Abstract
Dyslexia is a severe disorder in the acquisition of reading and writing. Several studies investigated the role of genetics for reading, writing and spelling ability in the general population. However, many of the identified SNPs were not analysed in case-control cohorts. Here, we investigated SNPs previously linked to reading or spelling ability in the general population in a German case-control cohort. Furthermore, we characterised these SNPs for functional relevance with in silico methods and meta-analysed them with previous studies. A total of 16 SNPs within five genes were included. The total number of risk alleles was higher in cases than in controls. Three SNPs were nominally associated with dyslexia: rs7765678 within DCDC2, and rs2038137 and rs6935076 within KIAA0319. The relevance of rs2038137 and rs6935076 was further supported by the meta-analysis. Functional profiling included analysis of tissue-specific expression, annotations for regulatory elements and effects on gene expression levels (eQTLs). Thereby, we found molecular mechanistical implications for 13 of all 16 included SNPs. SNPs associated in our cohort showed stronger gene-specific eQTL effects than non-associated SNPs. In summary, our results validate SNPs previously linked to reading and spelling in the general population in dyslexics and provide insights into their putative molecular pathomechanisms.
Dyslexia is a neurodevelopmental disorder which affects the ability of accurate and/or fluent word recognition and spelling1. These problems are thought to result from deficits in phonological decoding and are independent of general intelligence. With a prevalence of ~5% in Germany, it is one of the most common learning impairments for young school children2. Twin studies estimated the heritability of dyslexia at 50–70%3. The underlying genetics is complex and appears to consist of a large number of factors with rather small effect sizes. In recent years, several candidate genes and single nucleotide polymorphisms (SNPs) associated with dyslexia were found4. From those genes, DYX1C1, KIAA0319 and DCDC2 are the most prominent candidate genes for dyslexia. KIAA0319 is strongly expressed in the human cortex5 and knockdown in rats revealed disturbed neuronal migration in the neocrotex6. Similar findings were reported for DCDC2 and DYX1C1 where embryonic knockdown resulted in neuronal overmigration past the desired layer6. All three genes were reported to be linked with reduced white matter volume in the left hemisphere of dyslexics7, a well-known dyslexia brain phenotype. In addition to findings from dyslexia-control studies, several candidate genes were also found to be associated with reading or spelling ability in the general population8,9,10,11,12. These SNPs were mainly located in the DYX2 locus (DCDC28, KIAA031913 and TDP12 (also known as TTRAP)) but also variants in ROBO110, DYX1C19 and NKAIN211 were identified (Table 1).
Table 1. Reference studies for all 16 analyzed SNPs with the reported risk allele.
| SNP | Chromosome | Position[bp] | General population studies |
Case-control studies |
|||
|---|---|---|---|---|---|---|---|
| Study | Reportedrisk allele | Effect size | Reportingassociation | Not Reportingassociation | |||
| rs1091047-DCDC2 | 6 | 24295256 | Lind et al.8 | G | expl. variance = 0.64% | – | – |
| rs1419228-DCDC2 | 6 | 24178306 | Lind et al.8 | G | expl. variance = 0.87% | – | – |
| rs9467075-DCDC2# | 6 | 24205236 | Lind et al.8 | A | expl. variance = 0.35% | – | – |
| rs9467076-DCDC2# | 6 | 24209255 | Lind et al.8 | C | expl. variance = 0.34% | – | – |
| rs6922023-DCDC2 | 6 | 24348117 | Lind et al.8 | G | expl. variance = 0.40% | – | – |
| rs7765678-DCDC2 | 6 | 24330544 | Lind et al.8 | T | expl. variance = 0.45% | – | – |
| rs8037376-DYX1C1 | 15 | 55768321 | Paracchini et al.9 | C | −0.147 SD (spelling) per risk allele | – | – |
| rs8043049-DYX1C1# | 15 | 55777788 | Paracchini et al.9 | C | −0.163 SD (spelling) per risk allele | – | – |
| rs7174102-DYX1C1# | 15 | 55719687 | Paracchini et al.9 | T | −0.135 SD (spelling) per risk allele | – | – |
| rs8040756-DYX1C1 | 15 | 55798599 | Paracchini et al.9 | G | −0.196 SD (reading) per risk allele | – | – |
| rs2038137-KIAA0319 | 6 | 24645943 | Luciano et al.12; Paracchini et al.13 | G | 0.06 SD (reading) per copy of haplotype (rs4504469-rs2038137-rs2143340)12; Beta = −0.067 (adjusted reading values) per copy of haplotype (rs4504469-rs2038137-rs2143340)13 | Harold et al.19; Cope et al.20 | Couto et al.21; Scerri et al.27 |
| rs4504469-KIAA0319# | 6 | 24588884 | Luciano et al.12; Paracchini et al.13 | C | 0.06 SD (reading) per copy of haplotype (rs4504469-rs2038137-rs2143340)12; Beta = −0.003 (adjusted reading values) per copy of haplotype (rs4504469-rs6935076)13; Beta = −0.067 (adjusted reading values) per copy of haplotype (rs4504469-rs2038137-rs2143340)13 | Cope et al.20; Newbury et al.24; Becker et al.23 | Couto et al.21; Brkanac et al.22, Harold et al.19 |
| rs6935076-KIAA0319 | 6 | 24644322 | Luciano et al.12; Paracchini et al.13 | T | 0.06 SD (reading) per copy of haplotype (rs4504469-rs2038137-rs2143340)12; Beta = −0.003 (adjusted reading values) per copy of haplotype (rs4504469-rs6935076)13 | Cope et al.20; Couto et al.21 | Harold et al.19 |
| rs1842129-NKAIN2 | 6 | 124838090 | Luciano et al.11 | G | Not reported | – | – |
| rs1995402-ROBO1 | 3 | 79790407 | Bates et al.10 | A | Not reported | – | – |
| rs2143340-TDP2 | 6 | 24659071 | Luciano et al.12 ; Paracchini et al.13 | C | Expl. variance:1% (reading)12 ;Beta = −0.074 (adjusted reading values) per risk allele13; Beta = −0.067 (adjusted reading values) per copy of haplotype (rs4504469-rs2038137-rs2143340)13 | Newbury et al.24 | Cope et al.20; Couto et al.21, Scerri et al.27, Becker et al.23 |
Note that rs2038137 and rs4504469 were reported to associate as 3-marker-haplotype rs4504469-rs2038137-rs2143340 and rs4504469 as 2-marker-haplotype rs4504469-rs6935076 in a case-control study. SNP positions are based on HG19. Explained variances for SNPs identified by Lind et al.8 were calculated for a principal factor score computed from six reading and spelling related measures. For the other SNPs, phenotypes are given in brackets. Shown is the absolute value of the effect size as presented in the paper. SNPs with independent effects identified by pruning (R2 = 0.5) are bold, and SNPs without independent effects are marked with # and were tagged by the respective independent SNP shown above.
Genetic variants associating with a quantitative trait are highly relevant genetic candidate variants in a corresponding binary trait as seen in other diseases14. However, we identified that many of these SNPs (12 of 16 in four genes shown in Table 1) were not yet analysed in a dyslexia case-control setting, in these four genes only different SNPs were previously investigated (Supplemental Tables 1 and 2). As correlation between SNPs within the same gene can vary largely and SNPs may have independent effects due to locus heterogeneity, association results among different SNPs of the same gene may differ considerably. Furthermore, for the four of 16 SNPs case-control analyses already exist, information of additional association studies will provide valuable validation information helping to resolve partly contracting findings (Table 1).
Therefore, the primary aim of our study was to systematically investigate all these 16 SNPs in a German dyslexia case-control cohort. Previously, other reported dyslexia candidate SNPs were analysed in this cohort (or subsets of this cohort). Thereby, association was found for three of eight candidates from chromosome 1815, for a variant in FOXP216, and for a large 2445 bp deletion and certain SNPs in DCDC217 but not for a functional variant in KIAA031918 language gene.
A secondary aim of our study addresses the lack of functional hypotheses about potential molecular risk-mechanisms of these 16 SNPs. This information is valuable for an improved understanding of a potential pathomechanisms of an observed association. Moreover, it is also important as additional evidence for the validity of an observed association. Therefore, in order to address this secondary aim, we characterise all these 16 variants using in silico methods on large datasets from recent available high-throughput studies.
Results
We analysed 16 SNPs in five genes from six studies8,9,10,11,12,13 affecting reading and spelling in the general population that were genotyped in our cohort (see Table 1 for details). All SNPs were in Hardy-Weinberg-equilibrium (HWE) in controls (p-value > 0.05), the call rate for each SNP exceeded 98%, and the call rate per individual was in average 99% across all 16 SNPs.
Associations on single marker level
On single marker level, nominal significant allelic association was observed for the minor allele of rs7765678-DCDC2 (p-value = 0.023, odds ratio (OR) = 0.65 (0.5–0.9)) and rs6935076-KIAA0319 (p-value = 0.023, OR = 1.25 (1.0–1.5)) (Table 2). Nominal significant genotypic association was also observed for the major homozygous state of rs2038137-KIAA0319 (p-value = 0.036, genetic relative risk (GRR) = 1.35 (1.0–1.8)) and rs7765678-DCDC2 (p-value = 0.035, GRR = 1.52 (1.0–2.2)). Originally reported effect size of these associations was not markedly higher than for those without association. No associations reached significance when considering Bonferroni’s correction method. For all nominally associating SNPs, the observed risk alleles were in accordance to reported associations from the general population studies. Two of the three nominally associating SNPs were previously reported in case-control settings: For rs2038137-KIAA0319 the risk alleles were in accordance with the reported associations19,20 and for rs6935076-KIAA0319 one study reported same allele20 and one study reported the opposite allele as risk allele21.
Table 2. Association statistics.
| SNP | Reported risk allele and accordance | Major homozygous genotype | Minor homozygous genotype | Allelic association | |||
|---|---|---|---|---|---|---|---|
| p | GRR | p | GRR | p | OR | ||
| rs1091047-DCDC2 | Major (acc.) | 0.062 | 1.34 (1.0–1.8) | 0.720 | 0.88 (0.5–1.7) | 0.085 | 0.78 (0.6–1.0) |
| rs1419228-DCDC2 | Minor (acc.) | 0.491 | 0.90 (0.7–1.2) | 0.051 | 1.77 (1.0–3.1) | 0.292 | 1.15 (0.9–1.5) |
| rs9467075-DCDC2# | Major | 0.892 | 1.02 (0.7–1.4) | 0.481 | 1.29 (0.6–2.6) | 0.990 | 1.00 (0.7–1.3) |
| rs9467076-DCDC2# | Minor (acc.) | 0.985 | 0.99 (0.7–1.4) | 0.130 | 1.79 (0.8–3.8) | 0.767 | 1.05 (0.8–1.4) |
| rs6922023-DCDC2 | Major (acc.) | 0.268 | 1.19 (0.9–1.6) | 0.186 | 0.61 (0.3–1.3) | 0.186 | 0.83 (0.6–1.1) |
| rs7765678-DCDC2 | Major (acc.) | 0.035* | 1.52 (1.0–2.2) | 0.178 | 0.37 (0.1–1.6) | 0.023* | 0.65 (0.5–0.9) |
| rs8037376-DYX1C1 | Minor (acc.) | 0.951 | 1.01 (0.8–1.3) | 0.314 | 1.21 (0.8–1.7) | 0.741 | 1.04 (0.8–1.3) |
| rs8043049-DYX1C1# | Major | 0.576 | 1.08 (0.8–1.4) | 0.494 | 1.13 (0.8–1.6) | 0.970 | 0.99 (0.8–1.2) |
| rs7174102-DYX1C1# | Minor | 0.594 | 1.08 (0.8–1.4) | 0.439 | 1.15 (0.8–1.6) | 0.989 | 1.00 (0.8–1.2) |
| rs8040756-DYX1C1 | Major (acc.) | 0.305 | 1.19 (0.9–1.7) | 0.701 | 1.17 (0.5–2.6) | 0.38 | 0.87 (0.6–1.2) |
| rs2038137-KIAA0319 | Major (acc.) | 0.036* | 1.35 (1.0–1.8) | 0.887 | 0.97 (0.7–1.4) | 0.132 | 0.85 (0.7–1.0) |
| rs4504469-KIAA0319# | Major (acc.) | 0.987 | 0.99 (0.8–1.3) | 0.562 | 0.90 (0.6–1.3) | 0.802 | 0.97 (0.8–1.2) |
| rs6935076-KIAA0319 | Minor (acc.) | 0.088 | 0.78 (0.6–1.0) | 0.036* | 1.46 (1.0–2.1) | 0.039* | 1.25 (1.0–1.5) |
| rs1842129-NKAIN2 | Major | 0.100 | 0.78 (0.6–1.0) | 0.845 | 1.04 (0.7–1.5) | 0.261 | 1.13 (0.9–1.4) |
| rs1995402-ROBO1 | Minor (acc.) | 0.680 | 0.94 (0.7–1.3) | 0.708 | 0.94 (0.7–1.3) | 0.977 | 1.01 (0.8–1.2) |
| rs2143340-TDP2 | Minor | 0.442 | 1.14 (0.8–1.6) | 0.665 | 0.84 (0.4–1.8) | 0.406 | 0.88 (0.7–1.2) |
Shown are the respective p-values and genetic relative risks (GRR) for the homozygous major allele genotype and the homozygous minor allele genotype. Allelic associations relate to the effect of the minor allele reported in Supplemental Table 7. Shown is also the accordance (acc.) of the reported risk allele from literature with the observed risk allele of our study. SNPs with independent effects identified by priority pruning (R2 = 0.5) are bold, and SNPs without independent effects are marked with # and were tagged by the respective independent SNP shown above.
In literature, the haplotype rs4504469-rs2038137-rs2143340 was reported to associate with measures for reading12,13. We could not identify an association in our sample (Supplemental Table 3). No association was also observed of gene-wide haplotypes in DCDC2, KIAA0319, and DYX1C1 (Supplemental Table 3).
Meta-analysis
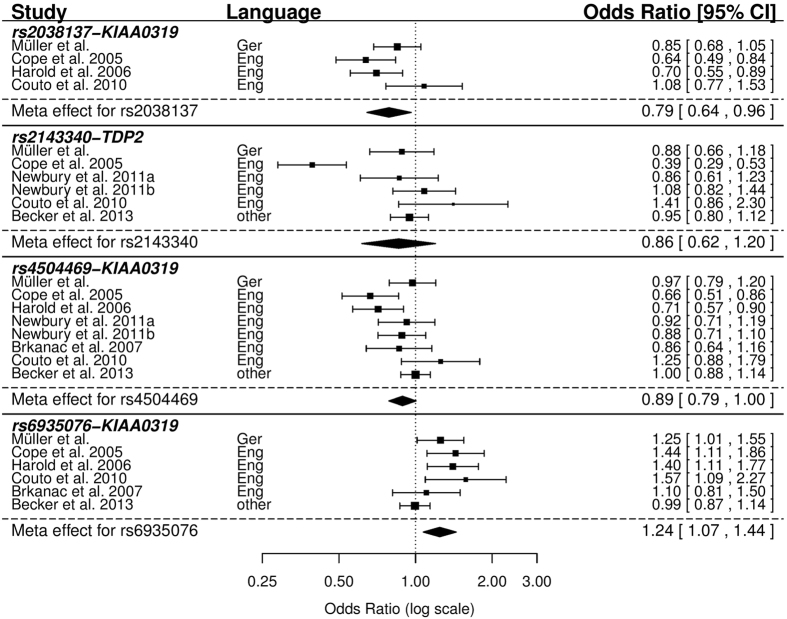
For four of the 16 investigated SNPs, case-control data of six different studies from a total of eight countries was available for meta-analysis19,20,21,22,23,24 (Table 1). These four SNPs included nominally associated variants rs2038137-KIAA0319 and rs693507-KIAA0319 but not rs7765678-DCDC2.
In line with our findings from our cohort alone, we found a significant meta-effect for SNPs rs2038137 and for rs6935076 (OR = 0.79 (0.64–0.96); p-value = 0.019 and OR = 1.24 (1.07–1.44); p-value = 0.004, respectively) across all studies (Fig. 1). For rs4504469-KIAA0319, only a trend-level significant meta-effect was found (p = 0.059). However, when stratifying the meta-analysis for languages, a significant meta-effect was found for this variant within English-speaking cohorts (OR = 0.84 (0.73–0.96); p-value = 0.009, Supplemental Fig. 1). No meta-effect was found for rs2143340-TDP2.
Figure 1. Forest plot representing the individual results of seven studies and the meta-effects for each SNP.
Note, that Brkanac et al.22 and Couto et al.21 performed TDT-studies and ORs were computed from transmitted allele counts.
Polygenic analyses
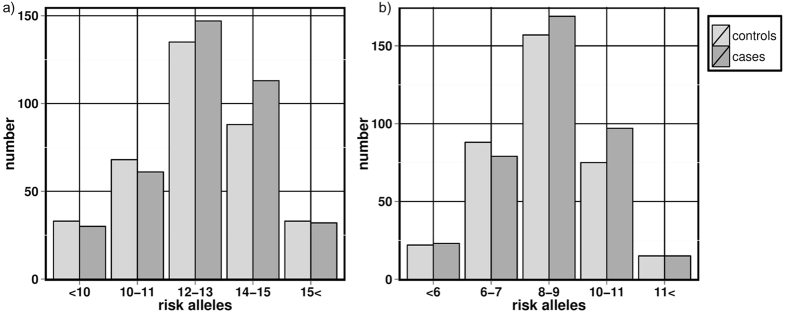
Next, we aimed to investigate effects that may not have been detectable in our single marker analysis. Therefore, we jointly analysed association of all 16 general-reading SNPs by creating a score summing individual-wise all risk alleles. In order to avoid bias due to linkage disequilibrium among these SNPs, we thereby considered the subset of 11 unlinked SNPs (see Material and Methods). We found that for nine of these 11 SNPs effect direction was concordant with the literature. Thereby, the reported risk alleles were increased in our cases. This was more than expected by chance (p-value = 0.033). Accordingly, an increased number of risk variants was observable within cases compared to controls (Fig. 2a). An increased number of reported risk variants in cases was also observable when including only the SNPs not associating on single marker level (Fig. 2b). However, a formal quantitative association analysis of the corresponding risk score did not meet significance (p-value > 0.2). Findings were similar when jointly considering these eleven SNPs and 14 additional genetic dyslexia candidate variants analysed in previous association studies of our cohort (Supplemental Fig. 2).
Figure 2. Distribution of risk alleles among dyslexia-cases and controls.
(a) Cases and controls with the total sum of all reported risk alleles from all included 11 independent SNPs. (b) Cases and controls with the total sum of all reported risk alleles but excluding all three SNPs associating at single marker level in our cohort.
Functional analyses
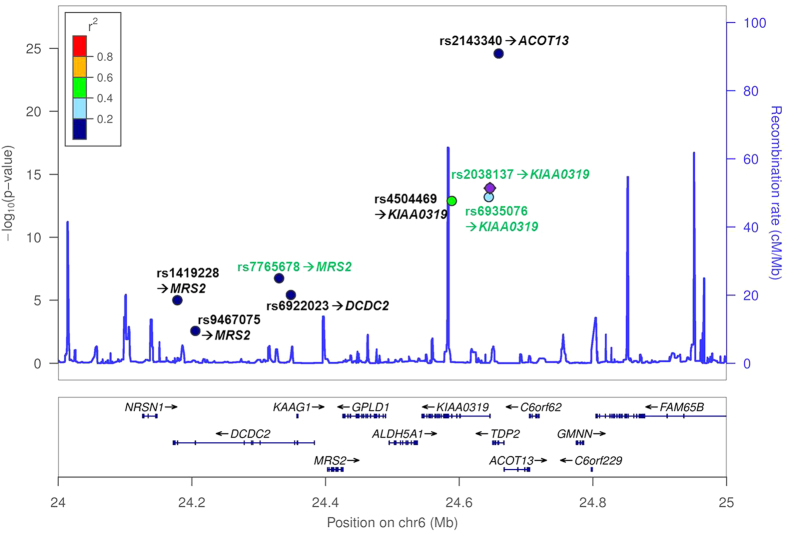
SNPs were evaluated for expression quantitative trait loci (eQTL)-effects by screening published eQTL-analyses. In total, twelve of all 16 SNPs showed a direct effect on expression levels of neighbouring genes (cis-eQTLs, Table 3). For 14 SNPs, proxy SNPs (R2 ≥ 0.5) were observed to be associated with the expression of a total of 16 genes in cis as well as in trans (see Table 3 and Supplemental Table 4). For DCDC2, KIAA0319, and DYX1C1, SNPs located within these genes also affected the expression of DCDC2, KIAA0319, and DYX1C1, respectively. This included nominally associated variants rs2038137 and rs693507 in KIAA0319 that actually affected gene expression of KIAA0319. However, nominally associated SNP rs7765678-DCDC2 was most strongly linked with the expression of MRS2. An overview of the eQTL effects of the SNPs from DYX2 (DCDC2, KIAA0319 and TDP2) is provided in Fig. 3. Consistently, this figure demonstrates that SNPs associated in our cohort showed stronger gene-specific eQTL effects than non-associated SNPs.
Table 3. EQTL and regulatory annotations for all analysed SNPs.
| SNP | eQTLs | Regulome DB | |
|---|---|---|---|
| eQTN (R2) | Affected gene | Predicted elements | |
| rs1091047-DCDC2 | rs17302582 (R2 = 0.79) | ALDH5A1 (p = 0.0013)67 | no data |
| rs1419228-DCDC2 | SNP = eQTN | MRS2 (p = 9.9E-06)67 | Motifs-PWM (FOXC1) |
| rs9467075-DCDC2 | SNP = eQTN | MRS2 (p = 0.0027)67 | Chromatin structure (DNase-seq, FAIRE) Protein binding-ChIPseq (RAD21, CTCF) |
| rs9467076-DCDC2 | rs9460977 (R2 = 1.00) | MRS2 (p = 0.0003)67 | no data |
| rs6922023-DCDC2 | SNP = eQTN | DCDC2 (p = 3.8E-06)69 | no data |
| rs7765678-DCDC2 | SNP = eQTN | MRS2 (p = 1.8E-07)67 | Chromatin structure (DNase-seq, FAIRE) Protein binding-ChIPseq (HNF4A, EP300, FOXA1, TCF4, FOXA2) |
| rs8037376-DYX1C1 | SNP = eQTN | PIGB (p = 1.9E-44)67CCPG1 (p = 5.2E-20)65DYX1C1 (p = 1.1E-05)69DYX1C1-CCPG1 (p = 1.4E-05)69RP11-139H15.1 (p = 1.7E-05)69RSL24D1 (p = 0.0005)67 | Chromatin structure (DNase-seq, FAIRE) Protein binding-ChIPseq (MEF2A) |
| rs8043049-DYX1C1 | SNP = eQTN | PIGB (p = 2.7E-47)67CCPG1 (p = 3.8E-19)67DYX1C1-CCPG1 (p = 2E-05)69RSL24D1 (p = 0.0025)67 | Chromatin structure (DNase-seq, FAIRE) |
| rs7174102-DYX1C1 | SNP = eQTN | PIGB (p = 5E-23)67CCPG1 (p = 1.2E-17)65DYX1C1 (p = 3.1E-06)69 | Motifs-PWM (Plagl1) Chromatin structure (DNase-seq, FAIRE) |
| rs8040756-DYX1C1 | SNP = eQTN | CCPG1 (p = 0.00018)55PIGB (p = 1.6E-05)67C15orf65 (p = 7.4E-08) | no data |
| rs2038137-KIAA0319 | SNP = eQTN | KIAA0319 (p = 1.3E-14)69RP1-30M3.5 (p = 3.6E-10)69ACOT13 (p = 9.9E-09)69KRT8P43 (p = 3.2E-06)69TDP2 (p = 8.5E-06)69 | Motifs-Footprinting(Staf) Motifs-PWM (Staf) Chromatin structure (DNase-seq, FAIRE) Protein binding-ChIPseq (RFX3, POLR2A, E2F6) |
| rs4504469-KIAA0319 | SNP = eQTN | KIAA0319 (p = 1.3E-13)65ACOT13 (p = 0.0003)67 | no data |
| rs6935076-KIAA0319 | SNP = eQTN | KIAA0319 (p = 6.4E-14)69ACOT13 (p = 7.6E-07)69 | Chromatin structure (DNase-seq, FAIRE) |
| rs1842129-NKAIN2 | no data | no data | Motifs-PWM (Lhx5, Lhx3, Lhx9) |
| rs1995402-ROBO1 | no data | no data | Motifs-PWM (Bbx) |
| rs2143340-TDP2 | SNP = eQTN | ACOT13 (p = 2.6E-25)67C6ORF62 (p = 2.5E-16)67RP1-30M3.5 (p = 2E-09)69 | Motifs-PWM (CSBP, ISGF-3, IRF-1) |
eQTL annotations include the gene whose expression levels were linked to the SNP and the study from which the evidence originates. The analyses were done for eQTLs identified in all tissues and for eQTLs identified in brain related tissue. Reported are the best-linked eQTNs (R2 ≥ 0.5), the linkage to the original SNP (R2), the affected gene and the strength of association (if an eQTL-association was reported in different studies, the strongest p-value is reported). EQTL-effects were identified in cis unless otherwise stated. Functional annotations classified as “predicted” and “known” from ‘RegulomeDB’ are also shown.
Figure 3. Local association plot of analysed DYX2-SNPs with reported eQTL-effect.
Depicted are the analysed SNPs of DYX2 whereby SNPs with nominal significant associations with dyslexia in our sample are coloured in green. Next to each SNP following the arrow we report the gene whose expression level was strongest associated with the SNP. P-values represent the association of the SNP with gene expression levels. Details can be found in Supplemental Table 4. The data points are coloured according to the linkage to the SNP with the strongest effect in the association and the meta-analysis (rs2038137).
To obtain complementary functional information for eQTLs, we analysed information regarding gene expression available in database ‘RegulomeDB’25 (Table 3). For SNP rs2038137-KIAA0319, we found the highest evidence of altered transcription factor binding. Ten of 16 SNPs revealed minor evidence to affect binding of transcription related factors, for five SNPs no data was available. All investigated genes, except TDP2, were expressed in brain according to the ‘Human Protein Atlas’26 (Supplemental Table 5).
Discussion
In this study, we investigated 16 candidate SNPs representing 11 independent genetic effects reported to alter reading or spelling ability in the general population. We analysed the relevance of these variants in a German-speaking dyslexia specific case-control cohort and meta-analysed our findings. Additionally, we characterised these SNPs for molecular evidence in order to get insights into potential molecular risk-mechanisms underlying these genetic variations.
Associations in case-control studies can provide evidence for additional, epidemiological features of risk variants described for the general population. Compared to studies of the general population, case-control studies are enriched for severe cases. Therefore, the relevance of such risk variants for rather extreme, but clinically highly relevant phenotypes is of interest. This investigation may prioritise variants that not only modify the normal range of reading/spelling but are also critical for the extreme ends.
To our knowledge, from all 16 investigated SNPs only four were yet analysed in case-control settings (rs4504469-KIAA0319, rs2038137-KIAA01319, rs6935076-KIAA0319, rs2143340-TDP2, Table 1). From those four SNPs, we could nominally replicate rs2038137-KIAA0319 and rs6935076-KIAA0319 in a genuine German dyslexia population and confirm this association in our meta-analysis (Fig. 1). Both SNPs were previously described for English speaking dyslexics and the latter also for a mixed central European population19,20,21,23,24,27. We did not find evidence that these associations are explained by rs9451046, a previously reported functional variant in KIAA0319 as linkage disequilibrium of our associated SNPs and this variant is rather weak (R2 = 0.22). In line with this, a direct association analysis of rs9451046 investigated previously in our cohort did not find a significant associations17,18. Contrarily, associating SNP rs2038137-KIAA0319 is in perfect LD (R2 = 1) with rs2179515 reported in the NeuroDys study23, therefore both associations refer to the same genetic phenomenon (Supplemental Table 1).
The third investigated variant in KIAA0319, rs4504469, was not associated in our cohort and reached only trend-level significance in the meta-analysis across all studies. Interestingly, when stratifying the meta-analysis for language, we found significant association of this variant among English speaking cohorts (Supplemental Fig. 1a). Therefore, we speculate that this SNP is more relevant in the development of dyslexia in English speaking humans. Indeed, regarding the language, the German language is a relatively transparent language with a clear grapheme-phoneme correspondence compared to English28. These differences may lead to the involvement of different brain regions, different neuronal networks and, consequently, different contributing genetic factors for speech processing29. For SNP rs4504469-KIAA0319, results from our meta-analysis helped to identify, whether non-replication in our cohort is more likely due to language differences or due to the described differences between case-control studies and studies in the general population. For the other SNPs not associated in our cohort, we do not have external data to perform a similar meta-analysis and therefore cannot resolve this ambiguity.
Our in silico characterisation on the molecular, functional level using ‘RegulomeDB’ further supported especially associated SNP rs2038137-KIAA0319. For this SNP strongest evidence for functionality was found in this database (Table 3): This SNP affected binding of the transcription factor RFX330, required for corpus callosum development31. In humans, corpus callosum deficits could lead to impaired motor coordination and balance problems, attributes known for dyslexics32. RFX3 was also associated with relative hand skill in individuals with dyslexia33, which is in line with the reported correlation of left-handedness and dyslexia34. Furthermore, RFX3 is involved in ciliogenesis which is suggested to play an important role in the aetiology of dyslexia4,35. When regarding eQTL findings for both associated SNPs rs2038137-KIAA0319 and rs6935076-KIAA0319, the strongest effect on gene expression was found on KIAA0319 itself. Effect sizes on KIAA0319 were consistent with association level observed in our case-control analysis (Fig. 3) as the two SNPs associated in our case-control and in our meta-analysis revealed the strongest effect on KIAA0319. This point to a mechanism of action of these SNPs including the regulation of expression levels of KIAA0319. Note that the strongest SNP affecting the expression levels of KIAA0319 is rs761100 (Supplemental Table 4). This SNP was previously associated with expressive language scores24 and, thus seems to be an additional reasonable candidate for future analyses.
To the best of our knowledge, SNP rs7765678-DCDC2 was identified for the first time in a case-control setting with our study. The effect size direction was also in accordance with the association in the general population. This variant is the second variant in DCDC2 with association in our sample as we previously found association with the intron 2 deletion of DCDC217,18. Note that we did not find linkage disequilibrium with other SNPs in DCDC2 for which association with dyslexia in a case-control cohort was previously reported, supporting novelty (Supplemental Table 1). Originally, SNP rs7765678-DCDC2 was associated with quantitative measures of regular word reading8. Interestingly, for SNP rs7765678-DCDC2 we did not found an association altered expression of DCDC2 itself, but with levels of MRS2 (Table 3 and Fig. 3). Consistently, the SNPs associated in our case-control cohort showed the strongest effect on expression levels of MRS2. MRS2 is associated with ‘hyperphosphatasia with mental retardation syndrome’ (HPMRS)36 and, besides other brain regions, highly expressed in the cerebellum26. Future studies have to show whether this relation might be part of a pathomechamism of this SNP. Not that in general, in early periods of neuronal development expression levels need to be tightly controlled, which is relevant for dyslexia37.
We are aware that due to the moderate sample size our results are limited and therefore require comparison with findings from additional studies as done for the SNPs with available external data in our meta-analysis. Nevertheless, advantages of our study also exist, e.g. the homogeneity of our cohort regarding language and local recruitment. It’s also worth mentioning, that we cannot completely exclude that population stratification and cryptic relationship might have a certain effect on the analyses. However, due to our recruitment strategy focussing on the local well defined population, this bias might be considerably low.
Still, given our limitations, we suggest that for an overall conclusion about general differences in the genetic architecture of reading in the general population and clinical dyslexia, additional studies have to be done.
Since dyslexia has a polygenic background and contributions of individual genetic markers might be low, it seems promising to not only analyse the SNPs on single marker level but jointly. Encouragingly, we found that the percentage of risk alleles was higher for more SNPs than expected by chance (p-value = 0.033). Supporting, we observed a general increase of the sums of all originally reported risk alleles in cases, Fig. 2a). Importantly, this increase was not only driven by the three SNPs that associated nominally at single level as this effect persisted when analysing only non-associated SNPs (Fig. 2b). When adding SNPs analysed in previous studies to the risk score, the characteristic risk allele pattern remains (Supplemental Fig. 2). Note that given the different measures used in the original studies, we could not apply a weighted score. Our observed shift between the risk score of cases and controls appears to be very similar to shifts found when summing up highly validated risk alleles from large genome-wide studies in other complex diseases38,39. It is in line with the finding that our study had 80% power to replicate the direction of a reported risk allele with very small effect sizes down to an OR of 1.093–1.152. Hence, our power to detect a reported trend is considerably more sensitive than the power to detect a reported association (i.e., here we have 80% power to detect an OR of 1.36–1.62). To the best of our knowledge our results provide first evidence in a relevant dataset that well-known strategies to sum risk alleles across multiple genes may be also promising in the field of dyslexia. Therefore, our findings may motivate future larger collaborative studies in dyslexia targeting variants with smaller genetic effect sizes.
In summary, our work corroborates the importance of rs7765678-DCDC2, rs2038137-KIAA0319, and rs6935076-KIAA0319 in the aetiology of dyslexia. The relevance of rs2038137-KIAA0319, and rs6935076-KIAA0319 was further supported by our meta-analysis. The increased number of originally reported risk alleles in our dyslexia cases compared to controls and the replicated effect directions above chance may support a contribution to dyslexia risk for at least some of the other SNPs without associations on single marker level. Additionally, we provided functional attributes for many of these SNPs which can be included in future studies aiming to elucidate links between these genetic variants and dyslexia.
Materials and Methods
Ethical approval
The study was approved by the Ethics Committee of the University of Leipzig. Involvement of children in schools was approved by the Saxon Ministry of Culture and Sports. Informed and written consent was obtained from probands’ parents. All experiments were conducted in accordance with the informed consent and the approved guidelines of the Ethics Committee of the University of Leipzig.
Sample
Our sample comprised 383 dyslexia cases and 357 controls. Dyslexia-cases were selected according to a two-stage process: In a first step, schools with special dyslexia classes were contacted. Children of these classes were tested with different psychometric tests at the end of the 2nd grade. According to the admission criteria of these classes only children with a discrepancy between IQ and reading performance of at least 1.25 SD or higher get access. In a second step, students underwent additional testing to avoid the inclusion of ADHD affected probands (test d2)40, to ensure an IQ ≥ 85 (CFT20)41 and to get a quantitative measure for reading/spelling performance (KNUSPEL-L)42. For further cohort and test descriptions see Wilcke et al.17 and Mueller et al.15.
SNP selection and genotyping
The literature was thoroughly screened for SNPs associated with reading or spelling in the general population. In total, six studies with 16 SNPs within five genes were identified (Table 1). The majority of these SNPs lack of replication in case-control studies. DNA was extracted from saliva or blood using standard protocols including Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), Macherey-Nagel NucleoMag 96 Blood Kit, the MACHEREY-NAGEL NucleoSpin® 8 and 96 Blood Kit (Macherey-Nagel, Dueren, Germany), and the Oragene DNA Genotek Saliva Kit (Kanata, Ontario, Canada).
In total, 11 SNPs (rs1419228, rs9467075, rs9467076, rs1091047, rs7765678, rs6922023, rs8037376, rs8043049, rs8040756, rs1842129, rs1995402) were genotyped by mass spectrometry via iPlex (Sequenom, Hamburg, Germany). Five SNPs (rs6935076, rs2143340, rs4504469, rs2038137, rs7174102) were genotyped by mass spectrometry using the single-base extension (SBE) method GenoSNIP43. Primer sequences are provided in Supplemental Table 6. In detail, PCR was performed in 10 μl reaction volume according to the following conditions: Initial denaturation at 95 °C for 15 min, 45 cycles with denaturation at 95 °C for 45 s, primer hybridization at 58 °C for 45 s, elongation at 72 °C for 45 s and a final extension step at 72 °C for 5 min. The resulting product was digested with exonuclease and shrimp alkaline phosphatase to remove leftover primers and to dephosphorylate dNTPs. A SBE reaction was performed with specific primers with biotin residue and a photocleavage site44. The SBE reaction was performed with the purified PCR product according to the following conditions: Initial denaturation at 95 °C for 4 min, 44 cycles with denaturation at 94 °C for 10 s, primer hybridization at 60 °C for 30 s, and elongation at 72 °C for 10 s. The final SBE products were genotyped by MALDI-TOF mass spectrometry.
Associations on single marker and haplotype level
The study was designed to detect an OR of 1.36–1.62 with a minor allele frequency (MAF) of 0.42–0.10 with β = 0.8. This power is in accordance with previously reported effect sizes20,23,24. Allelic associations were analysed using Chi2 statistics in order to detect group differences between dyslexia cases and controls. Genotypic associations were assessed by GRR estimations according to Lathrop45. Moreover, haplotypic effects were investigated using Haploview 4.146. Associations of haplotypes were analysed using the implemented Chi2 approach.
To identify independent genetic effects, we pruned the selected SNPs (R2 = 0.5) while priority was given to SNPs with higher effect sizes (i.e. stronger allelic odds ratios) this is also known as clumping.
Meta-analysis
We conducted a meta-analysis by integrating SNP data from dyslexia case-control and transmission/disequilibrium (TDT) studies. Studies were identified by systematically screening PubMed and Google Scholar with the keywords ‘dyslexia’ and the SNP-names.
Allele counts from cases and controls and numbers of transmitted alleles were used to estimate the individual effect sizes. ORs for TDT-studies were calculated according to Kazeem and Farrall47. Since between-cohort heterogeneity was expected, meta effect sizes were computed using a random effects model and effects were visualised by forest plots implemented in the metafor package48. Analyses were performed for all identified cohorts and in a second step stratified according to language. Newbury et al.24 analysed two different cohorts of cases with the same cohort of controls. Both analyses were included indicated with a) and b) as individual studies in the meta-analysis.
Polygenic analyses
We computed an additive risk score composed of the sum among all individually observed risk variants, and its distribution between cases and controls was analysed. To only consider independent genetic effects identified by pruning. This avoids unintendedly increasing the weight of SNPs with other SNPs in LD. Note that this procedure is similar with the analysis of an unweighted genetic risk score using the PRSice approach49. This approach was performed for three different sets of SNPs: All SNPs analysed in this study (Fig. 2a), all SNPs except the ones nominally associated (Fig. 2b) and all SNPs of this study with addition of the SNPs analysed in previous studies on the same cohort (rs793862-DCDC217*, rs807701-DCDC217*, rs807724-DCDC217, the intron 2 deletion of DCDC217*, rs12533005-FOXP216*, rs10502812-AK13101115*, rs11873029-DYM15*, rs11874896-EPB41L315*, rs11661879-KIAA042715*, rs1299348-MC5R15*, rs555879-MYO5B15*, rs12606138-NEDD4L15, rs8094327-NEDD4L15*, rs9461045-KIAA031918 (SNPs marked with * show independent effects (R2 ≥ 0.5) and were included for analysis).
The probability of finding the observed effect directions under the NULL for the pruned SNPs was calculated via the binomial distribution. The power to detect a reported trend was calculated as previously described50.
In a supplementary analysis, we additionally considered for the polygenic score all dyslexia candidate SNPs previously reported in our cohort into the score, regardless whether we had found association or not. These SNPs were rs793862-DCDC217*, rs807701-DCDC217*, rs807724-DCDC217, the intron 2 deletion of DCDC217*, rs12533005-FOXP216*, rs10502812-AK13101115*, rs11873029-DYM15*, rs11874896-EPB41L315*, rs11661879-KIAA042715*, rs1299348-MC5R15*, rs555879-MYO5B15*, rs12606138-NEDD4L15, rs8094327-NEDD4L15*, rs9461045-KIAA031918. SNPs marked with * show independent effects (R2 ≥ 0.5) and were included for analysis. Data for all these SNPS was available for 269 cases and 357 controls.
All statistical analyses were done in R 3.2.151 or perl using in-house scripts available upon request.
Functional in silico characterisation
We analysed the SNPs for effects on local and distant gene expression levels (cis and trans-eQTLs) using data from published large-scale experiments52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70. These studies cover a broad range of tissues, like blood-derived cells, skin, liver and different brain regions. To identify proxy SNPs in our eQTL analysis, we used a minimum level of R2 ≥ 0.5, linkage data was used from 1000 Genomes reference phase 1, release V3 and from HapMap Data (Release #28, lifted over to GRCh37/hg19). We report results of the proxy SNPs that showed strongest correlation and we only considered eQTLs meeting study-wide significance level. Most analysed SNPs were located in the DYX2 locus on chromosome 6. To provide a spatial overview of these SNPs and their eQTL effects we generated a local association plot using LocusZoom71.
SNPs were also annotated for known and predicted local regulatory elements via ‘RegulomeDB’25, and tissue-specificity of gene products was analysed by ‘Human Protein Atlas’26.
Additional Information
How to cite this article: Müller, B. et al. Association, characterisation and meta-analysis of SNPs linked to general reading ability in a German dyslexia case-control cohort. Sci. Rep. 6, 27901; doi: 10.1038/srep27901 (2016).
Supplementary Material
Acknowledgments
We would like to thank all members of the Legascreen consortium for their support during this study. Legascreen is funded by the Fraunhofer Society and the Max-Planck-Society as a project within the framework of the “Pakt für Forschung und Innovation”. Dr. Arndt Wilcke and Dr. Holger Kirsten received funding from the Federal Ministry of Education and Research (grant PtJ-Bio 0315883). The authors thank Elfi Quente for expert technical assistance.
Footnotes
Author Contributions H.K. and J.B. contributed equally to the work. B.M., H.K., P.A., A.W. and J.B. designed and interpreted the experiments. B.M. I.C. and H.K. conducted the experiments. B.M. and H.K. analysed the data and performed in silico experiments. B.M. wrote the paper, with contributions of H.K., A.W. and J.B. A.W. carried out the psychometric testing, with expert advice and methodical consultations of the LEGASCREEN consortium. All authors read and approved the final manuscript.
Contributor Information
The LEGASCREEN consortium:
Angela D. Friederici, Frank Emmrich, Jens Brauer, Arndt Wilcke, Nicole Neef, Johannes Boltze, Michael Skeide, Holger Kirsten, Gesa Schaadt, Bent Müller, Indra Kraft, Ivonne Czepezauer, and Liane Dörr
References
- Lyon G. R., Shaywitz S. E. & Shaywitz B. A. A definition of dyslexia. Ann. Dyslexia 53, 1–14 (2003). [Google Scholar]
- Scerri T. S. & Schulte-Körne G. Genetics of developmental dyslexia. Eur. Child Adolesc. Psychiatry 19, 179–97 (2010). [DOI] [PubMed] [Google Scholar]
- Harlaar N., Spinath F. M., Dale P. S. & Plomin R. Genetic influences on early word recognition abilities and disabilities: a study of 7-year-old twins. J. Child Psychol. Psychiatry. 46, 373–84 (2005). [DOI] [PubMed] [Google Scholar]
- Carrion-Castillo A., Franke B. & Fisher S. E. Molecular genetics of dyslexia: an overview. Dyslexia 19, 214–40 (2013). [DOI] [PubMed] [Google Scholar]
- Meng H. et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc. Natl. Acad. Sci. USA 102, 17053–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler W. T. et al. Position of Neocortical Neurons Transfected at Different Gestational Ages with shRNA Targeted against Candidate Dyslexia Susceptibility Genes. PLoS One 8, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darki F., Peyrard-Janvid M., Matsson H., Kere J. & Klingberg T. Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo-parietal white matter structure. Biol. Psychiatry 72, 671–6 (2012). [DOI] [PubMed] [Google Scholar]
- Lind P. a. et al. Dyslexia and DCDC2: normal variation in reading and spelling is associated with DCDC2 polymorphisms in an Australian population sample. Eur. J. Hum. Genet. 18, 668–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini S. et al. Analysis of dyslexia candidate genes in the Raine cohort representing the general Australian population. Genes. Brain. Behav. 10, 158–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T. C. et al. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav. Genet. 41, 50–7 (2011). [DOI] [PubMed] [Google Scholar]
- Luciano M., Montgomery G. W., Martin N. G., Wright M. J. & Bates T. C. SNP sets and reading ability: testing confirmation of a 10-SNP set in a population sample. Twin Res. Hum. Genet. 14, 228–32 (2011). [DOI] [PubMed] [Google Scholar]
- Luciano M. et al. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol. Psychiatry 62, 811–7 (2007). [DOI] [PubMed] [Google Scholar]
- Paracchini S. et al. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am. J. Psychiatry 165, 1576–84 (2008). [DOI] [PubMed] [Google Scholar]
- Welter D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, 1001–1006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B. et al. Genetic risk variants for dyslexia on chromosome 18 in a German cohort. Genes. Brain. Behav. 1–7, 10.1111/gbb.12118 (2013). [DOI] [PubMed] [Google Scholar]
- Wilcke A. et al. Imaging genetics of FOXP2 in dyslexia. Eur. J. Hum. Genet. 20, 224–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke A. et al. The role of gene DCDC2 in German dyslexics. Ann. Dyslexia 59, 1–11 (2009). [DOI] [PubMed] [Google Scholar]
- Kirsten H., Wilcke A., Ligges C., Boltze J. & Ahnert P. Association study of a functional genetic variant in KIAA0319 in German dyslexics. Psychiatr. Genet. 22, 216–7 (2012). [DOI] [PubMed] [Google Scholar]
- Harold D. et al. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol. Psychiatry 11, 1085–91, 1061 (2006). [DOI] [PubMed] [Google Scholar]
- Cope N. et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am. J. Hum. Genet. 76, 581–91 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto J. M. et al. Association of reading disabilities with regions marked by acetylated H3 histones in KIAA0319. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 153B, 447–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkanac Z. et al. Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B, 556–60 (2007). [DOI] [PubMed] [Google Scholar]
- Becker J. et al. Genetic analysis of dyslexia candidate genes in the European cross-linguistic NeuroDys cohort. Eur. J. Hum. Genet. 1–6, 10.1038/ejhg.2013.199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury D. F. et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav. Genet. 41, 90–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M. et al. Tissue-based map of the human proteome. Science (80−). 347, 1260419–1260419 (2015). [DOI] [PubMed] [Google Scholar]
- Scerri T. S. et al. DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol. Psychiatry 70, 237–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour P. H. K., Aro M. & Erskine J. M. Foundation literacy acquisition in European orthographies. Br. J. Psychol. 94, 143–74 (2003). [DOI] [PubMed] [Google Scholar]
- Bornkessel-Schlesewsky I. & Schlesewsky M. In Cogn. Biol. Basis Linguist. Struct. New approaches Endur. Themes. (Sanz M., Laka I. & Tanenhaus M. K.) 241–252 (Oxford University Press, 2013). [Google Scholar]
- Jolma A. et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20, 861–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benadiba C. et al. The ciliogenic transcription factor RFX3 regulates early midline distribution of guidepost neurons required for corpus callosum development. PLoS Genet. 8, e1002606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix Y. et al. Motor impairment in dyslexia: the influence of attention disorders. Eur. J. Paediatr. Neurol. 11, 368–74 (2007). [DOI] [PubMed] [Google Scholar]
- Brandler W. M. et al. Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS Genet. 9, e1003751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M., Reimers S. & Manning J. T. Hand preference for writing and associations with selected demographic and behavioral variables in 255,100 subjects: the BBC internet study. Brain Cogn. 62, 177–89 (2006). [DOI] [PubMed] [Google Scholar]
- Brandler W. M. & Paracchini S. The genetic relationship between handedness and neurodevelopmental disorders. Trends Mol. Med. 20, 83–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz P. M. et al. Mutations in PIGO, a member of the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 91, 146–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi M. & Lehner B. The effects of genetic variation on gene expression dynamics during development. Nature 505, 208–11 (2014). [DOI] [PubMed] [Google Scholar]
- Wolpin B. M. et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet. 46, 994–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C. H. T. et al. Use of Net Reclassification Improvement (NRI) Method Confirms The Utility of Combined Genetic Risk Score to Predict Type 2 Diabetes. PLoS One 8, e83093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickenkamp R. & Zillmer E. The d2 test of attention. (Hogrefe, 2002). [Google Scholar]
- Weiß R. H. Grundintelligenz Skala 2. (Hogrefe, 1998). [Google Scholar]
- Marx H. The d2 test of attentionKnuspels Leseaufgaben (KNUSPEL L). (Hogrefe, 1998). [Google Scholar]
- Kirsten H. et al. Robustness of single-base extension against mismatches at the site of primer attachment in a clinical assay. J. Mol. Med. (Berl). 85, 361–9 (2007). [DOI] [PubMed] [Google Scholar]
- Kirsten H., Dienst S., Emmrich F. & Ahnert P. CalcDalton: a tool for multiplex genotyping primer design for single-base extension reactions using cleavable primers. Biotechniques 40, 158–162 (2006). [DOI] [PubMed] [Google Scholar]
- Lathrop G. M. Estimating genotype relative risks. Tissue Antigens 22, 160–6 (1983). [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–5 (2005). [DOI] [PubMed] [Google Scholar]
- Kazeem G. R. & Farrall M. Integrating case-control and TDT studies. Ann. Hum. Genet. 69, 329–335 (2005). [DOI] [PubMed] [Google Scholar]
- Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 36, 7–10 (2010). [Google Scholar]
- Euesden J., Lewis C. M. & O’Reilly P. F. PRSice: Polygenic Risk Score software. Bioinformatics 31, 1466–1468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier S. et al. IRF5 rs2004640-T allele, the new genetic factor for systemic lupus erythematosus, is not associated with rheumatoid arthritis. Ann. Rheum. Dis. 66, 828–831 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team. R. R: A Language and Environment for Statistical Computing. (2015). at < http://www.r-project.org/>.
- Dimas A. S. et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325, 1246–1250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet. 87, 779–789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A. L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007). [DOI] [PubMed] [Google Scholar]
- Fehrmann R. S. N. et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 7, e1002197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenawalt D. M. et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E. et al. Population genomics in a disease targeted primary cell model. Genome Res. 19, 1942–1952 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Cho H., Lee D. & Webster M. J. Association between SNPs and gene expression in multiple regions of the human brain. Transl. Psychiatry 2, e113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D. et al. Impact of common regulatory single-nucleotide variants on gene expression profiles in whole blood. Eur. J. Hum. Genet. 21, 48–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. J. et al. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494–9 (2007). [DOI] [PubMed] [Google Scholar]
- Ramasamy A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, 1020–1032 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder a. et al. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 12–20, 10.1038/tpj.2011.44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras J.-B. et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4, e1000214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K. et al. SeeQTL: A searchable database for human eQTLs. Bioinformatics 28, 451–452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsten H. et al. Dissecting the genetics of the human transcriptome identifies novel trait-related trans-eQTLs and corroborates the regulatory relevance of non-protein coding loci†. Hum. Mol. Genet. 24, 4746–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen E. L. et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 6, e1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science (80−). 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol. Psychiatry 15, 779–784 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J. et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 27, 2336–2337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.