellulose and hemicellulose C–H deformations that were known to cleave under a fast pyrolysis treatment25, were broken between 240 °C and 250 °C (Fig. S2; Table S1). The grain size of torrefied biomass was over 1,400, 710, 355, and 106 μm or smaller, and the ratios were 11.40%, 25.30%, 28.45%, 23.06%, and 11.79%, respectively (Fig. S3). The chemical component comparison of raw and 240 °C torrefied biomass showed that some signals such as adipate, d-fructose, and l-glutamine disappeared, but many saccharide signals such as maltodextrin remained (Fig. S4; Table S2). Because denaturation of the supramolecular structures with the remaining main components such as cellulose at 240 °C may be able to provide adsorption of water and nutrient elements during incorporation into soil for plant growth, the torrefication temperature in these experiments was set at 240 °C. In addition, major toxic compounds (the profiles were already characterized in a previous study16) were under the detection limit in the NMR spectra of biomass torrefied at 240 °C, suggesting very low concentrations of these toxins in the biomass. Therefore, we presumed that the soil community should be less affected by toxins when biomass torrefied at 240 °C was used for soil amendment experiments. Moreover, most bacteria that existed in the biomass were eliminated because the torrefaction process has the effect of dry sterilization. Thus, we considered that further analyses using soil ecosystems would be unaffected by external bacteria that existed in the biomass.
Influence of torrefied biomass on soil
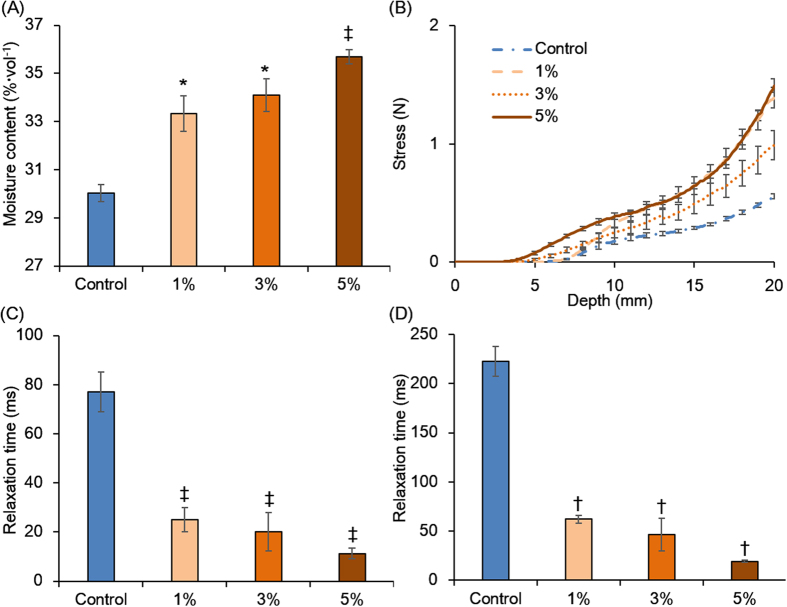
The physical properties of soils with or without torrefied biomass were evaluated by measuring the water-holding capability, soil compression stress, and relaxation time (Figs 2 and S5). The water contents of the control and 1%, 3%, and 5% torrefied biomass soils were significantly different at 30.0%, 33.3%, 34.1%, and 35.7%/volume, respectively (Fig. 2A). A similar trend in water contents was observed in soils with raw biomass (Fig. S6). Furthermore, the maximum mechanical stress values of the soils were 0.56, 1.39, 0.99, and 1.49 N, respectively (Fig. 2B). The 1% and 5% torrefied biomass soils were significantly different from the control at depths of 17.8 and 5.6 mm, respectively. The water T2 relaxation times of these soils for bound water with biomass in the soils were 76.99, 25.05, 20.16, and 11.27 ms, respectively, and significant differences between control and soils with torrefied biomass were observed (Fig. 2C). The T2 relaxation times of free water were 222.78, 62.02, 46.44, and 19.13 ms, respectively (Fig. 2D). The abundance ratios of binding vs. free water in the control and 1%, 3%, and 5% torrefied biomass soils were 63:37, 81:19, 66:34, and 79:21, respectively. The higher compression stress values and shorter T2 relaxation times in the torrefied biomass soils compared with the control soil suggested that torrefied biomass soils facilitated soil structural stability, resulting in higher water contents and retention capabilities. These results indicated that the physical character of soil was greatly changed by the addition of torrefied biomass and that soil water retention was improved. Although a similar effect of water retention capability was observed in soils mixed with raw biomass, the chemical properties were different between raw and torrefied biomass (Figs S1, S2 and S4). The torrefied biomass retained the cellulose component by degradation of the supramolecular structure known as lignocellulose. Therefore, one of the advantages of utilizing torrefied biomass compared with raw biomass is the easy access and exchange to energy by microbiota.
Figure 2. Physicochemical characterization of soils used for Jatropha cultivation with or without torrefied biomass.
The moisture contents of saturated absorption (A) and compression mechanical stress under wet soil conditions (B). T2 relaxation time of binding (C) and free water contents (D). The error bars show the standard error of the mean and the p value for comparison of the control with each sample calculated using Welch’s t test. *p < 0.05, †p < 0.01, and ‡p < 0.005.
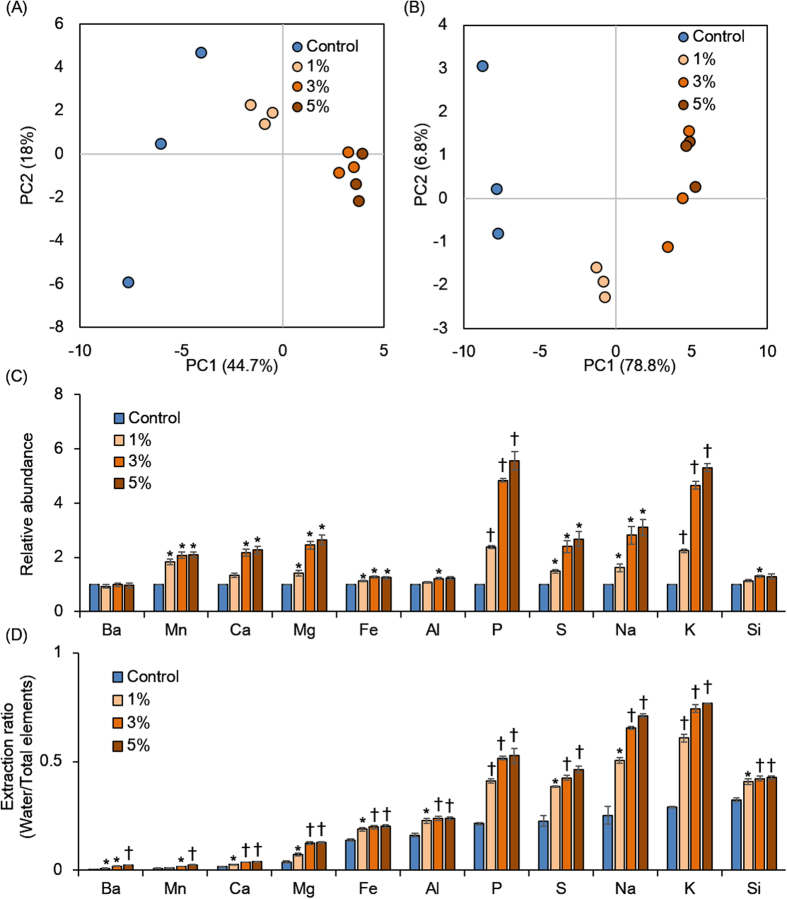
Soil elements with or without torrefied biomass were characterized by water extraction followed by HNO3 extraction using inductively coupled plasma-optical emission spectrometry (ICP-OES; Fig. 3). Principal component analysis (PCA) profiles showed similar trends between water and HNO3 extractions, meaning that the elemental profiles were different between control soils and those with added torrefied biomass (Fig. 3A,B). Some elements such as K, P, and S increased in the torrefied biomass soils compared with the control because these elements were derived from the torrefied biomass (Fig. 3C). In addition, the dissolution rates of some elements such as K, Na, and P in water compared with those in HNO3 were highly increased in the torrefied biomass soils compared with the control (Fig. 3D). Since water-soluble elements are readily accessible to plants, the elemental availability (especially of K, Na, and P) to plants was improved by the addition of torrefied biomass to the soils. However, many elements were largely dissolved in HNO3, suggesting that in nature these elements are trapped by supramolecular structures in the soil.
Figure 3. Evaluation of elemental components in soils.
Soil elemental profiles were evaluated from extractions of water (A) and HNO3 (B) using PCA score plots and the relative abundance of soil total elements compared the control with torrefied biomass (C) and extraction ratios compared water with total extracted elements (D). The error bars show the standard error of the mean and the p value for comparison of the control with each sample calculated using Welch’s t test. ∗p < 0.05 and †p < 0.01.
Soil maturation
The metabolic dynamics of microbiota during soil maturation were evaluated using 1H-NMR spectra (Fig. S7). Metabolic profiles from day 0 to 21 in the soils with torrefied biomass were varied, but each profile at day 28 was similar to that of the control. This result indicated that organic components such as polysaccharides were digested during the period from day 0 to 21, and that the available organic components were finally lost by day 28 (as in the control). Moreover, the metabolic profiles of soil with fishmeal were largely varied; degradation of creatine and trimethylamine N-oxide (TMAO) was accompanied by the production of acetate, methylamine, dimethylamine, and trimethylamine from day 0 to 13 (Fig. S8). Creatine and TMAO are the most abundant components in fish water-soluble fractions26,27. This result indicated that the soil microbiota metabolized and utilized these fish components and produced some metabolites such as methylamine, dimethylamine, and trimethylamine, which are derivatives of TMAO. However, a lot of nutrients remained in the fishmeal soil compared with the torrefied biomass, indicating that an excess of nutrients in the fishmeal soil prevented seedling and plant growth. Therefore, only the soils with torrefied biomass were used for further analyses.
Effect of torrefied biomass on the initial growth stage of J. curcas
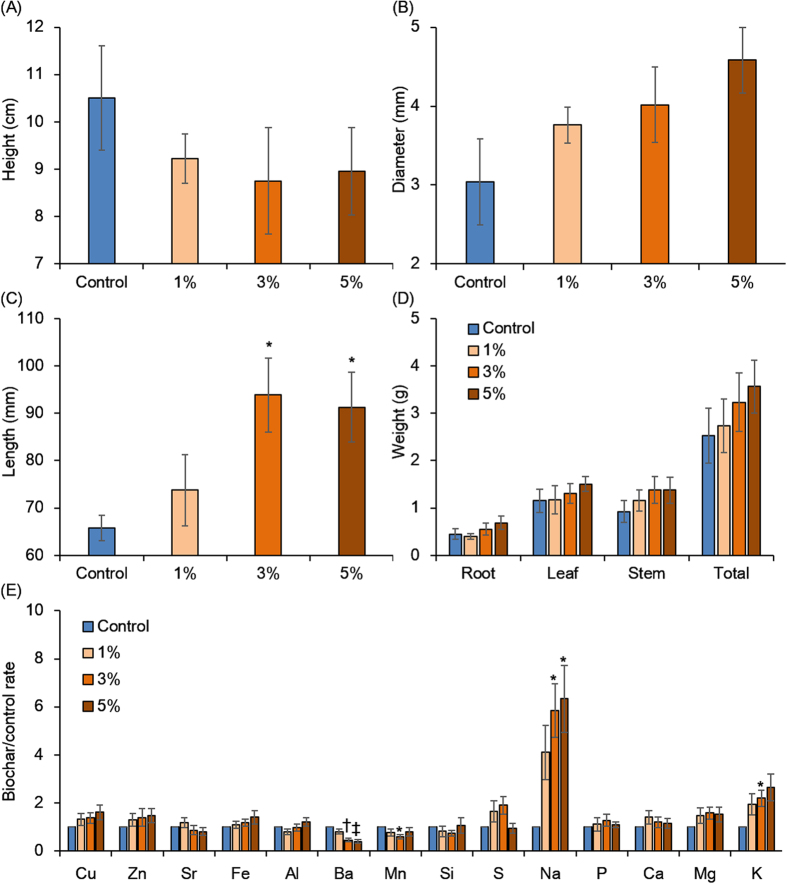
To evaluate the effect of torrefied biomass on Jatropha growth, germinated J. curcas were transplanted to matured soils and grown for 4 weeks. The plant heights of the control and of the 1%, 3%, and 5% torrefied biomass treatments were 10.50 and 9.23, 8.75, and 8.95 cm, respectively; the stem diameters were 3.04, 3.76, 4.02, and 4.58 mm, respectively (Fig. 4A,B); and the root lengths were significantly greater by 65.81, 73.75, 93.85, and 91.31 mm, respectively (Fig. 4C). The weights of the roots, stems, and leaves also tended to increase with increasing torrefied biomass treatments (Fig. 4D). The uptake of soil elements by plants in the initial growth stages was characterized by ICP-OES (Fig. 4E). K and Na were present in higher concentrations, but Si, Mn, and Ba were lower in plants grown in torrefied biomass-amended soils than in the control. Although Na is known to inhibit plant growth, the low concentration in the soils should not affect plant growth and less than 0.04% was incorporated into the plants. In addition, it is known that excessive Mn and Ba inhibit plant growth28,29. Moreover, the carboxyl functional group in organic compounds and polysaccharides such as hemicellulose in plants and alginic acid in algae is also known to be heavy metal adsorbents30,31,32. The result shows that torrefied biomass has the capacity to provide beneficial minerals such as K and to inhibit toxic element absorption. Thus, torrefied biomass can be utilized as a soil conditioner for soil amendment.
Figure 4. Evaluation of the effect of torrefied biomass on the initial plant growth stage.
The effect of torrefied biomass on the plant phenotype during the initial growth stage was evaluated by plant height (A), stem diameter (B), root length (C), whole weight (D), and elemental content ratios of torrefied biomass to the control (E). The error bars show the standard error of the mean and the p value for comparison of the control with each sample, calculated using Welch’s t test. *p < 0.05 and ‡p < 0.005.
Metabolic and microbial dynamics in soil during initial plant growth
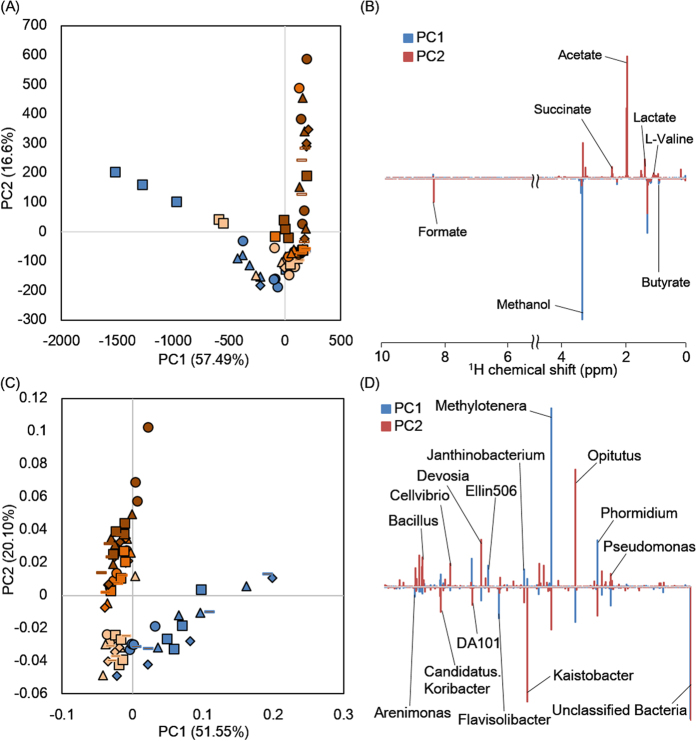
Metabolic soil dynamics during initial plant growth were evaluated using 1H-NMR spectra (Fig. 5A,B) in combination with 2D-J spectra for annotation (Fig. S9; Table S3). The metabolic profiles were clustered based on the differences between the control and torrefied biomass soils. In the 3% and 5% torrefied biomass soils, the profiles were PC1 positive with the factors being l-valine, lactate, acetate, and succinate. Organic acids produced by anaerobic microbes due to cellulosic degradation are known to be phosphate-solubilizing and plant growth-promoting substances33,34. Thus, the torrefied biomass was considered to effect soil fertilization and enhance plant growth. Formate decreased from week 0 to 1, and methanol and butyrate showed a similar trend increasing in weeks 1 and 3 and decreasing in week 4 in the control (Fig. S10A–C). In contrast, acetate, lactate, and succinate showed similar decreases from week 0 to 4 and l-valine increased from week 0 to 2 with 5% torrefied biomass (Fig. S10D–G).
Figure 5. Soil metabolic and microbial profiles during plant growth.
Metabolic (A,B) and microbial profiles (C,D) of soil during initial plant growth were evaluated using PCA score plots (A,C) and loading plots (B,D). Symbols in the score plot represent the control (blue), 1% (light orange), 3% (orange), and 5% torrefied biomass (brown) and weeks 0 (circle), 1 (triangle), 2 (diamond), 3 (square), and 4 (bar).
Microbial profiles during plant growth were analyzed with a MiSeq sequencer (Fig. 5C,D). The control contributed to PC1, and 3% and 5% torrefied biomass soils contributed to PC2. In the control, the most predominant microbe was Methylotenera sp. In contrast, some microbes including Opitutus sp. and Devosia sp. were associated with the 3% and 5% torrefied biomass soil. Methylobacterium sp. and Methylotenera sp., which exist on plant leaves and are associated with methanol consumption and production35,36, were highly abundant, and Methylotenera sp. greatly increased from week 0 to 1 in the control (Fig. S11A,B). Based on methanol variations, these microbes were inferred to be dominantly associated with a C1 metabolism in the control. Bacillus sp., Devosia sp., and Opitutus sp. were highly abundant, and Devosia sp. and Opitutus sp. showed similar trends during plant growth in 3% and 5% torrefied biomass soils (Fig. S11C–E). Devosia sp. and Opitutus sp. are known to use lactate as a carbon source37,38, and the time-course variations were associated with acetate, lactate, and succinate dynamics. Thus, these microbes were inferred to be associated with the metabolism of these organic acids and considered to be key players in torrefied biomass adjusted soil environments for promoting plant growth. In the future, the effects of soil amendments using torrefied biomass should be evaluated by field experiments.
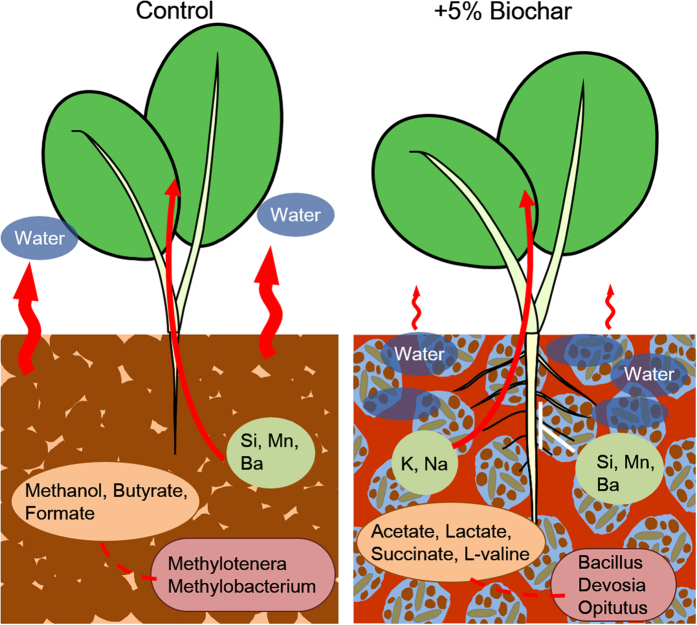
In conclusion, the effects of soil amended with torrefied biomass were evaluated with respect to their physical properties, initial plant growth, and metabolic and microbial dynamic soil profiles (Fig. 6). Torrefied biomass improved the physical and structural properties of soil such as water retentivity and structural stability. Soil amended with 3% and 5% torrefied biomass enhanced the initial growth of J. curcas in the form of increased stem diameter, root length, and element uptake ability. Although the metabolic and microbial dynamics of the control were associated with a C1 metabolism, those of the 3% and 5% torrefied biomass samples were associated with an organic acid metabolism. These results indicate that torrefied biomass is effective as a soil amendment by increasing water retentivity and structural stability, enhancing plant growth, and controlling soil metabolites and microbiota.
Figure 6. Schematic representation of the effects of incorporating torrefied biomass in soil on initial plant growth.
Although the metabolic and microbial dynamics of the control were associated with a C1 metabolism (left), those of the 3–5% torrefied biomass samples were associated with an organic acid metabolism (right). This is attributed to the fact that torrefied biomass can improve physical and structural soil properties such as water retentivity and structural stability (right). Therefore, a soil amended with 3–5% of torrefied biomass can enhance the initial growth of Jatropha curcas in the form of increased stem diameter, root length, and element uptake ability.
Methods
Sample preparation and experimental design
The overall experimental design to evaluate the effects of soil amendments based on physical, chemical, and biological characteristics is illustrated in Fig. 1. For the torrefaction analysis, stem and leaf mixtures of J. curcas were milled with a food cutter, divided into 50-g samples, and wrapped in aluminum foil. The samples were torrefied at 200 °C, 220 °C, 230 °C, 240 °C, 250 °C, and 300 °C for 10 min under 5-L/min N2 in an electric furnace (FO410; Yamato Scientific Co., Ltd., Tokyo, Japan).
For growth experiments using J. curcas, a soil sample was collected from a Jatropha agricultural field in Gaborone, Botswana, in 2014. The soil was separated into four parts each weighing 3 kg into which 0, 30, 90, and 150 g of biomass torrefied at 240 °C [control, 1%, 3%, and 5% (weight/weight), respectively], and 150 g of fishmeal [5% (weight/weight)] was incorporated. To stabilize the metabolic activities and microbial variations in soils, they were incubated in a chamber at 25 °C and 10% moisture for 1 month and sampled twice a week. Seeds of J. curcas IP2P accession39 were germinated in 0.8 wt% agar gel with no nutrients as described in a previous study40. The germinated seeds after 10 days of growth were transplanted into the matured soils using quadruplicate experiments for the control and 1%, 3%, and 5% of torrefied biomass. Jatropha growth experiments with and without torrefied biomass were conducted over 4 weeks in a chamber at 25 °C and 50% moisture. Samples of 50 g were taken from the soils once a week for 4 weeks during growth.
Characterization of torrefied biomass
A TG analysis was performed with an EXSTAR TG/DTA 6300 (SII Nanotechnology Inc., Tokyo, Japan) instrument following a previous study23. ATR FTIR was performed on a Nicolet 6700 FTIR (Thermo Fisher Scientific Inc., Waltham, MA, USA) instrument with a KBr disk following previous studies23,41. Grain size distribution was performed with a vibratory sieve shaker (Fritsch Japan Co., Ltd., Kanagawa, Japan) instrument, and the percentage was calculated for each weight.
Soil characterization
For the analysis of soil compression stress, 9 g of dried soil samples was analyzed with an EZ-LX autograph (Shimazu, Kyoto, Japan) and TRAPEZIUM 2 software (Shimazu) at 5 mm/min to a depth of 20 mm and 25 N of stress using a lunge test jig. Soil samples with an addition of 2.5 ml ultrapure water were also measured under wet conditions using the same method. To measure the water content in soils, soil with or without 1%, 3%, or 5% torrefied biomass and raw biomass were divided into 600 g fractions and placed in plant pots into which 200 ml of ultrapure water was added. Soil water contents were measured with an ML3-theta probe soil moisture sensor (Delta-T Devices Ltd., Cambridge, England).
Elemental analysis
For the ICP-OES analysis, 50 mg samples of Botswana soils with or without torrefied biomass and J. curcas in the initial growth stage were extracted with ultrapure water and then with HNO3 (6.9% v/v) following previous studies42,30 and analyzed using an ICP-OES instrument (SPS5510; SII Nanotechnology Inc., Tokyo, Japan).
One- and two-dimensional NMR analyses
Samples of maturing phase soils with Jatropha growth (40 g) with an addition of sterile water were homogenized with a sonicator for 30 min and heated at 55 °C for 5 min in a Thermomixer comfort (Eppendorf AG, Hamburg, Germany). After centrifugation, the supernatants were collected and dried in a centrifugal evaporator (CVE-3000, Tokyo Rikakikai Co. Ltd., Tokyo, Japan). The dried samples were dissolved in 1 ml of D2O/KPi (100 mM, pH 7.0) and transferred into a 5 mm NMR tube.
The torrefied biomass and soil after Jatropha growth were analyzed using 1H-13C HSQC to identify the components. NMR spectra were acquired at 25 °C using an AVANCE II 700 MHz Bruker Biospin (Rheinstetten, Germany) instrument equipped with an inverse (with proton coils nearest to the sample) 5 mm 1H/13C/15N cryoprobe. The peak of sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) was used as the internal reference (calibrated at δC 140, δH 0 ppm). NMR spectra were acquired from 11.704 to −2.296 ppm in F2 (1H) using 2048 data points for an acquisition time of 104 ms, recycling delay of 2 s, and 150−10 ppm in F1 (13C) using 256 data points of 48 scans. All one-dimensional Watergate and two-dimensional (2D) J-resolved (2D-J) spectra were acquired with the same NMR instrument to evaluate metabolic profiles of soil microbiota. Watergate spectra were measured from 14 to −3 ppm at 25 °C using 32 k data points. 2D-J spectra were acquired from 11.7568 to −2.2458 ppm in F2 (1H) using 16 k data points and from 20.0027 to −19.9973 Hz in F1 (J coupling) using 16 data points of eight scans. HSQC and 2D-J spectra were further analyzed for annotation of chemical components using SpinAssign (http://prime.psc.riken.jp)43,44, Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/)45, and Birmingham Metabolite Library (http://www.bml-nmr.org/)46.
Relaxation time analyses of the water content in soils were measured by solid-state NMR using a Bruker DRX-500 spectrometer operating at 500.13 MHz for 1H equipped with the Bruker 4 mm double-tuned MAS probe. For the NMR measurements, approximately 80 mg of a sample and 100 μl of sterilized water were placed in a ZrO2 rotor (outer diameter 4 mm) with a Kel-F cap. The magic angle (54.7°) pulse length for protons was set to 1.8 μs. The measurement program used 2D Carr Purcell Meiboom Gill and sampling of the decay/recovery curves was obtained at 2–80 ms.
Metasequencing
Microbial DNA extraction was performed using the PowerSoil™ DNA Isolation Kit (Mo Bio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. The polymerase chain reaction (PCR) protocol used for metasequencing was described previously47,48. Sequencing was performed on a MiSeq sequencer (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The data were analyzed using QIIME software (http://qiime.org/)49. Each sample was separated by a MiSeq barcode-attached 27F mod-534R primer and chimera check using Usearch software (http://drive5.com/usearch/manual/uchime_algo.html)50. The resulting operational taxonomic unit data defined over 97% similarity were assigned to sequences using the Ribosomal Database Project (http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp) classifier51.
Statistical analysis
All 1H-NMR data were processed using Topspin 3.1 software (Bruker Biospin), and raw data were exported as text files. Exported data were processed over a range of 11 to −1 ppm with approximately 27.5 k data points for 1H-NMR and binning using R 3.0.1 software (http://www.r-project.org/). The dataset was normalized using the sum of the DSS integral regions and analyzed by PCA using R software as previously described26,33,52.
Additional Information
How to cite this article: Ogura, T. et al. Improvement of physical, chemical, and biological properties of aridisol from Botswana by the incorporation of torrefied biomass. Sci. Rep. 6, 28011; doi: 10.1038/srep28011 (2016).
Supplementary Material
Acknowledgments
The authors wish to thank Minami Matsui, Tamotsu Kato, Sigeharu Moriya, and Kenji Sakata (RIKEN) for their useful advice and assistance with the analytical procedures. Furthermore, the authors would like to acknowledge the members of department of agriculture research in Botswana for their acceptance to investigate soil amendment effect in Japan. This research was supported in part by grants-in-aid for Scientific Research (to JK) and also partially supported by Science and Technology Research Partnership for Sustainable Development (SATREPS to JK and KA) from Japan Science and Technology Agency (JST) and the Japan International Cooperation Agency (JICA).
Footnotes
Author Contributions T.O., Y.D. and J.K. designed the study. T.O., Y.D., M.M. and T.C. performed experiments. T.O. and Y.D. analyzed the data and made the figures. T.O., Y.D. and J.K. wrote the paper. K.A. and J.K. supervised the study. All authors reviewed the manuscript and agreed with submission.
References
- Fairless D. Biofuel: The little shrub that could - maybe. Nature 449, 652–655, doi: 10.1038/449652a (2007). [DOI] [PubMed] [Google Scholar]
- Achten W. M. J. et al. Jatropha bio-diesel production and use. Biomass Bioenerg 32, 1063–1084, doi: 10.1016/j.biombioe.2008.03.003 (2008). [DOI] [Google Scholar]
- Thomas A. D. Impact of grazing intensity on seasonal variations in soil organic carbon and soil CO2 efflux in two semiarid grasslands in southern Botswana. Philos T R Soc B 367, 3076–3086, doi: 10.1098/rstb.2012.0102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inafuku-Teramoto S. et al. Production approaches to establish effective cultivation methods for Jatropha (Jatropha curcas L.) under cold and semi-arid climate conditions. International Journal of Agronomy and Plant Production 4, 3804–3815 (2013). [Google Scholar]
- Pennisi E. Ecology. Africa’s soil engineers: termites. Science 347, 596–597, doi: 10.1126/science.347.6222.596 (2015). [DOI] [PubMed] [Google Scholar]
- Glaser B., Lehmann J. & Zech W. Ameliorating physical and chemical properties of highly weathered soils in the tropics with charcoal - a review. Biol Fert Soils 35, 219–230, doi: 10.1007/s00374-002-0466-4 (2002). [DOI] [Google Scholar]
- Lehmann J. et al. Nutrient availability and leaching in an archaeological Anthrosol and a Ferralsol of the Central Amazon basin: fertilizer, manure and charcoal amendments. Plant Soil 249, 343–357, doi: 10.1023/A:1022833116184 (2003). [DOI] [Google Scholar]
- Pietikainen J., Kiikkila O. & Fritze H. Charcoal as a habitat for microbes and its effect on the microbial community of the underlying humus. Oikos 89, 231–242, doi: 10.1034/j.1600-0706.2000.890203.x (2000). [DOI] [Google Scholar]
- Steiner C. et al. Long term effects of manure, charcoal and mineral fertilization on crop production and fertility on a highly weathered Central Amazonian upland soil. Plant Soil 291, 275–290, doi: 10.1007/s11104-007-9193-9 (2007). [DOI] [Google Scholar]
- Hilber I. et al. Influence of activated charcoal amendment to contaminated soil on dieldrin and nutrient uptake by cucumbers. Environ Pollut 157, 2224–2230, doi: 10.1016/j.envpol.2009.04.009 (2009). [DOI] [PubMed] [Google Scholar]
- Whitman T. & Lehmann J. Biochar—One way forward for soil carbon in offset mechanisms in Africa? Environmental science & policy 12, 1024–1027 (2009). [Google Scholar]
- Barnes R. T., Gallagher M. E., Masiello C. A., Liu Z. L. & Dugan B. Biochar-Induced Changes in Soil Hydraulic Conductivity and Dissolved Nutrient Fluxes Constrained by Laboratory Experiments. Plos One 9, e108340 doi: 10.1371/journal.pone.0108340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslu A., Faaij A. P. C. & Bergman P. C. A. Pre-treatment technologies, and their effect on international bioenergy supply chain logistics. Techno-economic evaluation of torrefaction, fast pyrolysis and pelletisation. Energy 33, 1206–1223, doi: 10.1016/j.energy.2008.03.007 (2008). [DOI] [Google Scholar]
- Tumuluru J. S., Wright C. T., Hess J. R. & Kenney K. L. A review of biomass densification systems to develop uniform feedstock commodities for bioenergy application. Biofuel Bioprod Bior 5, 683–707, doi: 10.1002/Bbb.324 (2011). [DOI] [Google Scholar]
- Li J., Brzdekiewicz A., Yang W. H. & Blasiak W. Co-firing based on biomass torrefaction in a pulverized coal boiler with aim of 100% fuel switching. Appl Energ 99, 344–354, doi: 10.1016/j.apenergy.2012.05.046 (2012). [DOI] [Google Scholar]
- Watanabe T., Shino A., Akashi K. & Kikuchi J. Chemical profiling of Jatropha tissues under different torrefaction conditions: application to biomass waste recovery. Plos One 9, e106893, doi: 10.1371/journal.pone.0106893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolton M. et al. Impact of biochar application to soil on the root-associated bacterial community structure of fully developed greenhouse pepper plants. Appl Environ Microbiol 77, 4924–4930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.-J. et al. Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environmental science & technology 48, 9391–9399 (2014). [DOI] [PubMed] [Google Scholar]
- Anders E. et al. Biochar affects the structure rather than the total biomass of microbial communities in temperate soils. Agricultural and Food Science 22, 404–423 (2013). [Google Scholar]
- Fox A., Kwapinski W., Griffiths B. S. & Schmalenberger A. The role of sulfur-and phosphorus-mobilizing bacteria in biochar-induced growth promotion of Lolium perenne. FEMS microbiology ecology 90, 78–91 (2014). [DOI] [PubMed] [Google Scholar]
- Asakawa D. et al. Changes in elemental composition, molecular weight and 1H NMR spectra of the water-extractable hydrophobic acid fraction in Cambisol with season and soil depth. Soil science and plant nutrition 52, 361–370 (2006). [Google Scholar]
- Yuk J., Simpson M. J. & Simpson A. J. 1-D and 2-D NMR-based metabolomics of earthworms exposed to endosulfan and endosulfan sulfate in soil. Environ Pollut 175, 35–44, doi: 10.1016/j.envpol.2012.12.007 (2013). [DOI] [PubMed] [Google Scholar]
- Ogura T., Date Y. & Kikuchi J. Differences in Cellulosic Supramolecular Structure of Compositionally Similar Rice Straw Affect Biomass Metabolism by Paddy Soil Microbiota. Plos One 8, doi: 10.1371/journal.pone.0066919 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Date Y., Tsuboi Y. & Kikuchi J. Metabolic dynamics analysis by massive data integration: application to tsunami-affected field soils in Japan. ACS chemical biology 10, 1908–1915 (2015). [DOI] [PubMed] [Google Scholar]
- Ben H. & Ragauskas A. J. Comparison for the compositions of fast and slow pyrolysis oils by NMR characterization. Bioresour Technol 147, 577–584, doi: 10.1016/j.biortech.2013.07.151 (2013). [DOI] [PubMed] [Google Scholar]
- Yoshida S., Date Y., Akama M. & Kikuchi J. Comparative metabolomic and ionomic approach for abundant fishes in estuarine environments of Japan. Sci Rep 4, 7005, doi: 10.1038/srep07005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hoshino R., Date Y. & Kikuchi J. Visualization of Microfloral Metabolism for Marine Waste Recycling. Metabolites 6, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva S. et al. Silicon amelioration of manganese toxicity in Mn-sensitive and Mn-tolerant maize varieties. Environ Exp Bot 65, 189–197, doi: 10.1016/j.envexpbot.2008.11.006 (2009). [DOI] [Google Scholar]
- Suwa R. et al. Barium toxicity effects in soybean plants. Arch Environ Con Tox 55, 397–403, doi: 10.1007/s00244-008-9132-7 (2008). [DOI] [PubMed] [Google Scholar]
- Ito K., Sakata K., Date Y. & Kikuchi J. Integrated Analysis of Seaweed Components during Seasonal Fluctuation by Data Mining Across Heterogeneous Chemical Measurements with Network Visualization. Anal Chem 86, 1098–1105, doi: 10.1021/Ac402869b (2014). [DOI] [PubMed] [Google Scholar]
- Moon E. M. & Peacock C. L. Adsorption of Cu (II) to ferrihydrite and ferrihydrite–bacteria composites: Importance of the carboxyl group for Cu mobility in natural environments. Geochimica et Cosmochimica Acta 92, 203–219 (2012). [Google Scholar]
- Wei F. F., Ito K., Sakata K., Date Y. & Kikuchi J. Pretreatment and Integrated Analysis of Spectral Data Reveal Seaweed Similarities Based on Chemical Diversity. Anal Chem 87, 2819–2826, doi: 10.1021/Ac504211n (2015). [DOI] [PubMed] [Google Scholar]
- Date Y., Iikura T., Yamazawa A., Moriya S. & Kikuchi J. Metabolic sequences of anaerobic fermentation on glucose-based feeding substrates based on correlation analyses of microbial and metabolite profiling. Journal of proteome research 11, 5602–5610, doi: 10.1021/pr3008682 (2012). [DOI] [PubMed] [Google Scholar]
- Vyas P. & Gulati A. Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC microbiology 9, 174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustakhimov I., Kalyuzhnaya M. G., Lidstrom M. E. & Chistoserdova L. Insights into Denitrification in Methylotenera mobilis from Denitrification Pathway and Methanol Metabolism Mutants. J Bacteriol 195, 2207–2211, doi: 10.1128/Jb.00069-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita I., Nishikawa K., Nakamura S. & Fukui T. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions. Appl Microbiol Biotechnol 98, 3715–3725 (2014). [DOI] [PubMed] [Google Scholar]
- Galatis H., Martin K., Kämpfer P. & Glaeser S. P. Devosia epidermidihirudinis sp. nov. isolated from the surface of a medical leech. Antonie van Leeuwenhoek 103, 1165–1171 (2013). [DOI] [PubMed] [Google Scholar]
- Chin K. J., Liesack W. & Janssen P. H. Opitutus terrae gen nov, sp nov, to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int J Syst Evol Micr 51, 1965–1968 (2001). [DOI] [PubMed] [Google Scholar]
- Sakurai N. et al. Development of KaPPA-View4 for omics studies on Jatropha and a database system KaPPA-Loader for construction of local omics databases. Plant Biotechnol-Nar 29, 131–135, doi: 10.5511/plantbiotechnology.12.0508a (2012). [DOI] [Google Scholar]
- Komatsu T., Ohishi R., Shino A., Akashi K. & Kikuchi J. Multi-Spectroscopic Analysis of Seed Quality and 13C-Stable-Iotopologue Monitoring in Initial Growth Metabolism of Jatropha curcas L. Metabolites 4, 1018–1033, doi: 10.3390/metabo4041018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Shino A., Akashi K. & Kikuchi J. Spectroscopic investigation of tissue-specific biomass profiling for Jatropha curcas L. Plant Biotechnol-Nar 29, 163–170, doi: 10.5511/plantbiotechnology.12.0222a (2012). [DOI] [Google Scholar]
- Date Y. et al. In vitro evaluation method for screening of candidate prebiotic foods. Food Chem 152, 251–260, doi: 10.1016/j.foodchem.2013.11.126 (2014). [DOI] [PubMed] [Google Scholar]
- Chikayama E., Suto M., Nishihara T., Shinozaki K. & Kikuchi J. Systematic NMR analysis of stable isotope labeled metabolite mixtures in plant and animal systems: coarse grained views of metabolic pathways. Plos One 3, e3805, doi: 10.1371/journal.pone.0003805 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikayama E. et al. Statistical Indices for Simultaneous Large-Scale Metabolite Detections for a Single NMR Spectrum. Anal Chem 82, 1653–1658, doi: 10.1021/Ac9022023 (2010). [DOI] [PubMed] [Google Scholar]
- Ulrich E. L. et al. BioMagResBank. Nucleic acids research 36, D402–408, doi: 10.1093/nar/gkm957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C. et al. Birmingham Metabolite Library: a publicly accessible database of 1-D H-1 and 2-D H-1 J-resolved NMR spectra of authentic metabolite standards (BML-NMR). Metabolomics 8, 8–18, doi: 10.1007/s11306-011-0347-7 (2012). [DOI] [Google Scholar]
- Kim S. W. et al. Robustness of Gut Microbiota of Healthy Adults in Response to Probiotic Intervention Revealed by High-Throughput Pyrosequencing. DNA Res 20, 241–253, doi: 10.1093/dnares/dst006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T., Date Y. & Kikuchi J. Human metabolic, mineral, and microbiota fluctuations across daily nutritional intake visualized by a data-driven approach. Journal of proteome research 14, 1526–1534, doi: 10.1021/pr501194k (2015). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, doi: 10.1038/Nmeth.F.303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200, doi: 10.1093/bioinformatics/btr381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267, doi: 10.1128/Aem.00062-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D. M. et al. Biogeochemical typing of paddy field by a data-driven approach revealing sub-systems within a complex environment–a pipeline to filtrate, organize and frame massive dataset from multi-omics analyses. Plos One 9, e110723, doi: 10.1371/journal.pone.0110723 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.