Abstract
In the present study, we describe the cloning and functional characterization of chicken interleukin 26 (ChIL-26). ChIL-26, a member of the IL-10 cytokine family, induces the production of proinflammatory cytokines by T cells. The ChIL-26 cDNA encodes an 82-amino-acid protein whose amino acid sequence has 22.63, 46.31 and 43.15% homology with human IL-26, pig IL-26 and canary IL-26, respectively. ChIL-26 signals through a heterodimeric receptor complex composed of the IL-20R1 and IL-10R2 chains, which are expressed primarily in the CU91 T cell line as well as CD4+ and CD8+ T cells. Recombinant ChIL-26 protein induced Th1 cytokines (IL-16 and IFN-γ), Th2 cytokines (IL-4, IL-6 and IL-10), Th17 cytokines (IL-17A, IL-17D, and IL-17F), and chemokine transcripts (mainly CCL3, CCL4, CCL5, CCL20 and CXCL13) in the CU91 T cell line and in CD4+ and CD8+ T cells, however IL-18 was not expressed in the CU91 T cell line. Taken together, the data demonstrates that T cells express the functional ChIL-26 receptor complex and that ChIL-26 modulates T cell proliferation and proinflammatory gene expression. To the best of our knowledge, this is the first report of cloned ChIL-26. We evaluated its functional roles, particularly in the pathogenic costimulation of T cells, which may be significantly associated with the induction of cytokines.
Electronic supplementary material
The online version of this article (doi:10.1186/s13567-016-0342-0) contains supplementary material, which is available to authorized users.
Introduction
Interleukin 26 (IL-26) was originally discovered in humans [1] and zebrafish [2]. Human IL-26 (HuIL-26) was cloned as a novel cDNA clone, denoted as AK155, displaying weak but significant sequence homology (approximately 25% identity) to HuIL-10. The genes encoding the ligands of the IL-10 family are located on human chromosome 1 (Chr1) (IL-10, IL-19, IL-20, and IL-24) [3, 4] and Chr12 (IL-22 and -24), and genes for their receptors are located on Chr1 (IL-22R1), Chr3 (IL-20R2), Chr6 (IL-20R1, IL-22BP, and IFNGR1), Chr11 (IL-10R1) and Chr21 (IFNAR2, IL-10R2, IFNAR1 and IFNGR2) [2, 5].
The HuIL-26 gene is located on chromosome 12q15, between the genes for two other important class 2 cytokines, gamma interferon (IFN-γ) and IL-22. IL-26 is often coexpressed with IL-22 by activated T cells, especially Th17 cells [6, 7]. It signals through a heterodimeric receptor complex composed of the IL-20R1 and IL-10R2 chains [8]. HuIL-26 receptors are expressed primarily on non-hematopoietic cell types, particularly epithelial cells [7].
In chickens, only four avian members of the IL-10 family have been identified: IL-10, IL-19, IL-22, and IL-26. Similar to HuIL-26, chicken IL-26 (ChIL-26) is encoded in the same cluster with IL-10 on chromosome 26, in a syntenic region with human Chr1 [9, 10]. In humans, HuIL-26 has been reported to signal via the IL-10R2/IL-20R1 heterodimeric receptor [8, 11]. While IL-10R2 is broadly expressed, IL-20R1 is expressed in many epithelial cell types but not in hematopoietic cells [12, 13]. The only biological activity of IL-26 reported so far is the upregulation of IL-8, IL-10, tumor necrosis factor alpha (TNF-α) and CD54 expression in intestinal epithelial cell lines, in association with STAT3 and/or STAT1 phosphorylation [8, 12].
Recently, HuIL-26 was functionally characterized, and His-HuIL-26 was shown to induce IL-10 and IL-8 in the Colo-205 colon cancer cell line and IL-8 in the Lovo colon cancer and HaCaT cell lines [8]. HuIL-26 induces the production of proinflammatory cytokines and many chemokines (mainly CCL20) in myeloid cells and CD4+ T cells [14–16]. IL-26 is also produced by activated T cells and targets epithelial target cells for signal transduction [6, 17]. However, the molecular cloning and functional characterization of ChIL-26 have not yet been performed.
We therefore report here, for the first time, the cloning and functional characterization of ChIL-26. In addition, we examined the biological effects of recombinant ChIL-26 (rChIL-26) protein in the CU91 chicken T cell line, CD4+ T cells, and CD8+ T cells. We observed increased inflammatory responses, and production of proinflammatory molecules.
Materials and methods
Cloning and expression of rChIL-26
To clone full-length ChIL-26, the predicted ChIL-26 coding sequence (GenBank accession # XM_004937561) was amplified from total RNA of the intestinal mucosal layer using the following restriction enzyme-anchored primers: EcoRI-anchored forward primer, 5′-CGGAATTCATGAAAAATGTTTTCAGTCATCTTGG-3′; and HindIII-anchored reverse primer, 5′-CCAAGCTTTACTATGGTTTGGATGTAGGCCT-3′ (restriction sites are underlined). Total RNA was isolated using TRizol reagent (Invitrogen, Carlsbad, CA, USA) as described [18] from the intestinal mucosal layer of White Leghorn chickens, kindly provided by the Animal Biosciences and Biotechnology Laboratory (Beltsville, MD, USA) of the USDA-Agricultural Research Service. The first-strand cDNA was subsequently synthesized using a Maxima First Strand cDNA synthesis kit (Thermo Scientific Inc., Waltham, MA, USA). Polymerase chain reaction (PCR) was performed to amplify the full-length ChIL-26 cDNA, under the following conditions: initial denaturation at 94 °C for 3 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. The newly synthesized full-length ChIL-26 gene was directly inserted into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA, USA), followed by transformation into Escherichia coli TOP10 (Invitrogen). Transformed E. coli TOP10 cells were cultured overnight in Luria–Bertani media (Difco™ and BBL™, NJ, USA) at 37 °C. A transformant was selected by a combination of PCR screening and sequencing (Genotech Inc., Daejeon, Republic of Korea). For cloning into E. coli expression vectors, the ChIL-26-expressing plasmid was digested with the endonucleases EcoRI (Invitrogen) and HindIII (Promega, Madison, WI, USA). The digested ChIL-26 fragment was purified from an agarose gel using a QIAQuick gel extraction kit (QIAgen, Valencia, CA, USA) and then ligated into digested pET32a (EMD Millipore, Billerica, MA, USA). To express rChIL-26 in E. coli, the purified ChIL-26-pET32a plasmid was introduced into E. coli BL21 (Invitrogen).
The expression of rChIL-26 was induced by adding 1 mM IPTG (USB Corporation, Cleveland, OH, USA), and bacteria were cultured at 37 °C overnight at 250 rpm. The bacterial cells were pelleted by centrifugation at 3500 × g for 30 min at 4 °C. The supernatant was removed and the pellet treated with B-PER bacterial protein extraction reagent (Thermo Scientific). Recombinant ChIL-26 was purified using HisPur cobalt resin (Thermo Scientific) in the first step of purification as per the manufacturer’s instructions. To remove the endotoxin contaminants, we combined affinity chromatography with a non-ionic detergent washing step as previously described [19]. The purified protein was concentrated and the buffer changed by ultrafiltration using a 3000-molecular-weight-cutoff membrane (EMD Millipore). The samples were dialyzed in phosphate-buffered saline (PBS; pH 7.2) overnight, using SnakeSkinTM dialysis tubing (Thermo Scientific), with stirring, and were analyzed by SDS-PAGE.
Antibodies
The following antibodies were used: anti-chicken CD4 (IgG1, k) and anti-chicken CD8 monoclonal antibodies (CT-8), purchased from LifeSpan BioSciences, Inc. (Seattle, WA, USA), anti-His (C-Term)–horseradish peroxidase (HRP) antibody was purchased from Invitrogen and Biotin-labeled goat anti-mouse Ig and Plus-DM streptavidin particles were also purchased from BD Biosciences (San Jose, CA, USA).
Isolation of CD4+, CD8+ and splenic T cells
Spleens were obtained from 4 to 5 week-old disease-free Ross 308 chicks (YangJi Hatchery, Pyeongtaek, Republic of Korea). Total cells were strained through a cell strainer (mesh size, 70 µm; Thermo Scientific) into a 50 mL tube. CD4+ and CD8+ cells were isolated using a BD IMag magnetic bead cell separation system according to the manufacturer’s instructions (BD Biosciences Pharmingen) and described previously [20]. Briefly, the cell concentration was adjusted to 2 × 107 mL−1 and 50 µg/mL of anti-CD4 or anti-CD8 monoclonal antibody (LifeSpan BioSciences) was added as determined in preliminary titration experiments. The cells were then incubated on ice for 45 min and washed twice with Hank’s balanced salt solution (HBSS) (Invitrogen) containing 10% fetal bovine serum (FBS). Biotinylated goat anti-mouse IgG (BD Biosciences Pharmingen) was added at the appropriate concentration, and the cells were incubated for 15 min on ice and then washed with an excess volume of 1 × BD IMag buffer (BD Biosciences Pharmingen). Next, 50 µL of streptavidin-labeled magnetic particles (BD IMag Plus-DM streptavidin particles) per 1 × 107 total cells was added, and cells were incubated on ice for 30 min. Magnetic particles were isolated, and the positive and negative cell fractions were suspended in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS.
For stimulation of ChIL-26 and its receptors by concanavalin A (Con A, Sigma), primary T cells were isolated from the spleens of 4 to 5 week-old White Leghorn chickens as previously described [21]. Briefly, splenic T cells were prepared and purified using the Histopaque-1077 (Sigma) density gradient medium by centrifugation at 400 × g for 20 min to separation of single cell suspension. Splenic T cells (1 × 106 cells/well) were cultured in 6-well plates (Nunc™, Thermo Scientific) with 5 mL of DMEM (Invitrogen, Carlsbad, CA, USA) containing 100 IU/mL penicillin, 100 mg/mL streptomycin, and 10% heat-inactivated FBS (Invitrogen) in a humidified 5% CO2 atmosphere at 37 °C and 5 μg/mL of Con A was added. Cells were harvested after 24, 48 or 72 h of Con A stimulation, and total RNA was extracted from cells as described [18].
In vitro biological function analysis
The CU91 chicken T cell line [22] was cultured in DMEM (Invitrogen) containing 100 IU/mL penicillin, 100 mg/mL streptomycin, and 10% heat-inactivated FBS (Invitrogen) in a humidified 5% CO2 atmosphere at 37 °C. The CU91 T cell line and CD4+ and CD8+ T cells were cultured at 1 × 106 cells/well in 6-well plates and then treated with either medium alone, lipopolysaccharide (LPS) (5 µg/mL; from Salmonella enterica serotype Typhimurium; (Sigma), rChIL-26 (200 ng/mL) or LPS/rChIL-26). Cells were collected at 24, 48 and 72 h post treatment. In pilot experiments, rChIL-26 demonstrated a significantly greater effect at a concentration of around 200 ng/mL for the activation of certain kinases. After incubation, the cells were collected and total RNA was extracted from cells using TRizol reagent (Invitrogen) and cDNA was synthesized as described [18].
Western blotting
The ChIL-26 recombinant protein (100 ng/µL) was separated by SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (Invitrogen) using an iBlot® gel transfer device (Invitrogen). The transferred PVDF membrane was blocked with 5% nonfat milk (Sigma) dissolved in PBS containing 0.05% Tween 20 (PBST). The membrane was incubated with anti-His (C-Term)–horseradish peroxidase (HRP) antibody overnight at 4 °C, washed three times with PBST. Detection of immunoreactive bands was performed using Western Lightning Plus-ECL substrate (Thermo Scientific) as per the supplier’s instructions. A single ChIL-26 protein band with an apparent molecular mass of approximately 33.0 kDa was observed (Figure 1C). The increased size over the predicted 10.9 kDa was due to the three epitope tags (polyhistidine, S-protein, and thioredoxin) in the recombinant protein.
Figure 1.

Sequence comparison, phylogenetic tree of amino acid and western blot analysis of ChIL-26. A Chicken IL-10 (Accession # NP 001004414) and ChIL-26 amino acid sequences were aligned and secondary helices of IL-10 and IL-26 were shown in vertical and horizontal lined box, respectively. B Phylogenetic tree was constructed by Phylodendron program using amino acid sequences aligned with BioEdit and Mega4 and (C) Western blot analysis of ChIL-26 recombinant protein. E. coli BL21(DE3) transformed with the pET32a(+)-ChIL-26 plasmid was induced with 1.0 mM IPTG, purified with a HisPur Cobalt resin and analyzed by Western blotting using anti-His (C-Term)-HRP Antibody.
Quantitative real-time PCR (qRT-PCR) analysis of cytokine transcripts
For analysis of mRNA transcripts, 2 µg of total RNA isolated from cells was treated with DNase I (Sigma) according to the manufacturer’s instructions to eliminate possible genomic DNA contamination. Briefly, 2 µg of total RNA was treated with 1.0 unit of DNase I in 1.0 μL of 10× reaction buffer (Thermo Scientific) and incubated for 30 min at 37 °C. Total RNA was reverse transcribed using a Maxima First Strand cDNA synthesis kit (Thermo Scientific) according to the manufacturer’s recommendations. Briefly, 2 μg of RNA was combined with 0.2 μg of oligo (dT)18 primer and RNase-free water for a total volume of 12.0 μL. Next, 4.0 μL of 5× reaction buffer, 20 units of Ribolock RNase inhibitor, 10 mM dNTP mix, and 200 units of RevertAcid M-MuLV reverse transcriptase were added. The mixture was incubated at 42 °C for 60 min and then heated at 70 °C for 5 min to terminate the reaction. To analyze the transcripts of various cytokines, cytokine primers were designed using Lasergene software (DNASTAR Inc., Madison, WI, USA) (Table 1). Quantitative real-time PCR (qRT-PCR) was performed with 2× Power SYBR Green master mix (Roche, IN, USA) according to the manufacturer’s instructions, using a LightCycler 96 system (Roche). In brief, cDNA (100 ng) was added to a reaction mixture of 10 μL 2× Power SYBR Green master mix (Roche), 0.5 μL of each primer (10 pmol/µL) and RNase-free water for a total volume of 20 μL. GAPDH was used as an internal control to normalize the quantification of target genes. The relative quantification of gene-specific expression was calculated using the 2−ΔΔCt method after normalization with chicken GAPDH [23]. All qRT-PCR reactions were performed in triplicate.
Table 1.
Primer sequences for real-time PCR analyses of cytokine transcript expression
| Primer | F/R | Nucleotide sequencing (5′-3′) | Accession no |
|---|---|---|---|
| GAPDH | Forward | TGC TGC CCA GAA CAT CAT CC | NM_204305 |
| Reverse | ACG GCA GGT CAG GTC AAC AA | ||
| CCL3 | Forward | CAT TGC CTC CGC CTA CAT | EU999777 |
| Reverse | ACT CCT CGG GGT TTA CAC ATA | ||
| CCL4 | Forward | CCC CTT GTC ATC GGT CAC | NM_204720 |
| Reverse | AGA GGC AGG AGC AGA GCA | ||
| CCL5 | Forward | CAG CAA ATG CCC ACA GG | NM_001045832 |
| Reverse | TGC AGC TCC AGG AAG TTG AT | ||
| CCL20 | Forward | AGG CAG CGA AGG AGC AC | NM_204438 |
| Reverse | GCA GAG AAG CCA AAA TCA AAC | ||
| CXCL13 | Forward | GCC TGT GCC TGG TGC TC | XM_420474.4 |
| Reverse | TGC CCC CTT CCC CTA AC | ||
| IFN-γ | Forward | AGC TGA CGA CGG TGG ACC TAT TAT T | HQ739082 |
| Reverse | GGC TTT GCG CTG GAT TC | ||
| IL-4 | Forward | AGC ACT GCC ACA AGA ACC TG | NM_001007079 |
| Reverse | CCT GCT GCC GTG GGA CAT | ||
| IL-6 | Forward | CAA GGT GAC GGA GGA GGA C | JQ897539 |
| Reverse | TGG CGA GGA GGG ATT TCT | ||
| IL-8 (CXCLi2) | Forward | GGC TTG CTA GGG GAA ATG A | NM_205498 |
| Reverse | AGC TGA CTC TGA CTA GGA AAC TGC | ||
| IL-10 | Forward | CTG TCA CCG CTT CTT CAC CT | AJ621254 |
| Reverse | ACT CCC CCA TGG CTT TGT A | ||
| IL-16 | Forward | TGC CTC ACA AGA ATC AAC AAC T | AB104417.2 |
| Reverse | ATA GAG CCC TTC CCA CCT TC | ||
| IL-17A | Forward | TGT CTC CGA TCC CTT GTT CT | AM773756 |
| Reverse | GTC CTG GCC GTA TCA CCT T | ||
| IL-17D | Forward | ACC CCA CAA GAT ACC CTA AAT AC | EF570583 |
| Reverse | GTG CTG CGG AAG TGA AAA T | ||
| IL-17F | Forward | CTC CGA TCC CTT ATT CTC CTC | NM_204460 |
| Reverse | GTC CTG GCC GTA TCA CCT T | ||
| IL-18 | Forward | ATT TTC CCA TGC TCT TTC TCA | HM854281 |
| Reverse | GGA ATG CGA TGC CTT TTG | ||
| IL-26 | Forward | AAT GCC TGT CTT CCG TGT G | XM_004937561 |
| Reverse | TCA TTG ATG GCC TTG TAG ACC | ||
| TNFSF15 | Forward | CCT GAG TAT TCC AGC AAC GCA | NM_001024578 |
| Reverse | ATC CAC CAG CTT GAT GTC ATC AAC | ||
| IL-10R2 | Forward | GAG CAG ACC ACC CAT AAC G | NM_204857 |
| Reverse | ATA CCA AAA GGC AAA GAA ACA A | ||
| IL-20R1 | Forward | GTG GAT GAA TTG CTG GGT AAG | XM_419723 |
| Reverse | TCT GTG ATG CCG TGT GCT AT |
Bioinformatics and statistical analyses
Multiple-sequence alignments were analyzed using Lasergene software (DNASTAR Inc., Madison, WI, USA), and a phylogenetic tree was constructed using the neighbor-joining method, with a bootstrap value of 1000, in the MEGA4 program [24]. Signal peptides and N-like glycosylation were predicted using NetNGlyc software [25]. Statistical analysis was performed using IBM SPSS software (SPSS 21.0 for Windows; SPSS, Chicago, IL, USA). The results are expressed as mean ± standard error of mean (SEM) at three independent experiments for each group (n = 3) and were compared between groups using the Duncan’s multicomparison method.
Results
Chicken IL-26
ChIL-26 consists of 82 amino acids and has a predicted isoelectric point of 9.6 and a molecular mass of 10.9 kDa (Additional file 1). In contrast to HuIL-26, ChIL-26 does not contain signal peptides. Moreover, HuIL-26 has two glycosylation sites (NFIL and NKGI sites) and ChIL-26 has three glycosylation sites (NVFS, NACL and NELD sites). The ChIL-26 protein has 43.15% and 46.31% amino acid similarities to canary and pig IL-26, respectively, whereas the similarities to other mammalian IL-26 proteins are approximately 23% (Table 2). Analysis of the secondary structure showed four highly conserved helix structures between ChIL-10 and ChIL-26 (Figure 1A). A phylogenetic tree of predicted IL-26 proteins extracted from GenBank is shown in Figure 1B. The ChIL-26 protein comprises a grouping distinct from those of mammalian and amphibian species (Figure 1B).
Table 2.
Chicken IL-26 amino acid similarity and identity with other species
| Chicken | Accession no | ||
|---|---|---|---|
| Similarity (%) | Identity (%) | ||
| Human | 27.36 | 22.63 | NP_060872 |
| Fish | 22.1 | 20 | NP_001018635 |
| Baboon | 27.89 | 22.63 | XP_003906792 |
| Canary | 46.84 | 43.15 | XP_009093343 |
| Chimpanzee | 27.36 | 22.63 | XP_003808754 |
| Camelus | 23.68 | 21.57 | AIL25449 |
| Polar bear | 28.42 | 23.15 | XP_008682482 |
| Cat | 27.89 | 22.63 | XP_006933945 |
| Cattle | 28.42 | 23.15 | XP_005909110 |
| Pig | 48.94 | 46.31 | ABJ96481 |
The amino acid similarity and identity were analyzed using BLASTP with a BLOSUM62 scoring matrix.
ChIL-26 receptor complex in T cells
To determine whether ChIL-26 and the ChIL-26 receptor complex, consisting of IL-10R2 and IL-20R1, are expressed in T cells and to study this ligand-receptor system, we analyzed ChIL-26, IL-10R2 and IL-20R1 mRNA expression in the chicken CU91 T cell line and in CD4+ and CD8+ T cells. RT-PCR was performed to confirm IL-10R2 and IL-20R1 mRNA expression and demonstrated that the ChIL-10R2 and ChIL-20R1 genes were expressed and that ChIL-26 was not expressed in all T cells tested. A low expression of IL-10R2 in CD4+ and CD8+ T cell compared to CU91 T cell line may be caused by non-treatment of Con A because CU91 T cell line established using interleukin-containing conditioned medium or concanavalin A stimulated T cells (Figure 2A). Moreover, Con A treatment had an effect on ChIL-26, IL-10R2 and IL-20R1 mRNA expression in primary T cells (Figure 2B). IL-10R2 mRNA levels increased 3.4-, 4.19- and 5.9-fold compared with the control group after Con A treatment for 24, 48 (p < 0.01), and 72 h (p < 0.001), respectively. IL-20R1 mRNA levels also significantly increased (3.5-, 4.6- and 5.6-fold) compared with the control group, with varying levels of significance (Figure 2B). In addition, ChIL-26 expression increased considerably (3.8-, 6.9- and 8.4-fold) compared with the control group, also with varying levels of significance (Figure 2B). Therefore, mRNA expression of ChIL-26 and its receptors, IL-10R2 and IL-20R1, was affected by Con A treatment of splenocytes.
Figure 2.
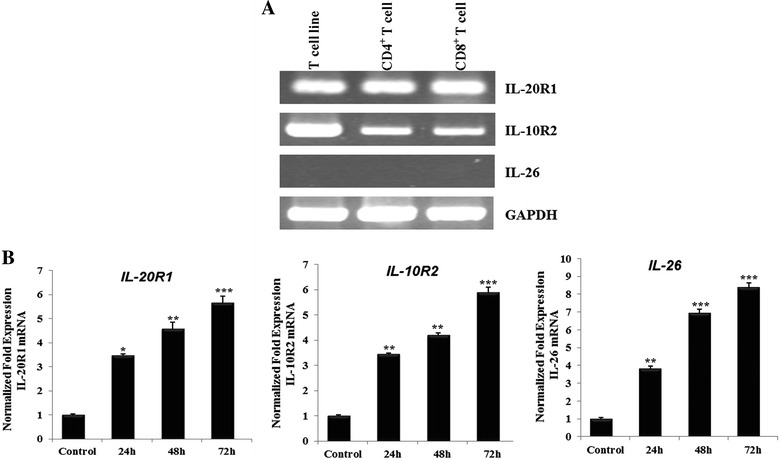
Receptors and GAPDH are expressed but not IL-26 are expressed in T cell. A IL-10 receptor 2 and IL-20 receptor 1 are expressed, but IL-26 is not expressed in T cell (T cell line, CD4+ T cell and CD8+ T cell). B IL-10R2, IL-20R1 and IL-26 expression in primary T cell induced by Con A (5 µg/mL), respectively. The expression of IL-10R2, IL-20R1 and IL-26 mRNA as analyzed by qRT-PCR analysis of mRNA derived from T cell as indicated. Data are presented as the mean ± SEM (n = 3) of three independent experiments: *p < 0.05, **p < 0.01, and ***p < 0.001.
Moreover, the expression of IL-10R2 and IL-20R1 mRNAs was confirmed in the CU91 T cell line and in CD4+ and CD8+ cells by qRT-PCR. The results indicated that the expression of IL-10R2 and IL-20R1 genes was upregulated in all T cells, particularly those cotreated with LPS and ChIL-26, including the CU91 T cell line, with 89.0-fold upregulation after 48 h and 88.9-fold upregulation after 72 h of treatment (Figure 3). Similar to the results obtained for the CU91 T cell line, IL-10R2 and IL-20R1 mRNA expression in CD4+ and CD8+ T cells was also highly upregulated by LPS/ChIL-26 cotreatment. In CD4+ T cells, IL-10R2 and IL-20R1 were upregulated 17.5- and 13.8-fold after 48 h treatment, and these receptors were increased 6.1-fold after 24 h and 5.5-fold after 72 h treatment with LPS/ChIL-26 in CD8+ T cells (Figure 3). This observation suggested that IL-10R2 and IL-20R1 are expressed on T cells and may be two chain for the chicken IL-26 receptor complex.
Figure 3.
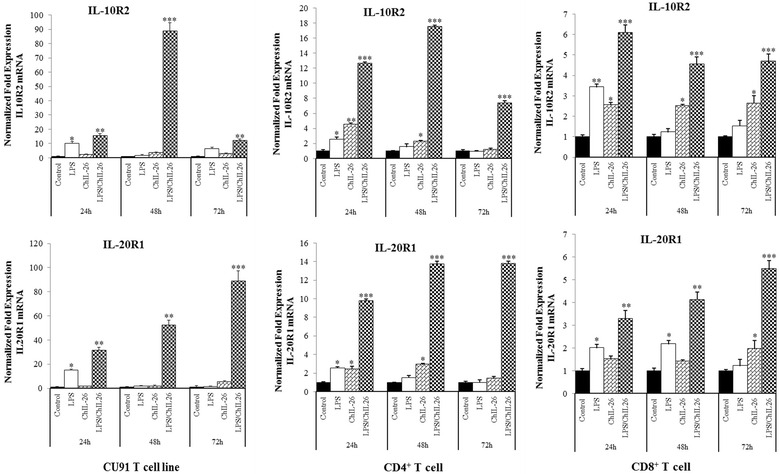
Interleukin 20 receptor 1 (IL-20R1) and interleukin 10 receptor 2 (IL-10R2) expressed in T cell. IL-20R1 and IL-10R2 mRNA expressed in CU91 T cell line, CD4+ T cell and CD8+ T cell as demonstrated by quantitative RT-PCR. Data are presented as the mean ± SEM (n = 3) of three independent experiments: *p < 0.05, **p < 0.01, and ***p < 0.001.
Costimulation with LPS/ChIL-26 leads to proinflammatory cytokine and chemokine expression in T cells
Previous reports demonstrated that HuIL-26 induces Th1 and Th17 as well as Th2 cytokines [14, 26]; therefore, we examined the transcriptional targets of this cytokine. The role of ChIL-26 in the transcriptional regulation of Th17 cytokines involved the induction of IL-17A, IL-17D and IL-17F cytokines. LPS and ChIL-26 treatment significantly enhanced the expression of IL-17A and IL-17D in all T cells at 72 h. In addition, IL-17F expression was also increased in the CU91 T cell line and in CD4+ and CD8+ cells at all time points, when cells were treated with LPS/ChIL-26 (Figure 4; Additional files 2 and 3). The IL-26-regulated expression of IL-17A, IL-17D and IL-17F by T cells and particularly in CU91 T cell line.
Figure 4.
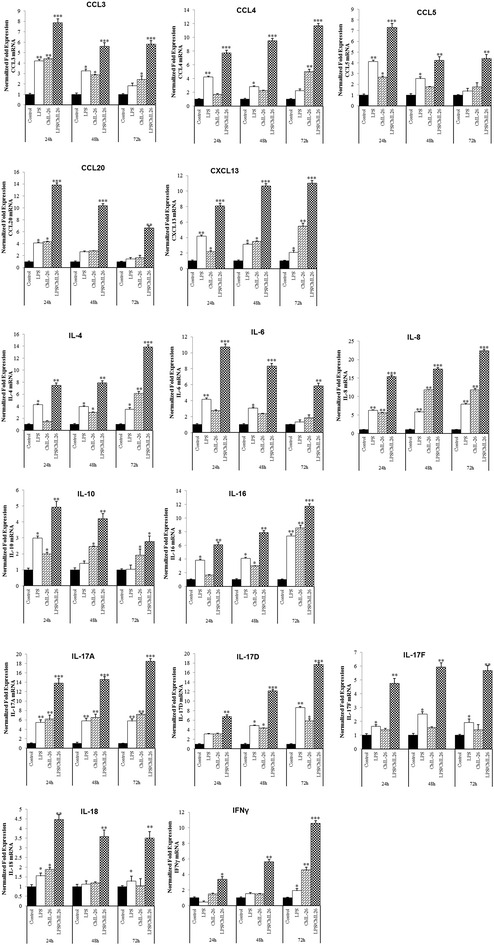
ChIL-26/LPS induces cytokine secretion by CD8 + T cells. The CD8+ T cell line was cultured in the presence or absence of 200 ng/mL ChIL-26, with or without 5 µg/mL LPS, and cytokine expression was analyzed by qRT-PCR. The results are indicated as fold increases of mRNA expression compared to that in unstimulated control cells. Data are presented as the mean ± SEM (n = 3) of three independent experiments: *p < 0.05, **p < 0.01, and ***p < 0.001.
The levels of the IL-4, IL-6, and IL-10 cytokines, which correspond to Th2 cytokines in birds, significantly increased following stimulation with LPS and ChIL-26 compared to the control. IL-4 mRNA was highly increased at 72 h, and its levels maximized at 131-, 16.4- and 13.8-fold in the CU91 T cell line and in CD4+ and CD8+ T cells, respectively compared to the control (p < 0.001) (Figure 4; Additional files 2 and 3). The expression of IL-6 and IL-10 mRNAs also increased significantly at 24 h post-LPS/ChIL-26 stimulation compared to the control in all T cells (p < 0.001) (Figure 4; Additional files 2 and 3).
IL-18 mRNA was not increased in the CU91 T cell line stimulated with LPS or ChIL-26, or both, however it increased 4.4-fold with LPS and ChIL-26 or with ChIL-26 only in CD4+ and CD8+ T cells (p < 0.05) (Figure 4; Additional files 2 and 3). The expression of IFN-γ, a Th1 cytokine, also increased, and reached its maximum expression level after 72 h of costimulation with LPS and ChIL-26 in all T cells (p < 0.01) (Figure 4; Additional files 2 and 3).
IL-16 mRNA was also highly expressed 24 h after costimulation in the CU91 T cell line and in CD4+ T cells and at 72 h in CD8+ T cells (p < 0.01) (Figure 4; Additional files 2 and 3). The LPS and ChIL-26 combination also had a positive effect on chemokine expression in T cells. When LPS and ChIL-26 were used in combination, the expression of CCL3, CCL4, CCL20, and CXCL13 mRNAs increased significantly in the CU91 T cell line at 24 h, in CD4+ cells at 24 h (CCL4 and CCL20) and 48 h (CXCL13), and CD8+ cells at 24 h (CCL3 and CCL20) and 72 h (CCL4 and CXCL13) compared with controls (p < 0.001) (Figure 4; Additional files 2 and 3). The expression of CCL5 and IL-8 (CXCLi2) mRNAs increased significantly compared with controls at 48 h (p < 0.001) (Additional files 2). In CD4+ and CD8+ T cells, the highest expression levels of all chemokines measured over all time points were seen with LPS and ChIL-26 treatments. Collectively, these results demonstrated that ChIL-26 upregulates the expression of proinflammatory cytokines and chemokines mRNA in the CU91 T cell line and in CD4+ and CD8+ T cells.
Discussion
This is the first report demonstrating a role for ChIL-26 in chickens. ChIL-26 is a novel member of the IL-10-like cytokine family and shares the IL-10R2 subunit with IL-10 [27] and the IL-20R1 subunit with IL-20 [28]. The IL-26 gene and its corresponding protein also share some sequence homology with HuIL-10 and ChIL-10. IL-10 has anti-inflammatory properties, however we demonstrated here that IL-26 has proinflammatory functions in T cells and induces cytokine upregulation upon costimulation with LPS. Other research groups first identified the receptors for HuIL-26 [8, 11]. HuIL-26 signals through a heterodimeric receptor complex composed of IL-20R1 and IL-10R2 chains. IL-20R1 functions as the specific ligand binding chain for IL-26, and IL-10R2 functions as an essential second chain to complete assembly of the active receptor complex. Similar to humans, signaling via chicken IL-10R2 and IL20R1 is also activated by LPS and ChIL-26 costimulation (Figure 3). Neutralizing antibodies against either the IL-20R1 or IL-10R2 chain can block induction of HuIL-26 signaling [8, 11]. In contrast to the anti-inflammatory properties of IL-10, we demonstrated that ChIL-26 modulates the proinflammatory functions of T cells. Similar to HuIL-26, ChIL-26 upregulates the gene expression of proinflammatory cytokines in T cells.
Depending on the T cell source, cytokines have been differentiated into Th1, Th2 and Th17 cytokines. Previous studies demonstrated an induction of HuIL-26 production particularly in Th1 cells [14]. Crohn’s disease is considered to represent Th1- and Th17-mediated intestinal inflammation, while ulcerative colitis has features, at least to some degree, of Th2-mediated colitis [29–31]. Our studies demonstrated that ChIL-26 strongly induces Th1 cytokines (IL-16 and IFN-γ; but with an absence of IL-18 in the CU91 T cell line), Th2 cytokines (IL-4, -6 and -10), Th17 cytokines (IL-17A, IL-17D and IL-17F), and chemokines (mainly CCL3, CCL4, CCL5, CCL20, CXCLi2 and CXCL13) in T cells (Figure 4; Additional files 2 and 3). Overall, the cytokine and chemokine genes were expressed more highly in the CU91 T cell line than in CD4+ and CD8+ T cells upon costimulation with LPS and ChIL-26. Researcher groups previously reported that resting T cells expressed lager diversity of genes and the patterns of gene expression showed markedly downregulation than transformed T cells cause by the different pathway involving antigen-presenting cell [32–34]. Therefore, the differences in gene expression for the CU91 T cell line and CD4+ and CD8+ T cells may result from disparities in the efficiency of antigen presentation via the MHC class I and II pathways, in the context of LPS and ChIL-26 cotreatment [35–37].
Avian IL-26 is another member of the IL-10 family, and chicken T cells express a functional IL-26 receptor complex (IL10R2 and IL-20R1). LPS binds to Toll-like receptor 4 (TLR4) on the cell surface and forms the LPS-TLR4 complex, which triggers both MyD88-dependent and MyD88-independent TRIF-dependent pathways. Toll-like receptor (TLR) family including TLR-4 expressed in chicken immune T cell subsets (CD4+ and CD8+ T cell) and cell lines (B and T cell lines) [38]. Signaling through the MyD88-dependent pathway is responsible for early-phase NF-κB and MAPK activation, which facilitates the induction of proinflammatory cytokines, chemokines, adhesion molecules, and MHC or costimulatory molecules [39]. As expected, the cytokines in T cells were significantly more highly expressed in cells receiving LPS/ChIL-26 costimulation than in cells treated with either LPS or ChIL-26 alone.
Here, we demonstrated the successful cloning and expression of ChIL-26 and its molecular functions in immune responses in the CU91 chicken T cell line and CD4+ and CD8+ T cells. In this study, the use of ChIL-26-deficient T cells revealed that ChIL-26 expression is necessary for optimal LPS induction of several cytokines. Therefore, we suggest that ChIL-26 as well as pathogens may influence the expression of Th1, Th2 and Th17 cytokines. Further investigation into the regulation of ChIL-26 gene expression with signaling pathway will undoubtedly reveal new insights into the regulation of T cell proinflammatory gene programs.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: ADT, YHH. Performed the experiments: ADT, BP and JB. Analysed the data: ADT, BP, JB and YHH. Contributed reagents/materials/analysis tools: YHH. Wrote the paper: ADT, YHH. Interpreted the data: ADT, YHH. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by the Golden Seed Project (PJ009925022016), the Ministry of Agriculture, Food and Rural Affairs (MAFRA), the Ministry of Oceans and Fisheries (MOF), the Rural Development Administration (RDA) and the Korea Forest Service (KFS) and by the Chung-Ang University Excellent Student Scholarship in 2014. We kindly thank Dr Hyun S. Lillehoj for providing the chicken samples.
Abbreviations
- MHC
major histocompatibility complex
- TCR
T cell receptor
- CTLs
cytotoxic T lymphocytes
- NKs
natural killer cells
- IL
interleukin
- TNF
tumor necrosis factor
- RT
reverse transcription
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- MAPK
mitogen-activated protein kinase
- TLR
toll-like receptor
Additional files
10.1186/s13567-016-0342-0 Nucleotide and deduced amino acid sequences of ChIL-26. The primers used to amplify the full-length ChIL-26 sequence are underlined.
10.1186/s13567-016-0342-0 ChIL-26/LPS induces cytokine secretion by the CU91 T cell line. The CU91 chicken T cell line was cultured in the presence or absence of 200 ng/mL ChIL-26, with or without 5 µg/mL LPS, and cytokine expression was analyzed by qRT-PCR. The results are indicated as fold increases of mRNA expression compared to that in unstimulated control cells. Data are presented as the mean ± SEM (n = 3) of three independent experiments: * p < 0.05, ** p < 0.01, and *** p < 0.001.
10.1186/s13567-016-0342-0 The CD4+ T cell line was cultured in the presence or absence of 200 ng/mL ChIL-26, with or without 5 µg/mL LPS, and cytokine expression was analyzed by qRT-PCR. The results are indicated as fold increases of mRNA expression compared to that in unstimulated control cells. Data are presented as the mean ± SEM (n = 3) of three independent experiments: * p < 0.05, ** p < 0.01, and *** p < 0.001.
Contributor Information
Anh Duc Truong, Email: truonganhduc84@gmail.com.
Boyeong Park, Email: hero173@naver.com.
Jihye Ban, Email: ring5648@gmail.com.
Yeong Ho Hong, Email: yhong@cau.ac.kr.
References
- 1.Knappe A, Hor S, Wittmann S, Fickenscher H. Induction of a novel cellular homolog of interleukin-10, AK155, by transformation of T lymphocytes with herpesvirus saimiri. J Virol. 2000;74:3881–3887. doi: 10.1128/JVI.74.8.3881-3887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Igawa D, Sakai M, Savan R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol Immunol. 2006;43:999–1009. doi: 10.1016/j.molimm.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, Vazquez N, Pestka S, Donnelly RP, Kotenko SV. Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10) Genes Immun. 2000;1:442–450. doi: 10.1038/sj.gene.6363714. [DOI] [PubMed] [Google Scholar]
- 4.Conti P, Kempuraj D, Frydas S, Kandere K, Boucher W, Letourneau R, Madhappan B, Sagimoto K, Christodoulou S, Theoharides TC. IL-10 subfamily members: IL-19, IL-20, IL-22, IL-24 and IL-26. Immunol Lett. 2003;88:171–174. doi: 10.1016/S0165-2478(03)00087-7. [DOI] [PubMed] [Google Scholar]
- 5.Kotenko SV. The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/S1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 6.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, Lecron JC, Kastelein RA, Cua DJ, McClanahan TK, Bowman EP, de Waal Malefyt R. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly RP, Sheikh F, Dickensheets H, Savan R, Young HA, Walter MR. Interleukin-26: an IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev. 2010;21:393–401. doi: 10.1016/j.cytogfr.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hor S, Pirzer H, Dumoutier L, Bauer F, Wittmann S, Sticht H, Renauld JC, de Waal Malefyt R, Fickenscher H. The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem. 2004;279:33343–33351. doi: 10.1074/jbc.M405000200. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser P, Poh TY, Rothwell L, Avery S, Balu S, Pathania US, Hughes S, Goodchild M, Morrell S, Watson M, Bumstead N, Kaufman J, Young JR. A genomic analysis of chicken cytokines and chemokines. J Interferon Cytokine Res. 2005;25:467–484. doi: 10.1089/jir.2005.25.467. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser P. The avian immune genome–a glass half-full or half-empty? Cytogenet Genome Res. 2007;117:221–230. doi: 10.1159/000103183. [DOI] [PubMed] [Google Scholar]
- 11.Sheikh F, Baurin VV, Lewis-Antes A, Shah NK, Smirnov SV, Anantha S, Dickensheets H, Dumoutier L, Renauld JC, Zdanov A, Donnelly RP, Kotenko SV. Cutting edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor 1 and IL-10 receptor 2. J Immunol. 2004;172:2006–2010. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- 12.Dambacher J, Beigel F, Zitzmann K, De Toni EN, Goke B, Diepolder HM, Auernhammer CJ, Brand S. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207–1217. doi: 10.1136/gut.2007.130112. [DOI] [PubMed] [Google Scholar]
- 13.Nagalakshmi ML, Murphy E, McClanahan T, de Waal Malefyt R. Expression patterns of IL-10 ligand and receptor gene families provide leads for biological characterization. Int Immunopharmacol. 2004;4:577–592. doi: 10.1016/j.intimp.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Corvaisier M, Delneste Y, Jeanvoine H, Preisser L, Blanchard S, Garo E, Hoppe E, Barre B, Audran M, Bouvard B, Saint-Andre JP, Jeannin P. IL-26 is overexpressed in rheumatoid arthritis and induces proinflammatory cytokine production and Th17 cell generation. PLoS Biol. 2012;10:e1001395. doi: 10.1371/journal.pbio.1001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sedwick C. IL-26 kick-starts rheumatoid arthritis. PLoS Biol. 2012;10:e1001398. doi: 10.1371/journal.pbio.1001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang HQ, Liu XG, Yang X, Chen T, Yu SG. Effect of different types of moxibustion intervention on expression of inflammatory cytokines IL-1 and TNF-alpha in rabbits with rheumatoid arthritis. Zhen Ci Yan Jiu. 2013;38:134–139. [PubMed] [Google Scholar]
- 17.Braum O, Klages M, Fickenscher H. The cationic cytokine IL-26 differentially modulates virus infection in culture. PLoS One. 2013;8:e70281. doi: 10.1371/journal.pone.0070281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong YH, Song W, Lee SH, Lillehoj HS. Differential gene expression profiles of beta-defensins in the crop, intestine, and spleen using a necrotic enteritis model in 2 commercial broiler chicken lines. Poultry Sci. 2012;91:1081–1088. doi: 10.3382/ps.2011-01948. [DOI] [PubMed] [Google Scholar]
- 19.Reichelt P, Schwarz C, Donzeau M. Single step protocol to purify recombinant proteins with low endotoxin contents. Protein Expr Purif. 2006;46:483–488. doi: 10.1016/j.pep.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 20.Hong YH, Lillehoj HS, Lee SH, Dalloul RA, Lillehoj EP. Analysis of chicken cytokine and chemokine gene expression following Eimeria acervulina and Eimeria tenella infections. Vet Immunol Immunopathol. 2006;114:209–223. doi: 10.1016/j.vetimm.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Annamalai T, Selvaraj RK. Chicken chemokine receptors in T cells isolated from lymphoid organs and in splenocytes cultured with concanavalin A. Poult Sci. 2010;89:2419–2425. doi: 10.3382/ps.2010-00968. [DOI] [PubMed] [Google Scholar]
- 22.Schat KA, Pratt WD, Morgan R, Weinstock D, Calnek BW. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 1992;36:432–439. doi: 10.2307/1591524. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.The MEGA4 homepage http://www.megasoftware.net/mega4/. Accessed 27 Dec 2014
- 25.The NetNGlyc homepage http://www.cbs.dtu.dk/services/NetNGlyc/. Accessed 30 Dec 2014
- 26.Alanara T, Karstila K, Moilanen T, Silvennoinen O, Isomaki P. Expression of IL-10 family cytokines in rheumatoid arthritis: elevated levels of IL-19 in the joints. Scand J Rheumatol. 2010;39:118–126. doi: 10.3109/03009740903170823. [DOI] [PubMed] [Google Scholar]
- 27.Yao Q, Fischer KP, Arnesen K, Lorne Tyrrell D, Gutfreund KS. The Pekin duck IL-10R2 common chain: cDNA cloning, genomic structure, molecular characterization and mRNA expression analysis. Int J Immunogenet. 2013;40:386–395. doi: 10.1111/iji.12042. [DOI] [PubMed] [Google Scholar]
- 28.Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121:1108–1111. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 29.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 30.Neurath MF. IL-23: a master regulator in Crohn disease. Nat Med. 2007;13:26–28. doi: 10.1038/nm0107-26. [DOI] [PubMed] [Google Scholar]
- 31.Dige A, Stoy S, Rasmussen TK, Kelsen J, Hvas CL, Sandahl TD, Dahlerup JF, Deleuran B, Agnholt J. Increased levels of circulating Th17 cells in quiescent versus active Crohn’s disease. J Crohns Colitis. 2013;7:248–255. doi: 10.1016/j.crohns.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Teague TK, Hildeman D, Kedl RM, Mitchell T, Rees W, Schaefer BC, Bender J, Kappler J, Marrack P. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc Natl Acad Sci U S A. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chtanova T, Newton R, Liu SM, Weininger L, Young TR, Silva DG, Bertoni F, Rinaldi A, Chappaz S, Sallusto F, Rolph MS, Mackay CR. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol. 2005;175:7837–7847. doi: 10.4049/jimmunol.175.12.7837. [DOI] [PubMed] [Google Scholar]
- 34.Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nat Rev Immunol. 2012;12:306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi Y, Operario DJ, Georas SN, Mosmann TR. The acute environment, rather than T cell subset pre-commitment, regulates expression of the human T cell cytokine amphiregulin. PLoS One. 2012;7:e39072. doi: 10.1371/journal.pone.0039072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribas A, Butterfield LH, Economou JS. Genetic immunotherapy for cancer. Oncologist. 2000;5:87–98. doi: 10.1634/theoncologist.5-2-87. [DOI] [PubMed] [Google Scholar]
- 37.Watts C. Capture and processing of exogenous antigens for presentation on MHC molecules. Annu Rev Immunol. 1997;15:821–850. doi: 10.1146/annurev.immunol.15.1.821. [DOI] [PubMed] [Google Scholar]
- 38.Iqbal M, Philbin VJ, Smith AL. Expression patterns of chicken Toll-like receptor mRNA in tissues, immune cell subsets and cell lines. Vet Immunol Immunopathol. 2005;104:117–127. doi: 10.1016/j.vetimm.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]


