Abstract
Figla is a germ-cell-specific transcription factor associated with ovary development and differentiation. In vertebrates, one transcriptional form of Figla is commonly found. However, besides the common form of this gene (named Figla_tv1), a new transcriptional form (named Figla_tv2) was identified in half-smooth tongue sole (Cynoglossus semilaevis). The full-length cDNA of Figla_tv1 was 1057 bp long with a 591-bp open reading frame encoding a predicted 196 amino acid protein, while Figla_tv2 encoded a 125 amino acid protein. Figla_tv1 and Figla_tv2 expression in various tissues was detected by qRT-PCR. Figla_tv1 was expressed mainly in ovary, skin and liver, while Figla_tv2 was expressed in all examined tissues. In the gonads, Figla_tv1 was expressed in ovary, while Figla_tv2 was predominately expressed in testis of pseudomales. Further, in situ hybridization located Figla_tv1 only in oocytes and Figla_tv2 mainly in germ cells of pseudomale testis. After knocking down Figla_tv2 in a pseudomale testis cell line, the expression of two steroid hormone-encoding genes, StAR and P450scc, was significantly up-regulated (P < 0.05). Our findings suggest that Figla_tv1 has a conserved function in folliculogenesis, as in other vertebrates, and that Figla_tv2 may have a role in the spermatogenesis of pseudomales by regulating the synthesis and metabolism of steroid hormones.
Basic helix-loop-helix (bHLH) proteins are members of a large superfamily that regulates a number of developmental and metabolic processes, including sex determination, cell differentiation, nervous system development, oncogenesis, and cholesterol metabolism1,2. Studies in many species, including Drosophila, Saccharomyces cerevisiae, Homo sapiens, and Arabidopsis, have provided a great deal of information about the structure and function of this family of proteins2,3,4,5,6,7,8.
The factor in the germline alpha (Figla) gene encodes a germ cell-specific bHLH transcription factor that mediates several developmental processes, including fertilization and early embryogenesis9. Many studies have focused on the important roles of Figla in gonad development and differentiation. In Mus musculus, Figla expression was first detected in the ovary at embryo day 13 (E13)10. Its expression increased dramatically at the end of embryo development and peaked at two days postpartum, when oocytes have become enclosed in primordial follicles10, suggesting the probable involvement of Figla in ovary follicle development. Furthermore, Figla was shown to regulate the expression of three zona pellucida genes (Zp1, Zp2, and Zp3) that encode proteins necessary for zona pellucida formation in growing oocytes11, indicating a crucial role for Figla in folliculogenesis. Moreover, targeted mutations of Figla in female M. musculus resulted in abnormal ovarian gonadogenesis, including failure to form primordial follicles, massive depletion of oocytes, and subsequent female sterility10. However, in mutated males, gonad development appeared to be normal, and these mice were fertile10. Together, these data indicate that Figla is indispensable only for ovary folliculogenesis and is not essential for testis development. In a subsequent study, it was suggested that Figla may balance sexually dimorphic gene expression in the postnatal ovary because Figla knockout resulted in the enhanced expression of many testis-specific genes in the oocytes of newborn M. musculus, while in male germ cells, the ectopic expression of Figla down-regulated a subset of these testis-specific genes12.
In teleosts, Figla has commonly been regarded as a marker gene of ovary development or early oocyte differentiation, but studies examining the regulation and roles of Figla in gonad development are limited13,14,15. Half-smooth tongue sole (Cynoglossus semilaevis) is an economically important marine flatfish in China. Because of their sex-dimorphic growth (females grow 2 to 4 times faster than males16), increasing the proportion of females in cultivated populations will be economically beneficial. To achieve this goal, understanding the sex-determination and differentiation mechanisms of C. semilaevis is particularly important. The primary sex of C. semilaevis is determined by the sex chromosomes: females (ZW) harbor a large W sex chromosome, while males possess two ZZ sex chromosomes17. Approximately 14% of ZW genetic females were shown to be sex-reversed to phenotypic males, the so-called pseudomales17. Interestingly, these pseudomales are fertile and can mate with the normal females to produce the viable offspring. A number of sex-related genes, including Dmrt1, wnt4a, Sox9, Foxl2, tesk1, piwil2, and Gadd45g, have been isolated from C. semilaevis; however, the molecular mechanisms associated with sex determination and differentiation are still poorly understood18,19,20,21,22,23. In a previous study, we identified two different transcripts of Figla (hereby named Figla, transcript variant 1 [Figla_tv1] and Figla, transcript variant 2 [Figla_tv2]) in our RNA-seq data from adult gonads of C. semilaevis and found that the methylation levels of these genes were closely related to gonad development24. However, the gonad expression patterns and functions of the two Figla transcripts during development are still unclear. In the present study, we cloned the full-length cDNAs of the two Figla isoforms in C. semilaevis by rapid amplification of cDNA ends (RACE) and used quantitative real-time polymerase chain reaction (qRT-PCR) and in situ hybridization (ISH) to detect the spatial and gonad expression of the two genes. Furthermore, the expression of genes that may be regulated by Figla_tv2 was analyzed after RNA interference (RNAi) knockdown of Figla_tv2. In the present study, we aimed to identify the sequences of the Figla_tv1 and Figla_tv2 transcripts, examine their sex-dimorphic expression profiles, and illustrate their functional diversity during gonad development.
Results
Sequence characteristics of two Figla homologues in C. semilaevis
In order to characterise the Figla homologues in C. semilaevis, we successfully isolated 5′- and 3′-RACE fragments of Figla_tv1 and Figla_tv2 from adult ovary and pseudomale testis of C. semilaevis and assembled two full-length cDNAs. The cDNA sequences have been deposited in GenBank with accession numbers KT966740 (Figla_tv1) and KT966741 (Figla_tv2).
The full-length cDNA of Figla_tv1 was 1050 bp long with an open reading frame of 591 bp encoding a 196 amino acid (aa) protein, and the 3′ and 5′ untranslated regions (UTRs) were 317 bp and 142 bp, respectively (Figure S1A). The putative Figla_tv1 protein was 22.2 kDa with a theoretical isoelectric point (pI) of 4.76. The full-length cDNA of Figla_tv2 was 1510 bp long with an open reading frame of 378 bp encoding a 125 aa protein, and the 3′ and 5′ UTRs were 317 bp and 815 bp, respectively. A single typical polyadenylation signal (AATAAA) site was found in the 3′ UTR of Figla_tv2 between nucleotides 1460 and 1465, 19 bp upstream of the poly (A) tail (Figure S1B). The putative Figla_tv2 protein was 14.0 kDa with a theoretical pI of 4.24.
To analyse the similarities/differences between the two Figla nucleotide sequences, a multiple sequence alignment was conducted by Clustal X. The 714-bp nucleotide sequences at the 3′-ends were nearly identical in the two transcripts, while the remaining sequences did not show much similarity (Figure S2). In order to illustrate the alternative splicing of the two transcripts, the cDNA sequences were successfully mapped to the genomic sequence. As shown in Fig. 1A, the genomic sequence of Figla was comprised of six exons (1–6) that were separated by five introns (A–E). The two transcripts were produced by alternatively splicing the different exons. Figla_tv1 was generated by splicing out the second exon, while Figla_tv2 was generated by splicing out the first exon, and the other exons were spliced together (Fig. 1B).
Figure 1. Schematic representation of the C. semilaevis Figla genomic structure and splice variants.
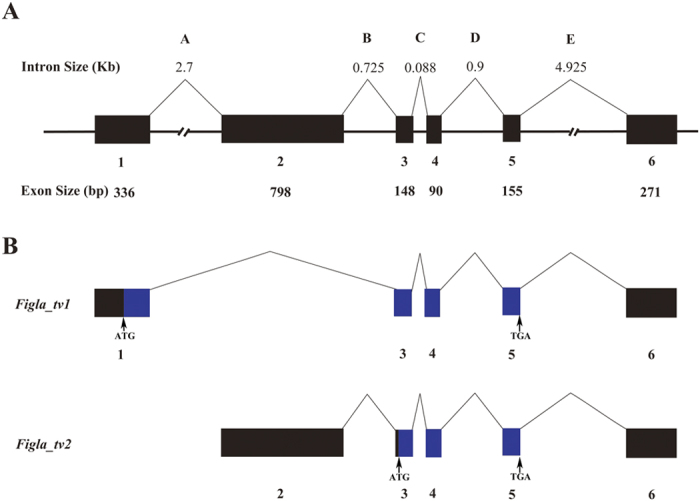
(A) The genomic structure of the C. semilaevis Figla gene. The six exons numbered under the solid boxes represent the Figla transcribed regions, and introns indicated by letters above the lines show the alternative splicing. (B) The alternatively spliced Figla mRNA isoforms. The transcribed nucleotides are shown with boxes, while the coding nucleotides are indicated with blue boxes. The locations of the initiation codons (ATG) and stop codons (TGA) are shown by vertical arrows.
To evaluate the similarities/differences of vertebrate Figla proteins, a multiple sequence alignment including Figlas from C. semilaevis and other vertebrates was conducted. All the vertebrate Figla proteins, including Figla_tv1 from C. semilaevis, had a typical conserved bHLH region composed of 57 aa (Fig. 2). However, Figla_tv2 from C. semilaevis lacked the basic region and six N-terminal amino acids from the HLH region (Fig. 2). The two translated Figla amino acid sequences from C. semilaevis shared high identities with proteins from other species as analyzed by MegAlign. The bHLH region from Figla_tv1 had especially high similarity to other amino acid sequences and shared 84.2%, 96.5%, 94.7%, 93.0%, 66.7%, 66.7%, 56.1%, and 56.1% identities with sequences from Acanthopagrus schlegelii, Dicentrarchus labrax, Oreochromis niloticus, Epinephelus coioides, Xenopus laevis, Chelonia mydas, Rattus norvegicus, and H. sapiens, respectively. Figla_tv2 shared 77.6%, 77.6%, 76.8%, 71.2%, 27.2%, 22.4%, 18.4% and 18.4% identities with Larimichthys crocea, O. niloticus, Pagrus major, D. labrax, X. laevis, C. mydas, R. norvegicus, and H. sapiens, respectively.
Figure 2. Multiple sequence alignment of Figla protein sequences from C. semilaevis and other vertebrates.
Sequences were aligned using Clustal X (Version 2.0) and the identical or similar amino acids are shaded by BOXSHADE. The presumed basic region is boxed, and the helix-loop-helix (HLH) region is underlined. Gaps introduced in the sequences to optimize the alignment are indicated by dashes. The abbreviated species names and their GenBank accession numbers are as follows: Cs Figla_tv2: Cynoglossus semilaevis Figla_tv2 (KT966741); Cs Figla_tv1: Cynoglossus semilaevis Figla_tv1 (KT966740); Lc Figla: Larimichthys crocea Figla (KKF33048.1); On Figla: Oreochromis niloticus Figla (NP_001298259.1); Dr Figla: Danio rerio Figla (NP_944601.2); Ol Figla: Oryzias latipes Figla (NP_001098215.1); Hs Figla: Homo sapiens Figla (NP_001004311.2); Mm Figla: Mus musculus Figla (NP_036143.1); Xl Figla: Xenopus laevis Figla (NP_001088667.1); Ac Figla: Anolis carolinensis Figla (XP_008120436.1).
Tissue expression of Figla_tv1 and Figla_tv2
To determine the expression patterns of the two Figla isoforms, we measured mRNA levels in 11 tissues of C. semilaevis. Figla_tv1 mRNA was predominately expressed in the skin, liver and brain of male and the gonad of female C. semilaevis and was negligibly expressed in the other tissues examined (Fig. 3). On the other hand, Figla_tv2 mRNA was expressed widely in the tissues of both sexes, including skin, liver, muscle and gill (Fig. 3), although there were some differences between the tissues from males and females.
Figure 3. Expression levels of Figla_tv1 and Figla_tv2 mRNAs in C. semilaevis tissues evaluated by qRT-PCR.
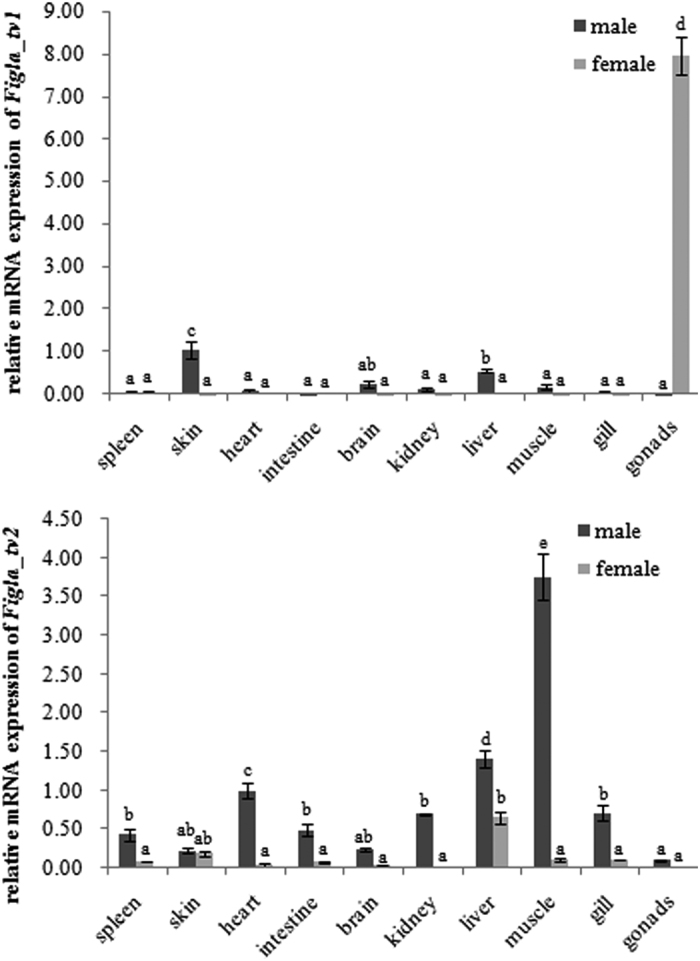
The expression levels were normalized using the geometric mean of the levels of two internal control genes (β-actin and Rpl13α). The mean ± SEM values from three separate individuals (n = 3) are shown. Bars with different letters indicate statistically significant differences (P < 0.05).
Sexual-dimorphic expression of Figla_tv1 and Figla_tv2 during gonad development
To measure the expression levels during the developmental stages of gonad differentiation, Figla_tv1 and Figla_tv2 expression was detected in ovary, testis and pseudomale testis at different stages by qRT-PCR. As shown in Fig. 4, Figla_tv1 mRNA was first detected at 120 days after hatching (dah) in the ovary and then continued to be expressed until it reached a maximum at 1 year after hatching (yah) before declining sharply at 2 yah. Conversely, Figla_tv1 mRNA was almost undetectable in the gonads of both male and pseudomale C. semilaevis during all the developmental stages tested (Fig. 4). Figla_tv2 mRNA was exclusively expressed in the pseudomale testis (Fig. 4), where it was initially detected at 120 dah and then rapidly increased to reach a maximum at 180 dah before declining sharply at 1 yah. Figla_tv2 mRNA persisted in the adult testis at 2 yah (Fig. 4).
Figure 4. Relative expression levels of Figla_tv1 and Figla_tv2 at different development stages in gonads of C. semilaevis.
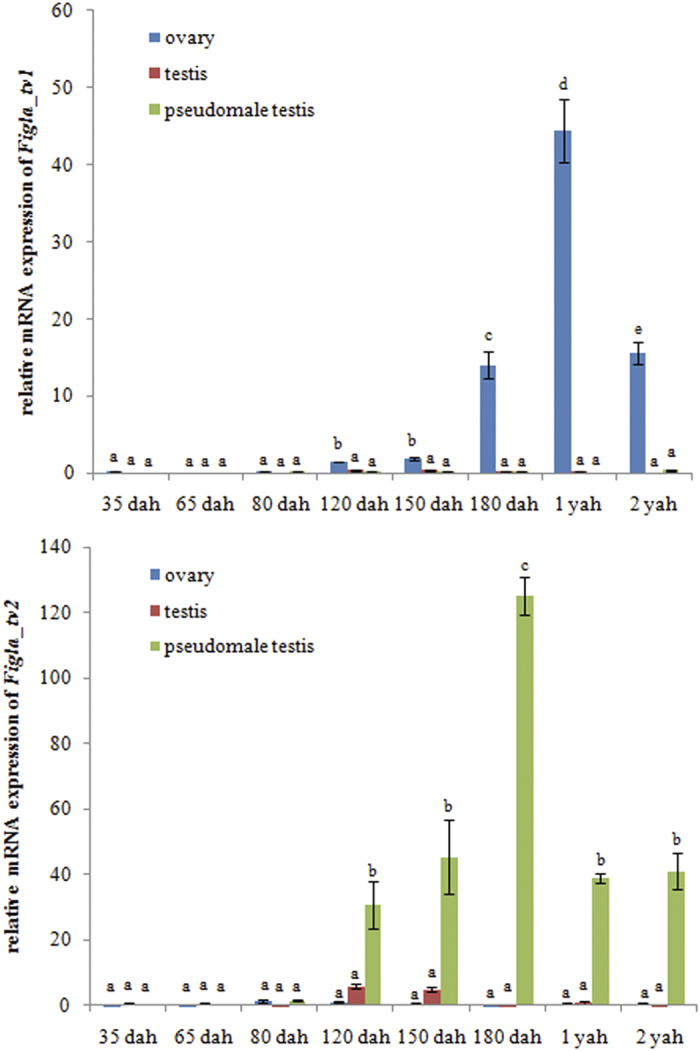
The expression level was normalized using the geometric mean of the levels of two internal control genes (β-actin and Rpl13α). The mean ± SEM values from three separate individuals (n = 3) are shown. Bars with different letters indicate statistically significant differences (P < 0.05).
Cyto-location of Figla_tv1 and Figla_tv2 in the gonads of C. semilaevis
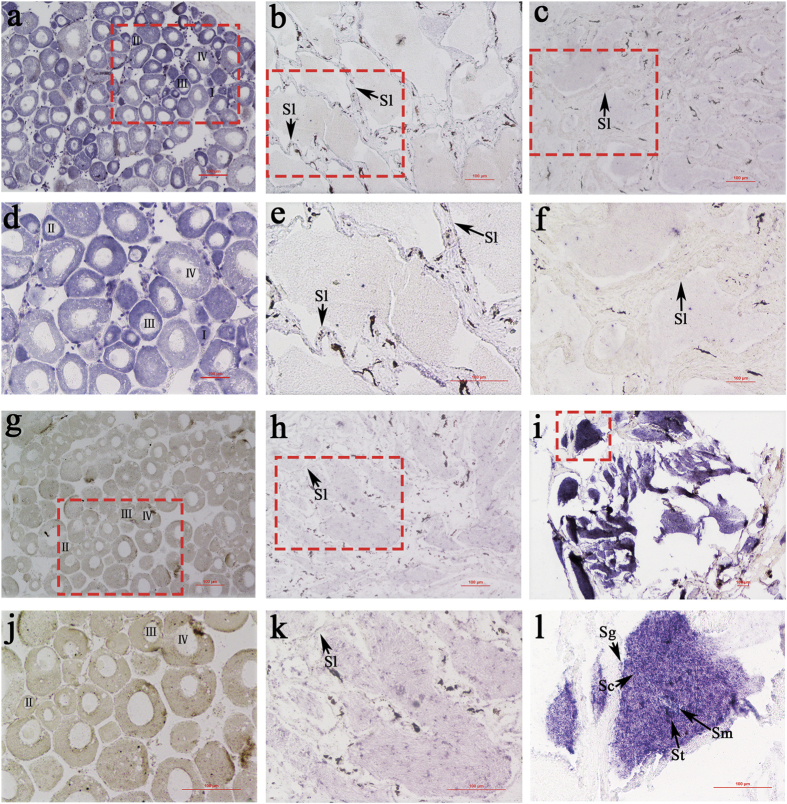
The cyto-location of Figla_tv1 and Figla_tv2 was detected by in situ hybridization (ISH). Ovaries from one-year-old fish contain somatic cells and oocytes at different developmental stages (stages I–IV). The ISH results showed that Figla_tv1 was expressed in the oocytes at all four developmental stages. Strong hybridization signals were detected in the oocytes at stages I, II and III, and only faint signals were detected in the oocytes at stage IV (Fig. 5a,d) compared with the controls (Figure S3a,d). In contrast to the intense signals in the ovary, no specific signals were detected in the testis of both males (Fig. 5b,e) and pseudomales (Fig. 5c,f) compared with controls (Figure S3b,e; Figure S3c,f), which was consistent with the weak or no Figla_tv1 expression in males and pseudomales at the 1 yah stage (Fig. 4). Testes from males and pseudomales contain germ cells from different developmental stages, including spermatogonia, spermatocytes, spermatids, and sperm. ISH revealed that Figla_tv2 mRNA was expressed mainly in the germ cells of pseudomale testis, with strong signals detected in spermatogonia, spermatocytes, spermatids, and sperm (Fig. 5i,l) compared with the controls (Figure S3i,l). No obvious hybridization signals were observed in ovary (Fig. 5g,j) and testis (Fig. 5h,k) compared with the controls (Figure S3g,j; Figure S3h,k).
Figure 5. Cyto-locations of Figla_tv1 and Figla_tv2 mRNAs in 1 yah gonads of C. semilaevis.
The figure shows the ovaries (left hand column), testes (middle column) and pseudomale testes (right hand column), labelled with Figla_tv1 antisense (a–f) and Figla_tv2 antisense (g–l) probes. (a–c) and (g–i) show the architecture with low magnification, while (d–f) and (j–l) indicate the red framed areas in (a–c,g–i) with large magnification. Oocytes at different developmental stages are marked by I, II, III and IV. Sg: spermatogonia; Sc: spermatocyte; St: spermatid; Sm: sperm; Sl: seminal lobule. Scale bars: 100 μm.
RNAi-mediated Figla_tv2 knockdown in C. semilaevis
Due to no suitable cell line for RNAi of Figla_tv1 established in C. semilaevis, we are unable to knock down the Figla_tv1 gene in vitro. The RNAi experiment was only conducted for Figla_tv2 gene in a C. semilaevis pseudomale gonad (CSPMG) cell line. To evaluate the silencing effects of RNAi, Figla_tv2 gene expression was detected at 48 h after siRNA transfection and was found to be reduced by approximately 54% in the si-cse-Figla 001-treated group (P < 0.05) and 76% in the si-cse-Figla 002-treated group (P < 0.05) compared with the control (Fig. 6A).
Figure 6. Relative mRNA expression levels of Figla_tv2, StAR and P450scc in cultured pseudomale gonad cells after RNAi treatment.
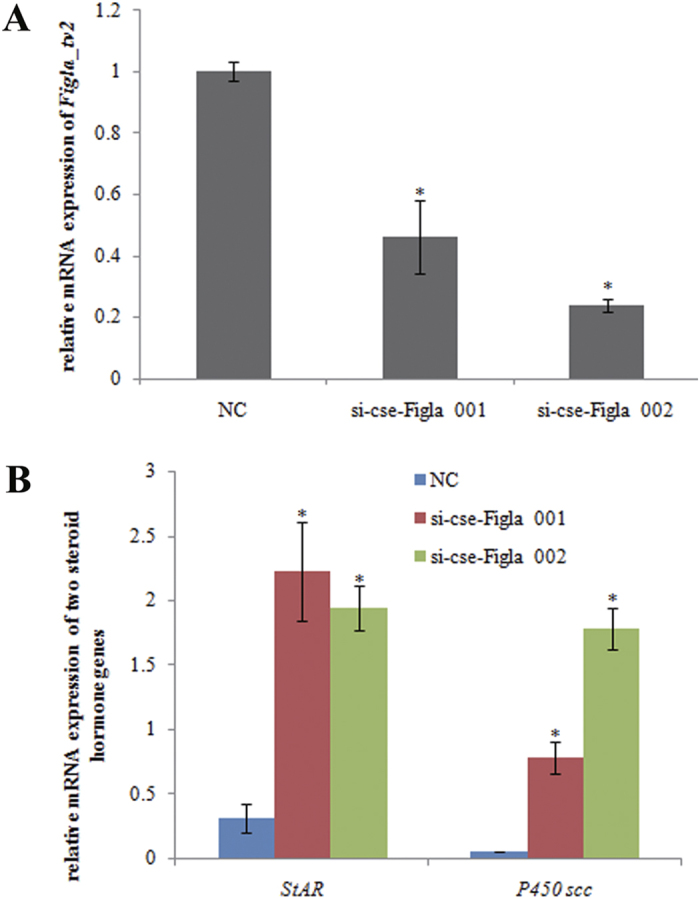
(A) Expression of Figla_tv2 after the transfection of the siRNAs for 48 h. (B) Expression of StAR and P450scc after the transfection of the siRNAs for 48 h. The transcripts of the β-actin and Rpl13α genes were used as internal controls to normalize the expression. NC, si-cse-Figla 001 and si-cse-Figla 002 indicate the gonad cells transfected with the siRNAs of the negative control (NC), si-cse-Figla 001 and si-cse-Figla 002, respectively. Asterisks above the bars indicate significant differences (P < 0.05) between the treated group and the control.
Effects of Figla_tv2 gene silencing on the expression of two steroid hormone encoding genes
To determine the effects of Figla_tv2 gene silencing on the expression of other sex-related genes, we measured the expression of Sox9a, wt1a and tesk1, three spermatogenesis-related genes in C. semilaevis20,21,25. However, no differential expression (P > 0.05) between the treated group and the control was observed for the three genes as detected by qRT-PCR (data not shown). Besides, the mRNA levels of two steroid hormone-encoding genes, steroidogenic acute regulatory protein (StAR) and cytochrome P450 side-chain cleavage (P450scc), were detected after RNAi silencing of Figla_tv2 expression. As shown in Fig. 6B, the expression levels of StAR and P450scc were strongly altered. StAR expression increased by approximately 6-fold in the si-cse-Figla 001-treated group and by 5-fold in the si-cse-Figla 002-treated group compared with the control (P < 0.05), and P450scc expression increased by approximately 15- and 35-fold (P < 0.05) in the si-cse-Figla 001- and the si-cse-Figla 002-treated groups, respectively, compared with the control (Fig. 6B).
Discussion
The Figla gene has been cloned in many mammalian and fish species, but until now only one transcript has been reported in each species. In the present study, in addition to the common form (Figla_tv1) of the gene, we isolated and characterized another transcript (Figla_tv2) from C. semilaevis. The highly conserved bHLH region is thought to be required for Figla binding to the E-box motif of the target protein10,11. Because Figla_tv2 lacked the conserved bHLH region (Fig. 2), we speculate that Figla_tv2 may be incapable of interacting with DNA directly and may function by associating with other molecules.
In vertebrates such as Bos taurus, M. musculus, O. latipes, O. niloticus and H. sapiens, Figla was expressed mainly in the gonads9,11,13,15,26. In this study, we also detected C. semilaevis Figla_tv1 expression mainly in the gonads of one-year-old female fish (Fig. 3), indicating the conserved role of Figla_tv1 in gonad tissue during development of the fish. We also noted that Figla_tv1 was expressed in other tissues, such as the skin and liver of male C. semilaevis (Fig. 3). However, the function of Figla in these tissues is not yet clearly understood. The robust expression of Figla in the skin and liver of C. semilaevis may indicate a role for Figla in immune functions, as previously reported in several teleosts27,28,29. It is very likely that the previous studies of Figla tissue distribution reported overlapping expression of both alternative splicing forms. For example, G. rarus Figla showed a similar composition to C. semilaevis Figla_tv1, but its expression pattern was the same as Figla_tv2 (Fig. 3)14. To evaluate whether there are multiple Figla isoforms in other teleosts too, we searched the transcriptome data in GenBank and Ensemble. We found that in most teleosts there was only one Figla isoform (Figla_tv1) except that Takifugu rubripes expressed three Figla isoforms. Therefore, we speculate the multiple Figla isoforms probably exist in some fish species or even are generally present in the teleosts, while they are undetectable due to the sequencing depth or coverage. More studies are needed to test these possibilities.
In C. semilaevis, the histological and cellular differentiation of the ovary is not synchronous; histological differentiation was reported to begin 56–62 dah, whereas cellular differentiation occurred at 120 dah with the appearance of the ovarian cavity18. In this study, Figla_tv1 mRNA was first detected at 120 dah in the ovary and persisted into adulthood (Fig. 4). This finding is reasonably consistent with the cellular differentiation period of the ovary, suggesting that Figla_tv1 may play a crucial role in ovary differentiation. In addition, qRT-PCR and ISH both detected dimorphic expression of Figla_tv1 in the gonads, with strong expression in the ovary during development and negligible expression in the testis (Figs 4 and 5). This result is consistent with previous reports in M. musculus, O. latipes, O. niloticus, and H. sapiens11,13,15,26. However, the ectopic expression of Figla in O. niloticus XY fish resulted in the depletion of germ cells and down-regulation of spermatogenesis-associated genes, suggesting Figla may play an important role in ovarian development by repressing the expression of spermatogenesis-associated genes15. Based on similar results with Figla_tv1 in C. semilaevis, we speculate that Figla_tv1 may be involved in ovary folliculogenesis.
In order to evaluate whether Figla_tv2 was involved in sex-reversal, we measured the levels of Figla_tv2 transcripts in the gonads of males, females and pseudomales. Figla_tv2 was expressed exclusively in the testis of pseudomales, with weak or no expression in the testis of males or in the ovary of females (Fig. 4), suggesting it is probably a pseudomale-biased gene that may play an important role in the sex-reversal process.
The ISH results showed that Figla_tv2 mRNA was mainly located in the germ cells of pseudomale testis (Fig. 5i,l), suggesting an important role in regulating spermatogenesis. However, the underlying regulatory mechanism is still unknown. Proteins that belong to the bHLH superfamily generally function as transcriptional enhancers or inhibitors of target genes1,2. Despite the lack of a complete bHLH region, we speculate that Figla_tv2 may regulate the development of pseudomale testis by repressing or activating its target genes. Steroid hormones are commonly present in the gonads of vertebrates, where they are known to play critical roles in gonad development30,31,32. StAR and P450scc are key regulatory proteins in the synthesis and metabolism of steroid hormones in teleosts33,34,35. StAR mediates the transport of cholesterol across the mitochondrial membrane, a rate-limiting step in steroid hormone synthesis32,33,34,35, and P450scc catalyzes the conversion of cholesterol to pregnenolone, a pivotal step in the initiation of steroidogenesis33,34,35. After the in vitro knockdown of Figla_tv2 in cultured pseudomale testis cells, qRT-PCR showed that StAR and P450scc mRNA levels were significantly up-regulated (Fig. 6B), suggesting C. semilaevis Figla_tv2 may be involved in spermatogenesis by regulating steroid hormones synthesis and metabolism in the gonads of pseudomales.
Conclusions
Two functional homologues, Figla_tv1 and Figla_tv2, were successfully isolated from C. semilaevis. The two transcripts had different tissue expression patterns in males and females, implying their probably functional diversity. Figla_tv1 exhibited predominant “vertebrate-like” ovarian expression, suggesting a conserved role in folliculogenesis. Figla_tv2 was expressed mainly in the testis of pseudomales and is likely to play a role in the spermatogenesis of pseudomales by influencing the synthesis and/or metabolism of steroid hormones.
Methods
Ethical statement
The collection and handling of the animals used in this study was approved by the Animal Care and Use Committee at the Chinese Academy of Fishery Sciences, and all the experimental procedures were performed in accordance with the guidelines for the Care and Use of Laboratory Animals at the Chinese Academy of Fishery Sciences.
Fish and sample collection
All half-smooth tongue sole (C. semilaevis) used in these experiments were purchased from Huanghai Aquaculture Ltd. (Haiyang, China). One-year-old fish (three individuals of each sex) were randomly sampled. Spleen, skin, heart, intestine, brain, head kidney, liver, muscle, gill, ovary, and testis tissues (11 in all) were collected and immediately transferred to liquid nitrogen and were then stored at −80 °C until RNA extraction. In addition, parts of the gonad were fixed in 4% paraformaldehyde (pH 7.4) at 4 °C for 24 h, then dehydrated through a methanol series (25, 50, 75, and 100%) and finally stored in 100% methanol at −20 °C for ISH analysis. Moreover, three independent gonadal samples at 35, 65, 80, 120, 150, and 180 dah and at 1 and 2 yah were sampled within one family. To determine the genetic sex of the fish, parts of the caudal fins were collected and fixed in 100% ethanol for DNA extraction.
cDNA synthesis and identification of the genetic sex
Total RNA was extracted from the frozen tissues with TRIzol reagents (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Then, the RNA was transcribed to cDNA using a PrimeScript™ RT reagent kit (TaKaRa Bio Inc., Otsu, Japan) with gDNA Eraser to avoid contamination by genomic DNA.
Genomic DNA was extracted from the fins using the phenol-chloroform method as described previously16. Female-specific primer pairs (CseF382F and CseF382R) (Table S1) were used to identify the genetic sex of the fish23. After PCR amplification with the conditions: 94 °C for 5 min, followed by 35 cycles (94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s), and 72 °C for 10 min, the products were run on a 1.2% agarose gel, and samples with a single 291 bp band were identified as genomic females, while samples with no bands were identified as genomic males.
Phenotypic sex was determined by histological analysis following procedures described previously36. Pseudomales are individuals for which the genomic sex is identified as female, but the phenotypic sex is male.
Isolation of Figla full-length cDNA from C. semilaevis tissues
RACE-ready first-strand cDNA was synthesized from total RNA using a SMART™ RACE cDNA amplification kit (Invitrogen) according to the manufacturer’s instructions. The specific primers for the outer and nest amplifications were based on the mRNA sequence that determined from whole-genome sequencing18 and the RNA-seq data24 of C. semilaevis. The sequences of all primers used for RACE amplifications are listed in Table S1. The universal primer UPM and the outer primers were used for the first 5′- and 3′-RACE amplifications. The products were then diluted 100 times with ddH2O and used as templates for the nested PCR reactions with the nest primers and the universal primer NUP. The outer and nest amplifications were both performed using the same touchdown PCR procedures: denaturation at 94 °C for 5 min, followed by 15 cycles (94 °C for 30 s, first five cycles 68 °C for 30 s, and subsequently the temperature was reduced by −2 °C per five cycles, and 72 °C for 2 min), 20 cycles (94 °C for 30 s, 60 °C for 30 s, and 72 °C for 2 min), and then incubation at 72 °C for 10 min. The nested PCR products were separated on a 1.2% agarose gel and purified with a DNA purification kit (TaKaRa Bio Inc.). The purified 5′- and 3′-RACE products were subcloned into a PMD-18T vector (TaKaRa Bio Inc.) and sequenced.
Bioinformatics analysis
The 5′- and 3′-RACE fragments were assembled using DNASTAR software (DNASTAR, Madison, WI) to obtain the full-length cDNA sequences of Figla_tv1 and Figla_tv2. Sequence identity and similarity of the two translated Figla proteins from C. semilaevis with known Figla proteins from other vertebrates were assessed using the MegAlign program (DNASTAR) with Clustal W method. Sequence alignments of two Figla nucleotide sequences were generated using the Clustal X37 and GeneDoc programs38. Multiple sequence alignments of Figla proteins from different species were conducted using the Clustal X37 and edited by BOXSHADE (http://www. ch.embnet.org/software/BOX_form.html).
qRT-PCR analysis
The expression levels of the two Figla genes in the tissues and gonads at differential developmental stages were determined by qRT-PCR as described previously23. Two pairs of primers (Table S1) were designed based on the specific gene regions of Figla_tv1 and Figla_tv2. The specificity of the primers was verified by a single distinct peak obtained in a melting curve analysis. The β-actin was identified previously as a reliable internal reference gene in various tissue samples of C. semilaevis39 and was used as the first reference gene. In addition, Rpl13α was validated as a suitable reference gene for quantifying gene expression in teleost species such as Danio rerio40. Our recent studies have found it was expressed with a relatively stable level in various tissues of C. semilaevis (data not shown). Therefore, it was employed as the second reference gene. The geometric means of β-actin and Rpl13α expression levels were used to normalize the data. Quantification was conducted using an ABI 7500 detection system (Applied Biosystems, Foster City, CA, USA) with SYBR Green Master Mix (TaKaRa Bio Inc.). The qRT-PCR procedure was as follows: 94 °C for 30 s, followed by 40 cycles of 94 °C for 5 s, and 60 °C for 34 s. Tissues and gonads were sampled from three individuals, and triplicate assays for each sample were conducted. The ABI 7500 system SDS software version 1.4 (Applied Biosystems) was used to analyze the qRT-PCR data, and baseline and cycle threshold values were set automatically. The relative mRNA expression of target genes was calculated by the 2−△△Ct method, as described previously41.
All data are presented as the mean ± SEM of three samples with three parallel repetitions. Differences between means were tested by one-way analysis of variance (ANOVA) followed by Duncan’s post-hoc test. (SPSS software version 18.0; SPSS Inc., Chicago, IL), and the significance level was set at P < 0.05. All assays in the qRT-PCR complied with the MIQE guidelines42.
In situ hybridization
To synthesize digoxigenin (DIG)-labeled RNA sense and antisense probes, the cDNA fragments of Figla_tv1 and Figla_tv2 were amplified by PCR using primer pairs Figla_tv1-ISH-F (BamHI site underlined) and Figla_tv1-ISH-R (EcoRV site underlined) and Figla_tv2-ISH-F (BamHI site underlined) and Figla_tv2-ISH-R (EcoRV site underlined) (Table S1), respectively. The PCR products were sub-cloned into the pEASY-T5-Zero vector and verified by sequencing. The recombinant plasmids with the correct insertion were extracted with a plasmid extraction kit (Tiangen Bio Inc., Beijing, China) and linearized by BamHI or EcoRV. The linearized plasmids were used as templates and the probes were synthesized with a DIG RNA Labeling Kit (Roche, Mannheim, Germany). For ISH analysis, the stored gonads were dehydrated in an ascending gradient of ethanol and embedded in paraffin wax, and 6-μm sections were cut. ISH was performed as described previously43 using three replicate samples. The sections were examined and photographed with a Nikon E80i microscope (Nikon Co., Tokyo, Japan).
RNAi silencing of Figla_tv2 and in vitro StAR and P450scc expression
Two Figla_tv2-specific small interfering RNAs (siRNAs), si-cse-Figla 001 and si-cse-Figla 002, and a nonspecific siRNA negative control (NC) were designed and synthesized by RayBiotech Co., Ltd. (Guangzhou, China). The cells used for the RNAi treatment were from the CSPMG cell line that was established previously44 and stored in liquid nitrogen. After thawing by incubating in a 42 °C water bath for 5 min, the CSPMG cells were harvested by centrifugation at 180 × g for 10 min. Then, the cells were incubated at 24 °C in 25-cm2 cell culture flasks with 20% FBS-DMEM/F12 medium. After an approximately 3 days of culturing, the cells formed a confluent monolayer. The cells were transferred to six-well plates and cultivated at 24 °C for 12 h to ensure complete attachment to the plates. The siRNA was transfected into the CSPMG cells using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s instructions. An NC siRNA labeled by Cy3 was used to assess the transfection efficiency, and the transfection efficiency was evaluated by calculating the ratio of cells expressing red fluorescence signals to all the cells used for transfection. The average transfection efficiency of the cells used for assay was calculated to be about 90%. The treated groups were transfected with siRNA (si-cse-Figla 001 and si-cse-Figla 002) at a final concentration of 30 nM. The controls were transfected with NC siRNA at the same concentration. The cells from the treated and control groups were cultivated at 24 °C for 48 h and were then harvested by centrifugation at 180 × g for 10 min. After washing three times with phosphate-buffered saline, the cells were stored in the same TRIzol reagents that were used for the RNA extraction. Three replicates were conducted for each group. Total RNA extraction and the re-transcriptions were performed according to the methods described above. The expression of Figla_tv2, StAR, and P450scc was measured by qRT-PCR using the primers listed in Table S1. The qRT-PCR data were analyzed as described above.
Additional Information
How to cite this article: Li, H. et al. Two Figla homologues have disparate functions during sex differentiation in half-smooth tongue sole (Cynoglossus semilaevis). Sci. Rep. 6, 28219; doi: 10.1038/srep28219 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Nature Science Foundation (31130057, 31402293, 31472269), the Taishan Scholar Project Fund of Shandong, China, the China Postdoctoral Science Foundation (2014M551990), the Post-Doctoral Innovation Project Fund of Shandong Province (201303035), and the Post-Doctoral Applied Research Project Fund of Qingdao City.
Footnotes
Author Contributions H.L., C.S. and S.C. conceived and designed the experiments. H.L., W.X., N.Z., Y.Z., Z.D. and H.X. performed the experiments. H.L., X.J. and N.W. analyzed the data. H.L., S.C. and W.X. wrote the main manuscript text. All authors reviewed the manuscript.
References
- Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol 5, 1–6 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir N. N., Ro¨nnstrand L. & Kazi J. U. The basic helix-loop-helix (bHLH) proteins in breast cancer progression. Med Oncol 30, 1–3 (2013). [DOI] [PubMed] [Google Scholar]
- Robinson K. A. & Lopes J. M. Saccharomyces cerevisiae basic helixloop-helix proteins regulate diverse biological processes. Nucleic Acids Res 28, 1499–1505 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. W., Barbel S., Jan L. Y. & Jan Y. N. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc Natl Acad Sci USA 97, 10436–10441 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrefitte S., Kahn D. & Haenlin M. New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mech Dev 104, 99–104 (2001). [DOI] [PubMed] [Google Scholar]
- Ledent V., Paquet O. & Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol 3, 1–18 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E. & Quail P. H. The Arabidopsis basic/helixloop-helix transcription factor family. Plant Cell 15, 1749–1770 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M. A. et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20, 735–747 (2003). [DOI] [PubMed] [Google Scholar]
- Tripurani S. K. et al. MicroRNA-212 post-transcriptionally regulates oocyte-specific basic-Helix-Loop-Helix transcription factor, factor in the germline alpha (FIGLA), during bovine early embryogenesis. PLoS ONE 8, e76114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal S. M., Amleh A. & Dean J. FIGα, a germ cell-specific transcription factor required for ovarian follicle formation. Development 127, 4645–4654 (2000). [DOI] [PubMed] [Google Scholar]
- Liang L. F., Soyal S. M. & Dean J. FIGα, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 124, 4939–4949 (1997). [DOI] [PubMed] [Google Scholar]
- Hu W., Gauthier L., Baibakov B., Jimenez-Movilla M. & Dean J. FIGLA, a basic helix-loop-helix transcription factor, balances sexually dimorphic gene expression in postnatal oocytes. Mol Cell Biol 30, 3661–3671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori A. et al. Comparative genomics approach to the expression of figα, one of the earliest marker genes of oocyte differentiation in medaka (Oryzias latipes). Gene 423, 180–187 (2008). [DOI] [PubMed] [Google Scholar]
- Yuan C. et al. Responsiveness of four gender-specific genes, figla, foxl2, scp3 and sox9a to 17a-ethinylestradiol in adult rare minnow Gobiocypris rarus. Gen Comp Endocrinol 200, 44–53 (2014). [DOI] [PubMed] [Google Scholar]
- Qiu Y. et al. Figla favors ovarian differentiation by antagonizing spermatogenesis in a teleosts, Nile Tilapia (Oreochromis niloticus). PLoS ONE 10, e0123900 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L. et al. Isolation of female-specific AFLP markers and molecular identification of genotypic sex in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol 9, 273–280 (2007). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Artificial gynogenesis and sex determination in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol 11, 243–251 (2009). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46, 253–260 (2014). [DOI] [PubMed] [Google Scholar]
- Hu Q. et al. Cloning and characterization of wnt4a gene and evidence for positive selection in half-smooth tongue sole (Cynoglossus semilaevis). Sci Rep 4, 7167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. L., Chen S. L. & Ji X. S. Molecular cloning, characterization and expression analysis of Sox9 and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Ocean Sin 30, 68–77 (2011). [Google Scholar]
- Meng L. et al. Cloning and characterization of tesk1, a novel spermatogenesis-related gene, in the Tongue Sole (Cynoglossus semilaevis). PLoS ONE 9, e107922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y. et al. Cloning, expression and methylation analysis of piwil2 in half-smooth tongue sole (Cynoglossus semilaevis). Mar Genom 18, 45–54 (2014). [DOI] [PubMed] [Google Scholar]
- Liu W. J. et al. Molecular characterization and functional divergence of two Gadd45g homologs in sex determination in half-smooth tongue sole (Cynoglossus semilaevis). Comp Biochem Physiol Part B 177–178, 56–64 (2014). [DOI] [PubMed] [Google Scholar]
- Shao C. et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res 24, 604–615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chen S., Liu Y., Wen H. & Zhu Y. WT1a gene molecular cloning and expression analysis during gender differentiation in half-smooth tongue sole (Cynoglossus semilaevis). J Fish China (in Chinese) 21(1), 26–36 (2014). [Google Scholar]
- Huntriss J. et al. Isolation, characterization and expression of the human factor in the germline alpha (FIGLA) gene in ovarian follicles and oocytes. Mol Hum Reprod 8, 1087–1095 (2002). [DOI] [PubMed] [Google Scholar]
- Matejusova I. et al. Gene expression profiles of some immune relevant genes from skin of susceptible and responding Atlantic salmon (Salmo salar L.) infected with Gyrodactylus salaris (Monogenea) revealed by suppressive subtractive hybridization. Int J Parasitol 36, 1175–1183 (2006). [DOI] [PubMed] [Google Scholar]
- Li H. X., Lu X. J., Li C. H. & Chen J. Molecular characterization of the liver-expressed antimicrobial peptide 2 (LEAP-2) in a teleost fish, Plecoglossus altivelis: Antimicrobial activity and molecular mechanism. Mol Immunol 65, 406–415 (2015). [DOI] [PubMed] [Google Scholar]
- Bayne C. J., Gerwick L., Fujiki K., Nakao M. & Yano T. Immune-relevant (including acute phase) genes identified in the livers of rainbow trout, Oncorhynchus mykiss, by means of suppression subtractive hybridization. Dev Comp Immunol 25, 205–217 (2001). [DOI] [PubMed] [Google Scholar]
- Baker P. J. et al. Expression of 3beta-hydroxysteroid dehydrogenase type I and type VI isoforms in the mouse testis during development. Eur J Biochem 2603, 911–917 (1999). [DOI] [PubMed] [Google Scholar]
- Barannikova I. A., Dyubin V. P., Bayunova L. V. & Semenkova T. B. Steroids in the control of reproductive function in fish. Neurosci Behav Physiol 328, 141–148 (2002). [DOI] [PubMed] [Google Scholar]
- Bauer M. P., Bridgham J. T., Langenau D. M., Johnson A. L. & Goetz F. W. Conservation of steroidogenic acute regulatory (StAR) protein structure and expression in vertebrates. Mol Cell Endocrinol 1681–1682, 119–125 (2000). [DOI] [PubMed] [Google Scholar]
- Arukwe A. Modulation of brain steroidogenesis by affecting transcriptional changes of steroidogenic acute regulatory (StAR) protein and cholesterol side chain cleavage (P450scc) in juvenile Atlantic salmon (Salmo salar) is a novel aspect of nonylphenol toxicity. Environ Sci Technol 3924, 9791–9798 (2005). [DOI] [PubMed] [Google Scholar]
- Arukwe A. Steroidogenic acute regulatory (StAR) protein and cholesterol side-chain cleavage (P450scc)-regulated steroidogenesis as an organ-specific molecular and cellular target for endocrine disrupting chemicals in fish. Cell Biol Toxicol 24, 527–540 (2008). [DOI] [PubMed] [Google Scholar]
- Aluru N., Renaud R., Leatherland J. F. & Vijayan M. M. Ah receptor-mediated impairment of interrenal steroidogenesis involves StAR protein and P450scc gene attenuation in rainbow trout. Toxicol Sci 842, 260–269 (2005). [DOI] [PubMed] [Google Scholar]
- Chen S. L. et al. Molecular marker-assisted sex control in half-smooth tongue sole (Cynoglossus semilaevis). Aquaculture 283, 7–12 (2008). [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K. B., Nicholas H. J. & Deerfiels D. W. GeneDoc: analysis and visualization of genetic variation. EMBnet News 4, 1–4 (1997). [Google Scholar]
- Li Z. et al. β-Actin is a useful internal control for tissue-specific gene expression studies using quantitative real-time PCR in the half-smooth tongue sole Cynoglossus semilaevis challenged with LPS or Vibrio anguillarum. Fish Shellfish Immun 29, 89–93 (2010). [DOI] [PubMed] [Google Scholar]
- Tang R., Dodd A., Lai D., Mcnabb W. C. & Love D. R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Bioch Bioph Sin 39, 384–390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth J. L. & Thomas D. S. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Kajiura-Kobayashi H. & Nagahama Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mech Develop 99, 139–142 (2000). [DOI] [PubMed] [Google Scholar]
- Sun A. et al. Establishment and characterization of a gonad cell line from half-smooth tongue sole Cynoglossus semilaevis pseudomale. Fish Physiol Biochem 41, 673–683 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.