Abstract
Green Fluorescent Protein (GFP) was originally found in cnidarians, and later in copepods and cephalochordates (amphioxus) (Branchiostoma spp). Here, we looked for GFP-encoding genes in Asymmetron, an early-diverged cephalochordate lineage, and found two such genes closely related to some of the Branchiostoma GFPs. Dim fluorescence was found throughout the body in adults of Asymmetron lucayanum, and, as in Branchiostoma floridae, was especially intense in the ripe ovaries. Spectra of the fluorescence were similar between Asymmetron and Branchiostoma. Lineage-specific expansion of GFP-encoding genes in the genus Branchiostoma was observed, largely driven by tandem duplications. Despite such expansion, purifying selection has strongly shaped the evolution of GFP-encoding genes in cephalochordates, with apparent relaxation for highly duplicated clades. All cephalochordate GFP-encoding genes are quite different from those of copepods and cnidarians. Thus, the ancestral cephalochordates probably had GFP, but since GFP appears to be lacking in more early-diverged deuterostomes (echinoderms, hemichordates), it is uncertain whether the ancestral cephalochordates (i.e. the common ancestor of Asymmetron and Branchiostoma) acquired GFP by horizontal gene transfer (HGT) from copepods or cnidarians or inherited it from the common ancestor of copepods and deuterostomes, i.e. the ancestral bilaterians.
Green fluorescent proteins (GFPs) are useful reagents for measuring molecular and cellular properties, such as gene expression, protein-protein interactions, and protein turnover1,2,3. They are structurally complex, being dimers of a monomer consisting of eleven beta sheets arranged in a cylinder with the fluorophore in the center and alpha helices at the top and bottom of the cylinder. This motif is so unique that GFP forms its own protein class with no other known protein with a similar structure4. While most GFPs fluoresce green, there are also some structurally related molecules that emit at other wavelengths5,6,7,8. Paradoxically, although the biotechnological applications of these molecules are well understood, much less is known about the functions of endogenous GFPs in animal cells. GFP was initially discovered in a luminous jellyfish (phylum Cnidaria) in which blue luminescent light is absorbed by the chromophore of the GFP which consequently gained electronic energy and then subsequently relaxed as photons in green fluorescence9. Endogenous GFPs have been found in at least three-dozen cnidarians (many of them non-luminous), six non-luminous copepods, and three non-luminous cephalochordate species in the genus Branchiostoma10,11,12, although GFP can be found in non-fluorescent species as well (GFP is then a chromoprotein)13. Clearly the occurrence of fluorescence does not necessarily indicate a relationship to GFP. Indeed, many compounds and proteins can trigger fluorescence in invertebrates and also vertebrates14,15,16.
There has been much discussion about the possible ecological relevance of light production in marine invertebrates, whether or not GFP is involved17. In bioluminescent cnidarians generally, the emitted light has been implicated in warning, defense, or attraction of prey. Since many pelagic organisms including jellyfishes perform diel vertical migration in the water column18,19, it has been suggested anecdotally for luminous jellyfishes that GFP might help to adjust the wavelength of light emitted to make it most visible at a given depth—for instance, blue at depth and green nearer the sea surface. The functions of GFP in non-luminescent organisms remain enigmatic. Suggested functions include photoprotection, spectral optimization for photosynthesis by mutualistic dinoflagellates, and protective antioxidation20,21,22. In addition, there is some evidence that prey are attracted to predators fluorescing when their GFP is excited by the blue wavelengths of sunlight penetrating relatively shallow water23.
When GFPs were initially discovered in copepods and cephalochordates, it was not clear whether the molecules were acquired from the diet, inherited from a common bilaterian ancestor or acquired via horizontal gene transfer (HGT)12,24. Sampling of more copepods indicated that GFPs had diversified within the group but did not answer the question of whether GFP had entered this lineage by horizontal gene transfer11. The same question was raised for cephalochordates (amphioxus, also known as lancelets), which have many GFPs12,25. The evolutionary conundrum about GFP (inheritance from common ancestor vs. HGT) largely derived from the fact that in all species studied thus far, GFPs share high similarity at both the structural and sequence levels, and yet the evolutionary lineages having GFPs (namely cnidarians, copepods and cephalochordates) are very sparsely distributed across the tree of life and are very distantly related to one another.
To address whether GFPs in the cephalochordate genus Branchiostoma were acquired by HGT, we also examined them from Asymmetron lucayanum, the most distant cephalochordate relative of Branchiostoma. There are three known genera of cephalochordates, where the Asymmetron genus branched off from the clade comprising Branchiostoma and Epigonichthys at least 120 mya26,27. Our comparison reveals that the genera Asymmetron and Branchiostoma share clearly homologous GFP-encoding genes. Thus, a recent acquisition of GFP-encoding genes in Branchiostoma via HGT is unlikely; instead GFP-encoding genes were probably present in the ancestral cephalochordates, although it remains to be determined whether they were horizontally transferred (e.g., via food intake, or symbiosis) to the ancestral cephalochordates or were inherited from the ancestral bilaterians.
Results
Asymmetron lucayanum fluorescent display and emission spectra
The notochord of A. lucayanum is iridescent under polarized light because the notochord cells are regularly spaced in a stack-of-coins arrangement that differentially refracts the incident light (Fig. 1A,B). In fluorescence mode (ex: 470 nm), dim green light was emitted diffusely throughout the body (in both genders) and was intense from the ripe ovaries (in females). Sometimes there was a red component at the distal end of the digestive tract, which was probably due to the chlorophyll in the algal diet (Fig. 1C). In fluorescence mode, the spawned eggs appeared bright green (Fig. 1D). The emission spectrum for the notochord, ovaries, and eggs had a sharp peak in the green (em: 525 nm), when excited at 470 nm, which was similar to Branchiostoma floridae (Fig. 2). In contrast, A. lucayanum also showed fluorescence when excited at 390 nm, with a broad blue-green spectrum, which was not observed for B. floridae (showing no fluorescence at all for that excitation). Excitation at 355 nm also triggered dim fluorescence in A. lucayanum (Fig. S1). Such blue fluorescence excitable at the shorter wavelength appears more specifically and more intensely in the eggs of A. lucayanum. This shorter-wavelength excitable fluorescence could originate from a variety of compounds other than GFP since broad fluorescence spectrum is typically not characteristic of any known GFP-family molecules. In cephalochordates however, some GFPs can produce fluorescence under a broader spectrum than the classic commercial GFP, such as described for the clade d GFPs in B. floridae25. One therefore cannot exclude that a GFP, or a maturation step of one of the Asymmetron GFPs (or its association with certain compounds) could lead to broader fluorescence under low excitation wavelength. This was also supported from the observation that in addition to the eggs having blue-green fluorescence, Asymmetron larvae also produce bright blue-shifted fluorescence (Fig. S1). This is consistent with the possible photoprotection against lower-wavelengths these life stages can be exposed to in the water column.
Figure 1. Imaging Asymmetron lucayanum.
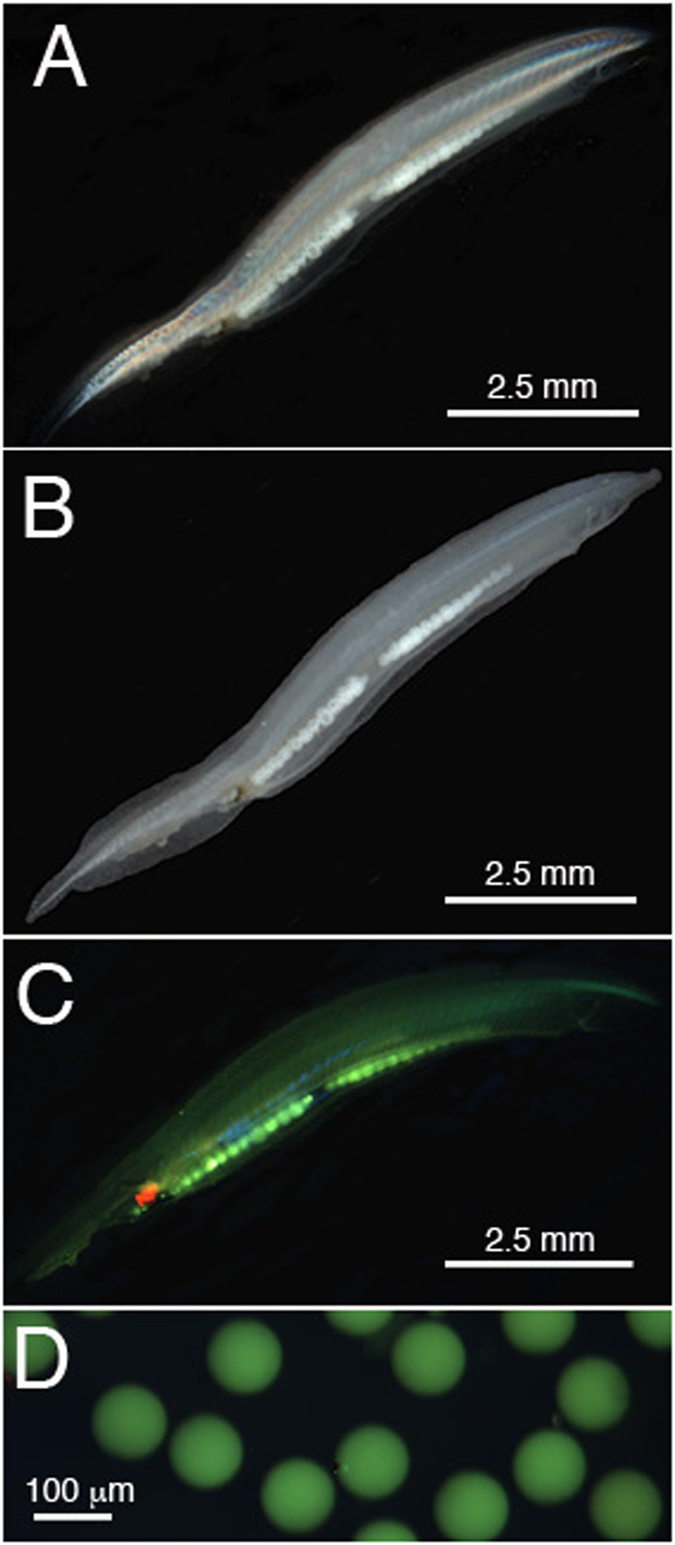
(A–C) Adult animal. (A) In bright field light with polarizer. (B) Under bright field light reflectance. (C) Under fluorescence excited at 470 nm. D. Egg fluorescence excited at 470 nm.
Figure 2. Fluorescence spectrum of cephalochordate GFPs excited at 470 nm (for both Asymmetron lucayanum and Branchiostoma floridae) and 390 nm (for A. lucayanum only).
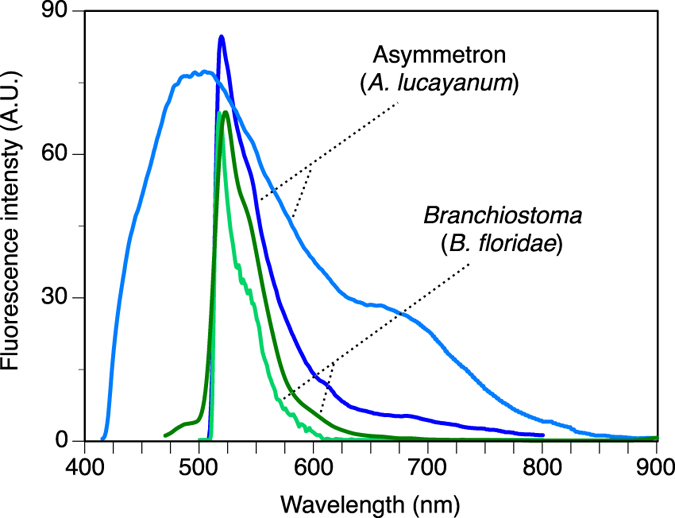
For the excitation at 470 nm, the A. lucayanum spectrum curve was shown in dark blue while the B. floridae specturm curve was shown in light green B. floridae GFPa) and darker green B. floridae GFPe). For the excitation at 390 nm, only the spectrum curve for A. lucyanum was shown (in light blue), since no fluorescence at that excitation wavelength was observed for B. floridae.
GFP-encoding genes identified in A. lucayanum and other cephalochordates
In this study, we identified one and two GFP-encoding genes from the A. lucayanum adult and larval transcriptomes, respectively. Our gene orthology identification analysis suggests the GFP-encoding gene in the adult transcriptome is the ortholog of one of the two GFP-encoding genes in the larva transcriptome. Therefore, we used the two GFP-encoding genes identified in the larva transcriptome as the non-redundant set for GFP-encoding genes in A. lucayanum. It is unlikely that additional GFP-encoding genes are present in A. lucayanum, since the transcriptome assembly we used was the most complete available27; however, we cannot rule out such a possibility. A definitive answer will be available until the A. lucayanum genome eventually becomes entirely sequenced.
In parallel, we found 13 GFP-encoding genes in both the Asian amphioxus Branchiostoma belcheri and the Florida amphioxus B. floridae. When we compared the 13 B. floridae GFP-encoding genes identified in this study with the 16 B. floridae GFP-encoding genes reported earlier25, we found the 3 “missing” GFPs had been deleted in the v2.0 assembly of the B. floridae genome when the two haploid genomes of the v1.0 assembly were collapsed into a single representative one; thus the 3 missing genes are likely to be redundant. In addition, 21 GFP-encoding genes have been cloned for the European amphioxus Branchiostoma lanceolatum (all deposited in NCBI GenBank database). In sum, we compiled a total of 49 cephalochordate GFP-encoding sequences from four cephalochordate species, two from A. lucayanum, 13 from B. belcheri, 13 from B. floridae and 21 from B. lanceolatum (Table 1). The nucleotide coding sequences (CDSs) and protein sequences of these genes are provided in Supplementary file 1 and 2, respectively.
Table 1. GFP-encoding genes identified in cephalochordates.
| Clade | Species | Gene name | Gene model ID | Chromophore region | Genomic coordinate (scaffold:start-end:strand) |
|---|---|---|---|---|---|
| GFPa | B. floridae | GFPa1 | 63256 | HL GYG YY | Bf_V2_107:1004669–1007396:+ |
| GFPa2 | 63262 | HL GYG YY | Bf_V2_107:912177–916945:+ | ||
| GFPb | B. floridae | GFPb1 | 75522 | HL GYA YY | Bf_V2_131:724134–735690:− |
| GFPb2 | 75521 | HL GYA FN | Bf_V2_131:710687–716998:− | ||
| GFPb3 | 75519 | QI GYG FH | Bf_V2_131:674613–680969:+ | ||
| GFPb4 | 75520 | HI GYG FY | Bf_V2_131:693853–703767:+ | ||
| B. belcheri | GFPb1 | 233630F | HF GYG YD | scaffold58:854039–856682:+ | |
| GFPb2 | 264830F | HF AYG YD | scaffold718:9844–13364:+ | ||
| GFPb3 | 240030F | HV GYG YH | scaffold6:4139317–4157041:+ | ||
| GFPc | B. floridae | GFPc1 | 75523 | NI GYG FH | Bf_V2_131:741722–750056:+ |
| B. belcheri | GFPc1 | 199320R | NL GYG FH | scaffold44:34684–36671:- | |
| GFPd | B. floridae | GFPd1 | 86184 | HL GYG HY | Bf_V2_226:1594162–1619868:− |
| GFPd2 | 126982 | HL GYG HY | Bf_V2_106:577911–582844:+ | ||
| B. belcheri | GFPd1 | 282900F | HL GYG FY | scaffold83:996645–1004389:+ | |
| GFPd2 | 005130R | HL GYG FY | scaffold1:5804381–5807161:− | ||
| GFPd3 | 005140R | HL GYG FY | scaffold1:5811232–5813910:− | ||
| GFPd4 | 145360F | HL GYG FY | scaffold291:172581–179080:+ | ||
| GFPd5 | 233640F | HL GFG FY | scaffold58:860037–874989:+ | ||
| GFPe | B. floridae | GFPe1 | 63257‡ | NL GYG FY | Bf_V2_107:979539–982204:+ |
| GFPe2 | 63260 | NL GYG FY | Bf_V2_107:950946–955643:+ | ||
| GFPe3 | 63258 | NL GYG FY | Bf_V2_107:971176–973558:− | ||
| B. belcheri | GFPe1 | 144690R | NL GYG FY | scaffold29:2454423–2458156:− | |
| GFPf | B. floridae | GFPf1 | 63259 | NL GYG YH | Bf_V2_107:961008–966261:− |
| B. belcheri | GFPf1 | 144780R | NL GYG YH | scaffold29:2657919–2677300:− | |
| B. belcheri | GFPf2 | 212280R | NL GYG YH | scaffold5:926780–961355:− | |
| A. lucayanum | GFP1 | asym20h* | NL GYG YH | ||
| Un- classified | B. belcheri | GFPx1 | 276530F | HL GYG FY | scaffold8:2499615–2549717:+ |
| A. lucayanum | GFP2 | asym20h** | HL GYG LY |
‡The internal sequence gap within the B. floridae gene model 63257 was filled based on our previous study23.
*Asym20h_comp74545_c2_seq1_m.28460.
**Asym20h_comp64813_c0_seq2_m.13123.
We compared the average sequence diversity of GFP-encoding genes within each cephalochordate species using four different measurements: nucleotide distance (DJC), protein sequence distance (DPois), nonsynonymous substitution rate (Dn) and synonymous substitution rate (Ds). In general, the Asymmetron GFP-encoding genes showed consistently higher sequence diversity than GFP-encoding genes from Branchiostoma species based on different measurements (Table 2). Within the genus Branchiostoma, GFP-encoding genes from B. belcheri seem to show higher sequence diversity than their B. floridae counterparts, whereas the B. lanceolatum GFP-encoding sequences showed much lower average sequence diversity. Since those B. lanceolatum GFP-encoding sequences were not identified from a systematic genome-wide survey such as we performed for the other two Branchiostoma species, it is highly likely that these sequences come from a biased GFP-encoding gene sampling in this species. This may, explain the extremely low sequence diversity among these sequences compared with their counterparts from the other two Branchiostoma species.
Table 2. Average molecular evolutionary rates for cephalochordate GFP-encoding genes within each species.
| DJC | DPois | Dn | Ds | Dn/Ds | |
|---|---|---|---|---|---|
| A. lucayanum | 0.649 | 0.809 | 0.562 | 1.033 | 0.544 |
| B. belcheri | 0.506 | 0.640 | 0.433 | 0.819 | 0.532 |
| B. floridae | 0.451 | 0.619 | 0.392 | 0.697 | 0.567 |
| B. lanceolatum | 0.202 | 0.280 | 0.163 | 0.357 | 0.421 |
Djc: Jukes-Cantor nucleotide substitution rate; DPois: Poisson amino acid substitution rate; Dn: nonsynonymous substitution rate; Ds: synonymous substitution rate.
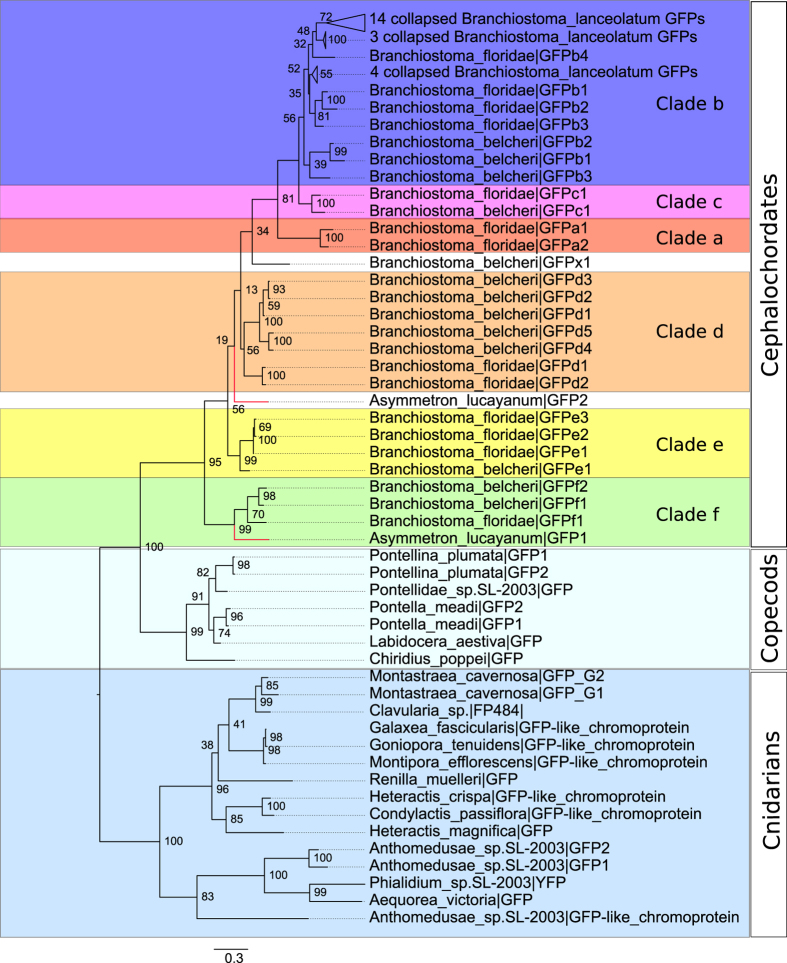
Phylogenetic relationship of cephalochordate GFP-encoding genes
In addition to the 49 cephalochordate GFP-encoding genes, we further incorporated 22 GFP-encoding genes from copepods and cnidarians as outgroups for the phylogenetic analysis (Fig. 3, Fig. S2 and Table S1). The trimmed sequence alignment used for this analysis is provided in Supplementary file 3. Overall, the tree showed that the cephalochordate GFPs have a closer affinity to those of copepods than to those of cnidarians (Figs 3 and S2). Within the cephalochordate GFP subtree, 47 out of 49 cephalochordate GFP-encoding genes were assigned to the six major clades (a–f) of cephalochordate GFPs previously demonstrated in B. floridae25, with the remaining two as unassigned (A. lucayanum GFP2 and B. belcheri GFPx1). One A. lucayanum GFP-encoding gene was positioned between clade d and clade e, whereas the other one fell unambiguously into clade f (Fig. 3). GFP-encoding genes from B. belcheri spread across clades b through f with only one member left as unassigned (Fig. 3), suggesting that clades b through f should have been established before the divergence of B. floridae and B. belcheri.
Figure 3. Phylogenetic tree of cephalochordate GFP-encoding genes, with cnidarian and copepod GFP-encoding genes as outgroups.
Branches representing the two GFP-encoding genes from A. lucayanum are highlighted in red. Branches corresponding to B. lanceolatum GFP-encoding sequences were collapsed and the collapsed nodes were represented by triangles in the tree. For each internal node, the local support value was calculated by 100 rapid bootstrapping via RAxML. The clades are highlighted with colors previously designating B. floridae GFP clades (following our earlier study)25. The full version of this tree without collapsing the B. lanceolatum sequences is provided as Fig. S2.
Interestingly, the unassigned B. belcheri GFP-encoding gene (B. belcheri GFPx1) encodes two other domains (class-A Low-density lipoprotein receptor domain and MFS/sugar transport protein domain) in addition to the GFP domain. One explanation is that this gene was mis-annotated during the original B. belcheri gene annotation by accidentally being joined to its flanking neighbors. However, our parallel phylogenetic analysis based on a modified gene model that does not contain those two additional domains placed this gene to the same phylogenetic position, suggesting the phylogenetic positioning of B. belcheri GFPx1 is not an artifact due to gene annotation error. All the 21 B. lanceolatum GFP-encoding sequences were tightly packed into clade b (Fig. 3), consistent with our previous conjecture that these closely sequences should come from strongly biased gene sampling in B. lanceolatum.
Lineage-specific expansion of GFP-encoding genes in the Branchiostoma genus
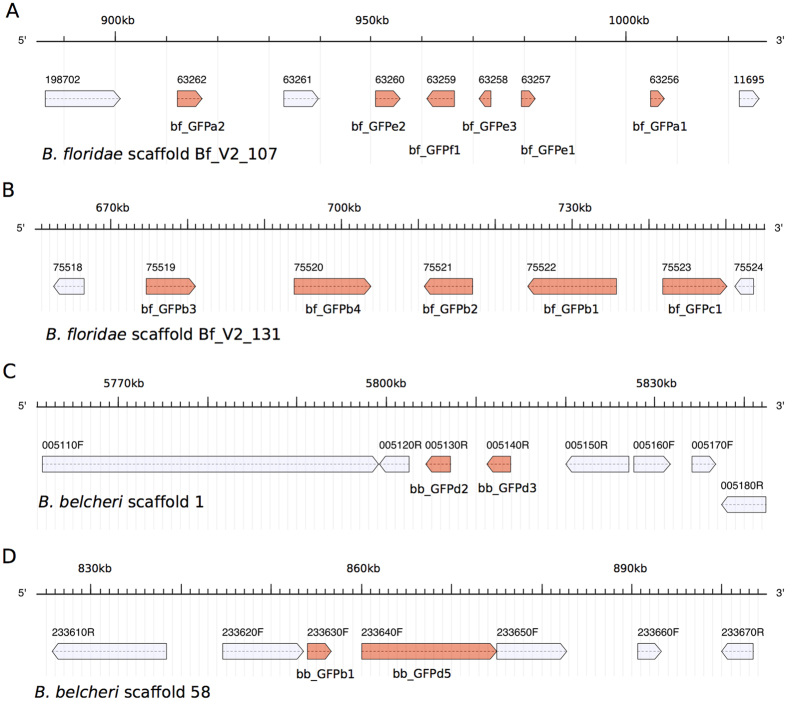
Compared with the earlier diverged A. lucayanum, copepod and even most cnidarians, the large number of GFP-encoding genes in B. floridae and B. belcheri seems to be the result of lineage-specific expansion. The lower average sequence diversity of Branchiostoma GFP-encoding genes compared with that of Asymmetron further supports this idea. Our phylogenetic analysis indicates that the starting condition for this expansion was the presence of at least five GFP genes corresponding to clade b through f (one for each clade) in the ancestral Branchiostoma species. The physical distribution of the Branchiostoma GFP-encoding genes (Table 1) suggests that at least some of the expansion resulted from tandem duplications. For example, there are 11 B. floridae GFP-encoding genes located in two tightly packed clusters: one 95.2 kb long (containing 6 genes) on scaffold Bf_V2_107 (Fig. 4A) and the other 75.4 kb long (containing 5 genes) on scaffold Bf_V2_113 (Fig. 4B). For B. belcheri, at least four GFP-encoding genes are attributable to tandem gene duplication (Fig 4C,D) while there could be even more since the current B. belcheri genome assembly is less complete than the B. floridae genome assembly.
Figure 4. The tandemly duplicated gene clusters of GFP-encoding genes in Branchiostoma floridae and Branchiostoma belcheri.
All the GFP-encoding genes were highlighted in red. (A) The genomic region with GFP-encoding gene tandem duplication on B. floridae scaffold Bf_V2_107. (B) The genomic region with GFP-encoding gene tandem duplication on B. floridae scaffold Bf_V2_131. (C) The genomic region with GFP-encoding gene tandem duplication on B. belcheri scaffold 1. (D) The genomic region with GFP-encoding gene tandem duplication on B. belcheri scaffold 58.
Purifying selection of GFP-encoding genes in cephalochordates
Expanding gene families can be shaped by diversifying selection when adaptive changes accumulate in different duplicated gene copies. Here, we assessed the selection imposed on cephalochordate GFP-encoding genes by measuring non-synonymous/synonymous substitution rate ratio (Dn/Ds). Neutral theory predicts Dn/Ds = 1 in the absence of natural selection, whereas deviations from this null model will suggest either diversifying selection (Dn/Ds > 1) or purifying selection (Dn/Ds < 1). For each cephalochordate species, we consistently observed Dn/Ds < 1 for all the GFP-encoding paralogs (Table 2), suggesting a general purifying selection scheme for GFP-encoding genes in cephalochordates despite the formation of different phylogenetic clades. Theoretically, it is possible that some specific codon sites could evolve under diversifying selection while purifying selection is shaping the evolution of the rest of the gene. However, no such sites were identified as statistically significant based on hypothesis testing of codeml’s site models (M1a vs. M2a and M7 vs. M8). We further examined selection scheme for different clades (b–f) based on B. floridae-B. belcheri (co-)orthologs28 within each individual clade. The clade a was excluded since not identified in GFP-encoding genes from B. belcheri. Apparently, Dn/Ds values were more elevated in clades b and d. In combination with their higher inter-specific sequence divergence (DJC and DPois), this indicates likely relaxation of purifying selection in these two clades (Table 3). Interestingly, they also have the most lineage-specific duplication events, suggesting that functional redundancy, due to recent gene duplication, might help relax the selection constraints in these two clades.
Table 3. Average molecular evolutionary rates between B. belcheri and B. floridae co-orthologs within each GFP clade.
| DJC | DPois | Dn | Ds | Dn/Ds | |
|---|---|---|---|---|---|
| Clade b | 0.301 | 0.424 | 0.249 | 0.507 | 0.493 |
| Clade c | 0.198 | 0.203 | 0.104 | 0.591 | 0.176 |
| Clade d | 0.312 | 0.387 | 0.230 | 0.649 | 0.354 |
| Clade e | 0.207 | 0.206 | 0.113 | 0.594 | 0.190 |
| Clade f | 0.277 | 0.279 | 0.191 | 0.656 | 0.290 |
Djc: Jukes-Cantor nucleotide substitution rate; DPois: Poisson amino acid substitution rate; Dn: nonsynonymous substitution rate; Ds: synonymous substitution rate. Data for clade a is not available since no B. belcheri GFPa gene was identified.
Evolutionary relationships between cephalochordate GFPs and those of other major clades
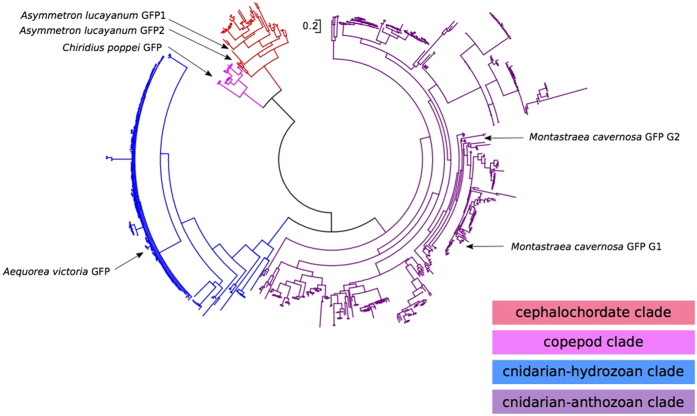
As Fig. 3 already shows, cephalochordate GFP-encoding genes formed their own phylogenetic group and are more closely related to those of copepods than to those of cnidarians. However, we cannot assess the possibility that there might be some unsampled copepod or cnidarian GFP-encoding genes that are closely related to cephalochordate ones, especially considering the small sample size of copepod and cnidarian outgroups considered in Fig. 3. To address this more comprehensively, we examined our data in the context of the entire National Center for Biotechnology Information (NCBI) non-redundant (nr) database of 1,832 GFP-encoding sequences. We manually reviewed the taxonomic information of these 1,832 GFP-encoding sequences and noticed that 1,128 of them are labeled as artificial constructs or recombinant vectors. These GFPs are lab-modified versions of natural GFPs (most of them are based on the same GFP: Aequorea victoria GFP). In addition, there are 22 GFPs in the nr database that were annotated with various origins (six from virus, seven from bacteria, one from oomycete parasite, five from protist parasites, one from mosquito, one from rat and one from human) but all of them are probably due to artificially introduced GFP vectors. There are 50 GFPs remaining with no traceable taxonomic origins. The 39 cephalochordate GFPs in the nr database were replaced with our 49 better-curated cephalochordate GFPs. This left us with a total of 642 GFP-encoding sequences with seemingly natural origins (174 from hydrozoan cnidarians, 403 from anthozoan cnidarians, 16 from copepods, and 49 from cephalochordates). The phylogenetic tree based on these sequences (Fig. 5) reveals three well-diverged clades: namely cephalochordates, copepods and cnidarians (with the last being divided into distinct hydrozoan and anthozoan subclades). This more comprehensive analysis emphasizes that the cephalochordate clade has clearly diverged from the copepod and cnidarian clades while the copepod and cephalochordate clades are closer to each other than either is to the cnidarian clade. The trimmed sequence alignment used for this analysis is provided in Supplementary file 4.
Figure 5. Phylogenetic relationship of cephalochordate GFPs relative to those in other evolutionary lineages.
The major taxonomic categories are cephalochordates (red), copepods (magenta), hydrozoan cnidarians (blue) and anthozoan cnidarians (purple). The phylogenetic position of GFP-encoding genes from representative species (one for each clade) of each clade was indicated.
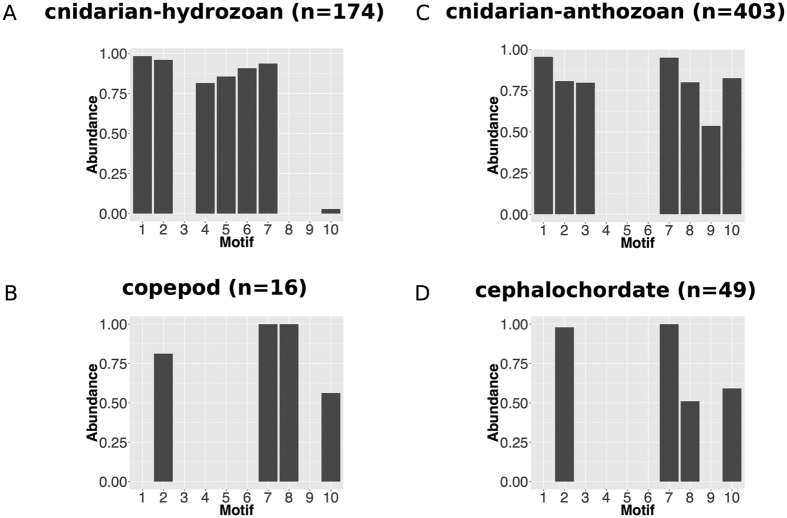
To compare the GFP domains from different evolutionary lineages in more detail, we further identified the top ten most conserved amino acid motifs (Table 4) within the GFP domain region and plotted their relative abundance (proportion of the sequences with this motif) in each evolutionary lineage (hydrozoans, anthozoans, copepods and cephalochordates) (Fig. 6A–D). In general, the GFP domains from all four evolutionary lineages show clear differences in overall conserved motif composition, reflecting lineage-specific changes of their GFP domains in their respective evolutionary histories. Consistent with what we observed from the phylogenetic tree, the cephalochordate and copepod lineages share more similarity in their motif composition compared with two cnidarian lineages, which once again suggests much closer evolutionary relationship of GFP-encoding genes from these two lineages relative to those cnidarian GFP-encoding genes.
Table 4. Top ten compositional motifs of the GFP domain.
| Motif ID | Regular expression for the motif* | E-value | Width |
|---|---|---|---|
| Motif 1 | D[YF]FK[QS][SA][FM]PEG[YF][SVT][WQ]ER | 5.7 e-6277 | 15 |
| Motif 2 | [ML][ED]G[DST]VNGH[KE]FS[IV][ES]GEGEG[KDN][PA][YTF][EY]G[KT][QL]T[LM]K[LF] | 1.2 e-9032 | 29 |
| Motif 3 | GVNFP[AP][ND]GPVMQKKTL[GK]WEPSTE[KR][ML] | 2.7 e-6330 | 25 |
| Motif 4 | NYNSHNVYI[MT]ADKQKNGIK[VA]NFKIRHNIEDGSVQLADHYQQ | 1.2 e-6048 | 41 |
| Motif 5 | PVLLPDNHYLSTQSALSKDPNEKRDHMVL | 1.0 e-4182 | 29 |
| Motif 6 | DGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEY | 2.0 e-6106 | 41 |
| Motif 7 | LP[FV][ASP][WF][DP][IT]L[SVT][TP][TA][FL]XYG | 3.9 e-4433 | 15 |
| Motif 8 | L[LK]L[EK][GD]GGH[YL]RC[DQ]F[KR][TS]TYKAKK | 1.1 e-4095 | 21 |
| Motif 9 | YH[FY]VDH[RK][IL][ED]I[TL]SH[DN][KE]DY[TN]KV[EK][LQ][YH]EHA[EV]A[RH] | 4.9 e-4025 | 29 |
| Motif 10 | M[TN][FY]EDG[GA][VI]CT[AV][TS][NQ]DI[ST]L[EQ]G[DNG]C | 1.7 e-3959 | 21 |
*Amino acids within the brackets are interchangeable.
Figure 6. The conserved motif composition of GFP domains from different evolutionary lineages.
The relative abundance of ten compositional motifs of the GFP domain was shown for hydrozoan cnidarians (A), anthozoan cnidarians (B), copepods (C), and cephalochordates (D). The numbers in the parenthesis indicate the total GFP-encoding sequences from the corresponding evolutionary lineage that were used in the analysis.
Discussion
The origin of GFP-encoding genes in cephalochordates
This study is the first demonstration of GFP-encoding gene evolution within the cephalochordate clade. A. lucayanum contains two GFP-encoding genes, in contrast to about a dozen such genes in each of two species in the genus Branchiostoma. Lineage-specific expansion of GFP-encoding genes was observed for the genus Branchiostoma, probably due at least in part to tandem duplication. Both of our phylogenetic and conserved motif analyses have emphasized that the sequences of the GFP-encoding genes of hydrozoans, anthozoans, and cephalochordates differ considerably among these four evolutionary lineages. Such marked sequence divergence does not favor the idea that there was recent HGT from one of these groups to the next—for instance, from cnidarians to cephalochordates.
Inheritance of GFP-encoding genes from a common ancestor is more likely, with three possible alternatives. First, GFP-encoding genes emerged from a common ancestor to cnidarians, copepods and cephalochordates, which should be indicative of GFP present in other early bilatarian lineages. We screened the proteomes of the sea urchin Strongylocentrotus purpuratus (Echinodermata), the acorn worm Saccoglossus kowalevskii (Hemichordata) and the sea snail limpet Lottia gigantea (Mollusca) and found no sequence with close similarity to GFP. It is still possible however that other representatives of these groups could have GFP-like genes, and looking at luminous species within these groups might resolve this question.
A second possibility is that HGT transfer did occur, but from some animal phyla containing GFP-encoding genes yet to be discovered. This speculation would then suggest that such an animal would have particular ecological relationships with the current groups of organisms with GFP-encoding genes—that is either being an important component of the diet, or a common symbiont or parasite.
Third, GFPs independently arose multiple times in different evolutionary lineages and then diversified within each of these three evolutionary lineages. The scattered taxonomic distribution of currently known GFP-encoding genes appears to favor this hypothesis. This scenario however seems less likely in light of the elaborate and unique structure of GFP proteins, which is specifically tuned to absorb and possibly re-emit light (not all GFP proteins produce fluorescence). If the specific functions of independently evolved GFP proteins had to be kept across different taxa, it is likely that the different group of organisms would have come up with different solutions to cover these functions. As an analogy, production of visible light through bioluminescence has appeared independently about 30–40 times across taxa during evolution, each time using different sets of proteins and molecules to produce light17. Clearly there is a need for a more thorough search for GFP-encoding genes throughout the animal kingdom, especially in luminescent but also non-luminescent taxa that have not previously attracted the attention of photobiologists.
The anatomical localization of fluorescence in cephalochordates could allow multiple ecological and/or biochemical functions
All cephalochordates so far assayed for fluorescence have conspicuous amounts of GFP proteins in their oocytes and spawned eggs. Then, during the subsequent embryonic and larval development, the GFP fluorescence is progressively lost, but in adults of Branchiostoma, remains strong in localized anatomical regions12. However in Asymmetron it remains diffusely distributed throughout the adult body (this study). The reason for the high concentrations of GFP signal in early developmental stages of cephalochordates is not known, although one possibility is photoprotection. Indeed, the embryos and larvae live planktonically in relatively shallow water, where they are sometimes captured during daylight hours29; under such conditions GFP would absorb blue (high-energy, possibly damaging) light and transform it to some extent into green (non-damaging) fluorescence. In any case, the main (most intense and sharper) spectrum of fluorescence for Asymmetron has a profile close to that of Branchiostoma GFPs from clade d or e, which is likely produced by A. lucyanum GFP2 although with a different amino acid sequence around the chromophore (Table 1). A previous study25 considered the Branchiostoma clade f GFP proteins as chromoproteins given that their spectra were too weak to be measured. In Asymmetron, it is possible that the broad blue-shifted weak spectrum is produced by A. lucayanum GFP1, which is also located in clade f in our phylogenetic analysis. This is not a rigorous demonstration, but is reasonable as the amino acid sequences are identical around their respective chromophore (Table 1).
Bomati et al.30 has shown that the amino acid sequence around the chromophore is critical for the fluorescence capacity of GFP, with a few changes changing the quantum efficiency from 0.1–100%30. The GFP chromophore sequence itself has been widely preserved through different taxa, and consistently reported as “GYG”, with the third glycine essential for the chromophore formation and fluorescence. The GYG reflects the electro- and stereo-chemical stability of this triplet allowing efficient energy transfer into fluorescence output, and found so far in all GFP templates31,32. In B. floridae, two GFPs with the GYA chromophore sequence were described in clade b (Bf GFPb1 and Bf GFPb2), although not associated with any detectable fluorescence. Also, there are two GFPs in B. belcheri with AYG (Bb GFPb2) and GFG (Bb GFPd5) chromophore sequences. These indicate that the general GFP protein motif could be preserved while performing different sets of biological/biochemical functions depending on the chromophore sequence, in association with fluorescence, but not necessarily.
In adults, differences in the fluorescent body regions between A. lucayanum and Branchiostoma spp are particularly striking—the chief site of fluorescence is diffuse distribution through the body in the former (present results) and restricted to the oral cirri of the latter12. All cephalochordates burrow shallowly in soft substrata with their anterior ends just within the burrow opening. There the mouth sucks in overlying sea water containing food particles that include motile planktonic organisms smaller than about 100 μm in diameter33. In the three species of Branchiostoma for which GFPs have been studied12, the oral cirri surrounding the mouth are highly fluorescent. The recent finding that jellyfish attracted prey with GFP fluorescence23 suggests that green light emanating from amphioxus cirri might attract motile planktonic prey, thus increasing their chance of being entrained in the feeding current entering the mouth. The stimulus for the fluorescence is ambient blue light in shallow sea water, which fits with species of Branchiostoma living from the shoreline to fairly moderate depths (max. ca. 100 m); by contrast, A. lucayanum, a species with non-fluorescent oral cirri, can be found in shallow water, but is more often captured at considerable depths, up to 1,000 m34.
It would be interesting to see if the oral cirri are fluorescent in a wider sample of Branchiostoma species and are non-fluorescent in additional species of Asymmetron, and in the single known species in the cephalochordate genus Epigonichthys, which is sister to Branchiostoma. Moreover, the idea that capture of small phototrophic prey items is enhanced by the fluorescence of the oral cirri in Branchiostoma species could be tested experimentally by manipulating the wavelengths of incident light impinging on the feeding animals. The data presented here, therefore, offer a new set of tools to address both the evolution and function of GFP in nature, which has largely been ignored.
Methods
Animal collection and fluorescent imaging
The Bahamas lancelet, Asymmetron lucayanum, was collected in Bimini, Bahamas35,36, and cultured and spawned in the laboratory according to previously described protocol37. Adults and unfertilized eggs were imaged in bright field and fluorescence under a Nikon SMZ 1500 stereoscope, equipped with a digital color QI camera. Fluorescence spectra were acquired using the PARISS hyperspectral imaging system (LightForm Inc.) mounted on a Nikon 80i microscope and spectra were generated in Excel and Deltagraph (Red Rock Inc.). All filters used were LP for all excitation wavelengths. These excitation wavelengths included 355, 390, 436, 470 nm, as per filter cubes commercially available from Nikon.
The identification of GFP-encoding genes in cephalochordates
Yue and colleagues27 constructed two non-redundant A. lucayanum transcriptome assemblies, respectively, from adult and larval libraries with protein-coding gene predictions. The details of the transcriptome assembly, redundancy removal, protein-coding gene annotation was described here27. The predicted coding DNA sequences (CDSs) and proteome sets based on these two A. lucayanum transcriptome assemblies was used in this study. We further added the CDSs and proteomes of B. floridae38 sequences (based on v2.0 assembly) and B. belcheri39 (based on v18h27.r3 assembly) for our GFP search. We used proteinortho (v5.11) with default settings40 to identify orthologous relationship among the cephalochordate proteomes that we used in this study. For each cephalochordate proteome, we used hmmsearch (option: -E 1e-4) from the hmmer (v3.1b2) package to search for all GFP-encoding genes based on the hidden Markov model of the GFP domain (PF01353) curated by the Pfam database (v27.0). The GFP-encoding genes that we identified from the two Asymmetron proteome sets were further collapsed based on the orthology identified by proteinortho. For each orthologous groups, the longest sequence was selected for the downstream analysis. In addition, existing B. lanceolatum GFP-encoding sequences deposited in NCBI GenBank was further added into our final cephalochordate GFP-encoding gene set after verifying their protein domains by the hmmer package.
Sequence alignment and phylogenetic reconstruction
In addition to the cephalochordate GFP-encoding genes that we compiled, we added 22 more GFP-encoding genes from copepods and cnidarians (including both hydrozoans and anthozoans) from GenBank as outgroups (Fig. S2 and Table S1). The protein sequences of these GFP-encoding genes were aligned by PROMALS3D41 with default settings. PROMALS3D searches against known protein structures and uses both structural and sequence constraints to generate highly accurate protein sequence alignment. The corresponding CDS sequence alignment was generated based on the PROMALS3D protein sequences alignment by PAL2NAL (v14)42 with default setting for later analysis. The PROMALS3D protein sequences alignment was further trimmed by trimAl (v1.4) (option: -gt 0.75)43 for phylogenetic analysis. We employed RAxML (v 8.2.6)44 for maximum likelihood (ML) phylogenetic tree construction with automatic model selection (model = PROTGAMMAAUTO) with 100 fast bootstrapping tests (option: -# 100) to assess topology stability. The final tree was visualized in FigTree (v1.4.2) (http://tree.bio.ed.ac.uk/software/figtree/). The different GFP clades were highlighted in different color to correspond to B. floridae GFP clades (a through f) defined in our earlier study25. In addition, during our phylogenetic analysis, we noticed that one B. belcheri GFP gene model (GFPx1: 276530F) has unusual domain composition and phylogenetic positioning. In order to test whether these anomalies are artifacts due to gene annotation error. We extracted the genomic sequence of this region together with 30 kb flanking region on both sides to run de novo gene annotation using FGENESH45 (organism specific gene-finding parameters: B. floridae). The resulting gene model was used to re-run our phylogenetic analysis described above for testing if such gene annotation change will affect the phylogenetic positioning of this gene.
Calculation of evolutionary rates
The Jukes-Cantor model46 and the Poisson model47 were used to calculate nucleotide (DJC) and amino acid substitution rate (DPois). The Nei-Gojobori model48 with Jukes-Cantor correction was used to calculate nonsynonymous substitution rate (Dn) and synonymous substitution rate (Ds). The CDS nucleotide alignment of cephalochordate GFP-encoding genes was used in this analysis. All such evolutionary rate calculation was performed in MEGA 9v6.06-mac49 with “pairwise deletion” option selected for alignment gap handling.
Detection of sites under diversifying (positive) selection
We used the codeml program from the PAML package50 (v.4.8a) to detect sites under diversifying (positive) selection based on the CDS nucleotide alignment of GFP-encoding genes within each cephalochordate species. The alternative codon model M2 and M8 were compared with null model M1 and M7 respectively. The statistical significance of potential positively selected sites was assessed by Bayes empirical Bayes (BEB) analysis51.
Screening and analyzing GFP-encoding sequences from the NCBI nr database and other bilaterian proteomes
The NCBI nr database was downloaded (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/) and all of its GFP-encoding sequences were identified by hmmsearch (option: -E 1e-4). The taxonomic origins of these sequences were mapped by MEGAN (v5.10.5)52 based on NCBI’s Gl (GenInfo identifier) number. By manual inspection, we eliminated all artificial constructs, recombinant vectors, as well as data with no traceable taxonomic information. After replacing the cephalochordate GFP-encoding sequences in the nr database with our better-curated sequences (two from A. lucayanum, 21 from B. lanceolatum, and 13 each from B. floridae and B. belcheri), sequence alignment, alignment trimming, and tree building were carried out by the method already described. We used FigTree (v1.4.2) to highlight the tree branches based on the taxonomic origin of the corresponding sequences.
In addition, we retrieved proteomes of several representative early-diverged bilaterian animals including sea urchin Strongylocentrotus purpuratus (Echinodermata), the sea snail limpet Lottia gigantea (Mollusca) and the acorn worm Saccoglossus kowalevskii (Hemichordata) from EnsemblMetazoa (http://metazoa.ensembl.org) (for S. purpuratus and L. gigantea) and Metazome v3.0 (www.metazome.net) (for S. kowalevskii). GFP-encoding gene was screened by hmmsearch (option: -E 1e-4) for these proteomes.
Characterizing conserved motif composition for the GFP domain
To characterize and compare conserved motif composition of the GFP domain in different evolutionary lineages. For each GFP-encoding gene investigated in this study, we performed hmmscan (option: -E 1e-4) to characterize its full domain composition and extracted the protein sequence of its GFP domain region accordingly. The protein sequences of all these GFP domains were scanned together by MEME (v4.11.1)53 (options: -protein -mod zoops -nmotifs 10 -evt 0.01 -maxsize 200000) to detect conserved motif composition shared among them. For each detected motif, we calculated the motif abundance (proportion of the test sequences with this motif) for all the GFP-encoding sequences within the corresponding evolutionary lineages (hydrozoan cnidarians, anthozoan cnidarians, copepods, and cephalochordates).
Additional Information
How to cite this article: Yue, J.-X. et al. The evolution of genes encoding for green fluorescent proteins: insights from cephalochordates (amphioxus). Sci. Rep. 6, 28350; doi: 10.1038/srep28350 (2016).
Supplementary Material
Acknowledgments
Research supported by the Air Force Office of Scientific Research, grant #AFOSR FA9550-14-1-0008 (to DDD). Jia-Xing Yue was supported by a postdoctoral fellowship from Fondation ARC pour la Recherche sur le Cancer (no. PDF20150602803). LZH was supported by NSF IOS-1353688. We are grateful to Deheyn lab technician Michael C. Allen for collection of some imaging and spectral data.
Footnotes
Author Contributions D.D.D. and J.-X.Y. designed the research. D.D.D. performed imaging and spectra collection while J.-X.Y. performed the bioinformatic analysis. N.D.H. and L.Z.H. collected and provided Asymmetron samples and helped with data interpretation; all authors contributed to writing the paper; all authors read and edited the manuscript.
References
- Tsien R. Y. The green fluorescent protein. Ann. Rev. Biochem. 67, 509–544 (1998). [DOI] [PubMed] [Google Scholar]
- Mocz G. Fluorescent proteins and their use in marine biosciences, biotechnology, and proteomics. Mar. Biotechnol. 9, 305–328 (2007). [DOI] [PubMed] [Google Scholar]
- Chudakov D. M., Matz M. V., Lukyanov S. & Lukyanov K. A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163 (2010). [DOI] [PubMed] [Google Scholar]
- Yang F., Moss L. G. & Phillips G. N. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14, 1246–1251 (1996). [DOI] [PubMed] [Google Scholar]
- Labas Y. A. et al. Diversity and evolution of the green fluorescent protein family. Proc. Natl. Acad. Sci. USA 99, 4256–4261 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M. V., Labas Y. A. & Ugalde J. in Green fluorescent protein: properties, applications, and protocols (eds Chalfie M. & Kain S. R.) 139–161 (John Wiley and Sons, 2006). [Google Scholar]
- Alieva N. O. et al. Diversity and evolution of coral fluorescent proteins. PLoS ONE 3, doi: 10.1371/journal.pone.0002680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann D. et al. A family of GFP-like proteins with different spectral properties in lancelet Branchiostoma floridae. Biol. Direct 3, doi: 10.1186/1745-6150-3-28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O., Johnson F. H. & Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59, 223–239 (1962). [DOI] [PubMed] [Google Scholar]
- Shagin D. A. et al. GFP-like proteins as ubiquitous metazoan superfamily: Evolution of functional features and structural complexity. Mol. Biol. Evol. 21, 841–850 (2004). [DOI] [PubMed] [Google Scholar]
- Hunt M. E., Scherrer M. P., Ferrari F. D. & Matz M. V. Very bright green fluorescent proteins from the Pontellid copepod Pontella mimocerami. PLoS One 5, doi: 10.1371/journal.pone.0011517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deheyn D. D. et al. Endogenous green fluorescent protein (GFP) in amphioxus. Biol. Bull. 213, 95–100 (2007). [DOI] [PubMed] [Google Scholar]
- Shkrob M. A., Mishin A. S., Chudakov D. M., Labas Y. A. & Lukyanov K. A. Chromoproteins of the green fluorescent protein family: Properties and applications. Russ. J. Bioorgan. Chem. 34, 517–525 (2008). [DOI] [PubMed] [Google Scholar]
- Kumagai A. et al. A Bilirubin-inducible fluorescent protein from eel muscle. Cell 153, 1602–1611, doi: 10.1016/j.cell.2013.05.038 (2013). [DOI] [PubMed] [Google Scholar]
- Sparks J. S. et al. The covert world of fish biofluorescence: A phylogenetically widespread and phenotypically variable phenomenon. Plos One 9, doi: 10.1371/journal.pone.0083259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorio M. G., Cordon G. B. & Iriel A. Reviewing the relevance of fluorescence in biological systems. Photoch. Photobio. Sci. 14, 1538–1559 (2015). [DOI] [PubMed] [Google Scholar]
- Haddock S. H. D., Moline M. A. & Case J. F. Bioluminescence in the sea. Ann. Rev. Mar. Sci. 2, 293–343, doi: 10.1146/annurev-marine-120308-081028 (2010). [DOI] [PubMed] [Google Scholar]
- Haddock S. H. D. & Case J. F. Bioluminescence spectra of shallow and deep-sea gelatinous zooplankton: ctenophores, medusae and siphonophores. Mar. Biol. 133, 571–582 (1999). [Google Scholar]
- Dupont N., Klevjer T. A., Kaartvedt S. & Aksnes D. L. Diel vertical migration of the deep-water jellyfish Periphylla periphylla simulated as individual responses to absolute light intensity. Limnol. Oceanogr. 54, 1765–1775, doi: 10.4319/lo.2009.54.5.1765 (2009). [DOI] [Google Scholar]
- Salih A., Larkum A., Cox G., Kuhl M. & Hoegh-Guldberg O. Fluorescent pigments in corals are photoprotective. Nature 408, 850–853 (2000). [DOI] [PubMed] [Google Scholar]
- Bou-Abdallah F., Chasteen N. D. & Lesser M. P. Quenching of superoxide radicals by green fluorescent protein. Biochim. Biophys. Acta 1760, 1690–1695 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S. G. et al. Host pigments: potential facilitators of photosynthesis in coral symbioses. Plant Cell Environ. 31, 1523–1533 (2008). [DOI] [PubMed] [Google Scholar]
- Haddock S. H. D. & Dunn C. W. Fluorescent proteins function as a prey attractant: experimental evidence from the hydromedusa Olindias formosus and other marine organisms. Biol. Open 4, 1094–1104, doi: 10.1242/bio.012138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Takenaka Y., Yamaguchi A., Nishikawa S. & Mizuno H. A novel yellowish-green fluorescent protein from the marine copepod, Chiridius poppei, and its use as a reporter protein in HeLa cells. Gene 372, 18–25 (2006). [DOI] [PubMed] [Google Scholar]
- Bomati E. K., Manning G. & Deheyn D. D. Amphioxus encodes the largest known family of green fluorescent proteins, which have diversified into distinct functional classes. BMC Evol. Biol. 9, 1–11, doi: 10.1186/1471-2148-9-77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T. et al. Phylogenetic position of a whale-fall lancelet (Cephalochordata) inferred from whole mitochondrial genome sequences. BMC Evol. Biol. 7, doi: 10.1186/1471-2148-7-127 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J. X., Yu J. K., Putnam N. H. & Holland L. Z. The Transcriptome of an amphioxus, Asymmetron lucayanum, from the Bahamas: A window into chordate evolution. Genome Biol. Evol. 6, 2681–2696, doi: 10.1093/gbe/evu212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338, doi: 10.1146/annurev.genet.39.073003.114725 (2005). [DOI] [PubMed] [Google Scholar]
- Rice H. J. Observation upon the habits, structure and development of Amphioxus lanceolatus. Am. Nat. 14, 73–95 (1880). [Google Scholar]
- Bomati E. K., Haley J. E., Noel J. P. & Deheyn D. D. Spectral and structural comparison between bright and dim green fluorescent proteins in Amphioxus. Sci. Rep. 4, doi: 10.1038/srep05469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M. a., Socolich M. & Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat. Struct. Biol. 7, 1133–1138 (2000). [DOI] [PubMed] [Google Scholar]
- Dedecker P., De Schryver F. C. & Hofkens J. Fluorescent Proteins: Shine on, You Crazy Diamond. J. Am. Chem. Soc. 135, 2387–2402, doi: 10.1021/ja309768d (2013). [DOI] [PubMed] [Google Scholar]
- Ruppert E. E., Nash T. R. & Smith A. J. The size range of suspended particles trapped and ingested by the filter-feeding lancelet Branchiostoma floridae (Cephalochordata: Acrania). J. Mar. Biol. Assoc. UK 80, 329–332, doi: 10.1017/S0025315499001903 (2000). [DOI] [Google Scholar]
- Poss S. G. & Boschung H. T. Lancelets (Cephalochordata: Branchiostomatidae): How many species are valid? Israel J. Zool. 42, S13–S66 (1996). [Google Scholar]
- Holland N. D. & Holland L. Z. Laboratory spawning and development of the Bahama lancelet, Asymmetron lucayanum (Cephalochordata): Fertilization through feeding larvae. Biol. Bull. 219, 132–141 (2010). [DOI] [PubMed] [Google Scholar]
- Holland N. D. Spawning periodicity of the lancelet, Asymmetron lucayanum (Cephalochordata), in Bimini, Bahamas. Ital. J. Zool. 78, 478–486 (2011). [Google Scholar]
- Holland N. D., Holland L. Z. & Heimberg A. Hybrids Between the Florida Amphioxus (Branchiostoma floridae) and the Bahamas Lancelet (Asymmetron lucayanum): Developmental morphology and chromosome counts. Biol. Bull. 228, 13–24 (2015). [DOI] [PubMed] [Google Scholar]
- Putnam N. H. et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008). [DOI] [PubMed] [Google Scholar]
- Huang S. F. et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat. Commun. 5, doi: 10.1038/ncomms6896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M. et al. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 12, doi: 10.1186/1471-2105-12-124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J. M., Kim B. H. & Grishin N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300, doi: 10.1093/nar/gkn072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D. & Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612, doi: 10.1093/nar/gkl315 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M. & Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973, doi: 10.1093/bioinformatics/btp348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313, doi: 10.1093/bioinformatics/btu033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V., Kosarev P., Seledsov I. & Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, 1–12, doi: 10.1186/gb-2006-7-s1-s10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H. & Cantor C. R. in Mamalian Protein Metabolism (ed Munro H. N.) 21–132 (Academic Press, 1969). [Google Scholar]
- Zuckerkandl E. & Pauling L. Molecules as documents of evolutionary history. J. Theor. Biol. 8, 357–366 (1965). [DOI] [PubMed] [Google Scholar]
- Nei M. & Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729, doi: 10.1093/Molbev/Mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591, doi: 10.1093/molbev/msm088 (2007). [DOI] [PubMed] [Google Scholar]
- Yang Z. H., Wong W. S. W. & Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118, doi: 10.1093/molbev/msi097 (2005). [DOI] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J. & Schuster S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386, doi: 10.1101/gr.5969107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. & Elkan C. in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology (ed UC San Diego Technical Report CS94-351) 28–36 (AAAI Press, 1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.