Abstract
Phosphorylation of eukaryotic initiation factor 2α (eIF2α) controls transcriptome-wide changes in mRNA translation in stressed cells. While phosphorylated eIF2α (P-eIF2α) attenuates global protein synthesis, mRNAs encoding stress proteins are more efficiently translated. Two eIF2α phosphatases, containing GADD34 and CReP, catalyze P-eIF2α dephosphorylation. The current view of GADD34, whose transcription is stress induced, is that it functions in a feedback loop to resolve cell stress. In contrast, CReP, which is constitutively expressed, controls basal P-eIF2α levels in unstressed cells. Our studies show that GADD34 drives substantial changes in mRNA translation in unstressed cells, particularly targeting the secretome. Following activation of the unfolded protein response (UPR), rapid translation of GADD34 mRNA occurs and GADD34 is essential for UPR progression. In the absence of GADD34, eIF2α phosphorylation is persistently enhanced and the UPR translational program is significantly attenuated. This “stalled” UPR is relieved by the subsequent activation of compensatory mechanisms that include AKT-mediated suppression of PKR-like kinase (PERK) and increased expression of CReP mRNA, partially restoring protein synthesis. Our studies highlight the coordinate regulation of UPR by the GADD34- and CReP-containing eIF2α phosphatases to control cell viability.
INTRODUCTION
The phosphorylation of eukaryotic initiation factor 2α (eIF2α) on serine-51 is a major point of translation control in cells experiencing environmental or metabolic stress (1, 2). Phosphorylated eIF2α inhibits eIF2B, attenuating its capacity to assemble the eIF2-GTP- ternary complex and thereby resulting in the global suppression of mRNA translation. While generally attenuating translation, eIF2α phosphorylation also enhances the translation of mRNAs to promote the expression of proteins required to execute the stress response (3–5). This mode of translational regulation is common to many stresses, including nutrient deprivation, iron deficiency, viral infection, and hypoxia (6), that employ four distinct eIF2α kinases to various degrees (7).
Counteracting the eIF2α kinases are two eIF2α phosphatases, each of which redirects protein phosphatase 1α (PP1α) to dephosphorylate phosphorylated eIF2α (P-eIF2α) (8–10). GADD34, encoded by the Ppp1r15a gene, displays increased expression, mediated by transcription and translation (11, 12), following eIF2α phosphorylation. Thus, GADD34 expression generates a feedback loop that reverses eIF2α phosphorylation (9). A second eIF2α phosphatase is assembled by CReP (encoded by Ppp1r15b), which shares structural homology with GADD34, particularly in the C-terminal PP1α-binding site (10, 13). However, unlike GADD34, CReP protein and mRNA levels are unchanged by stress (10). Thus, current models of stress signaling assign functions for GADD34 only in stressed cells, while CReP is thought to maintain low P-eIF2α levels in unstressed cells.
A major pathway that drives eIF2α phosphorylation is the unfolded protein response (UPR), an adaptive cellular response that is triggered by the accumulation of misfolded proteins in the endoplasmic reticulum (ER) (53, 54). In cells lacking GADD34, eIF2α phosphorylation remains chronically high and blocks the recovery of protein synthesis in later stages of UPR (14, 15). While the prolonged translational repression impairs the expression of key stress proteins, paradoxically, the pharmacological inhibition of GADD34/PP1α activity is remarkably cytoprotective (16, 17) and most likely functions by reducing the oxidative stress that results from the enhanced synthesis and folding of proteins (18). These studies suggest that eIF2α phosphatases play key roles in both the progression and resolution of UPR.
Analyses of GADD34 or CReP knockout mice provide critical insights into the roles of these eIF2α phosphatases. Disruption of the GADD34 gene yields live mice (14, 19, 20) that exhibit mild phenotypes, including modest deficits in hemoglobin synthesis (19) and metabolic dysregulation, namely, enhanced obesity when fed a high-fat diet (21). In contrast, the loss of the mouse CReP gene yields pups that die shortly after birth, displaying severe anemia (20). The combined GADD34/CReP gene knockout yields the most severe phenotype, with no detectable mouse embryos resulting from implantation defects (20). These data suggest that GADD34 and CReP play partially overlapping roles, such that the presence of only one of these eIF2α phosphatases is sufficient for mouse development. On the other hand, the different phenotypes of the GADD34 and CReP null mice also point to specialized roles for these eIF2α phosphatases. Current studies are focused on delineating how these eIF2α phosphatases, particularly the GADD34-containing enzyme, regulate mRNA translation in resting and stressed cells.
We utilized ribosome profiling in wild-type (WT) and GADD34−/− mouse embryonic fibroblasts (MEFs) to delineate the role of GADD34 in transcriptome-wide mRNA translation in resting and stressed cells. We report an unexpected role for GADD34 in the control of eIF2α phosphorylation and mRNA translation in resting cells. When cells were stressed, translation of GADD34 mRNA was rapidly and dramatically increased during early UPR. In the absence of GADD34, ER stress-induced eIF2α phosphorylation was elevated for prolonged periods and resulted in a stalled UPR, in that the molecular hallmarks of early UPR were maintained for many hours. Thus, in the cells that lacked GADD34, UPR progression was delayed due to the severe inhibition of protein synthesis, despite the continued recruitment of ribosomes to transcripts encoding the stress response proteins. At later stages of UPR, alternative mechanisms were activated in the GADD34 null cells suppressing PERK activity and increasing CReP mRNA levels, resulting in the partial reversal of eIF2α phosphorylation and reexpression of key UPR proteins. Together, our results highlighted the essential roles played by GADD34 and CReP in regulating mRNA translation during unstressed conditions and following ER stress.
MATERIALS AND METHODS
Cell culture.
Mouse embryonic fibroblasts (MEFs) were generated from wild-type (WT) and GADD34−/− mice (19) obtained from the Mutant Mouse Regional Resource Center (MMRRC) at the University of North Carolina, Chapel Hill, NC. MEFs were generated using 5 embryos at day 13.5 and immortalized by ectopic expression of simian virus 40 (SV40) large T antigen as previously described (http://ron.cimr.cam.ac.uk/protocols/ImmortalizeMEFs.pdf). CReP+/−, GADD34+/ΔC, and CReP−/−, GADD34+/ΔC MEFs were provided by David Ron, Cambridge Institute for Medical Research, University of Cambridge, United Kingdom.
MEFs were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen/Life Technologies) supplemented with 10% fetal bovine serum (HyClone/GE Healthcare), 100 U/ml penicillin-streptomycin (Gibco/Life Technologies), 1× minimal essential medium (MEM) nonessential amino acids (Gibco/Life Technologies), and 55 μM 2-mercaptoethanol (Sigma) at 37°C in a 5% CO2 incubator. Cells were cultured to 80% to 90% confluence and treated with the ER stress-inducing drugs thapsigargin (Tg) and tunicamycin (Tm) (purchased from Sigma-Aldrich) dissolved in dimethyl sulfoxide (DMSO).
For immunoblotting, cells were washed twice with cold phosphate-buffered saline (PBS) and lysed with radioimmunoprecipitation assay (RIPA) buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (wt/vol) NP-40, 0.1% (wt/vol) SDS, 0.1% (wt/vol) sodium deoxycholate, and 1 mM EDTA, supplemented with a cOmplete mini-protease inhibitor cocktail tablet (Roche) and a PhosSTOP phosphatase inhibitor cocktail tablet (Roche).
For ribosome profiling, cells were treated with 180 μM cycloheximide (CHX), washed with cold PBS, and fractionated between cytosol and endoplasmic reticulum (22, 23). Briefly, the plasma membrane was permeabilized by addition of a buffer containing 100 mM potassium acetate, 25 mM HEPES (pH 7.2), 15 mM MgCl2, 0.03% (wt/vol) digitonin (Calbiochem), 1 mM dithiothreitol (DTT), 50 μg/ml CHX, and 2 mM CaCl2. Digitonin-permeabilized cells were washed with the buffer described above containing 0.004% (wt/vol) digitonin. The ER was solubilized in a buffer containing 200 mM potassium acetate, 25 mM HEPES (pH 7.2), 15 mM MgCl2, 50 μg/ml CHX, 4 mM CaCl2, and 1% (vol/vol) NP-40 or 2% (wt/vol) N-dodecyl β-d-maltoside. All chemicals were purchased from Sigma-Aldrich.
SDS-PAGE and immunoblotting.
Cell lysates were subjected to centrifugation at 10,000 × g for 15 min at 4°C to clear the insoluble fraction. Protein quantification was performed using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). Lysates were heated at 95°C for 5 min in sample buffer containing 375 mM Tris-HCl (pH 6.8), 60% glycerol, 6% SDS, 0.03% (vol/vol) bromophenol blue, and 9% (vol/vol) 2-mercaptoethanol. Equal amounts of total protein were subjected to SDS-PAGE and subsequently transferred to Immun-Blot polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were incubated in Tris-buffered saline (TBS)-Tween containing 5% (wt/vol) bovine serum albumin (BSA) (Sigma-Aldrich) or 5% (wt/vol) nonfat milk (Sigma-Aldrich) prior to addition of primary antibodies followed by the secondary antibody conjugated to horseradish peroxidase (HRP) (Santa Cruz; 1:10,000 dilution). Antigen detection was performed using Pierce ECL Western blotting substrate or SuperSignal West Femto chemiluminescent substrate (Thermo Scientific), the blots were scanned, and band intensities were quantified using NIH ImageJ densitometry software.
The following antibodies were used in the current study: eIF2α (sc-11386; Santa Cruz; 1:500 dilution), phospho-eIF2α (Ser51) (3398; Cell Signaling Technologies; 1:1,500), ATF4 (10835-1-AP; ProteinTech; 1:1,500), CHOP (5554; Cell Signaling Technologies; 1:1,000), GADD34 (sc-825; Santa Cruz; 1:500), phospho-AKT (Ser473) (4060; Cell Signaling Technologies; 1:2,000), AKT (9272; Cell Signaling Technologies; 1:1,000), phospho-p70 S6 kinase (Thr389) (9206; Cell Signaling Technologies; 1:1,000), p70 S6 kinase (9202; Cell Signaling Technologies; 1:1,000), XBP-1 (sc-7160; Santa Cruz; 1:750), cleaved poly(ADP-ribose) polymerase (PARP) (9544; Cell Signaling Technologies; 1:1,000), cleaved caspase-3 (9661; Cell Signaling Technologies; 1:1,000), anti-phospho-PERK (Thr980) (16F8; Cell Signaling Technologies; 1:1,000), and tubulin (T5168; Sigma; 1:10,000).
RNA isolation and qRT-PCR.
Total RNA was extracted from cells and tissues using an RNeasy minikit (Qiagen). The yield and purity were assessed via UV spectrophotometry (NanoDrop 2000 UV-visible (UV-Vis) spectrophotometer; Thermo Scientific). RNA (1 μg) was used for cDNA synthesis performed with an iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR (qRT-PCR) was performed on a CFX96 Touch real-time PCR detection system (Bio-Rad) using SsoFast EvaGreen Supermix (Bio-Rad) and the following primer pairs: for ATF4, ATGGCCGGCTATGGATGAT and CGAAGTCAAACTCTTTCAGATCCATT; for CHOP, GCGACAGAGCCAGAATAACA and GATGCACTTCCTTCTGGAACA; for CReP, TGCTGGAGAAAGATACACCCATA and AATTCTTCCCATGGTCCTTTG; and for beta-actin, GATCTGGCACCACACCTTCT and GGGGTGTTGAAGGTCTCAAA. Transcripts were normalized to β-actin mRNA using the threshold cycle (ΔΔCT) method.
Protein synthesis measurement.
Cells were starved for 30 min by incubation in Met-Cys-deficient DMEM. Cells were incubated for 5 min with 50 μCi/ml of [35S]Met-Cys EasyTag Express protein labeling mix (PerkinElmer), followed by addition of 180 μM CHX. Cells were lysed in buffer containing 1% (wt/vol) CHAPS, 200 mM potassium acetate, and 15 mM HEPES (pH 7.2). Trichloroacetic acid (TCA) was added to a 10% (wt/vol) solution and kept on ice for 20 min. Incorporation of 35S label into TCA-precipitated protein was measured by liquid scintillation counting.
Ribosome profiling and data analyses.
Ribosome profiling was performed essentially as previously described (22, 24). Briefly, cell fractions were adjusted to 100 mM potassium acetate by dilution and were treated with 10 μg/ml micrococcal nuclease (Sigma-Aldrich) for 30 min at 37°C. Ribosomes were isolated by centrifugation over a 500 mM sucrose cushion at 100,000 × g for 30 min using a TLA 100.3 rotor (Beckman-Coulter). RNA from the ribosome pellet was extracted by phenol-chloroform extraction and was treated with 1 U/ml T4 polynucleotide kinase (PNK)–1× PNK buffer (New England BioLabs)–100 mM ATP for 1 h at 37°C. Ribosome footprints were isolated by electrophoresis on a 15% acrylamide gel containing 8 M urea–1× Tris-borate-EDTA (TBE) buffer. Gels were stained with SYBR gold (Invitrogen/Life Technologies). Ribosome footprints were excised, and RNA was extracted by crushing a gel slice in 375 mM sodium acetate (pH 5.2) and freezing to −80°C, followed by consecutive cycles of heating to 95°C and vortex mixing. The RNA samples were filtered through a SpinX column (Costar), and RNA was recovered by ethanol precipitation.
Deep-sequencing libraries were prepared using a NEBNext multiplex small RNA library prep set for Illumina (New England BioLabs) according to the manufacturer's protocol. In parallel, high-throughput RNA sequencing (RNA-seq) libraries were prepared by first depleting rRNAs using a RiboMinus kit (Ambion/Life Technologies), followed by the use of a TruSeq RNA sample preparation kit (Illumina), according to the manufacturer's instructions. Libraries were bar coded and sequenced using an Illumina HiSeq 2500 system by the Duke University's Genome Sequencing and Analysis Core Resource.
Reads were mapped to indexed mouse RefSeq mRNAs, using the longest coding sequence for each gene. The reads were aligned using Bowtie 1.0 (25), allowing for one mismatch, and translation and mRNA levels were quantified by read density within the coding sequence. All libraries were normalized by distribution of ribosomes in the ER or cytosol (22).
Contributions of translation efficiency and mRNA levels to changes in total translation levels were calculated as described previously (26). Coefficients of correlation between overall change in translation and the change in mRNA levels (to determine the relative contribution of mRNA levels) or the change in translation efficiency (to determine the relative contribution of translation efficiency) were then calculated. The percentage of change in total translation attributable to each variable was then calculated as the geometric mean of the data from all replicates of mRNA levels or translational efficiency divided by the geometric mean of the data from all biological replicates of the variables in question.
RNA-seq and ribosome footprinting were undertaken on duplicate cell samples representing each of two distinct genotypes (WT and GADD34−/−), following their treatment with either vehicle (DMSO) or Tg for increasing periods (0, 0.5, 1, 2, and 4 h), with mRNA translation also monitored in two separate subcellular compartments (namely, ER and cytosol). Thus, a total of 40 independent libraries were analyzed, and, where appropriate (e.g., for assessing translation efficiency), the quantitation of all ribosome-protected mRNA fragments was adjusted for the changes in the total levels of individual mRNAs.
Gene ontology enrichment analyses.
For each gene, the log2 change in expression between the control and treatment conditions was calculated. These values were used to calculate the mean change for each set of gene ontology values. Bootstrapping was used to calculate P values, where gene values were shuffled and means of gene ontologies were recalculated (27).
CellTiter-Glo luminescent cell viability assay.
Cells were plated in 96-well plates. After drug treatments, cells were lysed with CellTiter-Glo luminescent cell viability assay reagent (Promega) according to the manufacturer's instructions, and luminescence was read using a Tecan Infinite M200 microplate reader. Percent cell viability was calculated relative to DMSO-treated cells.
Tm administration in mice.
Male mice (13 to 15 weeks old) were housed in a temperature- and humidity-controlled room with light/dark cycles and given free access to standard chow diet and water. Tunicamycin (Tm) injections were performed on 4 to 8 animals per group as previously described (28, 29). Briefly, mice were injected intraperitoneally with Tm (1 mg/kg body weight) or vehicle DMSO dissolved in 200 μl of 150 mM dextrose and were monitored for up to 8 days after injection. All procedures were reviewed and approved by the SingHealth Institutional Animal Care and Use Committee (IACUC).
Histology.
Following CO2 narcosis, kidneys were dissected and fixed in 10% neutral buffered formalin (Sigma) overnight. Samples were embedded in paraffin, sectioned at 5 μm, and used for hematoxylin and eosin (H&E) staining or for immunohistochemistry (IHC) for cleaved caspase-3. For IHC, kidney sections were incubated with rabbit anti-active caspase-3 antibody (G7481; Promega; 1:250 dilution). Sections were stained with 3,3′-diaminobenzidine tetrahydrochloride (DAB), and the nuclei were counterstained with hematoxylin.
Tissue analyses.
Following CO2 narcosis, kidneys were dissected, snap-frozen in liquid nitrogen, and stored at −80°C until analysis or were stored in RNAlater RNA stabilization reagent (Qiagen) according to the manufacturer's instructions. Tissues were lysed in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.05% (wt/vol) SDS, and 1% (wt/vol) NP-40, supplemented with protease and phosphatase inhibitors. Tissue lysates were precleared by centrifugation at 10,000 × g for 15 min at 4°C. Protein quantification and immunoblotting were undertaken as described above.
XBP-1 splicing was assayed by the use of a modified protocol from Marcie Calfon and Heather Harding at Cambridge Institute for Medical Research, University of Cambridge. Briefly, 1 μg of total RNA was used for cDNA synthesis performed with a iScript cDNA synthesis kit (Bio-Rad). Amplification was performed with GoTaq Hot Start polymerase (Promega) using the following primer pair: AAACAGAGTAGCAGCGCAGACTGC and TCCTTCTGGGTAGACCTCTGGGAG. The amplified fragments were resolved on a 2.5% agarose gel, stained with SYBR-Safe (Invitrogen), and detected with ChemiDoc MP (Bio-Rad).
Puromycin labeling.
Cells were treated with 10 μg/ml puromycin for the final 30 min prior to cell lysis. Cells were then lysed in a mixture of 25 mM HEPES (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 5% glycerol, 1 mM Na3VO4, and 1% Triton X-100. Protein concentrations were normalized according to Bradford assay, and samples were processed for SDS-PAGE and immunoblotting with a mouse anti-puromycin antibody (Kerafast 3RH11). Puromycin labeling intensity was quantified using ImageJ.
Nucleotide sequence accession numbers.
All libraries are available on the Gene Expression Omnibus (accession no. GSE69800 and GSE53743).
RESULTS
GADD34 regulates mRNA translation in unstressed MEFs.
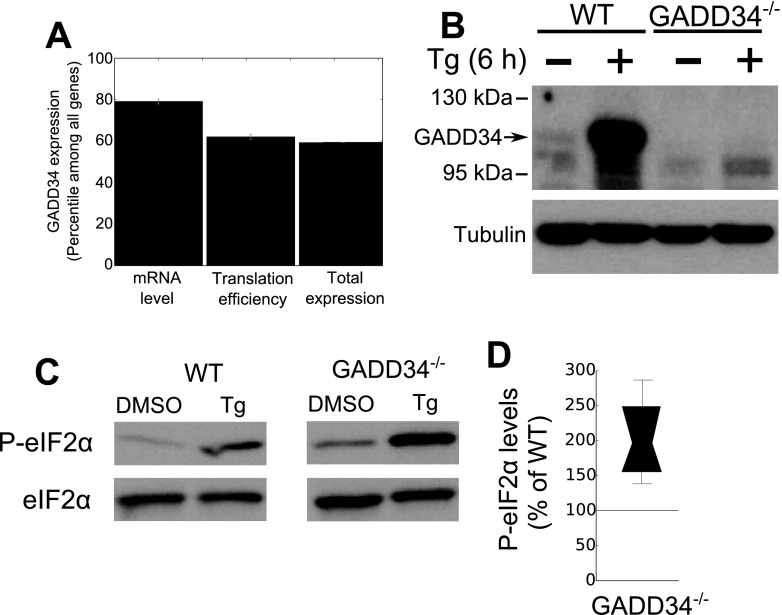
It has been widely reported that GADD34 is detected only in stressed cells. To assess the potential role for GADD34 in unstressed cells, we analyzed RNA-seq data from unstressed MEFs under normal growth conditions (Fig. 1A). GADD34 mRNA was detected at levels comparable to those seen with transcripts, such as mitochondrial ribosomal protein L35 and chloride channel Clcn5, that are constitutively expressed. Parallel experiments established that the GADD34 mRNA was undetectable in the GADD34−/− MEFs. Ribosome profiling also demonstrated that the GADD34 mRNA was actively translated in the unstressed WT cells (Fig. 1A) and, surprisingly, at a higher level than majority of the genes. Moreover, immunoblotting with an anti-GADD34 antibody established the presence of the GADD34 protein in the unstressed WT MEFs (Fig. 1B), albeit at much lower levels than in the cells exposed to ER stress (30). The approximately 100-kDa immunoreactive band represented GADD34 and was not observed in GADD34−/− MEFs, regardless of whether or not the cells were subjected to ER stress.
FIG 1.
Basal expression of GADD34 mRNA and protein. (A) mRNA levels, translational efficiency, and total translation of GADD34 mRNA in WT MEFs were assessed by ribosome profiling and RNA-seq. (B) GADD34 protein levels in unstressed WT MEFs and increased levels in MEFs subjected to ER stress (1 μM Tg for 6 h) were detected by immunoblotting with anti-GADD34 antibody. Lysates from unstressed and stressed GADD34−/− MEFs lacked the immunoreactive band. (C) Levels of eIF2α phosphorylation in WT and GADD34−/− MEFs were detected by immunoblotting with anti-P-eIF2α antibody. Total levels of eIF2α used as a loading control are also shown. (D) Box plot indicating the percentage of change in basal eIF2α phosphorylation in GADD34−/− MEFs relative to WT MEFs assessed by immunoblotting and quantification by ImageJ (n = 5).
To test whether the basal GADD34 protein functioned as an active eIF2α phosphatase, we compared eIF2α phosphorylation in unstressed WT and GADD34−/− MEFs. Basal P-eIF2α levels were approximately doubled (P = 0.014 [paired t test]) in the GADD34−/− MEFs (Fig. 1C and D). These data indicated that the GADD34 protein assembled an active eIF2α phosphatase in the unstressed MEFs.
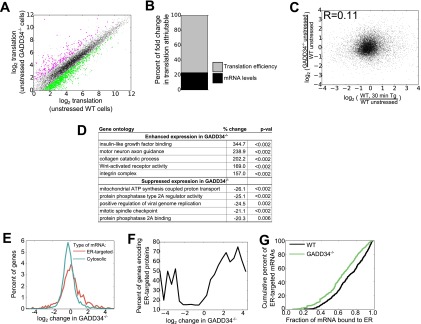
To examine the role of the GADD34-assembled eIF2α phosphatase in the transcriptome-wide gene expression in unstressed cells, we subjected unstressed WT and GADD34−/− MEFs to ribosome profiling and RNA-seq (see Table S1 in the supplemental material). In both cells, the ribosome profiling and RNA-seq data were highly reproducible (see Fig. S1). Ribosome profiling showed that the broad gene expression profile seen in WT MEFs was largely conserved in the GADD34−/− MEFs (Fig. 2A). There was, however, a cohort of mRNAs that showed different levels of translation in unstressed GADD34−/− MEFs compared to WT MEFs. Using a cutoff of a change in translation of at least 2-fold and a P value of <0.05 (Student's t test), we estimated that 414 genes showed enhanced translation whereas 470 genes demonstrated translational suppression selectively in the unstressed GADD34−/− MEFs. To determine whether these changes were driven by translational efficiency or changes in mRNA levels, we employed a previously described mathematical approach (26). Translational efficiency accounted for ∼80% of the change in total translation, while changes in mRNA levels contributed to the remaining ∼20%. The heavy reliance on translational efficiency is consistent with eIF2α phosphorylation being the primary driver of these changes rather than any downstream changes in gene transcription.
FIG 2.
Regulation of protein synthesis by GADD34 in unstressed MEFs. (A) Comparison of total levels of mRNA translation, analyzed by ribosome profiling, in unstressed WT and unstressed GADD34−/− MEFs (n = 2). The shaded area represents 5 standard deviations from the mean of results from WT biological replicates, with genes with significantly different results (at least 2-fold change and P < 0.05 by Student's t test [52]) highlighted in magenta (enhanced translation in the absence of GADD34) or green (suppressed translation in the absence of GADD34). (B) Percentages of changes in total translation in unstressed GADD34−/− cells relative to WT cells attributable to changes in mRNA levels and translation efficiency. Percentages were calculated as described in Materials and Methods. (C) Relationship between changes in translation in unstressed GADD34−/− cells relative to WT cells and changes in translation in WT cells after 30 min of induction of ER stress by the use of 1 μM Tg as reported previously (22). (D) Gene ontologies with changes in total translation in the absence of GADD34, with P values (p-val) determined by bootstrapping. (E) Histogram of differences in total translation between GADD34−/− and WT MEFs for mRNAs encoding ER-targeted proteins (containing signal sequence or transmembrane domain) (red) and cytosolic proteins (green). (F) Moving averages were calculated for the proportions of mRNAs encoding ER-targeted proteins as a function of GADD34-mediated changes in translation. Enrichment among the highly suppressed and highly enhanced genes highlights mRNAs encoding ER-targeted proteins as particularly sensitive to loss of GADD34. (G) Cumulative density plot representing the fraction of each mRNA encoding ER-targeted proteins associated with ER in WT and GADD34−/− cells.
To assess whether the translational changes in unstressed GADD34−/− cells resembled those seen following ER stress, we compared the ribosome profiling data in GADD34−/− cells and WT cells following their treatment with 1 μM thapsigargin (Tg) for 30 min (31). Surprisingly, there was only a weak correlation between the gene expression changes induced by the loss of GADD34 and those induced by UPR activation (Fig. 2C). This highlighted that the loss of GADD34 function generates a cellular response in unstressed cells that is quite distinct from that seen in response to acute ER stress and may result from long-term, low-level increases in P-eIF2α levels in the mutant cells.
Gene ontology analysis of the mRNAs whose translation was enhanced following the loss of GADD34 function highlighted specific classes of genes (Fig. 2D; see also Table S2 in the supplemental material). In these analyses, the mean change in total translation was calculated for all genes in any single ontology category, and P values were calculated by random permutation of gene values. Most notably, genes associated with insulin-like growth factor (IGF) binding showed significantly enhanced expression, with the most prominent among these being the mRNA encoding insulin-like growth factor-binding protein 2 (1,010-fold increase). Motor neuron axonal guidance genes were severely impacted by the absence of GADD34, with a 116-fold increase in the translation of mRNA encoding EPH receptor A4 (Eph4A). Other mRNAs encoding connective tissue growth factor protein (5-fold increase) and cysteine-rich motor neuron 1 protein (5-fold increase) showed more modest increases in their translation. Genes whose expression was reduced by the loss of GADD34 included genes encoding mitochondrial ATPase and genes whose protein products participated in the assembly and regulation of protein phosphatase 2A (PP2A).
GADD34 regulates translation of mRNAs encoding secretory and membrane proteins.
The translation of mRNAs encoding membrane and secretory proteins was particularly sensitive to the loss of GADD34 function. Although the mean changes in the steady-state levels of mRNAs encoding cytosolic and ER proteins (+3% for cytosolic proteins compared to −3% for ER-targeted proteins) were fairly similar, the fold change in translation of mRNAs encoding ER-targeted proteins was greater (Fig. 2E) (log2 variance of 0.89 for cytosolic proteins compared to 2.0 for ER-targeted proteins), with the ER-targeted proteins being represented by a significant fraction of mRNAs that showed either enhanced or reduced translation (Fig. 2F). This illustrated that GADD34 served a critical function in regulating the synthesis of membrane and secretory proteins in unstressed cells.
During acute ER stress, mRNAs encoding ER-targeted proteins were transiently released from the ER (22). This likely serves to reduce synthesis and the subsequent influx of ER-targeted proteins into the ER lumen. In this context, the degree of mRNA release correlated well with the increases in eIF2α phosphorylation, with around 50% of mRNAs released from the ER at the peak of eIF2α phosphorylation. Thus, we investigated whether the elevated basal eIF2α phosphorylation seen in the GADD34−/− MEFs promoted mRNA release from the ER. However, there was only modest (mean of 8.1%; P value = 10−25 [paired t test]) displacement of mRNAs encoding ER-targeted proteins from the ER to the cytosol in the unstressed GADD34−/− MEFs compared to the WT MEFs (Fig. 2G). This suggests that the higher levels of P-eIF2α seen in stressed cells are required to mobilize ER-localized mRNAs.
GADD34 mRNA is rapidly translated in response to ER stress.
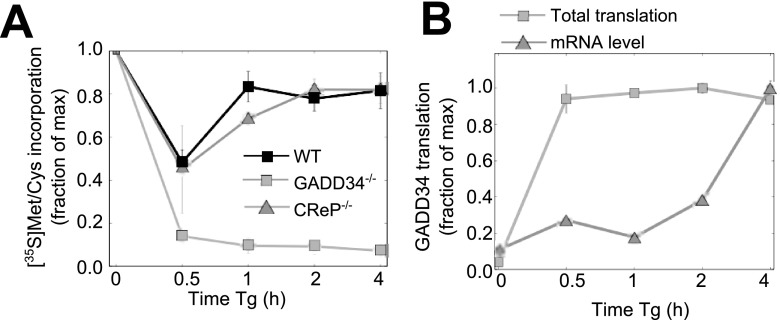
To define a role for basal GADD34 expression in ER stress, we analyzed the temporal changes in protein synthesis using [35S]Met-Cys labeling following Tg treatment (Fig. 3A). In WT cells, there was a rapid and substantial (∼50%) decrease in de novo protein synthesis after 30 min of Tg exposure that had partially recovered by 1 h. In contrast, cells lacking GADD34 displayed a profound (nearly 90% of control) decrement in protein synthesis at 30 min which failed to recover during the following 4 h, when eIF2α phosphorylation remained highly elevated. This hinted at a critical role for GADD34 in restoring protein synthesis in the early stages of UPR, significantly earlier than the well-described ER stress-induced transcription of the GADD34 gene. By comparison, the temporal changes in ER stress-modulated protein synthesis in MEFs lacking a functional CReP gene closely resembled those seen in WT MEFs. This strongly suggested that GADD34 functions as the predominant eIF2α phosphatase controlling translational recovery during the early stages of UPR.
FIG 3.
Functions of GADD34 and CReP in early stages of UPR. (A) Translational activity in WT, GADD34−/−, and CReP−/− cells during Tg-induced ER stress, as measured by pulse-labeling with [35S]Met-Cys (n = 3). Data for WT cells are reproduced from reference 22. max, maximum. (B) Time-dependent changes in GADD34 mRNA and translation in WT MEFs following exposure to 1 μM Tg, assessed by RNA-seq and ribosome profiling (n = 2).
The existence of GADD34 mRNA in unstressed cells and its known translational regulation via upstream open reading frames (uORFs) (12) raised the issue of whether translation of preexisting GADD34 mRNA was enhanced by ER stress. RNA-seq and ribosome profiling of WT MEFs following Tg exposure highlighted the rapid translation of GADD34 mRNA, with ∼20-fold enhancement in translation after 30 min even as the steady-state levels of GADD34 mRNA remained constant for up to 2 h following Tg treatment, after which they rose steadily, consistent with the ER stress-enhanced transcription of the GADD34 gene (Fig. 3B). These data are consistent with earlier studies (12) that combined Tg with the transcriptional inhibitor actinomycin D to suggest that ER stress promoted the translation of the GADD34 mRNA, which in turn made a significant contribution to the cellular levels of GADD34 protein.
Absence of GADD34 stalls the UPR translational program.
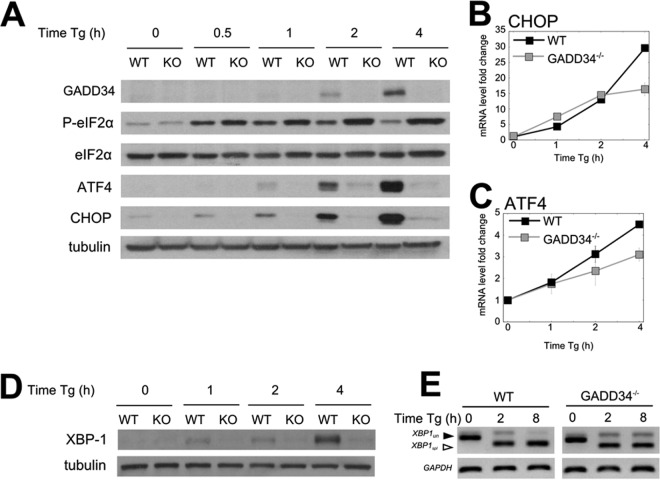
Prior studies suggested that GADD34 was required for the expression of selected stress response proteins (14, 15). To confirm and extend these findings to the entire transcriptome, both WT and GADD34−/− MEFs were treated with Tg. In WT MEFs, a transient increase in eIF2α phosphorylation was observed that peaked at between 30 min and 1 h (Fig. 4A). Significant levels of GADD34 protein were induced by 2 h, resulting in a subsequent decrease in eIF2α phosphorylation that returned to near-baseline levels at 4 h despite the continued Tg exposure. These data supported the well-established role for GADD34 in a feedback loop that dephosphorylates eIF2α and allows translation recovery following ER stress. After 1 h of ER stress, a time-dependent increase in expression of the two hallmark UPR proteins, ATF4 and CHOP, was noted in WT MEFs. In contrast, much higher and sustained levels of eIF2α phosphorylation were observed in Tg-treated GADD34−/− MEFs and were accompanied by the nearly complete loss in expression of these UPR proteins (Fig. 4A). However, levels of mRNAs encoding CHOP (Fig. 4B) and ATF4 (Fig. 4C) increased in response to ER stress in both WT and GADD34−/− MEFs in similar manners. Expression of Xbp-1 protein, another UPR-activated transcription factor, was also significantly impaired in the GADD34−/− MEFs compared to the WT MEFs (Fig. 4D), even as the XBP-1 mRNA levels and splicing results were nearly identical in the WT and GADD34−/− MEFs. Despite the inability of GADD34−/− cells to express these stress response proteins, the profiling data suggested that the ribosomes were heavily recruited to these mRNAs in the GADD34−/− MEFs (see Fig. S2 in the supplemental material). This suggested that the lack of expression of these stress response proteins was primarily due to reduced ribosome scanning or reduced translational activity, attributable to higher P-eIF2α levels in the GADD34−/− MEFs.
FIG 4.
UPR gene expression is suppressed in the absence of GADD34. (A) Levels of UPR-induced proteins analyzed by immunoblotting following Tg treatment of WT and GADD34−/− MEFs. Tubulin is shown as a loading control. KO, knockout. (B and C) mRNA levels of CHOP (B) and ATF4 (C) were analyzed by quantitative PCR (qPCR) in MEFs following Tg exposure (n = 3). (D) Time-dependent changes in the expression of XBP1 protein in WT and GADD34−/− MEFs following Tg treatment. (E) XBP1 splicing following Tg treatment of WT and GADD34−/− MEFs was analyzed by exon-spanning PCR as described in Materials and Methods.
As genetic deletions can result in adaptations with altered expression of other proteins or activation of other pathways (32, 33), we examined the impact of elevating cellular P-eIF2α levels using small-molecule inhibitors of eIF2α phosphatases (see Fig. S3 in the supplemental material). Brief treatment of WT MEFs with Sal003, an analogue of salubrinal that inhibits both GADD34- and CReP-associated eIF2α phosphatases (17), and guanabenz (GBZ), which selectively targets GADD34 (16), enhanced P-eIF2α levels and downstream signaling. Both drugs enhanced expression of ATF4, CHOP, and GADD34, albeit less effectively than Tg, which activates the eIF2α kinase PERK and triggers ER stress. The higher levels of P-eIF2α seen with combined treatments with Tg and GBZ or Tg and Sal003 significantly reduced the expression of ATF4, CHOP, and GADD34. The greater efficacy and unique specificity of GBZ argued that inhibiting GADD34 to increase levels of P-eIF2α, as in the GADD34−/− MEFs, attenuates the expression of UPR genes.
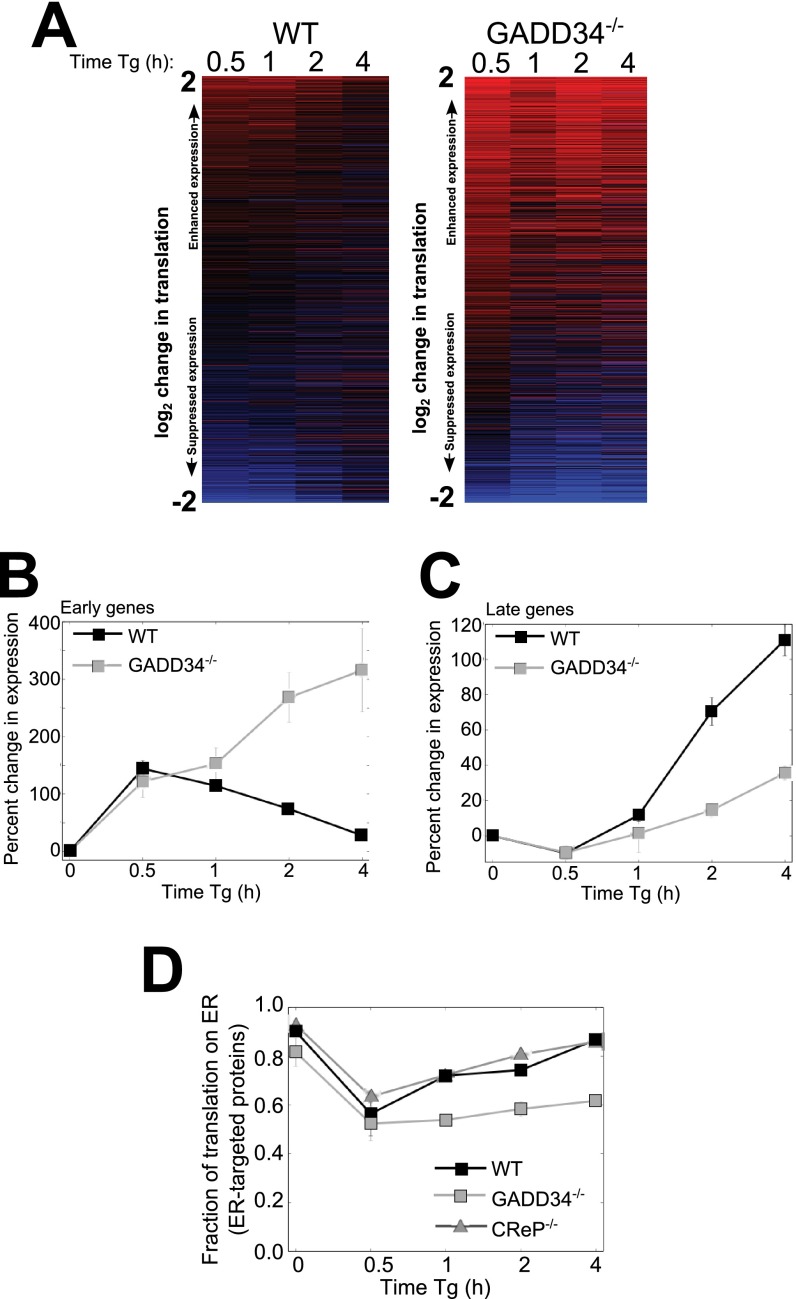
To assess the broader genome-wide impact of loss of GADD34 function on mRNA translation, we analyzed ribosome profiling and RNA-seq in WT and GADD34−/− MEFs over a time course following Tg treatment. It is noteworthy that, while the ribosome profiling data reflect significantly less total protein synthesis in the stressed GADD34−/− cells (Fig. 3A), they also highlight the activity of ribosomes and the stress response programs that remain active in the mutant cells. In the initial analyses, we generated heat maps that provide a snapshot of the temporal changes in mRNA translation. In WT MEFs, there was a major reprogramming of translation over the initial 30 min (Fig. 5A). As UPR progressed, this program changed significantly such that the alterations in mRNA translation after 4 h barely resembled those at early time points. This early-to-late progression in mRNA translation is a hallmark of the UPR stress response (22). Our studies suggested that the early UPR is focused on the expression of proteins that catalyze translational suppression and degradation of misfolded proteins, while during late UPR, ER protein folding capacity is enhanced through the increased synthesis of chaperones (22). In contrast to WT MEFs, GADD34−/− MEFs displayed a gene expression pattern that was constant over time. While the initial reprogramming in mRNA translation seen in WT cells was also noted in the mutant cells following Tg exposure, this response stalled in the absence of GADD34 function.
FIG 5.
GADD34 is required for timely progression of the UPR program. (A) A heat map of changes in total translation following Tg treatment of WT and GADD34−/− MEFs is shown. The mRNAs were sorted based on mean changes in translation levels across all time points and cell types. (B) Median changes in translation of 5% of the genes most highly translationally enhanced after 30 min of Tg treatment in WT cells (n = 2) are shown. (C) Translation responses for 5% of the genes most highly translationally enhanced in WT cells following 4 h of Tg treatment relative to the mean for untreated cells and cells treated with Tg for 30 min (n = 2). (D) Temporal changes in ER enrichment for translation of mRNAs encoding ER-targeted proteins are shown for Tg-treated WT, GADD34−/− (n = 2), and CReP−/− (n = 1) cells.
We then analyzed the translation of early and late UPR genes (listed in Table S3 in the supplemental material) in GADD34−/− cells to understand the nature of the stalled UPR. For genes that were translationally activated after 30 min of Tg treatment, such as those encoding ATF4 and CHOP, their translation peaked early in WT cells and then diminished over time. In contrast, the expression of the same genes continued to increase over time in the GADD34−/− cells (Fig. 5B), presumably due to the sustained eIF2α phosphorylation. On the other hand, genes expressed late in UPR in WT cells were largely suppressed in the GADD34−/− cells (Fig. 5C). These data indicated that in the absence of GADD34, the UPR translational program stalled in a state resembling the early UPR.
However, in the GADD34−/− MEFs, the high, sustained eIF2α phosphorylation resulted in a severe reduction in de novo protein synthesis (Fig. 3A). Therefore, despite the increasing levels seen with ATF4 mRNAs (Fig. 4B) and CHOP mRNAs (Fig. 4C) as well as the increased targeting of ribosomes to these mRNAs (Fig. 5B), the synthesis of core UPR proteins, like ATF4 and CHOP, was largely inhibited (Fig. 4A).
GADD34 regulates ER stress-induced relocalization of ER-bound polysomes.
Our prior studies showed that ER-bound polysomes were released into the cytosol during early UPR and later returned to their ER-bound location (22). To determine the contribution of GADD34 in this process, we analyzed mRNA release from ER in WT and GADD34−/− MEFs following Tg treatment. In WT MEFs, mRNAs encoding ER-targeted proteins were maximally displaced from ER at 30 min but largely recovered during the ensuing 4 h despite the continued exposure of cells to Tg (22). By comparison, in the GADD34−/− MEFs, the released mRNAs failed to return to the ER over the same period (Fig. 5D), consistent with a stall in early UPR. Preliminary analyses in CReP−/− MEFs showed that the time course of displacement of ER-bound polysomes closely resembled that seen in WT cells. These data established that GADD34 function was also essential for returning mRNAs encoding ER-targeted proteins to the ER during UPR.
GADD34-null mice are protected from tunicamycin-induced renal toxicity.
Deficits in UPR signaling were noted in MEFs derived from mutant mice (14, 15), generated by targeting exon 2 (14) or exon 3 (15) in the mouse GADD34 gene. Yet these mice were described as “normal” or “indistinguishable” from WT mice. However, when stressed by a high-fat diet, the exon 2-targeted GADD34 mutant mice showed increased susceptibility to obesity, fatty liver, and insulin resistance (21), although the authors reported that “dephosphorylation of P-eIF2α was not the main cause” of these metabolic alterations. The current report presents the first analysis of UPR signaling in MEFs derived from mice lacking exons 2 and 3, eliminating the entire GADD34 coding sequence (19). These mice display a mild phenotype in the absence of stress, namely, reduced hemoglobin synthesis. When subjected to iron deficiency, the exon 2/exon 3-deleted mutant mice recovered more slowly than WT mice. However, the contribution of reprogramming mRNA translation in mouse tissues to the altered phenotypes has not investigated.
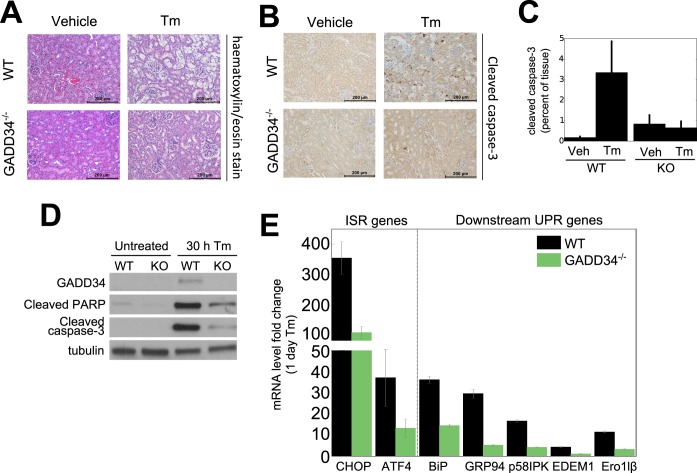
To ascertain whether the UPR stalling or delay seen in MEFs occurred in tissues, we subjected WT and GADD34−/− mice to intraperitoneal injection of tunicamycin (Tm), a documented model of ER stress. As previously noted (28, 29), Tm induced renal lesions, similar to those seen in human acute tubular necrosis, in WT mice in 4 to 6 days (Fig. 6A). In contrast, the GADD34−/− mice were significantly protected from Tm-induced toxicity, with little evidence of renal lesions in 8 days. This was emphasized by immunostaining of renal sections, 4 days after Tm treatment, for cleaved caspase-3, a hallmark of apoptosis. Staining for cleaved caspase-3 was elevated 3-fold to 5-fold in kidney sections from Tm-treated WT mice but was unchanged in the GADD34−/− mice (Fig. 6B and C). Immunoblotting lysates from MEFs exposed to Tm for 30 h also showed enhanced caspase-3 and PARP cleavage in WT MEFs, with little evidence of apoptosis in the GADD34−/− MEFs (Fig. 6D). qRT-PCR analysis of mouse kidneys exposed to Tm for 24 h showed reductions in the levels of a broad array of UPR genes, including ATF4 and CHOP, chaperones and cochaperones (e.g., BiP, GRP94, and p58IPK), ERAD genes (e.g., EDEM1), and the gene encoding the oxidoreductase, Ero1lβ (Fig. 6E), consistent with a stalled or delayed UPR in the mutant mouse kidney. These data established that the molecular hallmarks of delayed UPR were common to GADD34−/− MEFs and mutant mouse kidney, supporting the notion of a role for GADD34 in the propagation of UPR signals and induction of cell death from prolonged ER stress.
FIG 6.
Loss of GADD34 function protects mice against tunicamycin-induced renal toxicity. WT and GADD34−/− mice were injected intraperitoneally with Tm (1 mg/kg body weight) or DMSO as described in Materials and Methods. (A) Representative images (magnification, ×200) of kidney slices from WT and GADD34−/− mice 4 days after Tm administration and after staining with H&E are shown. Scale bar, 200 μm; n = 4 to 12. (B) Paraffin-embedded sections of fixed mouse kidneys were stained with anti-cleaved caspase-3 antibody. Representative images (×200) show cells positive for cleaved caspase-3 (dark brown) at sites of renal lesions observed 4 days after Tm injection. Scale bar, 200 μm; n = 2 to 4. (C) Quantification of cleaved caspase-3 in mouse kidneys as a percentage of the image stained for cleaved caspase-3 using ImageJ. Error bars represent standard deviations (SD) (n = 4). Veh, vehicle. (D) An immunoblot of apoptotic markers in WT and GADD34−/− MEFs 30 h after Tm (2 μg/ml) treatment is shown. (E) RNA was extracted from kidney samples and assayed by qPCR, normalizing against β-actin mRNA. ISR, integrated stress response.
Activation of mechanisms that promote partial recovery from UPR stalling in the absence of GADD34 function.
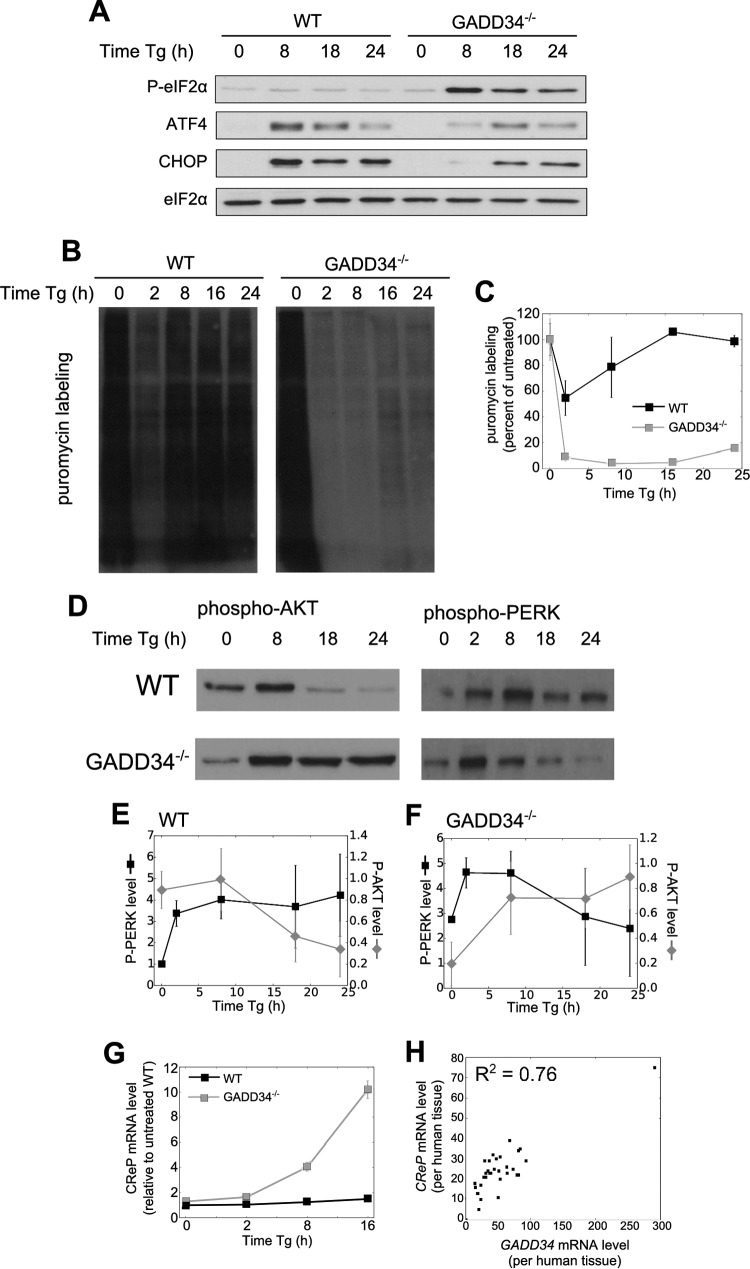
To investigate the long-term consequences of a stalled UPR, we analyzed UPR signaling over a longer time following Tg treatment of WT and GADD34−/− MEFs. In WT cells, eIF2α phosphorylation was largely reversed by 8 h following Tg exposure. At 18 to 24 h, levels of the downstream effectors, ATF4 and CHOP, had significantly receded (Fig. 7A). A markedly different UPR profile was observed in GADD34−/− cells. Most importantly, eIF2α phosphorylation remained high beyond 8 h following Tg exposure but, surprisingly, declined at later time points (Fig. 7A). This decline in eIF2α phosphorylation was associated with the successful execution of UPR signaling. Specifically, expression of ATF4 and CHOP proteins, which was undetectable in GADD34−/− cells in early stages of UPR (Fig. 4A), was clearly visible after 18 h. These data suggested that, rather than causing a terminal stalling of the UPR, the absence of GADD34 delays UPR signaling and hints at the activation of mechanisms that reduce eIF2α phosphorylation and reactivate UPR signaling at later times in the absence of GADD34.
FIG 7.
Late recovery of UPR suppression in GADD34-null MEFs. (A) eIF2α phosphorylation and selected UPR proteins were analyzed by immunoblotting following Tg treatment in WT and GADD34−/− MEFs. (B) Protein synthesis in MEFs treated with Tg was measured by puromycin labeling. MEFs were treated with Tg, and after 30 min with 10 μg/ml puromycin, lysates were analyzed by immunoblotting with an antipuromycin antibody. (C) Quantification of puromycin labeling used ImageJ (n = 3; error bars represent SD). (D) AKT and PERK phosphorylation in WT and GADD34−/− MEFs is shown. (E and F) AKT and PERK phosphorylation was quantified by ImageJ in WT (E) and GADD34−/− (F) MEFs (n = 3; error bars represent SD). (G) CReP mRNA levels were quantified by qRT-PCR following Tg treatment of WT and GADD34−/− MEFs (n = 3; error bars represent SD). (H) Expression levels of GADD34 and CReP mRNA in human tissues from the Human Protein Atlas (38) are shown. Each point represents a different human tissue.
To first demonstrate that the reduction in eIF2α phosphorylation at later times restored protein synthesis, we analyzed WT and GADD34−/− MEFs after Tg treatment by puromycin labeling (Fig. 7B and C). In WT cells, protein synthesis was significantly inhibited after 2 h Tg treatment but had largely recovered by 8 h. In contrast, GADD34−/− cells showed prolonged suppression of protein synthesis. However, a discernible increase in protein synthesis was observed by 24 h, paralleling the reduction in P-eIF2α levels (Fig. 7A). Multiple independent experiments established that protein synthesis at 24 h recovered to approximately 16% of that in control untreated cells and significantly above the 4% seen at 8 h of Tg treatment (P < 0.05 [Student's t test]). This partial recovery in protein synthesis was sufficient to drive the expression of ATF4 and CHOP at 18 and 24 h.
To elucidate mechanisms that contribute to reversal of the P-eIF2α phenotype in GADD34−/− MEFs, we analyzed ER stress-induced activation of AKT, which has been implicated in PERK inhibition (34–36). In WT cells, AKT activity, monitored by phosphorylation of serine-473, was transiently increased following 8 h of Tg treatment (Fig. 7D and E), thereafter declining to low levels at 18 and 24 h. The temporal changes in ER stress-stimulated AKT activity paralleled changes in P-eIF2α, as previously reported (36). Immunoblotting for phospho-PERK showed that PERK activity was rapidly increased by Tg treatment in WT cells and that the increase was maintained up to 24 h (Fig. 7D and E). In contrast, in GADD34−/− cells, AKT phosphorylation, which was elevated by Tg, remained persistently high up to 24 h. This contrasted with PERK activation by Tg in the GADD34−/− MEFs, which peaked at 2 h but declined after 8 h to reach resting levels by 24 h. These results are consistent with the AKT-mediated phosphorylation and inactivation of PERK (35), likely contributing to the reduction in levels of P-eIF2α and expression of ATF4 and CHOP in later stages of UPR (Fig. 7A).
While earlier studies showed that CReP protein levels did not change with the exposure of cells to stress (10), recent work has suggested that ER stress-mediated IRE1α activation enhances degradation of the CReP mRNA (37). This in turn reduces CReP protein, increases eIF2α phosphorylation, and attenuates protein synthesis. Thus, we analyzed CReP mRNA in Tg-treated cells by RT-PCR. Our data showed that CReP mRNA levels were constant in Tg-treated WT MEFs over 16 h (Fig. 7G). However, in Tg-treated GADD34−/− cells, a 10-fold increase in the level of CReP mRNA was observed over the same period. Using available antibodies against human CReP, we were unable to detect a band specific for mouse CReP in WT or GADD34−/− MEFs and thus were unable to address the changes in CReP protein levels in the GADD34−/− MEFs, but ribosome profiling suggests that CReP mRNA was actively translated in GADD34−/− MEFs over the period analyzed. Regardless, our data hint at enhanced transcription of the CReP gene or stabilization of CReP mRNA by ER stress, particularly in the absence of GADD34, and suggest coordinate regulation of the two eIF2α phosphatases.
To examine GADD34 and CReP expression in mammalian tissues, we queried the Human Protein Atlas for GADD34 and CReP mRNAs (38). According to the current UPR model, GADD34 is encoded by an inducible gene whereas CReP is constitutively expressed. Thus, we anticipated that CReP mRNA levels would be constant while GADD34 mRNA levels would differ widely across tissues. Surprisingly, we observed a positive correlation (R2 = 0.76) between GADD34 and CReP mRNA levels, although the levels of the GADD34 transcript were higher in most tissues. This supports the notion that the two genes encoding essential components of eIF2α phosphatases are coordinately regulated.
DISCUSSION
Based on the common observations that GADD34 is a stress-induced protein that is virtually undetected in unstressed cells and that CReP is a constitutively expressed repressor of eIF2α phosphorylation, most current discussions of UPR segregate CReP functions to maintaining low P-eIF2α levels in unstressed cells while assigning a role for GADD34 in reversing stress-induced increases in P-eIF2α levels. In this regard, the current studies expanded knowledge of the role of GADD34, whose mRNA was actively translated in unstressed cells. Our data showed that while at low levels, the GADD34 protein maintained low P-eIF2α levels in resting or unstressed cells. This contrasts with the lack of change in basal P-eIF2α levels following small interfering RNA (siRNA)-mediated knockdown of CReP, although peak P-eIF2α levels in stressed cells were significantly elevated by reducing cellular CReP levels (10). Our data also showed that GADD34 plays a critical role in the recovery from transient repression of de novo protein synthesis and restored ER-bound mRNAs that had been released into cytosol during the early stages of the UPR. As we also observed increases in CReP mRNA levels during late stages of UPR in GADD34−/− MEFs, our studies argued for a revision of the current UPR model to ascribe functions for both GADD34 and CReP in regulating P-eIF2α and mRNA translation in resting and stressed cells.
Ribosome profiling highlighted nearly 900 mRNAs whose translation is regulated by GADD34 in the absence of stress. Thus, the loss of basal GADD34 results in substantial translational reprogramming, with a particular focus on the secretome. The mRNAs whose translation was sensitive to loss of GADD34 function encoded proteins implicated in numerous diseases associated with the UPR, including metabolic disease (39), neurodegenerative disorders (40), and mitochondrial dysfunction. Thus, the absence of GADD34 enhanced translation of the mRNA encoding IGF-binding protein 2 over 1,000-fold and prior work showed that elevated expression of IGF-binding protein 2 protected mice from obesity and insulin resistance (41). Translation of the mRNA encoding Eph4A, a disease modifier whose elevated expression correlates with vulnerability of motor neurons to amyotrophic lateral sclerosis (42), was also enhanced over 100-fold by loss of GADD34. These data pointed to altered UPR signaling and possibly changes in GADD34 expression as factors contributing to aging-related chronic diseases. Analysis of mRNA translation in unstressed cells also highlighted the number of mRNAs encoding proteins involved in assembly or regulation of protein phosphatase-2A (PP2A). These represented a class of mRNAs whose translation required the low levels of GADD34 seen in unstressed cells, and the higher levels of GADD34 observed in stressed cells did not further enhance their translation, highlighting a role for GADD34 solely in the basal expression of these proteins.
The transcriptome-wide translational profile of early UPR in WT MEFs was extended for long periods in the GADD34−/− MEFs. Similar hallmarks of UPR stalling were also seen with the pharmacological inhibition of GADD34 in WT cells, suggesting that high P-eIF2α levels and severe suppression of protein synthesis attenuated UPR progression. The stalled UPR seen in GADD34−/− cells did not persist indefinitely but triggered changes in other UPR-activated mechanisms to reduce levels of P-eIF2α later during UPR, and global protein synthesis recovered from a low of 4% seen with the control at 8 h to approximately 16% by 24 h in stressed GADD34−/− MEFs. The expression of ATF4 and CHOP recovered to a much greater extent, equivalent to the results seen with Tg-treated WT MEFs at 24 h. This suggested that the translational recovery in late UPR was weighted in favor of restoring stress response proteins.
In considering mechanisms for translational recovery, we analyzed AKT, which was activated by ER stress (34–36) and, by phosphorylating threonine-799, inhibited PERK activity (35). In WT cells, the transient AKT activation had no impact on cellular phospho-PERK levels, which remained high over the 24 h of Tg exposure. In contrast, the AKT activation was higher and more prolonged in the GADD34−/− MEFs and was accompanied by the inhibition of PERK and reductions in P-eIF2α levels during the late UPR.
Consistent with previous studies (10), there was no change in CReP mRNA levels over 24 h of Tg exposure in the WT MEFS. However, loss of GADD34 function resulted in a stress-induced (more than 10-fold) increase in CReP mRNA levels above those seen with WT cells. These and other data mandate a redefinition of CReP as a constitutive repressor of eIF2α phosphorylation, as ER stressors also increased CReP mRNA levels in rat INS-1E cells in a PERK-dependent manner (43). In contrast, tunicamycin-induced IRE-1 activation decreased CReP mRNA and protein levels over 12 h in other cells (37). Finally, a mutation (R658C) in human CReP that attenuates protein phosphatase-1 binding was accompanied by a compensatory increase in CReP mRNA and protein levels in lymphoblasts of affected individuals (44). As GADD34 was also highly induced by ER stress in the CReP−/− MEFs (20), we speculate that there is coordinated control of cellular GADDD34 and CReP levels. Indeed, analysis of GADD34 and CReP expression in human tissues supported the idea of the conditional coregulation (32, 33) of these eIF2α phosphatases.
Despite our findings that GADD34 regulated mRNA translation in unstressed and stressed cells, mice lacking GADD34 are remarkably devoid of serious physiological defects (14, 19, 20). Moreover, when subjected to stress, the GADD34−/− mice show both adverse effects, such as accelerated obesity on a high-fat diet (21) and delayed recovery from iron deficiency (19), and beneficial outcomes, such as resistance to tunicamycin-induced renal toxicity (29; this study). These paradoxical findings reflect the heterogeneity in UPR signaling in mammalian tissues, which is highlighted by mutations in other mouse UPR genes (45). Striking heterogeneity in UPR signaling is seen in mouse models of amyotrophic lateral sclerosis (ALS), where motor neurons can be segregated as vulnerable or resistant based on activation of the UPR, specifically, of markers such as PERK, P-eIFα, and ATF4, despite all neurons expressing the mutant superoxide dismutase (SOD1) and possessing ubiquitin-conjugated protein deposits (44). One potential factor distinguishing the vulnerable and resistant neurons is their secretory load, or the production and export of synaptic vesicles. Consistent with identification of GADD34 as a major regulator of the secretome, haploinsufficiency of GADD34 ameliorated motor neuron disease in mutant SOD1 (mSOD1) transgenic mice (46). Guanabenz also reduced motor neuron loss in this ALS mouse model (47) with hallmarks of a stalled UPR, namely, reduced expression of Bip and CHOP. While numerous studies linked chronic activation of the PERK–P-eIF2a–ATF pathway with cell death, there is some controversy over the downstream events that signal cell death. Genome-wide chromatin immunoprecipitation-sequencing (ChIP-seq) studies of ATF4 and CHOP (18) found no evidence for the expression of proapoptotic genes and accredited cell death to oxidative stress associated with enhanced protein synthesis, mediated in part by GADD34 (29). In agreement with these studies, we observed no differences in translation of mRNAs encoding pro- or antiapoptotic proteins in WT and GADD34−/− cells. Thus, we hypothesize that carefully staged control of protein synthesis with attenuation of mRNA translation during the early UPR and partial recovery, specifically, expression of UPR proteins, during the late UPR may account for the resistance of GADD34−/− MEFs and mouse kidney to ER stress-induced cell death.
In contrast, the homozygous R658C mutation in human CReP impaired eIF2α phosphatase and was associated with skeletal defects, microcephaly, and early-onset diabetes (43, 48), a phenotype reminiscent of Wolcott-Rallison syndrome, which was associated with mutations in human PERK (49). Thus, increased P-eIF2α levels resulting from CReP mutation and reduced eIF2α phosphorylation resulting from PERK mutations yielded overlapping disease profiles. This emphasizes the precise balance in mRNA translation that is critical for cell viability. Thus, understanding the translational program(s) controlled by CReP, combined with current work, should provide crucial insights into the benefits and liabilities associated with drugs that target GADD34 (16, 50) or CReP (51) or both eIF2α phosphatases (17) to treat human disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank S.S. and C.V.N. laboratory members for helpful discussions and acknowledge the Advanced Molecular Pathology Laboratory, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore, for undertaking histology. We thank Ralph Bunte for the analysis of renal histology.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00190-16.
REFERENCES
- 1.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5:897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 2.Prostko CR, Brostrom MA, Brostrom CO. 1993. Reversible phosphorylation of eukaryotic initiation factor 2 alpha in response to endoplasmic reticular signaling. Mol Cell Biochem 127–128:255–265. [DOI] [PubMed] [Google Scholar]
- 3.Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 4:e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 5.Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11:619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 7.Samuel CE. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem 268:7603–7606. [PubMed] [Google Scholar]
- 8.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. 2001. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol 21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novoa I, Zeng H, Harding HP, Ron D. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jousse C, Oyadomari S, Novoa I, Lu P, Zhang Y, Harding HP, Ron D. 2003. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J Cell Biol 163:767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Hendershot LM. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem 278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 12.Lee YY, Cevallos RC, Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J Biol Chem 284:6661–6673. doi: 10.1074/jbc.M806735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brush MH, Weiser DC, Shenolikar S. 2003. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojima E, Takeuchi A, Haneda M, Yagi A, Hasegawa T, Yamaki K, Takeda K, Akira S, Shimokata K, Isobe K. 2003. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J 17:1573–1575. [DOI] [PubMed] [Google Scholar]
- 15.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J 22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsaytler P, Harding HP, Ron D, Bertolotti A. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 17.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. 2005. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA, Kaufman RJ. 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson AD, Hollander MC, Miller GF, Fornace AJ Jr. 2006. Gadd34 requirement for normal hemoglobin synthesis. Mol Cell Biol 26:1644–1653. doi: 10.1128/MCB.26.5.1644-1653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, Ron D. 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc Natl Acad Sci U S A 106:1832–1837. doi: 10.1073/pnas.0809632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishio N, Isobe K. 2015. GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance. Sci Rep 5:13519. doi: 10.1038/srep13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. 2014. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158:1362–1374. doi: 10.1016/j.cell.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagannathan S, Nwosu C, Nicchitta CV. 2011. Analyzing mRNA localization to the endoplasmic reticulum via cell fractionation. Methods Mol Biol 714:301–321. doi: 10.1007/978-1-61779-005-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid DW, Nicchitta CV. 2012. Primary Role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem 287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jovanovic M, Rooney MS, Mertins P, Przybylski D, Chevrier N, Satija R, Rodriguez EH, Fields AP, Schwartz S, Raychowdhury R, Mumbach MR, Eisenhaure T, Rabani M, Gennert D, Lu D, Delorey T, Weissman JS, Carr SA, Hacohen N, Regev A. 2015. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science 347:1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efron B. 1982. The jackknife, the bootstrap, and other resampling plans. Society for Industrial and Applied Mathematics, Philadelphia, PA. [Google Scholar]
- 28.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev 12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brush MH, Shenolikar S. 2008. Control of cellular GADD34 levels by the 26S proteasome. Mol Cell Biol 28:6989–7000. doi: 10.1128/MCB.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thastrup O, Cullen PJ, Drobak BK, Hanley MR, Dawson AP. 1990. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci U S A 87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafri R, Bar-Even A, Pilpel Y. 2005. Transcription control reprogramming in genetic backup circuits. Nat Genet 37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- 33.Kafri R, Levy M, Pilpel Y. 2006. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A 103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu P, Han Z, Couvillon AD, Exton JH. 2004. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem 279:49420–49429. doi: 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- 35.Mounir Z, Krishnamoorthy JL, Wang S, Papadopoulou B, Campbell S, Muller WJ, Hatzoglou M, Koromilas AE. 2011. Akt determines cell fate through inhibition of the PERK-eIF2alpha phosphorylation pathway. Sci Signal 4:ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan B, Krokowski D, Majumder M, Schmotzer CL, Kimball SR, Merrick WC, Koromilas AE, Hatzoglou M. 2014. Translational Control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α. J Biol Chem 289:12593–12611. doi: 10.1074/jbc.M113.543215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.So JS, Cho S, Min SH, Kimball SR, Lee AH. 2015. IRE1alpha-dependent decay of CReP/Ppp1r15b mRNA increases eukaryotic initiation factor 2alpha phosphorylation and suppresses protein synthesis. Mol Cell Biol 35:2761–2770. doi: 10.1128/MCB.00215-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlén M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. 2015. Proteomics tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 39.Scheuner D, Kaufman RJ. 2008. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev 29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hetz C, Mollereau B. 2014. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci 15:233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- 41.Wheatcroft SB, Kearney MT, Shah AM, Ezzat VA, Miell JR, Modo M, Williams SCR, Cawthorn WP, Medina-Gomez G, Vidal-Puig A, Sethi JK, Crossey PA. 2007. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes 56:285–294. doi: 10.2337/db06-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, Peeters E, Philips T, Goris A, Dubois B, Andersen PM, Al-Chalabi A, Thijs V, Turnley AM, van Vught PW, Veldink JH, Hardiman O, Van Den Bosch L, Gonzalez-Perez P, Van Damme P, Brown RH Jr, van den Berg LH, Robberecht W. 2012. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat Med 18:1418–1422. doi: 10.1038/nm.2901. [DOI] [PubMed] [Google Scholar]
- 43.Abdulkarim B, Nicolino M, Igoillo-Esteve M, Daures M, Romero S, Philippi A, Senee V, Lopes M, Cunha DA, Harding HP, Derbois C, Bendelac N, Hattersley AT, Eizirik DL, Ron D, Cnop M, Julier C. 2015. A missense mutation in PPP1R15B causes a syndrome including diabetes, short stature and microcephaly. Diabetes 64:3951–3962. doi: 10.2337/db15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saxena S, Cabuy E, Caroni P. 2009. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci 12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 45.Fullwood MJ, Zhou W, Shenolikar S. 2012. Targeting phosphorylation of eukaryotic initiation factor-2α to treat human disease. Prog Mol Biol Transl Sci 106:75–106. doi: 10.1016/B978-0-12-396456-4.00005-5. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Popko B, Tixier E, Roos RP. 2014. Guanabenz, which enhances the unfolded protein response, ameliorates mutant SOD1-induced amyotrophic lateral sclerosis. Neurobiol Dis 71:317–324. doi: 10.1016/j.nbd.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang HQ, Ren M, Jiang HZ, Wang J, Zhang J, Yin X, Wang SY, Qi Y, Wang XD, Feng HL. 2014. Guanabenz delays the onset of disease symptoms, extends lifespan, improves motor performance and attenuates motor neuron loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neuroscience 277:132–138. doi: 10.1016/j.neuroscience.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 48.Kernohan KD, Tétreault M, Liwak-Muir U, Geraghty MT, Qin W, Venkateswaran S, Davila J, Care4Rare Canada Consortium, Holcik M, Majewski J, Richer J, Kym M., Boycott KM. 2015. Homozygous mutation in the eukaryotic translation initiation factor 2alpha phosphatase gene, PPP1R15B, is associated with severe microcephaly, short stature and intellectual disability. Hum Mol Gen 24:6293–6300. doi: 10.1093/hmg/ddv337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delépine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. 2000. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet 25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 50.Das I, Krzyzosiak A, Schneider K, Wrabetz L, D'Antonio M, Barry N, Sigurdardottir A, Bertolotti A. 2015. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348:239–242. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Gassart A, Bujisic B, Zaffalon L, Decosterd LA, Di Micco A, Frera G, Tallant R, Martinon F. 2016. An inhibitor of HIV-1 protease modulates constitutive eIF2α dephosphorylation to trigger a specific integrated stress response. Proc Natl Acad Sci U S A 113:E117–E126. doi: 10.1073/pnas.1514076113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Winter JFC. 2013. Using the Student's t-test with extremely small sample sizes. Pract Assess Res Eval 18:1–12. [Google Scholar]
- 53.Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 54.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.