Abstract
Here we analyzed in leaves the effect of FT overexpression driven by meristem-specific KNAT1 gene homolog of Arabidopsis thaliana (Lincoln et al., 1994; Long et al., 1996) on the transcriptomic response during plant development. Our results demonstrated that meristematic FT overexpression generates a phenotype with an early flowering independent of photoperiod when compared with wild type (WT) plants. Arabidopsis FT-overexpressor lines (AtFTOE) did not show significant differences compared with WT lines neither in leaf number nor in rosette diameter up to day 21, when AtFTOE flowered. After this period AtFTOE plants started flower production and no new rosette leaves were produced. Additionally, WT plants continued on vegetative stage up to day 40, producing 12–14 rosette leaves before flowering. Transcriptomic analysis of rosette leaves studied by sequencing Illumina RNA-seq allowed us to determine the differential expression in mature leaf rosette of 3652 genes, being 626 of them up-regulated and 3026 down-regulated. Overexpressed genes related with flowering showed up-regulated transcription factors such as MADS-box that are known as flowering markers in meristem and which overexpression has been related with meristem identity preservation and the transition from vegetative to floral stage. Genes related with sugar transport have shown a higher demand of monosaccharides derived from the hydrolysis of sucrose to glucose and probably fructose, which can also be influenced by reproductive stage of AtFTOE plants.
Keywords: Arabidopsis, Flowering locus T, RNA-seq analysis, Plant development, Transgenic plants
1. Introduction
In plants, floral transition initiates reproduction changes from vegetative to floral meristem. This transition is controlled by multiple genetic pathways in response to various developmental and environmental cues such as temperature, photoperiod and nutrient availability (Lee and Amasino, 1995; Andres and Coupland, 2012; Romera-Branchat et al., 2014). Moreover, endogenous signals influencing the timing of the floral transition lead to the conversion of the shoot apical meristem into an inflorescence meristem and subsequently to the formation of flowers (Pidkowich et al., 1999; Wang, 2014). However, the way these cues interact with the pathways is not yet fully understood (Graciet and Wellmer, 2014). Arabidopsis plants initiate flower development through six known routes: age-, vernalization-, gibberellin (GA)-, temperature-, photoperiod-dependent, and autonomous pathways. These converge to regulate a small number of “floral integrator genes”, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Fornara et al., 2010; An et al., 2004; Samach et al., 2000; Yoo et al., 2005).
The FT protein is a component of the florigenic signaling system that translocates long- distance through the phloem (Corbesier et al., 2007; Tamaki et al., 2007; Lin et al., 2007). Plants perceive differences in day length in mature leaves; when a threshold is reached, phloem-mobile FT triggers the floral transition in the shoot apical meristem (SAM) to initiate floral morphogenesis via interaction with 14-3-3 proteins (Turck et al., 2008; Taoka et al., 2011; Yoo et al., 2013). Arabidopsis is a facultative long-day plant that flowers earlier under long days (LDs) of 16 h of light than under short days (SDs) of 8 to 10 h of light. Under LD, a cascade results in the activation of the FT and its homolog TWIN SISTER OF FT (TSF) genes by the transcriptional regulator CONSTANS (CO) in the leaf vasculature (An et al., 2004; Suarez-Lopez et al., 2001; Valverde et al., 2004). FT is expressed in the companion cell (CC) of source leaves, selectively enters the phloem translocation stream and is then carried to the apex by cell-to-cell movement (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007). FT is unloaded into the surrounding tissue by selective trafficking through plasmodesmata, after which it reaches the shoot apical meristem (SAM) where it triggers floral transition by forming a complex with FD, a bZIP transcription factor (Abe et al., 2005; Wigge et al., 2005; Amasino, 2010; Yoo et al., 2013). FT activates SUPPRESSOR OF OVEREXPRESSION OF CO (SOC1), FRUITFULL (FUL) and later APETALA1 (AP1), all of which encode MADS-box transcription factors (Turck et al., 2008; Torti et al., 2012; Yoo et al., 2013; Balanzà et al., 2014; Smaczniak et al., 2012). It has been proposed that high levels of FT accumulates in the postphloem pathway providing the FT reservoir required to support its selective cell-to-cell trafficking into the axillary meristem (AM) (Yoo et al., 2013). SAM is located beyond the limits for FT trafficking and a threshold level is required to bind the bZIP transcription factor FD to activate the floral developmental pathway in the SAM (Amasino, 2010; Yoo et al., 2013). The floral transition involves a dramatic transcriptional reprogramming of the shoot meristem; however, many of the global changes originated by gene expression occurring specifically in the SAM are not yet fully understood (Torti et al., 2012).
Constitutive FT expression driven by the CaMV 35S promoter causes early flowering, leading to the production of terminal flowers immediately upon germination (Kardailsky et al., 1999; Kobayashi et al., 1999). Additionally, it has been demonstrated that FT overexpression induces early flowering in both short-day and long-day conditions (Xu et al., 2012); on the other hand, loss-of-function alleles have a late-flowering phenotype (Koornneef et al., 1991). Aside form its well-established function in flowering induction; FT also regulates development of seeds, pods, and other tissues (Xu et al., 2012; Mouradov et al., 2002).
The objective of the present study was to analyze the effect of FT overexpression driven by the Arabidopsis meristem-specific promoter KNAT1, a homolog of KNOTTED-1 (Lincoln et al., 1994; Long et al., 1996) on the whole-plant transcriptome. Our results demonstrate that meristematic FT overexpression generate a phenotype with an early flowering independent of photoperiod when compared with wild type (WT) plants. Despite that FT overexpression was confined to the apical meristem and cambium, a massive change in gene expression was observed in rosette leaves. These changes were analyzed in the context of functional categories in the Arabidopsis metabolism.
2. Materials and methods
2.1. Plant growth
Wild type Arabidopsis Columbia-0 ecotype was employed in the present study. Plants were grown in greenhouse and in controlled growth chambers. Hydroponic cultures were employed as follows: Arabidopsis seeds were stratified, kept at 4 °C for 3 days in the dark and then germinated and grown in hydroponic system (Conn et al., 2013) at 22 °C under controlled conditions, initially in short days (SD) (8 h light, 16 h dark) and then transferred to long-day photoperiod (LD) (16 h light, 8 h dark), under 100 to 120 mmol m−2 s− light. Plants were monitored daily for flowering, while rosette development was measured with software ImageJ (http://rsb.info.nih.gov).
2.2. Design of genetic constructs
The 2356 bp upstream region of the KNAT1 gene (Long et al., 1996) was PCR-amplified from Arabidopsis genomic DNA using the specific primers pAT4G08150 F (5′-TTCTTAACATTTGACCATTGATTGAAA-3′) and pAT4G08150 R (5′-ACCCAGATGAGTAAAGATTTGAGAGAG-3′), which includes the 5′UTR. The FT ORF was PCR- amplified from cDNA previously synthetized using the specific primers AtFT1F (5′-ATGTCTATAAATATAAGAGACCCTCTT-3′) and AtFT2R (5′-CTAAAGTCTTCTTCCTCCGCAGCC-3′). The NOS terminator was amplified using plasmid DNA from pB7W320 with the primers T-Nos (F) (5′-TGACCCCTAGAGTCAAGCAGATCGTTCAAACAT-3′) and T-nos (R) (5′-ATCAGCTTGCATGCCGGTCGATCTAGTA-3′). The size of the amplified products (KNAT1 promoter, FT ORF- and T-Nos) was 2356 bp, 528 bp and 294 bp, respectively. The PCR products were independently cloned into the pDRIVE® vector (Qiagen, Santa Clarita, CA). In order to express FT driven by the KNAT1 promoter and NOS terminator, the assembly PCR technique was employed (https://www.geneoracle.com/documents/GeneIOS_Manual.pdf). The expected size of the gene expression unit was 3178 bp. The sequence of this PCR product was cloned into the PCR8/GW/TOPO TA vector, (Invitrogen, Carlsbad, CA) and verified by sequencing. The expression unit was subcloned into the destination vector pBGWFS7.0 (https://gateway.psb.ugent.be/search/index/transcriptional_reporters/any). The recombinant destination vector harboring PKNAT1::FT::T-NOS was introduced into Agrobacterium tumefaciens strain C58C1 by electroporation.
Transformation of Arabidopsis was performed using the floral-dip method (Zhang et al., 2006) employing A. tumefaciens C58C1 strain, harboring the specific construct P-KN::FT::T-NOS. Seeds obtained from the transformation were collected and then germinated in soil containing ammonium glufosinate (FINALE, Bayer, Germany). Transformation was verified by PCR employing the specific primers OGMFT (R) (5′-ACCAAAGTATAGAAGTTCCTGAGGTCTTCT-3′) and transgene knotted (F) (5′-CCTTGACGAATTCTATATACCTAGTTCGTT-3′).
2.3. Determination of T-DNA copy number
Droplet digital PCR (Bio-Rad) was employed to determine transgene (FT) copy variation number (CVN) in Arabidopsis transgenic plants. For this purpose, 2.5 ng of genomic DNA previously digested with HindIII was employed as template. Droplets were generated for PCR reaction with the specific primers AtFT-qPCR (F) (5′-TCCGTTTAATAGATCAATCAC-3′), FT-qPCR (R) (5′-CCACCATAACCAAAGTATAG-3) and probe TaqMan ddPCRFT [5′FAM] TCCTGAGGTCTTCTCCACCA [3′BHQ1]. The PCR-amplified product was 152 bp. Arabidopsis HMGB1 (AT3G51880) was used as an internal, reference gene. The primers used for the reference were HMGB1 probe [5′HEX]AGGCACCGGCTGAGAAGCCT[3′BHQ1], HMGB1F (5′-CAGAAAGGTGGGAAAGAGGA-3′) and HMGB1-R (5′-AAGGACCCAAACAAACCAAA-3′). The HMGB1 PCR-amplified product was 96 bp. After cycling, the PCR nanodroplets were counted using the droplet reader Bio-Rad QX100 (Bio-Rad 2012).
2.4. FT mRNA quantification by real time quantitative RT-PCR
Total RNA was isolated using the RNeasy Plant Kit (Qiagen, Valencia, CA). Quantitative RT-PCR was performed with KAPPA SYBR® FAST qPCR kitmaster mix (2×) ABI Prism™. The qRT-QPCR analysis was carried out using the StepOnePlus™ Real-Time PCR System (Life Technologies). Gene-specific primers employed were AtFT-ddPCR (F) (5′-TCCGTTTAATAGATCAATCAC-3′) and At-qPCR (F) (5′-CCACCATAACCAAAGTATAG-3′). To normalize the mRNA expression, Arabidopsis 18S rRNA was utilized as internal, constitutive gene, and was amplified with the specific primers 18S-F (5′-GTGATGGGGATAGATCATTGCAATTGTTGG-3′) and 18S-R (5′-TGGACTTCTCGCGACGTCGCGGGCGGCG-3′. Relative expression levels were calculated by the (2−ΔΔCt) method (Livak and Schmittgen, 2001). Three technical replicates were made per sample and averaged to calculate relative expression.
2.5. RNA purification
For RNA-Seq, total RNA from rosette leaves of 35-day old AtFT2.1 overexpressors and Wild Type (WT) plants was extracted with the RNeasy Plant Kit (Qiagen). Seeds of both genotypes were kept at 4 °C for 3 days and then germinated in a hydroponic system (Conn et al., 2013). Seedling growth was performed under controlled conditions, initially in short days (8 h light, 16 h dark) and after 21 days the seedlings were transferred to long-day (16 h light, 8 h dark) photoperiod under 100 to 120 mmol m−2 s−1. The RNA-seq experiments were conducted with RNA isolated from three biological replicates per overexpressing line and two biological replicates for WT.
2.6. Massive mRNA-sequencing, bioinformatics analysis and data processing
Illumina sequencing was performed at Otogenetics (Georgia, USA), Quality analysis and quantification of extracted RNA was carried out with a NanoDrop ND-2000 Spectrophotometer (NanoDrop Technologies, Wilminton, DE, USA) and a Bioanalyzer 2100 (Agilent Technologies Palo Alto, CA; http://www.agilent.com). The 260/280 nm ratios of all the samples ranged from 1.99 to 2.11. The RNI value of each sample reached >7.0. Illumina HiSeq Sequencing with PE50 yielded 20 million reads by triplicate. The raw data files are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) accession numbers SRR2094583 and SRR2094587 for AtFTOE replicates 1–3 and AtWT for control replicates 1–2 respectively. The quality control was performed using FasQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). The paired-end reads were aligned to the reference Arabidopsis genome using Tophat (version 2.0.10) (Trapnell et al., 2009) and Bowtie2 (version 2.1.0) (Langmead and Salzberg, 2012). The reference Arabidopsis genome and gene model annotation files (TAIR10 < ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Arabidopsis_thaliana/NCBI/TAIR10/Arabidopsis_thaliana_NCBI_TAIR10.tar.gz>) were downloaded from the Illumina iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.html). Differential expression was determined by Cufflinks (version 2.1.1) as described by Trapnell et al., 2012 and then was visualized by CummeRbound, an edgeR package (Goff et al., 2013).
FT gene expression levels in WT and overexpressing (OE) lines were determined by calculating the FPKM (Fragment per Kilobase of transcript (exon model) per Million mapped reads) values (Mortazavi et al., 2008, Mizrachi et al., 2010). To analyze the variation in expression between two replicates from WT and three replicates from OE it was calculated the absolute difference of the log2 fold change and adjusted to P-value ≤ 0.05.
2.7. Assignment of gene ontology terms
Gene Ontology (GO) analysis and GO enrichment was performed to annotate the genes with differential expression using the TAIR gene ontology annotation (https://www.arabidopsis.org/portals/genAnnotation/functional_annotation/go.jsp) and also the bioinformatics resource DAVID version 6.7, which is suitable for RNA-seq data, (Huang et al., 2009a, 2009b). Identified differentially expressed genes were subsequently mapped to the Mapman databases (http://mapman.gabipd.org/web/guest/mapman; Ath_AGI_LOCUS_TAIR10_Aug2012-3.m02; Thimm et al., 2004) in order to visualize the genes metabolic pathway diagrams.
2.8. Validation of differential gene expression through Quantitative RT-PCR
Differential gene expression of both AtFTOV2.1 and WT samples was validated through qRT-PCR for the following transcripts: MAF5, SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), SPL4, STP13 and ST3. Total RNA was isolated using the RNeasy Plant Kit (Qiagen, Valencia, CA) and quantitative RT-PCR was performed with KAPPA SYBR® FAST qPCR kit master mix (2×) ABI Prism™. The qRT-QPCR analysis was carried out using the StepOnePlus™ Real-Time PCR System (Life Technologies). Gene-specific primers were designed in order to amplify a segment of each gene (see Table 1 in Duplat-Bermúdez et al., in press). To normalize the mRNA expression, Arabidopsis 18S rRNA was used as internal, constitutive gene, and was amplified with the specific primers 18S-F (5′-GTGATGGGGATAGATCATTGCAATTGTTGG-3′) and 18S-R (5′-TGGACTTCTCGCGACGTCGCGGGCGGCG-3′. Relative expression levels were calculated by the (2−ΔΔCt) method (Livak and Schmittgen, 2001). Three technical replicates were made per sample and averaged to calculate relative expression.
Table 1.
Overexpressed genes involved in flower induction in AtFTOE plants.
| Gene | Locus ID | Localization | Description | FC |
|---|---|---|---|---|
| FRUITFULL(FUL)/AGL8 | At5g60910 | Nucleus | MADS-box protein | 2.52 |
| MADS-BOX PROTEIN SOC1 | At2g45660 | Nucleus, cytoplasm |
MADS-box protein. Controls flowering and is required for CO to promote flowering. | 1.89 |
| Agamous-like MADS-box protein AGL3, SEP4, AGAMOUS- LIKE 3 |
At2g03710 | Nucleus | MADS-box protein involved in the development of sepals, petals, stamens and carpels. It also plays a central role in the determination of flower meristem and organ identity. |
2.31 |
| AGAMOUS-LIKE 68,/MAF5 | At5g65080 | Nucleus | Is upregulated during vernalization and regulates flowering time. Encodes MADS-domain protein. |
14.82 |
| AGL42 (FOREVER YOUNG FLOWER) (FYF) |
At5g62165 | Nucleus | MADS-box protein | 3.04 |
| AP2/ERF and B3 domain containing transcription factor RAV1 |
At1g13260 | Nucleus | Encodes an AP2/B3 domain transcription factor | 2.01 |
| FLOWERING LOCUS T | At1g65480 | Nucleus, cytoplasm |
Promotes flowering and acts as a long-range signal | 15.36 |
| Squamosa promoter-binding-like protein 4 SPL4 |
At1g53160 | Nucleus, cytoplasm |
Encodes a member of the SPL (squamosa-promoter binding protein-like) DNA binding proteins and putative transcription factors. Involved in regulation of flowering and vegetative phase change. |
6.8 |
| AP2/B3-like | At3g53310 | Nucleus | AP2/B3-like transcriptional factor family protein | 3.09 |
| CYTOCHROME P450 | At3g20100 | oxidation-reduction process | 3.56 | |
| CELLULOSE SYNTHASE LIKE E1 | AT1g55850 | Plasma membrane |
Encodes a protein similar to cellulose synthase | 3.24 |
| DUF581 | At1g22160 | Mitochondrion | Protein of unknown function | 2.36 |
| At3g28500 | 60S acidic ribosomal protein family. |
Cytoplasm, cytosol, cytosolic ribosome, plasma membrane, ribosome | 17.09 | |
| ATNPF2.13, NITRATE TRANSPORTER 1.7, NPF2.13, NRT1.7, NRT1/PTR FAMILY 2.13 |
At1g69870 | Encodes a low affinity nitrate transporter NRT1.7. Expressed in phloem. Responsible for source-to-sink remobilization of nitrate. |
1.89 | |
| At4g23680 | Polyketide and lipid transport superfamily protein | 6.47 |
3. Results
3.1. Overexpression of FT driven by the KNAT1 promoter induces early flowering under non-inductive short days (SD)
Three independent transformed lines overexpressing FT in vegetative meristem were employed for further studies. The presence of the construct in the tested lines was assessed by PCR, yielding in positive cases a 560 bp (see Fig. 1A in Duplat-Bermúdez et al., in press) corresponding to a fragment that includes the KNAT1 promoter and the FT ORF. Copy number in genome, determined by ddPCR, identified only one copy of the construct per independent line see Fig. 1B in Duplat-Bermúdez et al., in press) The flowering time of these three independent AtFT overexpressing lines was experimentally determined, grown in parallel with wild type Arabidopsis Columbia-0 plants. When plants were grown under non-inductive SD conditions, flowering occurred earlier than wild type (Col-0) plants. Quantification of timing of flowering revealed that >80% of AtFT overexpressor plants (AtFTOE) flowered at day 21 in SD conditions (Fig. 2A). In contrast, wild type plants flowered until day 40. Thus, AtFTOE reduced their flowering time by 19 days, which is half of the normal vegetative growth. T-student test was employed to assess the data, indicating that there is a significant difference (α = 0.05) in the three independent experiments performed.
Fig. 1.
Phenotypic comparison of AtFTOE vs Wild Type (WT) plants. (A) Plants of three independent lines AtFTOE and WT at 21 day-old growing under short days (SD) (8 h light, 16 h dark) (B) Plants AtFTOE 2.1 vs WT plants at 35 day-old under long days (LDs) of 16 h of light (C) Plants WT at 40 day-old starting flowering under long days (LDs) of 16 h of light. (D) Comparison of rosette leaves at 40 days of growth, rosette diameter in wild type plants vs AtFTOE lines under long days (LDs) of 16 h of light.
Fig. 2.
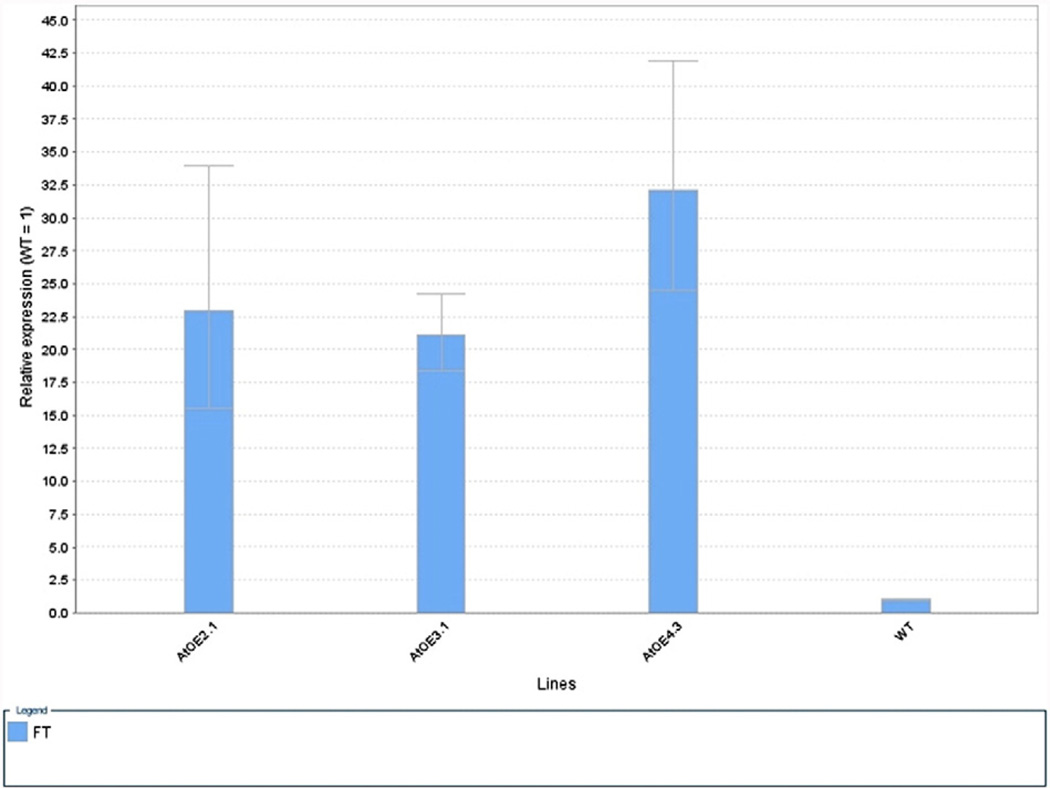
Relative expression profiles of FT in three AtFTOE overexpressors determined by quantitative q-PCR. At 35 day-old plants. Values plotted were normalized to 18S rRNA and are the mean of three biological replicates ± SE.
Morphological differences were quantified during plant development by measuring the number of rosette leaves and in rosette diameter of SD-grown plants (see Fig. 2 in Duplat-Bermúdez et al., in press) No significant differences were observed from germination to day 21 in both WT and AtFTOE lines (Fig. 1A). However, from days 21 to 35 all AtFTOE plants had already transitioned to flowering with 8–9 > 1 mm rosette leaves, while most WT plants were still in the vegetative stage (Fig. 1B) and still producing leaves, until 12–14 leaves developed at day 40. Flowering in wild type Arabidopsis started at 40 days. Stem initial, as indicative of flowering initiation is observed in the plants (Fig. 1C). Rosette leaves paused growth when FT-overexpressing plants started flowering, in contrast with WT, which continued leaf expansion. After 40 days of growth, rosette diameter in wild type plants was significantly larger than AtFTOE lines (Fig. 1D).
3.2. Identification of differentially expressed genes
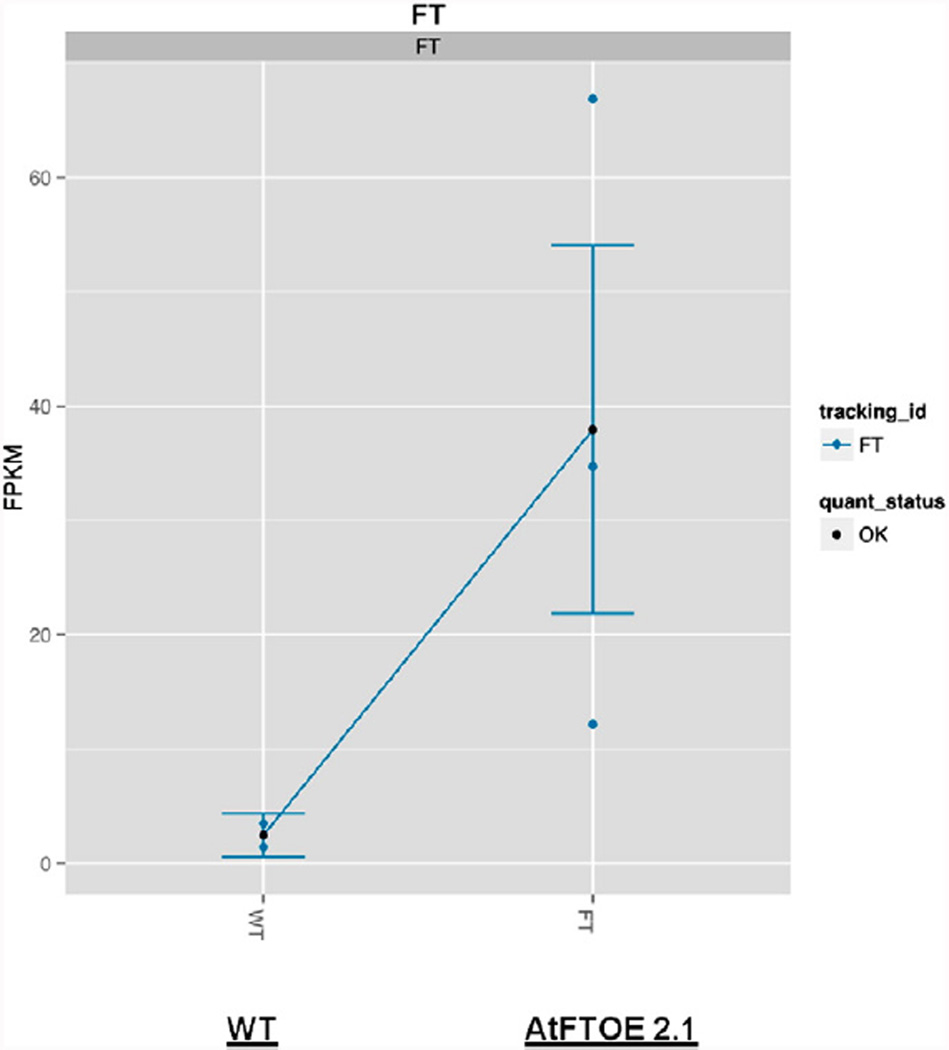
Hyperaccumulation of FT mRNA in three independent AtFTOE lines was determined by qPCR (Fig. 2), confirming the association between the observed early flowering phenotype and the accumulation of FT mRNA. However, to better understand the molecular basis for the early flowering phenotype of AtFTOE, the AtFTOE 2.1 line was selected for further massive RNA sequence analysis (RNA-seq). In order to compare transcript profiles of wild type with AtFTOE, three biological samples were assayed to generate paired-end libraries for the sequencing step. A total of 118,584,576 reads (average length = 106 bp) were generated using Illumina HiSeq2000/2500 PE100 sequencer. Each sample was represented by an average de 21–26 millions reads (see Table 2 in Duplat-Bermúdez et al., in press) The alignment performed to the to the reference Arabidopsis genome using Tophat version 2.0.10 (Trapnell et al., 2009) and Bowtie2 version 2.1.0 (Langmead and Salzberg, 2012) software resulted in read alignment of 60.8% average between replicates of WT and 59.2% for AtFTOE 2.1. Cufflinks program (Trapnell et al., 2010) provided relative abundance values by calculating fragments per kilobase of exon per million fragments mapped (FPKM) (Mortazavi et al., 2008, Mizrachi et al., 2010). The average of FT mRNA FPKM for the three biological replicates for AtFTOE was 38 vs a FPKM of 2, in WT plants, confirming the overexpression of the FT gene in the AtFTOE line (Fig. 3). We identified a total of 3699 differentially expressed genes in rosette leaves of AtFT OE vs WT plants; these data were visualized in a Heatmap. Furthermore a number of isoforms (5107), promoters (3699), CDS (4677), TSS (4504) or sites of alternative splicing (4504) was determined. The differential expression was statistically different with a p < 0.05 in a Student's t-test.
Table 2.
Induced genes in AtFTOE plants involved in sugar transport.
| Genes | Locus ID | Localization | Description | FC |
|---|---|---|---|---|
| Sugar transporter ERD6-like 11 | At3g05165 | Membrane, plasma membrane | Substrate-specific transmembrane transporter activity, carbohydrate transmembrane transporter activity, transporter activity, sugar: hydrogen symporter activity. |
1.68 |
| ATSTP13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
At5g26340 | Cytoplasm, integral component of plasma membrane, membrane, plasmodesma |
Encodes a protein with high affinity, hexose-specific/H+ symporter activity. | 4.59 |
| Sugar transport protein 3, STP3 | At5g61520 | Membrane | Carbohydrate transmembrane transporter activity, sugar: hydrogen symporter activity. |
2.42 |
| A/N-INVC, ALKALINE/NEUTRAL INVERTASE C |
At3g06500 | Extracellular region, mitochondria | Encodes an alkaline/neutral invertase which localizes in mitochondria. | 1.83 |
| POLYOL TRANSPORTER 5 | At3g18830 | Membrane, plasma membrane | Glucose import | 1.63 |
| POLYOL TRANSPORTER 6 | At4g36670 | Membrane, plasma membrane | Glucose import, hexose transmembrane transport | 2.07 |
Fig. 3.
Differential expression of FT measured in fragments per kilobase of transcript per million mapped reads (FPKM) determined by Cufflinks in WT and in the AtFTOE 2.1 line.
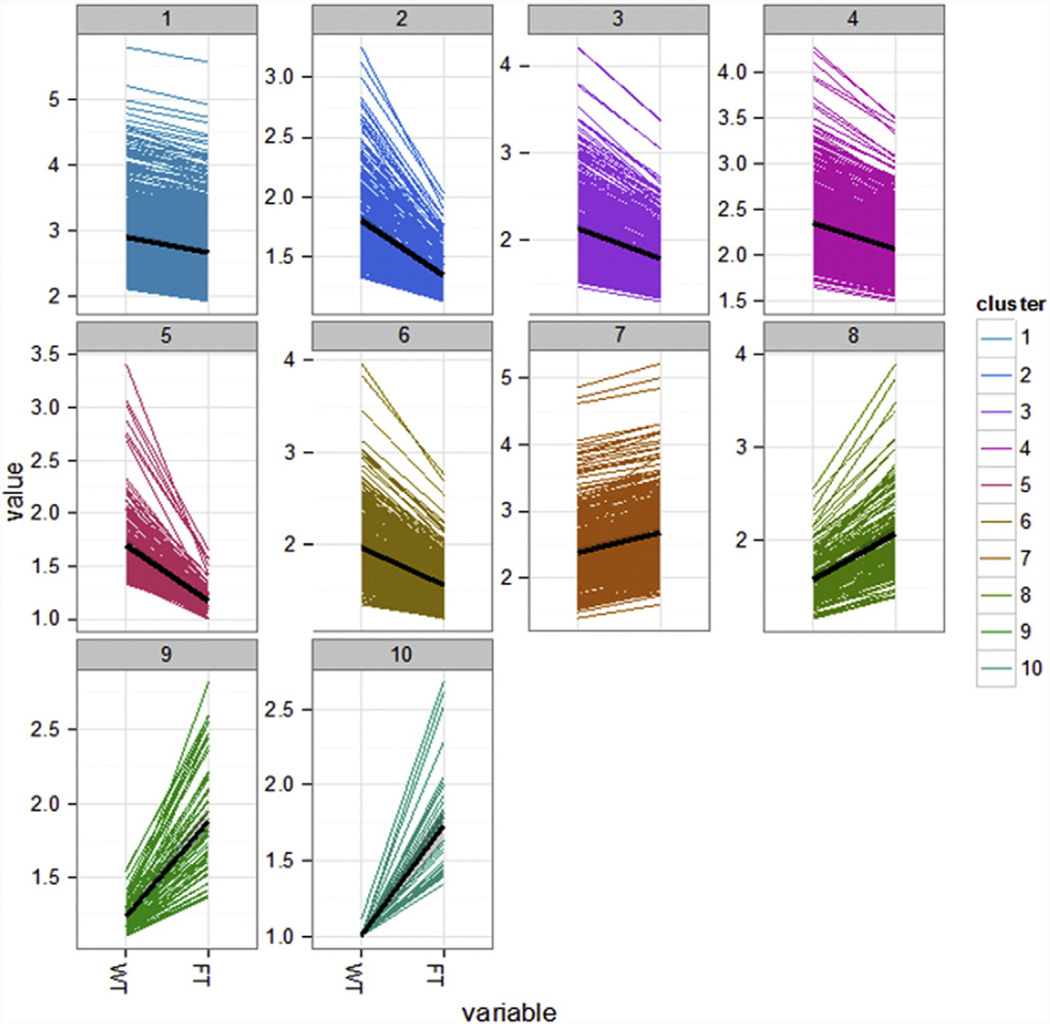
The identification of gene expression clusters calculated by K-means (CummeRbound) yielded 10 clusters four of which were upregulated and 6 downregulated in AtFTOE in comparison with WT (Fig. 4). The genes showing a fold change (FC) with infinite numbers were removed for further analysis (such as cluster 10), resulting in 3652 differentially expressed genes.
Fig. 4.
Clusters of differentially expressed genes obtained by K-means.
3.3. Functional classification by GO
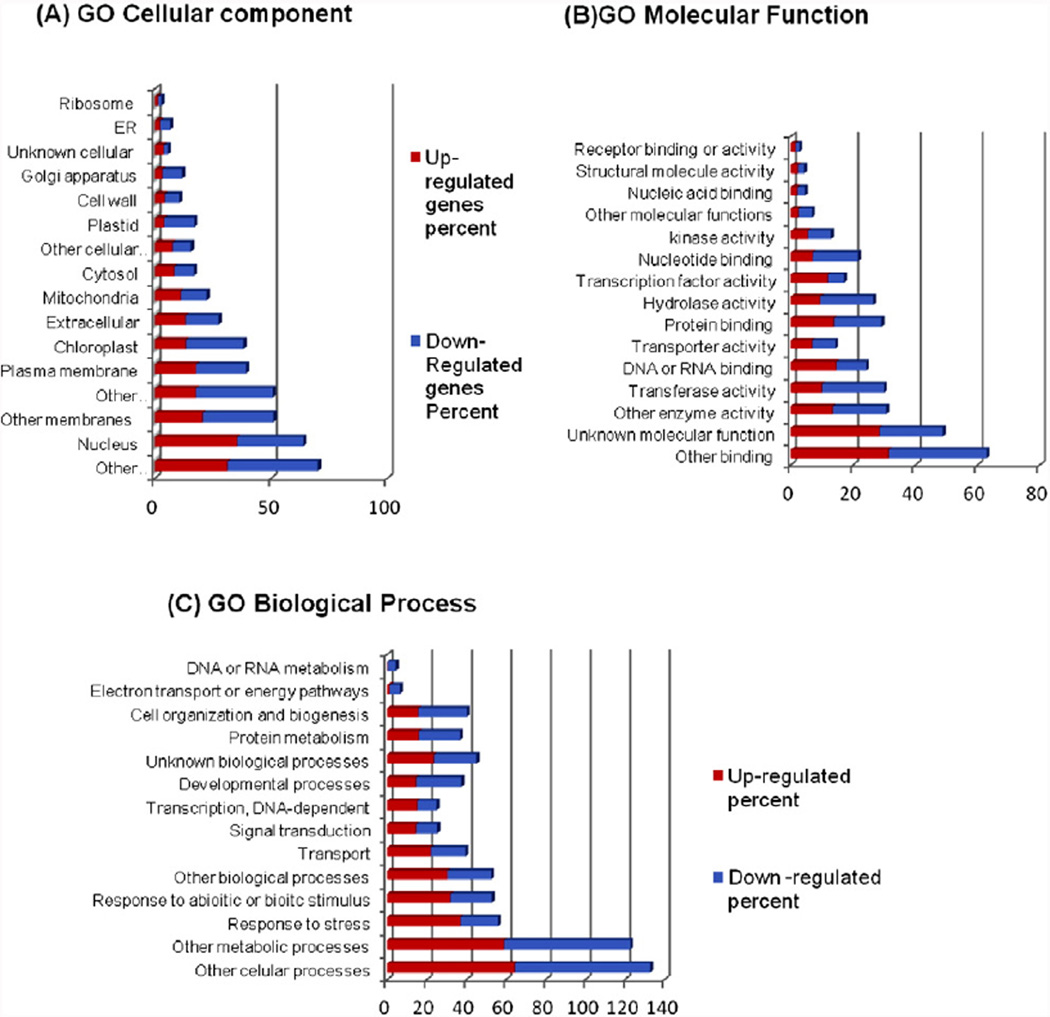
Gene ontology (GO) annotations from the TAIR database (http://www.Arabidopsis.org/tools/bulk/go/index.jsp) were used to analyze the functional annotation of statistically significant differentially expressed genes. To compare the effect of overexpression of FT in rosette leaves of AtFTOE vs WT plants, we analyzed groups of genes for which the Fold Change (FC) was ≥ 1.5 for upregulated and downregulated genes. The RNA-seq analysis showed that overexpression of FT caused changes (p < 0.05) in 3652 differentially expressed genes in rosette leaves of Arabidopsis, which 3026 were down regulated; of these, 2913 were assigned to cellular components, 2782 were assigned to Biological processes, and 2678 genes were assigned to molecular function. 626 genes were upregulated, of which 579 were assigned to cellular components, 572 were assigned to molecular functions and 583 were assigned to biological processes. The functional classification of the differentially expressed genes is shown in Fig. 5A, B, and C, in terms of percentage of the number of significantly changed annotated genes. Interestingly, different transcript accumulation patterns were observed in each cluster. Thus, more genes were downregulated than upregulated in AtFTOE relative to WT plants in the three categories. A total of 579 upregulated and 2913 downregulated genes were grouped the “cellular component” cluster. As mentioned above, there were more downregulated than upregulated transcripts in most of the categories with the exception of “nucleus” and “unknown cellular components”, suggesting changes in transcriptional regulators (Fig. 5A). In the “molecular Function” cluster 572 upregulated genes and 2678 downregulated genes were grouped, being most of them in “other binding”. There are more upregulated than downregulated transcripts in the category “unknown molecular function”, “DNA, RNA binding, “transcription factor activity”, “Structural molecule activity” and “Receptor binding activity” (Fig. 5B). In the GO clustering of “Biological process” most upregulated and downregulated annotated genes were in the categories “other cellular processes” and “other metabolic processes”, and a smaller proportion of these were found grouped in “stress Response”, “response to abiotic or biotic stimulus”, “other biological process”, “transport”, “signal transduction”, “transcription DNA-dependent”, and “unknown biological processes” (Fig. 5C).
Fig. 5.
Distribution of differentially-expressed genes in AtFTOE compared with WT from whole Arabidopsis in several gene ontology (GO) categories. (A) Cellular Component (N = 579 upregulated genes and N = 2913 down-regulated). (B) Molecular Function (N = 572 up-regulated and N = 2678 down-regulated genes) (C) Biological Process (N = 583 up-regulated and N = 2782 down-regulated genes) using GO annotations from the TAIR base.
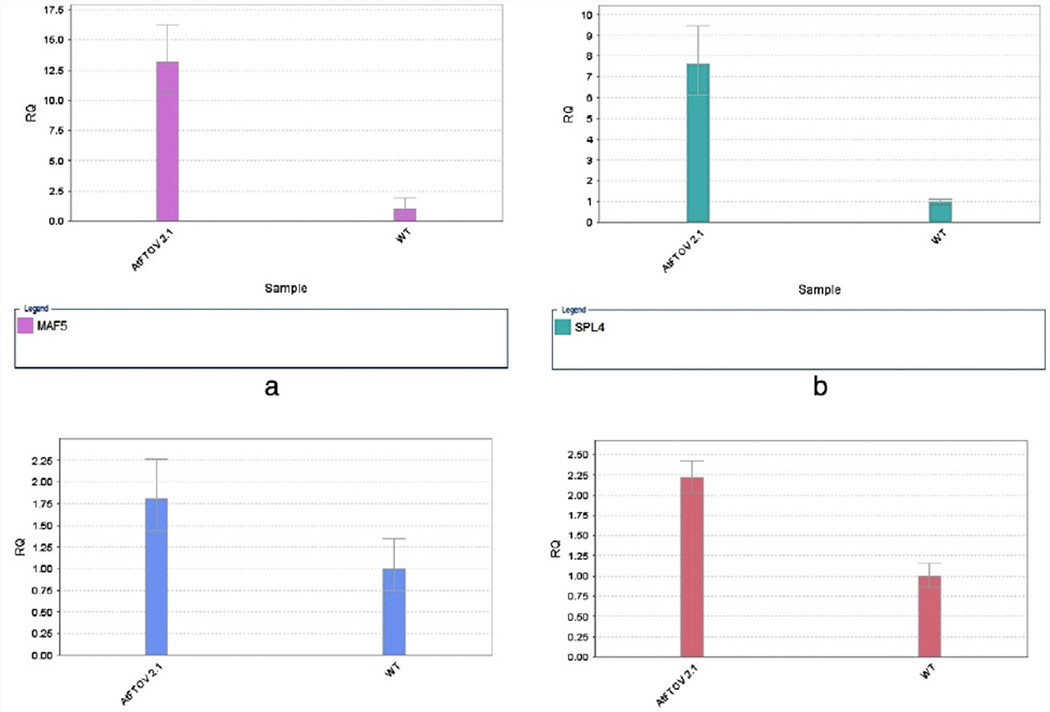
In order to identify the genes grouped on each category, additional analyses were performed with the bioinformatics resource DAVID. The clusters obtained by K-means were analyzed (Fig. 6), focusing first on the clusters with overexpressed transcripts belonging to groups 7, 8 and 9. The last group showed the highest values with 70 transcripts overexpressed with a FC ≥ 4. We determined 83 clusters when we analyzed by DAVID groups 7, 8 and 9 with a total of 626 genes overexpressed with a FC ≥ 1.5. The cluster with genes in the category of flowering processes is represented in Fig. 6 and their identity is presented in Table 1. In this cluster transcripts for five MADS-box proteins were identified: (FUL/AGL8, SOC1, SEP4 (AGL3), AGL42 and MAF5), AP2/B3-like protein (AP2/ERF and B3 domain containing transcription factor RAV1 (AT1g13260), SPL (squamosa-promoter binding protein-like gene family), SPL4, (MAF5) and AP2/ERF and a B3 domain containing transcription factor RAV1. An experimental validation of three differentially expressed genes (MAF5, SPL4 and SOC1) was performed by quantitative real time RT-PCR (Fig. 7A, B, C).
Fig. 6.
Cluster of differentially expressed genes related with flowering obtained by David Bioinformatics Resources 6.7. In this 2D graph it can be observed the genes related with flowering (X axis) and their associated annotation (Y Axis). Green colour corresponds to positively reported gene-term association and black colour corresponds to gene-term association not reported yet.
Fig. 7.
Quantitative Real Time RT-PCR analysis of selected transcripts on AtFTOE relative to WT. (A) MAF5, (B) SPL4, (C) SOC1 and (D) STP3.
A comparison of AtFTOE 2.1 transcript data with that reported by Schmid et al. (2003) allowed us to investigate the response to photoperiod induction at the shoot apex. Twelve common overexpressed genes were identified, of which there are the aforementioned 5 MADS-box containing protein genes, which are involved in floral transition and meristem identity (Dorca-Fornell et al., 2011, Ditta et al., 2004; Smaczniak et al., 2012). We also found other overexpressed genes such as At3g53310, At3g20100, At1g22160, At3g28500, At4g23680, At1g69870 and At1g55850, the function of which is described in Table 1. We found that several genes known to be induced in the shoot meristem upon floral induction were overexpressed in rosette leaves of AtFTOE lines such as (FRUITFULL (FUL)/AGL8), MADS box protein SOC1, and SPL4 (Borner et al., 2000; Hempel et al., 1997). FRUITFULL (FUL)/AGL8) and MADS box protein SOC1 play essential roles in this commitment process and in the induction of flowering downstream of FT signal (Torti et al., 2012; Balanzà et al., 2014; Jung et al., 2012).
Since it is likely that flower induction implies repression of genes involved in maintenance of the vegetative state, it was speculated that potential flowering repressors such as At3g27200 and At5g43270 (SPL2) would be downregulated in AtFTOE (Teaster et al., 2012; Schmid et al., 2003). Indeed, upon comparison of the present RNA co-transcript data with data obtained by Schmid et al. (2003) in the Global Transcriptional Profiling, twenty-eight common floral repressor genes were found to be downregulated (see Table 3 in Duplat-Bermúdez et al., in press). Further studies could help understand the molecular basis of this phenomenon, i.e., the mechanisms by which genes that repress flower induction are upregulated during the vegetative growth stage.
Table 3.
Genes involved in sugar transport repressed in AtFTOE plants.
| Gene | Locus ID | Localization | Description | FC |
|---|---|---|---|---|
| SUC 2 | At1g22710 | Plasma membrane | High-affinity transporter essential for phloem loading and long-distance transport. | 0.60 |
| SUC 3 | At2g02860 | Plasma membrane | Sucrose transporter in sieve elements and a number of sink tissues and cell types. | 0.57 |
| SUC 4 | At1g09960 | Plasma membrane, vacuole | Low affinity (10 mM) sucrose transporter in sieve elements (phloem). | 0.38 |
It is known that genes related with sugar transport have an important role in flowering, the demand of which is related with the physiological stage of the plant (Wang, 2014; Bolouri-Moghaddam and Van den Ende, 2013). Indeed, among transcripts induced in the AtFTOE 2.1 line there are some that encode for hexose transport, specifically glucose and fructose. These genes are STP13, STP3 and Sugar transporter ERD6-like11. Genes At3g06500, corresponding to an invertase and two-polyol transporter 5 and 6 were also overexpressed (Table 2). A validation of differentially expressed gene STP3 was performed by quantitative real time RT-PCR (Fig. 7D). In addition, genes related to sugar transport were downregulated in AtFTOE, namely three sucrose transporter mRNAs: SUC2, SUC3 and SUC4 (Table 3).
In order to visualize the 3652 differentially expressed genes onto cell function diagrams they were subsequently mapped to the Mapman databases (http://mapman.gabipd.org/web/guest/mapman; Ath_AGI_LOCUS_TAIR10_Aug2012–3.m02; Thimm et al., 2004). In the obtained graph (see Fig. 3 in Duplat-Bermúdez et al., in press) are represented 2032 mapped genes from the 3652 genes differentially expressed, being visible 1806 data points. Differential expression can be observed in cell functions such as cell division and cell cycle, DNA synthesis, biotic and abiotic stress, regulation of transcription, RNA processing, protein synthesis and AA activation, development, hormone regulation, protein modification, protein degradation, unclassified no ontology and unknown, enzyme families, redox and transport.
4. Discussion
Floral induction requires a large number of genes; changes in gene expression are expected to be more relevant in the shoot apex than in other tissues. However, it is clear that effects in other tissues upon flower induction do occur. Indeed, overexpression of the floral induction gene FT driven by a constitutive promoter results in premature flowering, which is the result of activation of gene networks in vegetative tissues (Kardailsky et al., 1999; Kobayashi et al., 1999). It is also worth mentioning that certain genes required for floral transition are expressed in leaf and other tissues; for example, potato FT has been recruited during evolution to function in triggering tuber formation (Navarro et al., 2011). In the present research, the meristem-specific promoter KNAT1 (Lincoln et al., 1994, Long et al., 1996, Heyer et al., 2004) was employed to drive the expression of the FT gene. Transformed Arabidopsis plants showing a phenotype with early flowering were compared with WT plants, under non-inductive short day growth conditions. Although morphology, size and number of leaves did not differ from during the first 21 days, afterwards AtFTOE lines started bolting, which paused leaf expansion compared to WT. Indeed, WT plants continued vegetative growth up to day 40, producing 1–14 rosette leaves before flowering. Leaf formation in WT plants occurred as has been reported by Boyes et al. (2001). Afterwards, rosette leaves of WT plants started to grow while AtFTOE leaves stopped growing. Considering that the flower meristem is a strong sink tissue, leaf expansion arrest could constitute a strategy to balance the demand of energy exerted by the floral meristem, thus explaining the smaller size of leaves in AtFTOE plants compared with WT.
Aiming to understand the basis of the molecular responses associated with FT overexpression in leaves driven by a meristem-specific promoter, a transcriptomic analysis by RNA-seq was attempted; its analysis using DAVID software identified a cluster of overexpressed genes in rosette leaves related to flowering, data that was also in agreement with the study of Schmid et al. (2003). Most of the predicted gene products localize to the nucleus, as expected for transcription-associated proteins. Among overexpressed genes, there are five MADS-box domain transcription factors which are known to be important regulators of cellular processes related with flowering time, determination of floral meristem identity, floral organogenesis, fruit formation and endothelium development (Parenicová et al., 2003). However, the mechanism by which MADS-domain proteins activate or repress the expression of their target genes and the nature of their cofactors are still largely unknown (Parenicová et al., 2003; Smaczniak et al., 2012). Interestingly, SUPPRESOR OF OVEREXPRESSION OF CO1 (SOC1), which encodes a MADS box transcription factor which integrates multiple flowering signals derived from photoperiod, temperature, hormone and age-related signals was found to be overexpressed in AtFTOE plants. It is known that SOC1 is activated mainly by FT in the flowering stage and is necessary to establish and maintain flower meristem identity (Lee and Lee, 2010, Schmid et al., 2003). WT contains high levels of SOC1 transcript in leaves under long day relative to short day conditions, and SOC1 mutants show a delay in flowering (Samach et al., 2000; Schmid et al., 2005). This could suggest that the accumulation of SOC1 mRNAs in rosette leaves in AtFTOE lines is also important to maintain the identity of the floral meristem. Another upregulated transcript in rosette leaves is FRUITFULL (FUL/AGL8), a MADS box gene closely related to SOC1 (Schmid et al., 2003; Balanzà et al., 2014). SOC1 and FUL downstream function of FT (Torti et al., 2012) form a heterodimer that may mediate the vegetative and meristem identity transitions (Balanzà et al., 2014). There is an FT-dependent increase in FUL expression in rosette leaves, suggesting that this gene may be involved in changing the fate of leaf primordia during the transition to flowering. It has also been reported the FUL-dependent reduction in leaf size caused by FT and FD (Teper-Bamnolker and Samach, 2005). The overexpression of AP2/ERF and the B3 domain-containing transcription factor RAV1 generates less lateral roots and rosette leaves (Hu et al., 2004). The accumulation of these transcripts in AtFTOE plants could explain in part the smaller size of rosette leaves relative to WT plants.
The differentially accumulated mRNA for the MADS-box transcription factor AGL42 (closely related phylogenetically to SOC1 and directly regulated by this gene) is also involved in the floral transition (Dorca-Fornell et al., 2011). According to available microarray data, AGL42 is expressed in roots, rosette, cauline leaves and inflorescences (Schmid et al., 2005). Another MADS-box overexpressed transcript was AGL3/SEP2, which plays an important role in flower meristem identity (Ditta et al., 2004) and MAF5, which acts as a floral activator (Ratcliffe et al., 2003). The overexpression of one Squamosa promoter binding-like protein SPL4, which has a role in floral transition and belongs to those negatively regulated genes by microRNA156 and microRNA157 was also observed (Schwab et al., 2005). It is possible then that this response is influenced by the miRNA 153-SPL4 module which, at the same time, may regulate the overexpression of floral markers in leaves that maintain the plant in productive stage.
The induction of floral transition markers such as MADS-box and Squamosa Promoter Binding protein Like (SPL) in leaves, which are normally expressed in the shoot apex, is intriguing. However, two facts may account for these observations: 1) the KNAT1 promoter is also active in cambium (and thus leaf tissue) (Lincoln et al., 1994); 2) FT protein has the ability to move cell-to-cell and long distance; it is possible that if this is the case in AtFTOE plants, FT could well be transported outside the cambium into other cell types whereupon induction of the aforementioned genes could occur.
Regarding the downregulated genes in AtFTOE lines, several have been previously suggested to be potential flowering repressors, such as At3g27200 (Teaster et al., 2012) and At5g43270 (SPL2) (Teaster et al., 2012 and Schmid et al., 2003). There are twenty-seven downregulated genes in common with those reported by Schmid et al. (2003) It will be of interest to determine the mechanism through which these genes act as flowering repressors. Interestingly, the products of these downregulated genes in flowering plants are potentially localized mainly in chloroplast and in extracellular components.
Is interesting to highlight that bioinformatics analyses did not show differential gene expression of genes such as CONSTANS (CO), a B-box-containing protein that acts in the promotion of flowering by long days and promote the expression of FT and SOC1, regulating at both the mRNA and protein levels (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000; Suarez-Lopez et al., 2001; Hepworth et al., 2002; Valverde et al., 2004. We found no evidence of the differential expression of genes that code for photoreceptors such as PHYTOCHROME A (PHYA) and CRYPTOCHROMEs (CRY1 and CRY2), which are necessary for the stabilization of CO protein (Valverde et al., 2004; Kim et al., 2008). These results demonstrate that the FT expression is independent of genes associated with photoperiod, as it occurs naturally.
In the present work, the mRNA massive analysis with bioinformatics tools allowed the identification of pathways signaling in plant leaves overexpressing AtFTOE, which showed a phenotype with early flowering when compared with WT Arabidopsis; however, we also consider necessary to perform complementary analyses regarding the energetic cost, because increasing demand of the newly differentiated floral meristem has implications on a heterotrophic tissue that depends on nutrients and sugars. It is also important to consider the fact that when sugars act like signaling molecules or are transported to sink organs they may affect the control of floral transition by activating or inhibiting genes. (Corbesier et al., 1998; Roldan et al., 1999; Ohto et al., 2001; Seo et al., 2011; Yang et al., 2013). Arabidopsis is a plant with major apoplastic movement of photoassimilates, as suggested by the big family of mono- and di-saccharide symporters identified in its genome. In order to understand the demand of this sink tissue to source distant tissues, we identified which genes have been up regulated and down regulated in agreement with apoplastic transport, mono and disaccharide symporters and polyol transporter. Our results show the upregulation of the genes related with monosaccharide transport such as STP13 and STP3 (Table 2) which belong to the family of the Arabidopsis sugar transporter (AtSTP) that mediate the uptake of hexoses from the apoplastic space across the plasma membrane in the cell (Büttner, 2007, 2010). STP3 and STP13 transport glucose but interestingly STP13 also transport fructose (Norholm et al., 2006), the two hexoses that result from the hydrolysis of sucrose by cell wall invertases. La upregulation of these genes and the gene At3g06500 (Table 2) that codifies for an invertase indicate that there is a demand of glucose and fructose possibly generated by the early presence of floral meristems which are strong sink and heterotrophic tissues in AtFTOE plants. Other two upregulated genes were polyol transporter 5 (At3g18830) and 6 (At4g36670) (Table 2) which have the function of transporting polyols and hexoses, mainly glucose and pentoses (ribose). Analysis of transport properties and expression in Arabidopsis indicate that polyol transporters, transport a wide range of sugars into specific sink tissues in the plant (Reinders et al., 2005; Klepek et al., 2005) showing that the AtFTOE plants are in a hexose-demanding stage possibly generated by early flowering and accelerated plant growth. The finding of overexpressed symporters in Arabidopsis is in agreement with the sugar translocation mediated by transporter through the plasma membrane; however, it is not possible to discard the sugar symplasmic movement, mediated by plasmodesmata, a bona fide translocation route of mobile molecules such as FT, responsible of the dramatic change in plant development documented here, to produce flowering in Arabidopsis.
Considering that sucrose is both a metabolite and a signaling molecule and that manipulating the rate of the synthesis, transport or degradation of it affects plant growth, development an physiology (Wind and Smeekens, 2010) is interesting the fact that three sucrose transporters (SUC 2, 3 and 4) (Table 3) were down regulated in AtFTOE plants when compared with WT plants which could indicate that WT plants still require sucrose for leaf growth in this developmental stage (Proveniers, 2013), in contrast with the demand of hexoses in AtFTOE plants.
The bioinformatics tool Mapman performed analyses of differentially regulated genes. For a general overview of cellular function, 2032 genes were mapped and in the graph 1806 data points were visualized (see Fig. 3 in Duplat-Bermúdez et al., in press). Differential expression was observed in genes associated with cell division and cell cycle; considering that we analyzed mRNA from matures leaves, cell division is not a major function in AtFTOE plants as it is in WT plants. The DNA synthesis differential expression is in agreement with this interpretation. We would like to highlight the differential expression of genes associated to biotic and abiotic stress, plant development and regulation. Differential expressions of genes associated to hormone biosynthesis in leaves most likely indicate a supracellular regulation to distant tissues, based in plant regulators or hormones. A differential expression major downregulation was observed in protein synthesis, modification, degradation and transport, probably suggesting the normal turnover is impaired to provide longer half-life to newly synthesized proteins. The redox balance comprised different examples of differential expression, indicating the crucial role of maintaining redox metabolism for the overall metabolic pathways.
5. Conclusion
In the present work, transgenic lines overexpressing Flowering Locus T (AtFTOE) were obtained driven by the meristem-specific promoter KNAT1. AtFTOE displayed an early-flowering phenotype when compared with WT plants. The number of leaves (7–9 > 1 mm) and rosette size did not differ from WT plants up to day 21, when AtFTOE plants started to flower. However, after this time, these plants did not produce more leaves and committed to develop floral structures with equivalent seed productivity, while WT plants produced more expanded leaves before flowering. Transcriptomic analysis of rosette leaves by RNA-seq allowed the identification of 3652 differentially expressed genes in rosette leaves of Arabidopsis including different floral markers such as transcription factors of the type MADS and SPL, located in the nucleus and thus affecting the transcription of selected genes. Differentially expressed genes initiate a cascade of events leading to FT-dependent transcriptional changes in hundreds of genes, the result of which is entrance to reproductive stage in a non-reversible way. Further research is still required to understand the regulatory networks that integrate signals from meristem to rosette leaves to ensure the reproductive success in plants.
Acknowledgments
This work was supported by departmental funds from CINVESTAVIPN, by CONACyT grant nos. 156162 to RR-M and 105985 to BX-C, and a SENASICA-SAGARPA grant to BX-C and RR-M. LD-B was supported by a doctoral fellowship from CONACyT no 388937. This research was partially supported by the Intramural Research Program of the NIH, NLM, NCBI. We are grateful to the members of the laboratory of Plant Biotechnology for critical comments to this manuscript.
Abbreviations
- wt
wild type
- FT
Flowering locus T
- AtFTOE
Arabidopsis FT-overexpressor lines
- SOC1
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- SAM
shoot apical meristem
- LD
long days
- SD
short days
- TSF
TWIN SISTER OF FT
- CC
companion cell
- FUL
FRUITFULL
- AP1
APETALA1
- AM
axillary meristem
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Araki T. FD, a bZIP Protein Mediating Signals from the Floral Pathway Integrator FT at the Shoot Apex. Science. 2005;309(5737):1052–1056. doi: 10.1126/science.1115983. Retrieved from http://science.sciencemag.org/content/309/5737/1052.abstract. [DOI] [PubMed] [Google Scholar]
- Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. http://dx.doi.org/10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Coupland G. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development (Cambridge, England) 2004;131(15):3615–3626. doi: 10.1242/dev.01231. http://dx.doi.org/10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. http://dx.doi.org/10.1038/nrg3291 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Balanzà V, Martínez-Fernández I, Ferrándiz C. Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J. Exp. Bot. 2014;65(4):1193–1203. doi: 10.1093/jxb/ert482. ( http://doi.org/10.1093/jxb/ert482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri-Moghaddam MR, Van den Ende W. Sugars, the clock and transition to flowering. Front. Plant Sci. 2013 Feb;4:22. doi: 10.3389/fpls.2013.00022. http://dx.doi.org/10.3389/fpls.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleißner R, Wisman E, Apel K, Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. http://dx.doi.org/10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill J, Hoffman NE, Davis KR, Görlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M. The monosaccharide transporter -like gene family in Arabidopsis. FEBS Lett. 2007;581(12):2318–2324. doi: 10.1016/j.febslet.2007.03.016. http://dx.doi.org/10.1016/j.febslet.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Büttner M. The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol. 2010;12:35–41. doi: 10.1111/j.1438-8677.2010.00383.x. http://dx.doi.org/10.1111/j.1438-8677.2010.00383.x. [DOI] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Gilliham M. Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods. 2013;9(1):4. doi: 10.1186/1746-4811-9-4. http://dx.doi.org/10.1186/1746-4811-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana, comparison between the wild type and a starchless mutant. Planta. 1998;206:131–137. doi: 10.1007/s004250050383. http://dx.doi.org/10.1007/s004250050383. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Coupland G. FT protein movement contributes to long-distance signaling in floral Induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. http://dx.doi.org/10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Ditta Gary, Pinyopich Anusak, Robles Pedro, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. http://dx.doi.org/10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Dorca-Fornell C, Gregis V, Grandi V, Coupland G, Colombo L, Kater MM. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 2011;67(6):1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x. http://dx.doi.org/10.1111/j.1365-313X.2011.04653.x. [DOI] [PubMed] [Google Scholar]
- Duplat-Bermúdez L, Ruiz-Medrano R, Landsman D, Mariño-Rámirez L, Xoconostle-Cázares B. Bioinformatic data of differentially-expressed genes in Arabidopsis expressing FT in vascular tissues. Gene. 2016 (Data in Brief. in press) [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. SnapShot: control of flowering in Arabidopsis. Cell. 2010;141(3):3–5. doi: 10.1016/j.cell.2010.04.024. http://dx.doi.org/10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Goff L, Trapnell C, Kelley D. cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R package version 2.8.2. 2013 [Google Scholar]
- Graciet E, Wellmer F. Gene networks controlling Arabidopsis thaliana flower development. Tansley review. 2014 doi: 10.1111/nph.12444. http://dx.doi.org/10.1111/nph.12444. [DOI] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel Ma, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development (Cambridge, England) 1997;124(19):3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Antagonistic, Coupland G. Regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002;21(16):4327–4337. doi: 10.1093/emboj/cdf432. http://dx.doi.org/10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer AG, Raap M, Schroeer B, Marty B, Willmitzer L. Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana. Plant J. 2004;39(2):161–169. doi: 10.1111/j.1365-313X.2004.02124.x. http://dx.doi.org/10.1111/j.1365-313X.2004.02124.x. [DOI] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004;14(1):8–15. doi: 10.1038/sj.cr.7290197. http://dx.doi.org/10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37(1):1–13. doi: 10.1093/nar/gkn923. (2009b) http://dx.doi.org/10.1093/nar/gkn923 (Epub 2008 Nov 25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;4(1):44–57. doi: 10.1038/nprot.2008.211. http://dx.doi.org/10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 2007;17(12):1050–1054. doi: 10.1016/j.cub.2007.05.008. http://dx.doi.org/10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jung J-H, Ju Y, Seo PJ, Lee J-H, Park C-M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012;69:577–588. doi: 10.1111/j.1365-313X.2011.04813.x. http://dx.doi.org/10.1111/j.1365-313X.2011.04813.x. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. http://dx.doi.org/10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kim SY, Yu X, Michaels SD. Regulation of CONSTANS and FLOWERING LOCUS T expression in response to changing light quality. Plant Physiol. 2008;148(1):269–279. doi: 10.1104/pp.108.122606. http://dx.doi.org/10.1104/pp.108.122606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepek Y-S, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+–symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell Online. 2005;17(1):204–218. doi: 10.1105/tpc.104.026641. http://dx.doi.org/10.1105/tpc.104.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. http://dx.doi.org/10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991;229:57–66. doi: 10.1007/BF00264213. http://dx.doi.org/10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. http://dx.doi.org/10.1038/nmeth.1923 (Retrieved from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 1995;108:157–162. doi: 10.1104/pp.108.1.157. http://dx.doi.org/10.1104/pp.108.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010;61(9):2247–2254. doi: 10.1093/jxb/erq098. http://dx.doi.org/10.1093/jxb/erq098. [DOI] [PubMed] [Google Scholar]
- Lin M-K, Belanger H, Lee Y-J, Varkonyi-Gasic E, Taoka K-I, Miura E, Lucas WJ. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19(5):1488–1506. doi: 10.1105/tpc.107.051920. http://dx.doi.org/10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6(12):1859–1876. doi: 10.1105/tpc.6.12.1859. http://dx.doi.org/10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods (San Diego, Calif.) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. http://dx.doi.org/10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long J, Moan E, Medford J, Barton K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Lett. Nat. 1996;379:66–69. doi: 10.1038/379066a0. http://dx.doi.org/10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 2007;17(12):1055–1060. doi: 10.1016/j.cub.2007.05.009. http://dx.doi.org/10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Mizrachi E, Hefer CA, Ranik M, Joubert F, Myburg AA. De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genomics. 2010;11:681. doi: 10.1186/1471-2164-11-681. http://dx.doi.org/10.1186/1471-2164-11-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. http://dx.doi.org/10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell. 2002;14(Suppl):S111–S130. doi: 10.1105/tpc.001362. http://dx.doi.org/10.1105/tpc.001362.S112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oro E, Cuellar CA, Tamaki S, Silva J, Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–122. doi: 10.1038/nature10431. http://dx.doi.org/10.1038/nature10431 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Norholm MH, Nour-Eldin HH, Brodersen P, Mundy J, Halkier BA. Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett. 2006;580:2381–2387. doi: 10.1016/j.febslet.2006.03.064. http://dx.doi.org/10.1016/j.febslet.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001;127:252–261. doi: 10.1104/pp.127.1.252. http://dx.doi.org/10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Colombo L. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15(7):1538–1551. doi: 10.1105/tpc.011544. http://dx.doi.org/10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidkowich MS, Klenz JE, Haughn GW. The making of a flower: control of floral meristem identity in Arabidopsis. Trends Plant Sci. 1999;4:64–70. doi: 10.1016/s1360-1385(98)01369-7. http://dx.doi.org/10.1016/S1360-1385(98)01369-7. [DOI] [PubMed] [Google Scholar]
- Proveniers M. Sugars speed up the circle of life. elife. 2013;2:e00625. doi: 10.7554/eLife.00625. http://dx.doi.org/10.7554/eLife.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family:MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15(5):1159–1169. doi: 10.1105/tpc.009506. http://dx.doi.org/10.1105/tpc.009506.mous. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Panshyshyn JA, Ward JM. Analysis of transport activity of Arabidopsis sugar alcohol permease homolog AtPLT5. J. Biol. Chem. 2005;280(2):1594–1602. doi: 10.1074/jbc.M410831200. http://dx.doi.org/10.1074/jbc.M410831200. [DOI] [PubMed] [Google Scholar]
- Roldan M, Gomez-Mena C, Ruiz-Garcia L, Salinas J, Martinez-Zapater JM. Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. 1999;20:581–590. doi: 10.1046/j.1365-313x.1999.00632.x. http://dx.doi.org/10.1046/j.1365-313X.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M, Andrés F, Coupland G. Flowering responses to seasonal cues: what's new? Curr. Opin. Plant Biol. 2014;21:120–127. doi: 10.1016/j.pbi.2014.07.006. http://dx.doi.org/10.1016/j.pbi.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. http://dx.doi.org/10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development (Cambridge, England) 2003;130(24):6001–6012. doi: 10.1242/dev.00842. http://dx.doi.org/10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. http://dx.doi.org/10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. http://dx.doi.org/10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Ryu J, Kang SK, Park C-M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011;65:418–429. doi: 10.1111/j.1365-313X.2010.04432.x. http://dx.doi.org/10.1111/j.1365-313X.2010.04432.x. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muino JM, Blanvillain R, Busscher M, Busscher-Lange J, Kaufmann K. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. 2012;109(5):1560–1565. doi: 10.1073/pnas.1112871109. http://dx.doi.org/10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–1120. doi: 10.1038/35074138. http://dx.doi.org/10.1038/35074138 (Retrieved from) [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. http://dx.doi.org/10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Chihiro Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors or rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. http://dx.doi.org/10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Teaster ND, Keereetaweep, Kilaru JA, Wang Y-S, Tang Y, Tran N-QC, Ayre BG, Chapman KD, Blancaflor EB. Overexpression of fatty acid amide hydrolase induces early flowering in Arabidopsis thaliana. Front. Plant Sci. 2012;3:32. doi: 10.3389/fpls.2012.00032. http://dx.doi.org/10.3389/fpls.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17(10):2661–2675. doi: 10.1105/tpc.105.035766. http://dx.doi.org/10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. http://dx.doi.org/10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Coupland G. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell. 2012;24(2):444–462. doi: 10.1105/tpc.111.092791. http://dx.doi.org/10.1105/tpc.111.092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. http://dx.doi.org/10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, VanBaren MJ. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. http://dx.doi.org/10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. http://dx.doi.org/10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. http://dx.doi.org/10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. http://dx.doi.org/10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Wang J. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014;65(17):4723–4730. doi: 10.1093/jxb/eru246. http://dx.doi.org/10.1093/jxb/eru246. [DOI] [PubMed] [Google Scholar]
- Wigge Pa, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–1059. doi: 10.1126/science.1114358. http://dx.doi.org/10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Wind J, Smeekens S. Sucrose: metabolite and signaling molecule. Phytochemistry. 2010;7:1610–1614. doi: 10.1016/j.phytochem.2010.07.007. http://dx.doi.org/10.1016/j.phytochem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Xu F, Rong X, Huang X, Cheng S. Recent advances of flowering locus T gene in higher plants. Int. J. Mol. Sci. 2012;13(3):3773–3781. doi: 10.3390/ijms13033773. http://dx.doi.org/10.3390/ijms13033773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife. 2013;2 doi: 10.7554/eLife.00260. http://dx.doi.org/10.7554/eLife.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Yoo SY, Lee JS, Ahn JH. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering. Plant Physiol. 2005 Oct;139:770–778. doi: 10.1104/pp.105.066928. http://dx.doi.org/10.1104/pp.105.066928.promotion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-C, Chen C, Rojas M, Daimon Y, Ham B-K, Araki T, Lucas WJ. Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J. Cell Mol. Biol. 2013;75(3):456–468. doi: 10.1111/tpj.12213. http://dx.doi.org/10.1111/tpj.12213. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1(2):641–646. doi: 10.1038/nprot.2006.97. http://dx.doi.org/10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]