Abstract
Antiangiogenic therapies, such as sunitinib, have revolutionized renal cell carcinoma (RCC) treatment. However, a precarious understanding of how resistance emerges and a lack of tractable experimental systems hinder progress. We evaluated the potential of primary RCC cultures (derived from tumors and tumor grafts) to signal to endothelial cells (EC) and fibroblasts in vitro and to stimulate angiogenesis ex vivo in chorioallantoic membrane (CAM) assays. From 65 patients, 27 primary cultures, including several from patients with sunitinib-resistant RCC, were established. RCC cells supported EC survival in coculture assays and induced angiogenesis in CAM assays. RCC-induced EC survival was sensitive to sunitinib in half of the tumors and was refractory in tumors from resistant patients. Sunitinib sensitivity correlated with vascular endothelial growth factor (VEGF) production. RCC induced paracrine extracellular signal-regulated kinase (ERK) activation in EC which was inhibited by sunitinib in sensitive but not in resistant tumors. As determined by fibroblast growth factor receptor substrate 2 (FRS2) phosphorylation in fibroblasts, RCC broadly induced low-level fibroblast growth factor receptor (FGFR) signaling. Whereas ERK activation in EC was uniformly inhibited by combined VEGF/platelet-derived growth factor (PDGF)/FGF receptor inhibitors, paracrine ERK activation in fibroblasts was blocked in only a fraction of tumors. Our data show that RCC activates EC through VEGF-dependent and -independent pathways, that sunitinib sensitivity correlates with VEGF-mediated ERK activation, and that combined inhibition of VEGF/PDGF/FGF receptors is sufficient to inhibit mitogenic signaling in EC but not in fibroblasts.
INTRODUCTION
Therapies against tumors may be directed at tumor cells or nonmalignant components. Drugs like imatinib or gefitinib were developed to target aberrantly activated growth factor signaling kinases in tumor cells such as Abl or epidermal growth factor receptor (EGFR). In contrast, sunitinib (Su) was developed to inhibit growth factor receptor tyrosine kinases in endothelial cells (EC) and pericytes, which are implicated in angiogenesis.
The most common malignant kidney tumors are renal cell carcinomas (RCCs), and the most common subtype is clear cell renal cell carcinoma (ccRCC). In ccRCC, angiogenesis is directly activated as a consequence of mutations in the tumor cells. ccRCC is characterized by von Hippel-Lindau gene (VHL) inactivation, which is found in over 80% of tumors (1, 2). The VHL protein (pVHL) is a component of a multisubunit ubiquitin ligase complex (3–9) that targets hypoxia-inducible factor alpha (HIFα) subunits for degradation (10–12). The loss of pVHL in tumors stabilizes HIFα, leading to constitutive HIF activation and the upregulation of HIF target genes, including the vascular endothelial growth factor gene (VEGF), encoding an important angiogenic factor (13). This may account, at least in part, for the vascular nature of ccRCC and may explain the unusual sensitivity of this tumor type to antiangiogenic agents.
Tumor-induced angiogenesis may be specifically disrupted by antagonizing ligands or receptors. VEGF activity is blocked by neutralizing antibodies such as bevacizumab (14), while VEGF receptors (VEGFRs) are inhibited by small-molecule ATP analogues, including sunitinib. Antiangiogenic drugs such as bevacizumab and sunitinib are routinely used in the clinic to treat advanced RCC. However, the benefit is modest and resistance uniformly develops. Therapy that includes the use of bevacizumab (in combination with alpha interferon) delays tumor progression by approximately 3 months compared to alpha interferon use alone (15, 16), whereas sunitinib therapy delays progression by approximately 6 months (17, 18). Unfortunately, the majority of patients develop resistance within a year of treatment, and in a small proportion of patients (10% to 20%) resistance occurs de novo, prior to treatment (17, 18).
Little is known about how resistance to antiangiogenic agents, which typically target EC, develops. In contrast, substantial advances have been made in our understanding of resistance to drugs targeting cancer cells. Cancer cells evade targeted therapies by multiple mechanisms. Drugs targeting receptor tyrosine kinases may be rendered ineffective through mutations in (i) the drug target, precluding drug binding, (ii) effector proteins downstream of the drug target, and (iii) signaling proteins in parallel pathways (19). Understanding resistance has been greatly facilitated by the ability to model the process in vitro. Exposure of tumor cells in culture to an inhibitor may result in the emergence of resistant clones, and mechanisms of resistance seen in vitro often reproduce those in patients (20–22).
Unlike cancer cells, EC are unlikely to undergo genetic alterations and acquire mutations conferring resistance. EC respond to paracrine stimulation from other cells, including cancer cells. Thus, cancer cells may induce resistance in EC by altering the microenvironment in which EC reside. The complex interaction of EC and cancer cells poses a challenge for the study of antiangiogenic resistance in vitro.
Various experimental approaches have been developed to probe into the process of resistance to antiangiogenic agents. Welti et al. screened a panel of growth factors for those that overcame sunitinib-mediated inhibition of EC (23). Those authors found that addition of exogenous fibroblast growth factor 2 (FGF2) to cultures of EC treated with sunitinib restored EC proliferation. FGF2 was also identified as a putative mediator of resistance to anti-VEGF signaling agents in a transgenic tumor model involving the expression of a simian virus oncoprotein in pancreatic β-cells (24). Casanovas et al. showed that resistance to anti-VEGFR2 antibodies correlated with FGF2 production and that combined inhibition of VEGF signaling and fibroblast growth factor (FGF) signaling was more effective. Both of those studies showed that FGF2 confers resistance to VEGFR2 inhibitors, but the role of FGF2 in sunitinib resistance in humans is unknown.
MATERIALS AND METHODS
Cell lines.
786-O, A498, and Caki-2 cell lines were obtained from the American Type Culture Collection. Caki-1 cells were obtained from Osamu Ogawa from the Department of Urology, Kyoto University, Japan. Monoclonal 786-O cell lines stably transfected with empty vector (RCC3) or the wild-type VHL gene (WT8) were gifts from William G. Kaelin, Jr., Dana-Farber Cancer Institute, Boston, MA. 786-O cells were maintained in RPMI 1640 (Life Technologies) supplemented with 20 mM HEPES (pH 7.4) and 0.5% glucose; A498 in minimal essential medium (MEM) with Earle's salts and 2 mM glutamine (Life Technologies); and Caki-1 and Caki-2 in McCoy's 5A medium (Life Technologies). HEK293 cells and mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 4,500 mg/ml glucose, l-glutamine, NaHCO3, and pyroxidine (Sigma). All media were supplemented with 10% fetal bovine serum (FBS) (HyClone) and 1% penicillin/streptomycin (P/S) (Sigma) unless otherwise specified. WT8 and RCC3 cells were grown in the same medium used for 786-O supplemented with G418 (Sigma) (1 mg/ml). Human umbilical vein endothelial cells (HUVEC) and human dermal microvascular endothelial cells (HDMEC) were purchased from ScienCell Research Laboratories (Carlsbad, CA) and were maintained in endothelial cell medium (ECM) (ScienCell Research Laboratories; catalog no. 1001) on coated plates. Plates were coated with 0.5% gelatin (Sigma) for 30 min and then with 20% FBS–phosphate-buffered saline (PBS) (Sigma) for 30 min at 37°C. All cells were kept in a humidified incubator (37°C, 5% CO2).
Primary RCC cell line generation and culture.
Primary renal tumors (or metastasis) from patients or mice were kept in transport medium (MEM with Earle's salts and 2 mM glutamine supplemented with 50 μg/ml of gentamicin [Life Technologies], 1% P/S, and 1% amphotericin B [Fungizone] [Life Technologies]) and stored at 4°C for less than 24 h before processing. Tissues were washed twice with 3 to 5 volumes of transport medium, transferred to a 15-cm-diameter petri dish, and minced to pieces <0.5 mm3 in size. Subsequently, 15 ml of tissue digestion medium (transport medium supplemented with 0.1 mg/ml collagenase [Sigma], 0.1 mg/ml hyaluronidase [Sigma], and 20 μg/ml DNase I [Sigma; D5025]) was added. Tissues were incubated in a humidified incubator (37°C, 5% CO2) for 1 to 2 h. Samples were transferred to 15-ml tubes and spun down at 600 × g for 2 min. Pellets were washed with 5 ml of transport medium and spun again. Pellets were resuspended and plated in primary RCC cell medium. Primary RCC cell medium consisted of MEM with Earle's salts and 2 mM glutamine, 10 ng/ml EGF (Life Technologies), 1% MEM nonessential amino acid (CellGro), 0.4 μg/ml hydrocortisone (Sigma), 1% P/S, and 10% FBS. All cells were kept in a humidified incubator (37°C, 5% CO2). The medium was changed every 3 days, and cells were split 1:2 as necessary.
Pleural fluid was processed as soon as it became available. Fluid was divided into aliquots in 50-ml tubes and spun at 600 × g for 5 to 10 min. Cell pellets were resuspended in transport medium and then pooled in one tube and spun again. Cells were plated in primary RCC cell medium. The next day, medium was changed to remove red blood cells, and cell maintenance procedures were similar to those used for the cells generated from solid tissues.
Tumor graft treatment.
Tumor graft experiments were performed as previously described (25) except that mice were treated for 3 days to examine the effect of sunitinib on different tumor cell types. Briefly, when tumors reached 250 to 300 mm3 in size, mice were treated with vehicle (Ve), rapamycin (LC Laboratories), or sunitinib (in the form of sunitinib malate [LC Laboratories]). Rapamycin was administered by intraperitoneal (i.p.) injection every 48 h at 0.5 mg/kg of body weight in a mixture consisting of 5% ethanol, 5% polyethylene glycol 400 (PEG 400) (Sigma), 5% Tween 80 (Sigma), and 85% D5W (5% dextrose in water). Vehicle (5% ethanol, 5% PEG 400, 5% Tween 80, 85% D5W) was administered by i.p. injection every 48 h. Sunitinib was administered by oral gavage every 12 h at 10 mg/kg in 0.5% carboxymethyl cellulose (CMC; Sigma)–D5W. Mice were sacrificed ∼3 h after the last treatment, and tumors were collected and flashed frozen in liquid nitrogen or fixed in 10% buffer formalin phosphate (Fisher Scientific).
In vitro drug treatments and FGF2 neutralization.
RCC cells were plated at 2,000 cells per well in 96-well white clear-bottom plates (Fisher Scientific). The next day, the medium was removed and replaced with fresh medium containing vehicle (dimethyl sulfoxide [DMSO]; Sigma) or sunitinib (1 μM). From the day 3 to day 6, cell proliferation was assessed daily using a CellTiter-Glo kit (Promega) and a PolarStar plate reader (BMG Labtech) according to the instructions of the manufacturers. A similar experimental setup was used for HUVEC and HDMEC. ECM was replaced by EC proliferation assay medium (M199 [Life Technologies] with 10% FBS and 1% P/S)-containing vehicle, sunitinib (100 nM), or dovitinib (LC Laboratories) (500 nM) without or with VEGF (Life Technologies) (100 ng/ml) and/or FGF2 (PeproTech) (50 ng/ml) as indicated. To determine the 50% inhibitory concentration (IC50) of dovitinib for RCC cells, dovitinib was serially diluted in DMSO and cells were incubated for 4 days and assayed as described above. To determine the IC50 of dovitinib for HUVEC, cells were incubated with drug serially diluted in DMSO in the proliferation assay medium supplemented with 50 ng/ml FGF2 for 4 days and assayed as described above.
For FGF2 neutralization experiment, HUVEC were plated as described above. However, only 10 ng/ml of FGF2 was used. Rabbit anti-FGF2 antibodies (PeproTech) or normal rabbit IgG (Santa Cruz Biotechnology) was added according to the recommendation of the manufacturer at the same time as the growth factors and sunitinib.
Coculture assays.
On day 1, HUVEC (EC) were plated at 1.5 × 105 cells/well in 6-well plates, and RCC cells (or HEK293 cells) were plated at 105 cells/Transwell permeable support (Fisher Scientific) (0.4-μm pore size). The next day, both EC and RCC cells were washed once with PBS, and 2.5 ml of coculture medium (M199 with 1% P/S and 5% FBS) was added to the EC. Transwell inserts were placed on 6 wells, and 1.5 ml of coculture medium was added onto Transwell inserts. For drug treatment experiments, coculture medium was supplemented with vehicle, sunitinib (100 nM), or dovitinib (500 nM) as indicated. Cells were kept in a humidified incubator (37°C, 5% CO2) for 3 days. At the time of harvest, cell culture supernatants were collected for enzyme-linked immunosorbent assay (ELISA) and HUVEC numbers were counted using a hemocytometer. The sensitivity was expressed as the ratio of HUVEC numbers in cocultures treated with drug to those in cocultures treated with vehicle. Sensitivity was determined using the t test as described in the Statistics section below.
ELISA.
Levels of secreted VEGF and FGF2 in culture supernatants were determined with Quantikine human VEGF and human basic FGF immunoassay kits (R&D Systems) and a Spectra Max M5 plate reader (Molecular Devices) according to the instructions of the manufacturers.
IHC, immunofluorescence (IF), and IHC followed by IF (IHC-IF).
Immunohistochemistry (IHC) analyses were performed as described previously (26), and images were taken at ×200 magnification using a Nikon camera.
Primary RCC cells were grown in coverslips (Fisher Scientific), fixed with 10% buffered formalin phosphate for 10 min, permeabilized, blocked with 0.5% Tween 20 and 10% bovine serum albumin (BSA) in PBS for 30 min, and incubated with the indicated primary antibodies diluted with 1% BSA in PBS overnight (ON) at 4°C. All washes were performed using PBS. Cy3-conjugated donkey anti-mouse or -rabbit (Jackson ImmnoResearch Laboratories) antibodies were used as secondary antibodies. CAM5.2 (cytokeratin) (1:50) and PAX-2 (1:100) antibodies were purchased from Becton Dickinson and Life Technologies, respectively. Images were taken at ×200 magnification.
A similar protocol was followed for IHC-IF with the following changes: antigens were retrieved by boiling in retrieving buffer (10 mM sodium citrate, pH 6.0) for 30 min and cooling at room temperature for 30 min. After washing and incubating in hydrogen peroxide, tissues were blocked using 5% normal goat serum in Tris-buffered saline (TBS) with 0.1% Tween 20 (TBST) (RPI Research Products). To detect phosphorylation of S6, tissues were incubated with rabbit anti-phospho-S6 S240/244 (P-S6 S240/244; Cell Signaling) at a 1:100 dilution for 1 h at room temperature. To detect phosphorylation of extracellular signal-regulated kinase (ERK), tissues were incubated with rabbit anti-phospho-ERK T202/Y204 (Cell Signaling) at a 1:50 dilution overnight (ON) at 4°C. After counterstaining with hematoxylin was performed, tissues were blocked with 5% BSA in TBST and then a rat anti-mouse CD31 monoclonal antibody (Dianova, Germany) was added at a 1:50 dilution ON at 4°C. Washes were performed using TBST. A Cy3-conjugated donkey anti-rat antibody (Jackson ImmunoResearch Laboratories) was used to detect EC, and DAPI (4′,6-diamidino-2-phenylindole) (Sigma) (0.5 μg/ml) was diluted into the mixture of secondary antibodies to detect nucleus. Images were taken at ×400 magnification using a Nikon camera. Adobe Photoshop CS2 software was used for merging images and Image J for analysis. The level of phosphorylated S6 was expressed as a ratio of phospho-S6+CD31+/CD31+ cells. A similar approach was used for Phospho-ERK. Two mice were used for each treatment, and for each mouse, 10 pictures of random fields were taken. Typically, ∼20 to 40 EC could be identified from each field. Thus, for each treatment, 200 to 400 EC were typically examined.
Collection of conditioned media and EC stimulation.
RCC cells were grown until ∼90% confluence, and then cells were washed with PBS once and MEM with Earle's salts and glutamine was added (no FBS [base medium]). Cells were incubated for 2 to 3 days, and culture supernatants were collected and filtered using a 0.2-μm-pore-size filter and then frozen at −80°C until needed for stimulation of MEFs or HUVEC.
HUVEC were plated at 2 × 105 cell/well in 6-well plate and allowed to grow to ∼85% confluence. To perform RCC-conditioned medium (RCC-CM) stimulation, cells were starved in the base medium supplemented with vehicle or the indicated drug, sunitinib (100 nM), dovitinib (500 nM), or PD173074 (LC Laboratories) (50 nM), for ∼4 h and then stimulated with RCC-CM containing the corresponding drugs for 20 min. As a negative control, cells were stimulated with base medium supplemented with the indicated drugs.
MEFs were plated at 1.5 × 105 cells/well in 6-well plates for ∼24 h. Cells were starved in base medium in the presence of vehicle, sunitinib (200 nM), dovitinib (1 μM), or PD173074 (LC Laboratories) (100 nM) for ∼20 h and then stimulated with RCC-CM containing the corresponding drugs for 20 min. Negative controls for these experiments were similar to those described for HUVEC above.
To perform stimulation using growth factors (insulin, VEGF, or FGF2 or both VEGF and FGF2), MEFs or EC (HUVEC or HDMEC) were plated as described above. For EC, cells were starved for 1 h using stimulating medium (M199 supplemented with 1% FBS and 1% P/S) containing either vehicle or sunitinib. Then, cells were stimulated with VEGF alone or in combination with FGF2 as indicated. To stimulate MEFs, cells were plated and starved as described above and then stimulated with FGF2 (25 ng/ml), VEGF (50 ng/ml), or insulin (Sigma) (300 nM) for 20 min.
Western blot analysis.
Western blot analyses of cell lysates were performed as described previously (27). To make lysates from tumor tissue, frozen tumor pieces (2 mm3) were alternately homogenized in lysis buffer on dry ice and wet ice until no tissue could be seen (28). The homogenates were passed through QIAshedder mini spin columns (Qiagen) twice, and protein concentrations were determined using Bradford assay (Bio-Rad). Lysates were boiled in the presence of sample loading buffer, and Western blot analysis was performed. The following antibodies were purchased from Cell Signaling Technology: phospho-S6 kinase (S6K) (T389), phospho-S6 (S235/236 and S240/244), total S6, phospho-AKT (S473), total AKT, and phospho-FRS2 (Y196). The following antibodies were purchased from Sigma: phospho-ERK (T183/Y185), total ERK, and tubulin.
Quantitative RT-PCR (qRT-PCR).
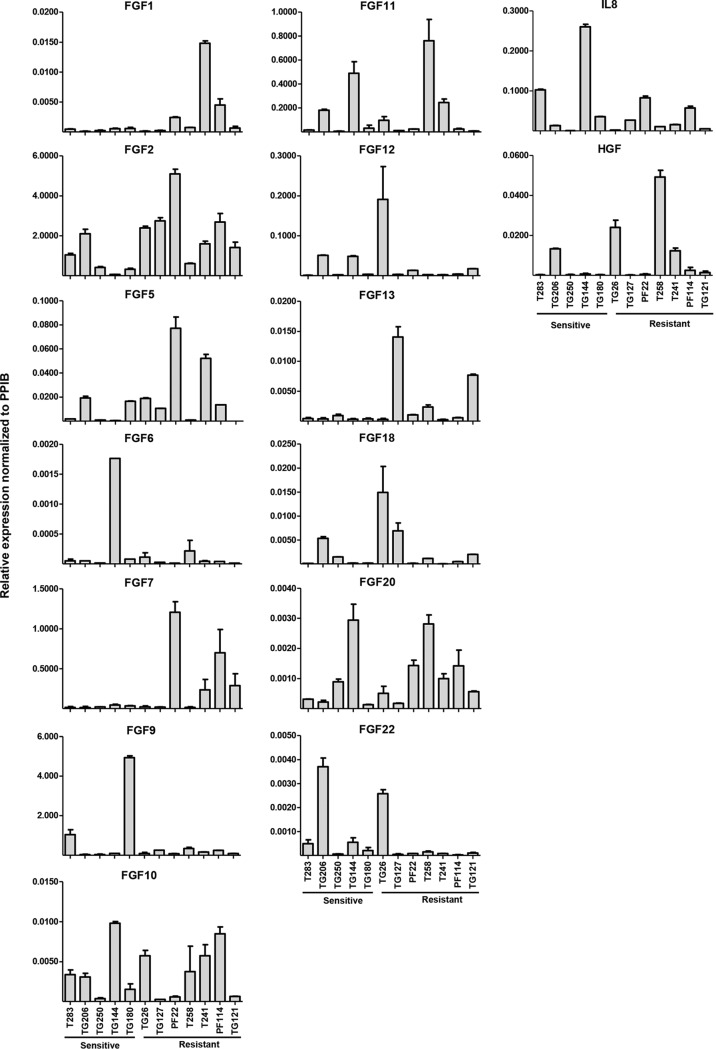
Total RNA from indicated cells was isolated using an RNAeasy kit (Qiagen) according to the instructions of the manufacturer. cDNA synthesis was performed using random hexamers from 1 μg of total RNA and Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT; Life Technologies). Because we did not know which cell line expressed all of the growth factors of interest, we pooled the total RNA from all cell lines to reach a final amount of 2 μg and used the pooled RNA to synthesize cDNA as described above. This cDNA pool was used as the template to generate standards for all GFs by PCR. GFs that could not be detected were evaluated using 2 or 3 additional primer pairs. The sequences of the primers used for the reactions that gave rise to the data shown (see Fig. 10) were as follows: for FGF1-F, CACATTCAGCTGCAGCTCAG; for FGF1-R, TGCTTTCTGGCCATAGTGAGTC; for FGF2-F, CTTCTTCCTGCGCATCCACC; for FGF2-R, CACATACCAACTGGTGTATTTC; for FGF5-F, GCTGTGTCTCAGGGGATTGT; for FGF5-R, TGAAAACGCTCCCTGAACTT; for FGF6-F, CTACTGCAACGTGGGCATCG; for FGF6-R, TGCTCAGGGCAATGTAGGTC; for FGF7-F, GAAGACTCTTCTGTCGAACAC; for FGF7-R, TATTGCCATAGGAAGAAAGTGG; for FGF9-F, GCAGCTATACTGCAGGACTG; for FGF9-R, AATGCAACATAGTATCGCCTTC; for FGF10-F, GCATGTGCGGAGCTACAATCAC; for FGF10-R, CTCTCCTTGGAGCTCCTTTTCC; for FGF11-F, GAGGATACCAGCTCCTTCAC; for FGF11-R, CTGCCTTGGTCTTCTTAACTC; for FGF12-F, GCACCCAGATGGTACCATTG; for FGF12-R, TTCTTGCTGGCGGTACAGTG; for FGF13-F, CTGTACTTGGCAATGAACAGTG; for FGF13-R, CGTGAGATCGTGCAGTGATG; for FGF18-F, ACTTCCTGCTGCTGTGCTTC; for FGF18-R, CTTACGGCTCACATCGTCC; for FGF20-F, GGTATCTTGGAATTCATCAGTG; for FGF20-R, TCTGATGCCTCTTGGACCTG; for FGF22-F, TTCTTCCTGCGCGTGGATC; for FGF22-R, TAGGTGTTGTGGCCGTTCTC; for HGF-F, AGCATGTCCTCCTGCATCTC; for HGF-R, TGGTTTTTATCTTCAGTGCTGG; for IL-8-F, CGGAAGGAACCATCTCACTG; for IL-8-R, AGCACTCCTTGGCAAAACTG; for PPIB-F, GAGGAAAGAGCATCTACGGTG; and for PPIB-R, GCTTCTCCACCTCGATCTTG.
FIG 10.
Expression of detectable FGFs, IL-8, and HGF in primary RCC cell lines. Results of qRT-PCR analysis of indicated growth factors normalized to PPIB. Data are means ± SEM (n = 2). PPIB, protein coding, peptidylprolyl isomerase B.
Labeling of primary RCC cells and VHL reconstitution of 786-O cells.
Primary RCC cells were transduced with a lentivirus encoding cytoplasmic zsGreen (pLVX-ZsGreen1-C1, Clontech) and selected with puromycin (Sigma) (2 μg/ml) to generate fluorescently green versions of each cell line prior to chorioallantoic membrane (CAM) implantation. 786-O cells were reconstituted with either empty vector (EV) (pBabe-hygro; plasmid database reference no. 392) or VHL (pBabe-hygro-HA-VHL; reference no. 618) using retroviruses followed by hygromycin selection (Life Technologies) (250 μg/ml). 786-O EV cells were labeled with green fluorescent protein (GFP) by transfection with pIRES-eGFP-H2B (catalog no. 585) using Lipofectamine Plus reagent (Life Technologies) followed by fluorescence-activated cell sorter (FACS) analysis and selection using 2 μg/ml puromycin. 786-O VHL cells were labeled with mCherry using a method similar to the GFP labeling method used for 786-O EV cells except that the plasmid used for transfection was pIRES-mCherry-H2B (catalog no. 584).
Chicken chorioallantoic membrane implantation and drug treatment.
Ex ovo avian embryos were prepared and maintained as described previously (29, 30), and cells were implanted at day 9 of embryonic development. For primary cell implantation of 40 animals, 4 to 6 T175 cell culture flasks of cells grown to 90% confluence were washed with PBS twice, trypsinized (0.05% Trypsin-EDTA; Life Technologies), and pelleted at 200 × g for 10 min at room temperature. Supernatant was removed and mixed with an equal volume of normal-growth Matrigel (Life Technologies) and kept on ice until implantation into the CAM.
To generate tumors, an autoclaved Whatman No. 1 filter paper disc (5-mm diameter) was placed onto the CAM and allowed to adsorb to the CAM for 1 min. The filter paper disc was quickly snap removed using forceps, and then 10 μl of cell-Matrigel mix was quickly applied to affected area of CAM, and the CAM was immediately placed back in the animal incubator. Tumors were treated every other day by applying 10 μl (10 μM) of indicated drug directly over the implanted tumor. For 786-O cells, they were pretreated with sunitinib or vehicle overnight before implantation using the same postimplantation treatment schedule as described above. After 7 to 8 days postimplantation, the tumor take rate was determined by the presence of tumor growth in the CAM using a fluorescent stereoscope (Lumar; Zeiss Inc.). To determine if vessels formed within the tumor graft, 100 μl of dextran-Alexa Fluor 555 (Life Technologies) (0.1 μg/μl) was injected intravenously as described previously (30, 31).
Statistics.
Pooled or Satterthwaite t testing was used to calculate P values when data showed equal or unequal variances, respectively. Correlation coefficients were calculated using the Pearson or Spearman method for data distributed normally or nonnormally, respectively. Fisher's exact test was used to calculate the P value for CAM experiments. SAS 9.3 for Windows was used for statistical analysis.
Study approval.
Tumor samples were obtained from patients providing written consent to a University of Texas Southwestern Medical Center Institutional Review Board (IRB)-approved protocol allowing the use of discarded surgical specimens for research. Tumor graft studies were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
RESULTS
RCC tumor cells are resistant to sunitinib in vitro and in vivo.
While sunitinib was originally designed to inhibit VEGF and platelet-derived growth factor (PDGF) receptors (32), sunitinib also targets other receptor tyrosine kinases, such as KIT, RET, and CSF1R (33). Thus, in addition to obstructing EC function and neovascularization, sunitinib also directly inhibits tumor cells dependent on these receptors. However, in contrast to other tumor types, kinases are rarely mutated in ccRCC (34), and whether sunitinib directly affects ccRCC cell growth is unclear (35–37).
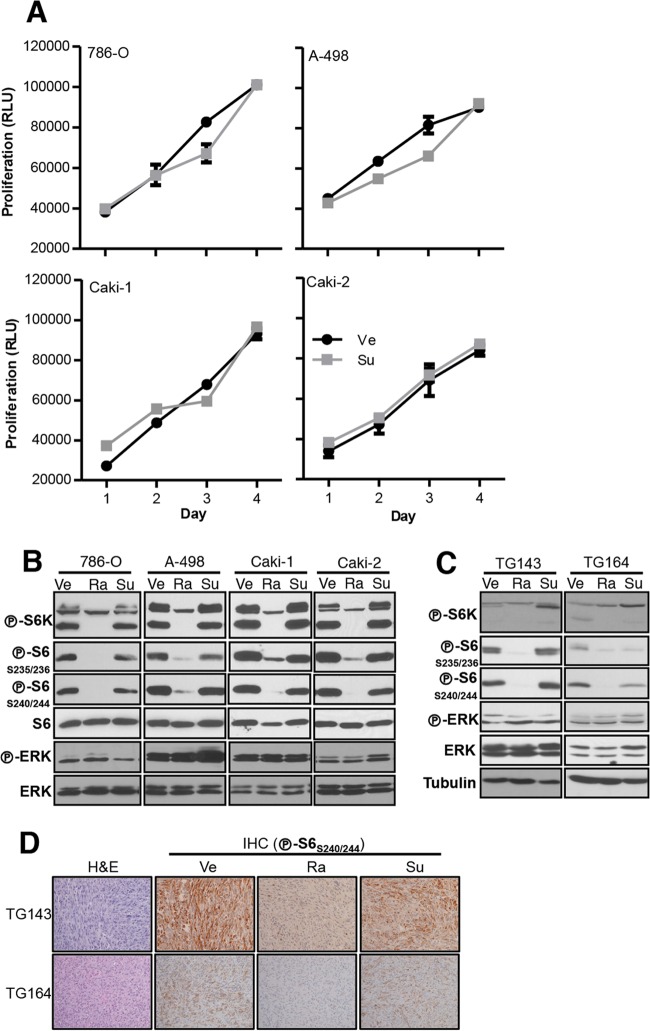
To determine if sunitinib inhibits RCC cell proliferation, we treated four established RCC cell lines (786-O, A-498, Caki-1, and Caki-2) with sunitinib and evaluated their proliferation for 4 days. Despite sunitinib administration at 1 μM, 10-fold-higher levels than the maximum concentration of drug in patients' serum (Cmax) (38), RCC cell proliferation was unaffected by the drug (Fig. 1A). In addition, as assessed by the phosphorylation of ERK, S6 kinase (S6K), and S6, there was no evidence of mitogen-activated protein kinase (MAPK) or mTORC1 inhibition by sunitinib (Fig. 1B). As a control, mTORC1 was readily inhibited by rapamycin (also called sirolimus, the active metabolite of temsirolimus) at concentrations reached in plasma in patients (39) (Fig. 1B). Thus, sunitinib does not inhibit proliferation or growth factor signaling in RCC cells in vitro.
FIG 1.
Sunitinib does not inhibit RCC cell proliferation or signaling. (A) Proliferation assays of the indicated RCC cell lines treated with vehicle (Ve) or sunitinib (Su; 1 μM). Data are means ± standard errors of the means (SEM); n = 4. RLU, relative luminescence units. (B and C) Western blot analysis of RCC cell lines treated with Ve, rapamycin (Ra; 25 nM), or Su (10 μM) overnight (ON) (B) or of the indicated tumor grafts in mice treated with Ve, Ra, or Su for 3 days (C). (D) Hematoxylin and eosin (H&E) staining and IHC analysis of the same tumors as those used for Western blotting in panel C (×100 magnification).
The use of cell lines is limited, however, by the acquisition of mutations and copy number alterations (40). Some of these alterations may directly affect angiogenic pathways, such as homozygous deletions of the HIF1A gene (41). To study the effects of sunitinib in a more physiologically accurate setting, we turned to tumor grafts (tumors from patients directly implanted in mice; also called patient-derived xenografts [PDX]). As we showed recently, RCC tumor grafts preserve the architecture and molecular signature of patient tumors, including gene expression levels, DNA copy number alterations, and mutations (25). Using this experimental system, we previously reported that, as in humans, sunitinib inhibits RCC growth in mice (25). In these experiments, tumor graft-bearing mice were treated with sunitinib (or erlotinib, as a control) for ∼28 days. Analyses of tumors at the end of this time period showed mTORC1 inhibition in tumor cells (25). However, due to the long treatment period, we could not distinguish whether sunitinib directly inhibited tumor cells or whether this was an indirect consequence of targeting the vasculature.
To evaluate the effects of sunitinib on tumor cells in vivo, we (orthotopically) implanted ccRCCs from 2 patients in the kidney of mice and studied the effects of sunitinib treatment after a short interval. Tumor graft-bearing mice were treated with sunitinib every 12 h by gavage at 10 mg/kg to achieve drug exposures similar to those observed in humans (25). For these experiments, tumors were collected at the end of 3 days. ERK or mTORC1 was not inhibited in any of the tumors as shown by Western blot analysis (Fig. 1C). Similar results were observed by IHC, which allowed us to evaluate the effects specifically on tumor cells (Fig. 1D). In contrast, rapamycin, which was used as a control, suppressed mTORC1 in all the tumors (Fig. 1C and D). These data suggest that sunitinib does not directly inhibit tumor cells and that the effects of sunitinib on tumor cells may be indirect.
Sunitinib inhibits EC proliferation and signaling in vitro and in primary orthotopic tumor grafts.
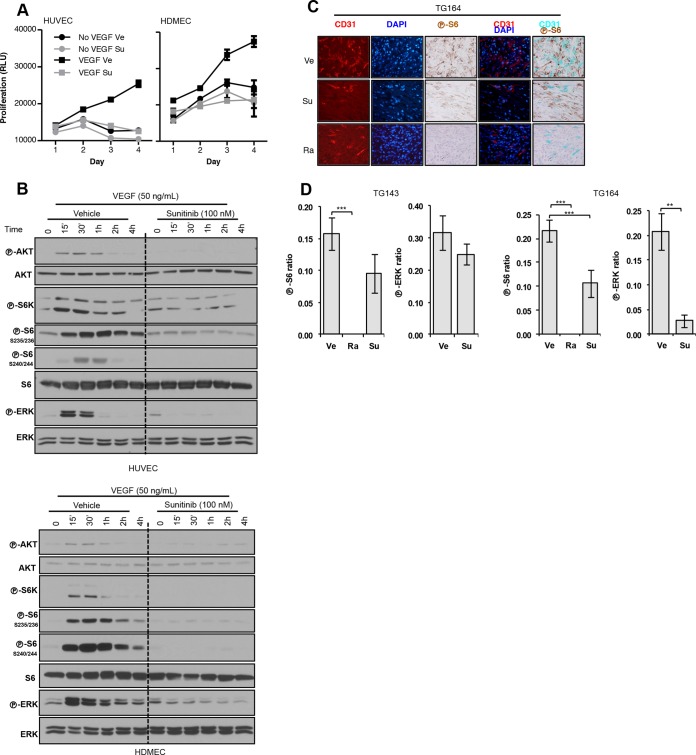
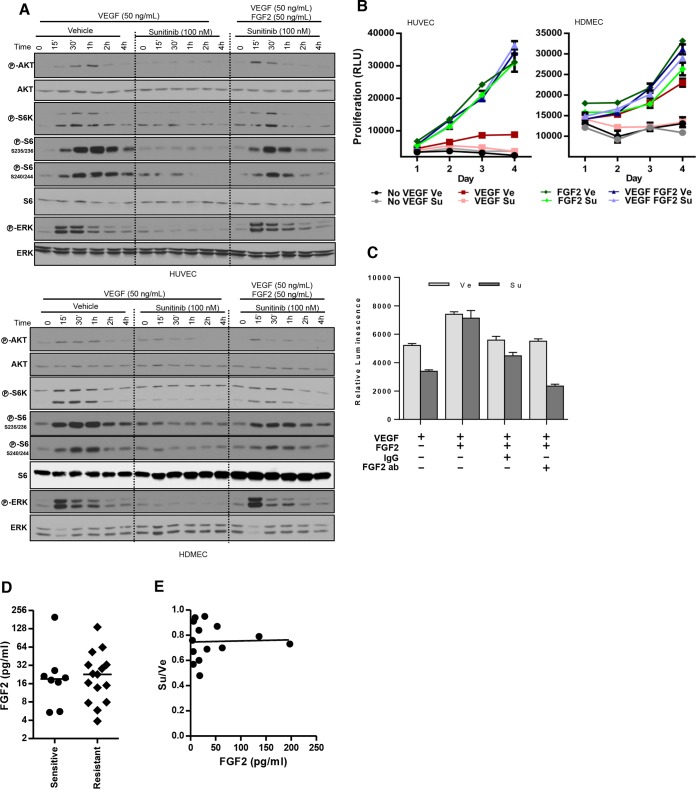
We studied the effect of sunitinib on EC using human umbilical vein endothelial cells (HUVEC) and human dermal microvascular endothelial cells (HDMEC). It is well established, that growth factors are essential for EC survival in culture (42, 43). VEGF stimulated EC to proliferate, but sunitinib blocked this effect (Fig. 2A). Sunitinib also inhibited AKT, ERK, and mTORC1 activation by VEGF (Fig. 2B).
FIG 2.
Sunitinib inhibits endothelial cell (EC) proliferation and signaling. (A) Proliferation assay of EC treated with vehicle (Ve) or sunitinib (Su; 100 nM) (VEGF, 100 ng/ml). (B) Western blot analysis of EC pretreated with Ve or Su and stimulated with VEGF for the indicated periods of time. (C) Tumor graft sections from tumors treated with Ve, Su, or Ra and stained for CD31, DAPI, and phospho-S6 (×400 magnification). (D) Quantification of phospho-S6- or phospho-ERK-positive EC in tumor grafts treated with Ve, Su, or Ra. Ratios refer to the numbers of EC that stained positive for either phospho-S6 or phospho-ERK divided by the total EC number. Data are means ± SEM. n = 2. **, P < 0.01; ***, P < 0.001.
Next, we examined the effects of sunitinib on EC in vivo in our tumor graft model. To specifically evaluate the effects of sunitinib on EC, tumor grafts (after 3 days of treatment) were subjected to IHC-immunofluorescence (IHC-IF). Tumor sections were costained for EC markers and either phosphorylated ERK or S6, and the number of positive EC per field was determined to assess the extent of inhibition (Fig. 2C). Sunitinib inhibited ERK and S6 phosphorylation in EC in both tumor grafts, albeit to different extents (Fig. 2D). S6 phosphorylation was also inhibited by rapamycin in EC (Fig. 2D). Thus, whereas rapamycin inhibits both tumor and EC, sunitinib appears to affect only EC.
Establishment of an in vitro assay to evaluate the cross talk between RCC and EC.
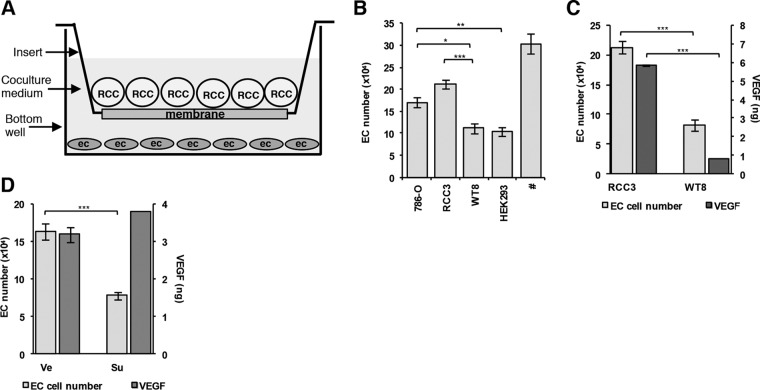
We attempted to model tumor-induced angiogenesis in vitro in coculture assays. Interestingly, established RCC cell lines can contribute to the support of EC in coculture assays (44–46). To further evaluate the role of RCC cell lines in supporting EC, we set up coculture assays using a tumor cell line, 786-O, derived from a ccRCC with a frameshift mutation in the VHL gene (47). EC and 786-O cells were grown in the same chamber separated by a permeable membrane, in growth factor limited medium (Fig. 3A). As a control, we used HEK293 cells, which were derived from a normal kidney. Over the course of 3 days, approximately 40% of EC cocultured with HEK293 cells died (Fig. 3B). In contrast, EC numbers were preserved in cocultures of 786-O cells (Fig. 3B).
FIG 3.
786-O cells support EC survival in a VHL gene-dependent and sunitinib-sensitive manner. (A) Coculture assay schematic. (B) Histogram of EC numbers in cocultures with the indicated cell lines. #, EC number in cultures supplemented with recombinant VEGF (100 ng/ml). (C) Histogram of the effects of VHL gene reconstitution on EC numbers and VEGF levels using RCC3 (VHL gene deficient) or WT8 (VHL gene reconstituted) (n = 4). (D) Histogram of the effects of sunitinib (Su; 100 nM) on EC numbers and VEGF levels in cocultures of 786-O cells. Data are means ± SEM. *, P < 0.05; **, P < 0.01, ***, P < 0.001. In panel C, n = 21 for 786-O, n = 8 for RCC3, n = 13 for WT8, and n = 10 for HEK293; in panel D, n = 21 for EC numbers and n = 11 for VEGF levels.
To probe further, we evaluated the effects of VHL gene reconstitution in 786-O cells. We compared WT8 cells, clones of 786-O cells reconstituted with the wild-type VHL gene, to RCC3 cells reconstituted with an empty vector (48). Interestingly, restoration of pVHL in WT8 cells compromised their ability to support EC (Fig. 3B and C). WT8 cells supported EC survival to an extent similar to that seen with HEK293 cells (Fig. 3B). As expected, this effect was accompanied by a substantial reduction in VEGF production (Fig. 3C).
Next, we tested whether the survival effects of 786-O cells on EC could be blocked by sunitinib. Consistent with a previous report (49), 786-O cells failed to support EC survival in the presence of sunitinib (Fig. 3D). As anticipated, this inhibition was not due to a downregulation of VEGF secretion (Fig. 3D).
Establishment of an ex vivo assay to further evaluate the cross talk between RCC and EC.
To further evaluate the interaction between RCC and EC, we established a second experimental system using the chorioallantoic membrane (CAM) of chicken embryos. On day 4 of chicken embryo development, the extraembryonic membrane allantois fuses with the chorion to form the CAM (50). This double-layered membrane has a rich vascular network that is connected to the embryonic circulation (51). Chicken CAM assays have been extensively used to study angiogenesis (51), including tumor-induced vascularization (52, 53). However, to our knowledge, this system has never before been deployed for the study of tumor resistance to angiogenesis inhibitors.
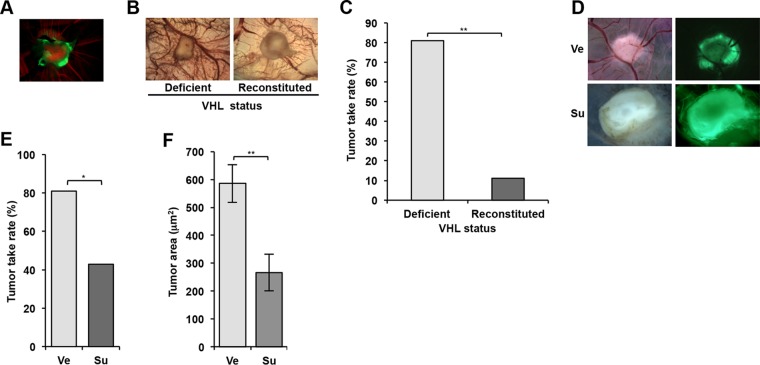
To evaluate this system, we first assayed 786-O cells. First, we determined if 786-O cells implanted on the CAM would form tumors. To distinguish tumor tissue from host tissue, 786-O cells were transduced to express cytoplasmic GFP (using a lentivirus; see Materials and Methods). Implanted 786-O cells formed well-vascularized tumors (Fig. 4A).
FIG 4.
786-O cells form tumors on the chorioallantoic membrane (CAM) of chicken embryos in a VHL gene- and sunitinib-dependent manner. (A) Fluorescence image of a tumor generated from 786-O cells (labeled with GFP) implanted on CAM. Green, tumor cells; red, Alexa Fluor 555-dextran labeling blood vessels. (B) Bright-field images of tumors on CAM using 786-O reconstituted with empty vector (Deficient) or VHL gene (Reconstituted). (C) Tumor take rates of VHL gene-deficient and reconstituted 786-O cells. (D) Bright-field and fluorescence images of 786-O-derived tumors on CAM treated with vehicle (Ve) or sunitinib (Su). (E) Tumor take rates of 786-O cells treated with Ve or Su. (F) Area of 786-O tumors treated with Ve or Su. Data are means ± SEM (n = 11 for Ve and n = 6 for Su). *, P < 0.05; **, P < 0.01.
We next determined the dependency on VHL gene loss and evaluated the vascularization and tumor engraftment rate of 786-O cells reconstituted with either empty vector (EV) or VHL gene. Consistent with the coculture assays, VHL gene reconstitution significantly reduced tumor vascularization (Fig. 4B). We also observed a significant reduction in the tumor take rate (Fig. 4C and Table 1).
TABLE 1.
Tumor take rates of 786-O cells depending upon on VHL status and sunitinib treatment
| VHL status | Treatment | Tumor take rate (%) (no. of samples with tumors/total no. of samples) |
|---|---|---|
| Deficient | Vehicle (n = 21) | 81.0 (17/21) |
| Reconstituted | Vehicle (n = 9) | 11.1 (1/9) |
| Deficient | Sunitinib (n = 21) | 57.1 (12/21) |
Next, we examined whether sunitinib would suppress 786-O-induced vascularization and tumor growth. Similarly to coculture assays, sunitinib administration inhibited vascularization (Fig. 4D) and resulted in a lower take rate (Fig. 4E and Table 1). This was associated with a significant reduction in the extent of the tumor area (Fig. 4F). Together, these results established our coculture assay as a second assay suitable for the study of angiogenic drug sensitivity in renal cancer.
Primary RCC cultures support EC survival in a tumor-dependent manner.
Next we asked whether tumors from patients supported EC survival in vitro. We collected tumor samples from 65 patients and successfully generated 27 short-term primary cultures. Most samples were derived from kidney tumors. There were 26 renal cell carcinomas: 21 with clear cell histology, 2 papillary, 2 unclassified, and 1 chromophobe. In addition, one cell line was generated from a primitive malignant neoplasm (Table 2).
TABLE 2.
Tumor characteristics of samples giving rise to primary culturesa
| ID | Pathological staging | Source | Histology | % of sarcomatoid | Fuhrman grade | Pretreatment(s) | Resistance to sunitinib in patients |
|---|---|---|---|---|---|---|---|
| T285 | T3aNxMx | Kidney | Clear cell | 0 | 3 | No | |
| T283 | T3aN0Mx | Kidney | Clear cell | 0 | 3 | No | |
| T279 | T3bN0Mx | Vein thrombus | Clear cell | 0 | 3 | No | |
| TG206 | Brain met | Clear cell | NA | No | |||
| TG250 | T2aN0Mx | Kidney | Clear cell | 0 | 3 | No | |
| TG158 | T3aNxMx | Kidney | Clear cell | 5 | 3 | No | |
| T264 | T3bN0Mx | Kidney | Clear cell | 0 | 3 | No | |
| TG144 | T2aN1Mx | Kidney | Clear cell | 0 | 4 | No | |
| TG185 | Kidney | Unclassified | 0 | No | |||
| TG142 | T3aNxMx | Kidney | Clear cell | 0 | 3 | No | |
| TG191 | T3aNxMx | Kidney | Clear cell | 0 | 3 | No | |
| TG143 | T4NxMx | Kidney | Clear cell | 95 | 4 | No | |
| TG180 | Bone met | Clear cell | NA | No | |||
| T249 | T3aN0Mx | Vein thrombus | Clear cell | 0 | 3 | No | |
| TG26 | Adrenal gland | Clear cell | NA | No | |||
| TG169 | T4N1Mx | Kidney | Unclassified | NA | Su | Yes | |
| TG127 | T3aNxMx | Kidney | Clear cell | 0 | 3 | No | Yes* |
| PF22 | Pleural fluid | Clear cell | NA | Su, Ev, Pa, So | Yes | ||
| T258 | T3aN0M1 | Kidney | Clear cell | 15–20 | 4 | No | Yes* |
| T241 | T3aN0M1 | Kidney | Clear cell | 0 | 3 | No | |
| TG194 | T3aNxMx | Kidney | Clear cell | 0 | 3 | No | |
| T239 | T1aNxMx | Kidney | Clear cell | 0 | 2 | No | |
| T256 | Kidney | Prim. Mal. Neo. | 0 | No | |||
| T230 | T2bN1Mx | Kidney | Papillary | 0 | No | ||
| PF114 | Pleural fluid | Clear cell | 0 | Su, Pa | Yes | ||
| TG121 | T1aNxMx | Kidney | Papillary | 0 | No | ||
| T255 | T2aNxMx | Kidney | Chromophobe | 0 | No |
List of samples giving rise to primary cultures. Samples are sorted according to their degree of sensitivity to sunitinib in coculture assays (see Table 3); samples resistant to sunitinib are indicated with boldface characters. Cell lines exhibiting sunitinib resistance in patients were defined as (i) a line derived from a tumor in a patient whose tumor had progressed on sunitinib or (ii) a line derived from a patient subsequently treated with sunitinib (after generation of the line) that exhibited tumor progression within 3 months of treatment onset (indicated with an asterisk). ID, identifier; NA, not available; met, metastasis; Prim. Mal. Neo., primitive malignant neoplasm; Su, sunitinib; Pa, pazopanib; So, sorafenib; Ev, everolimus; No, no pretreatment. When multiple treatments are indicated, they are displayed in the order in which they were administered.
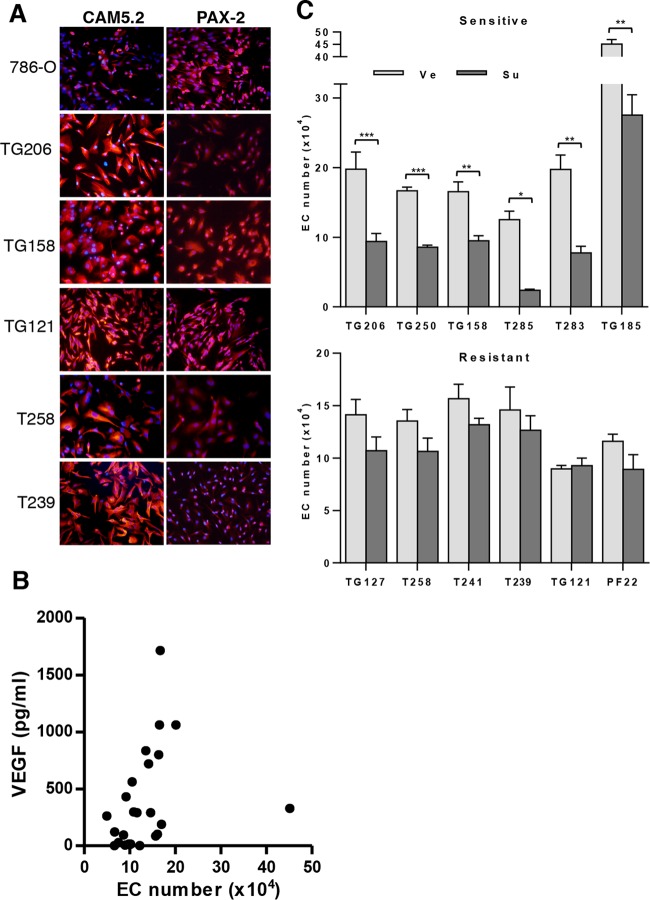
More than half of the RCC tumors were of high grade, and a few contained sarcomatoid elements (Table 2). Twenty cell lines were generated from tumors in the kidney; 2 from tumor thrombi; 1 from an adrenal gland metastasis; 1 from a brain metastasis; 1 from a bone metastasis; and 2 from malignant pleural effusions (Table 2). Some tumor samples were implanted orthotopically in mice to generate tumor grafts. These tumor grafts reproduce the characteristics of human tumors (25), and both tumors and tumor grafts were used to generate primary cultures. Cell lines generated from tumor grafts are denoted with “TG” followed by a number. Cell lines generated directly from patient tumors (either primary tumors or metastases) are labeled with a “T” followed by a number. Cell lines derived from malignant pleural fluid were generated directly from the fluid without passage in mice, and the corresponding samples are labeled with the letters “PF” followed by the patient number. To assess whether the cells generated were of renal epithelial origin, they were stained with two RCC markers, CAM5.2 (low-molecular-weight cytokeratin) (54) and the transcription factor PAX-2 (55) (Fig. 5A).
FIG 5.
Evaluation of the effects of sunitinib on EC survival in coculture with primary RCC. (A) CAM5.2 and PAX-2 staining of 786-O and indicated primary RCC cells; ×200 magnification. (B) Tumor-induced EC survival correlates with secreted VEGF levels; R = 0.57 and P = 0.0028. (C) EC survival in cocultures. Data are means ± SEM. n = 13 for TG206, n = 3 for TG250, n = 4 for TG158, n = 3 for T285, n = 3 for T283, n = 4 for TG185, n = 10 for TG127, n = 4 for T258, n = 6 for T241, n = 8 for TG121, n = 4 for PF22, and n = 6 for T239. *, P < 0.05; **, P < 0.01, ***, P < 0.001.
Among the patients whose tumors/tissues were collected for this study, 3 had been previously treated and had had tumor progression on sunitinib, and their samples were regarded as sunitinib resistant. In addition, among the remaining 24 patients, 2 other patients subsequently treated with sunitinib (after their samples were collected) were resistant to it (progression within 3 months). Overall, samples were obtained from 5 tumors regarded as sunitinib resistant (Table 2). In summary, sunitinib-resistant lines were defined as (i) derived from a tumor in a patient progressing to sunitinib (TG169, PF22, and PF114) or (ii) derived from a tumor that had not been exposed to sunitinib but from a patient who was subsequently treated with sunitinib and whose tumor had progressed within 3 months of treatment onset (TG127 and T258).
In preliminary experiments, we asked whether primary cultures could support EC survival, and we compared the results to those of cocultures with 786-O and HEK293. The degree to which primary cultures supported EC survival varied from sample to sample. Overall, 10 lines supported EC to an extent similar to or greater than that seen with 786-O cells (data not shown) whereas the rest failed to support EC survival to the same extent. The most active tumor was an unclassified RCC (TG185). Otherwise, the lines most supportive of EC were from ccRCC tumors. Perhaps this should not have been unexpected, given the role of the VHL gene in the regulation of angiogenic pathways.
Next, we explored whether a correlation existed between EC support and VEGF secretion. Interestingly, VEGF secretion correlated well with tumor-induced EC survival (R = 0.57 and P = 0.0028) (Fig. 5B). Of note, the levels of VEGF detected by enzyme-linked immunosorbent assay (ELISA) in cocultures were much (at least 50-fold) lower than the levels of VEGF used in the EC cell proliferation assays presented in Fig. 2A. Lower VEGF levels further diluted in culture medium may explain the observation that RCC cells could support only EC survival but not proliferation in the coculture assays.
Sunitinib inhibits tumor-induced EC survival in a RCC-dependent manner.
We asked whether the effects of tumor cells on EC survival were sunitinib sensitive. A tumor was defined as sensitive if there was a statistically significant reduction in the EC numbers in the sunitinib-treated versus vehicle-treated groups. To normalize for differences in EC numbers across cultures, where appropriate, results are expressed as a ratio (Su/Ve). Sunitinib inhibited EC survival in 13 cocultures (Fig. 5C and Table 3). The degree of inhibition ranged between 30% and 80% (Table 3). Notably, EC survival was inhibited by sunitinib in both highly EC-supportive and less-supportive tumors (data not shown). Overall, these data show that primary RCC cultures support EC survival and that tumors can be divided into sunitinib-sensitive and -insensitive tumor categories. Interestingly, among 14 lines that were resistant, we found all 5 corresponding to RCC tumors that were resistant in patients.
TABLE 3.
Response to sunitinib in coculturesa
| Cell line sunitinib sensitivity category | ID | Su/Ve ratio | P value |
|---|---|---|---|
| Sensitive | T285 (n = 3) | 0.19 | 0.013 |
| T283 (n = 3) | 0.39 | 0.0019 | |
| T279 (n = 3) | 0.42 | 0.010 | |
| TG206 (n = 13) | 0.48 | 0.0005 | |
| TG250 (n = 3) | 0.51 | <0.0001 | |
| TG158 (n = 4) | 0.57 | 0.0047 | |
| T264 (n = 4) | 0.58 | 0.014 | |
| TG144 (n = 10) | 0.60 | 0.0002 | |
| TG185 (n = 4) | 0.61 | 0.0022 | |
| TG142 (n = 4) | 0.62 | 0.0014 | |
| TG191 (n = 8) | 0.67 | 0.027 | |
| TG143 (n = 3) | 0.67 | 0.037 | |
| TG180 (n = 10) | 0.73 | 0.0009 | |
| Resistant | T249 (n = 4) | 0.68 | 0.051 |
| TG26 (n = 2) | 0.69 | 0.095 | |
| TG169 (n = 8) | 0.70 | 0.17 | |
| TG127 (n = 10) | 0.76 | 0.10 | |
| PF22 (n = 4) | 0.77 | 0.14 | |
| T258 (n = 4) | 0.79 | 0.11 | |
| T241 (n = 6) | 0.84 | 0.15 | |
| TG194 (n = 4) | 0.85 | 0.17 | |
| T239 (n = 6) | 0.87 | 0.47 | |
| T256 (n = 4) | 0.88 | 0.27 | |
| T230 (n = 2) | 0.91 | 0.75 | |
| PF114 (n = 4) | 0.94 | 0.79 | |
| TG121 (n = 8) | 1.03 | 0.71 | |
| T255 (n = 4) | 1.09 | 0.80 |
List of cell lines evaluated for sunitinib sensitivity in coculture assays with EC. Cell lines are separated into categories of sensitive and resistant (based on P value) and ranked within each group based on the Su/Ve ratio (ratio of EC numbers in RCC cocultures in the presence of sunitinib or vehicle). Sunitinib-resistant cell lines are indicated with boldface characters.
Testing sunitinib sensitivity using CAM assays.
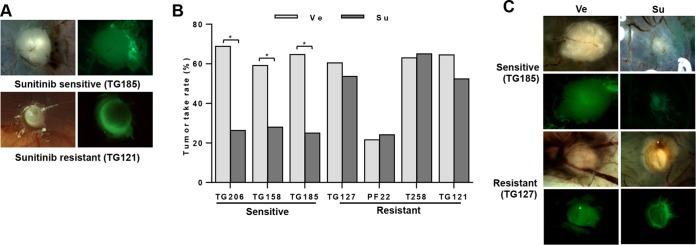
A subset of the primary RCC cells (both sunitinib sensitive and sunitinib resistant) was used to test for sunitinib sensitivity in CAM assays. Like 786-O cells, these primary RCC cells were labeled with zsGreen (to distinguish tumor cells from host tissue), and they formed vascularized tumors (Fig. 6A). Overall, the tumor engraftment rates were around 60%, with the exception of PF22 (21.6%) (Table 4). PF22 cells were isolated from pleural fluid, and that may have reduced their ability to engraft and form solid tumors in the CAM assays. Notably, sunitinib treatment significantly reduced tumor engraftment rates for all the cell lines that had been shown to be sensitive in coculture assays (Fig. 6B and Table 4). Those that were resistant in coculture assays were also resistant in CAM assays. When tumors from sunitinib-sensitive lines formed in the presence of the drug, those tumors were also smaller (Fig. 6C). Overall, there seemed to be good reproducibility across assays for sunitinib sensitivity and resistance. These data also underscore the critical role of neoangiogenesis for tumor development on CAM assays. Specifically, the reductions in the tumor take rates and in the size of tumors in the presence of sunitinib from cells that support EC survival in a sunitinib-dependent manner suggest that tumor development depends upon the ability of RCC cells to sustain EC proliferation and productive angiogenesis. These data further show that RCC development on CAM is intrinsically linked to efficient angiogenesis.
FIG 6.
Testing sunitinib sensitivity using CAM assay. (A) Bright-field and fluorescence images of sunitinib-sensitive and -resistant tumors. Green, tumor cells. (B) Tumor take rates of indicated primary RCC cells treated with vehicle or sunitinib. (C) Bright-field and fluorescence images of sunitinib-sensitive and -resistant tumors following treatment with vehicle (Ve) or sunitinib (Su). *, P < 0.05.
TABLE 4.
Response to sunitinib treatment in CAM assaysa
| RCC cell line | Treatment | Tumor engraftment rate (%) (no. of samples with tumors/total no. of samples) | Su/Ve | P value |
|---|---|---|---|---|
| TG206 | Vehicle (n = 16) | 68.8 (11/16) | 0.40 | 0.037 |
| Sunitinib (n = 18) | 27.8 (5/18) | |||
| TG158 | Vehicle (n = 22) | 59.1 (13/22) | 0.47 | 0.042 |
| Sunitinib (n = 25) | 28.0 (7/25) | |||
| TG185 | Vehicle (n = 17) | 64.7 (11/17) | 0.39 | 0.022 |
| Sunitinib (n = 20) | 25.0 (5/20) | |||
| TG127 | Vehicle (n = 43) | 60.4 (26/43) | 0.89 | 0.628 |
| Sunitinib (n = 28) | 53.6 (15/28) | |||
| PF22 | Vehicle (n = 37) | 21.6 (8/37) | 1.12 | 1.000 |
| Sunitinib (n = 29) | 24.1 (7/29) | |||
| T258 | Vehicle (n = 27) | 63.0 (17/27) | 1.04 | 1.000 |
| Sunitinib (n = 23) | 65.2 (15/23) | |||
| TG121 | Vehicle (n = 31) | 64.5 (20/31) | 0.81 | 0.405 |
| Sunitinib (n = 21) | 52.4 (11/21) |
List of cell lines evaluated for sunitinib sensitivity in CAM assays. Cells are sorted based on sunitinib sensitivity (see Table 3). Sunitinib-resistant cell lines (as determined based on coculture assays) are indicated with boldface characters.
Sunitinib sensitivity correlates with VEGF production.
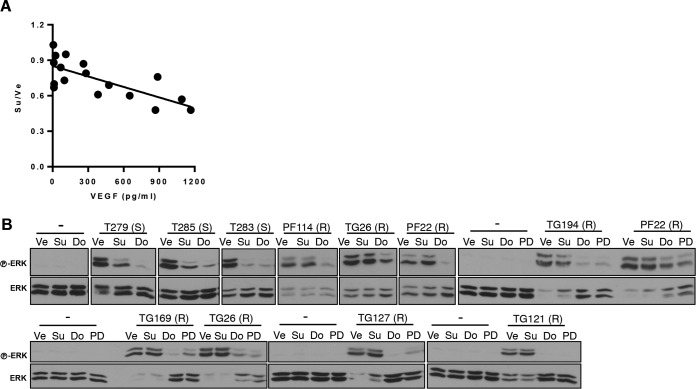
Next, we explored whether a correlation existed between VEGF secretion and sunitinib sensitivity. We evaluated the levels of secreted VEGF as a function of sunitinib sensitivity. We found a significant correlation between secreted VEGF levels and sunitinib sensitivity (R = −0.73 and P = 0.0008) (Fig. 7A). These results are consistent with a model in which VEGF production by tumor cells promotes EC survival in a sunitinib-dependent manner. We speculate that tumors resistant to sunitinib produce other growth factors that stimulate EC and consequently exhibit a lesser dependency on VEGF.
FIG 7.
VEGF dependency for mitogenic signaling in EC from sunitinib-sensitive but not sunitinib-resistant tumors. (A) Correlation of VEGF levels and sunitinib sensitivity (R = −0.73 and P = 0.0008). (B) Western blot analysis of HUVEC pretreated with Ve, Su (100 nM), dovitinib (Do; 500 nM), or PD173074 (PD; 50 nM) and then stimulated with conditioned media from the indicated RCC. “(S)” and “(R)” indicate sunitinib-sensitive and -resistant RCC lines, respectively. −, cells in the same base medium that was used to collect RCC-conditioned media.
We predicted that, if our hypothesis was correct, conditioned medium (CM) from sunitinib-resistant RCC lines should be able to sustain growth factor signaling in EC despite the presence of sunitinib. First, we established that CM from all RCC lines induced ERK phosphorylation (used as a readout for growth factor receptor activation) in EC (Fig. 7B). Provocatively, sunitinib pretreatment blocked ERK activation in EC cultured with CM from the sensitive but not from the resistant RCC lines (Fig. 7B). These data suggest that mitogenic signaling in EC is dependent on VEGF in sunitinib-sensitive lines but is independent of VEGF in the resistant lines. Together with the findings that sunitinib sensitivity correlated with VEGF secretion and that VEGF-induced ERK activation in EC is sunitinib sensitive (Fig. 7B), these data suggest that whereas some RCC tumors induce angiogenesis through a VEGF-dependent and sunitinib-sensitive pathway, others can activate signaling independently of this mechanism.
Recombinant FGF2 is sufficient to overcome sunitinib effects on EC.
One potential mechanism of resistance to sunitinib involves fibroblast growth factor receptor (FGFR) signaling. FGFRs are expressed in EC (56) and are not inhibited by sunitinib (57, 58). FGFRs can be activated by FGFs, including FGF2, a potent inducer of angiogenesis (56, 59). Building upon previous results (23), we found that recombinant FGF2 restored ERK activation in EC treated with sunitinib (Fig. 8A). Similarly, FGF2 restored proliferation of sunitinib-treated EC (Fig. 8B). This effect was specific to FGF2 and could be blocked with FGF2 neutralizing antibodies (but not an IgG control) (Fig. 8C). Thus, FGF2 is sufficient to sustain mitogenic signaling and EC proliferation despite the presence of sunitinib.
FIG 8.
FGF2 restores EC proliferation and signaling despite the presence of sunitinib, but FGF2 production does not correlate with sunitinib resistance in RCC. (A) Western blot analysis of HUVEC or HDMEC pretreated with Ve or Su (100 nM) and stimulated with VEGF (50 ng/ml) or VEGF plus FGF2 (50 ng/ml). (B) Proliferation curves of HUVEC and HDMEC treated with Ve or Su in the absence or presence of VEGF (100 ng/ml) or FGF2 (50 ng/ml) or both. (C) Neutralizing assay using anti-FGF2 antibodies to suppress the effect of FGF2. Data are means ± SEM. n = 4 (for panels B and C). (D) FGF2 levels in coculture supernatant from sensitive and resistant RCC lines. Horizontal bars indicate medians. (E) Correlation between FGF2 levels and sunitinib sensitivity (R = 0.035 and P = 0.91).
However, while FGF2 may contribute to the VEGF-independent effect of RCC on EC, a correlation could not be found between sunitinib resistance and FGF2 production (Fig. 8D and E). In addition, the effect of FGF2 on sunitinib resistance disappeared when FGF2 concentrations were reduced to levels detected by ELISA in cocultures with RCC cells (see Fig. 8D; data not shown). We could not rule out, however, the possibility that the effective concentration of FGF2 in the tumor microenvironment was substantially higher than that detected by ELISA. Furthermore, a role for FGF2 in mediating sunitinib resistance was also suggested by our finding that sunitinib induced FGF2 production, though the changes were similar in the sunitinib-sensitive and -resistant RCC cell lines (data not shown). Overall, these data show that FGF2 is able to overcome sunitinib effects on EC, but its role in mediating this effect in resistant RCC tumors remains to be fully elucidated. FGF2 is not the only ligand that activates FGFRs, and we then turned our focus to the receptors.
RCCs broadly induce FGFR signaling.
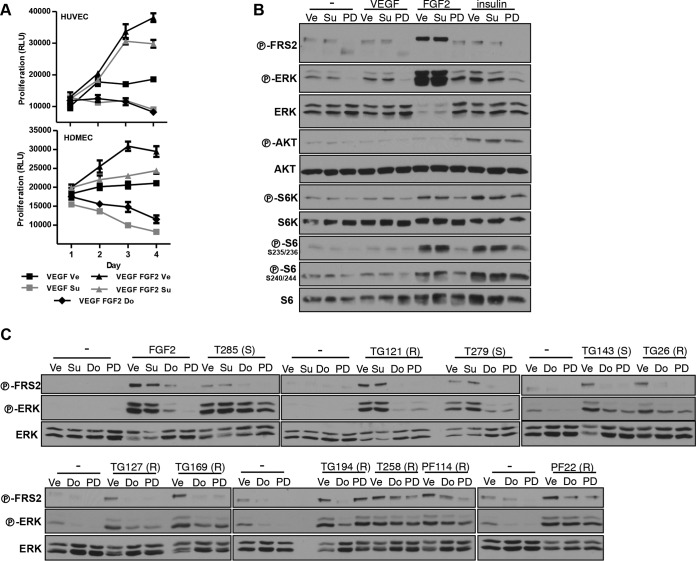
To address the role of FGFR in sunitinib resistance, we evaluated the effects of dovitinib (a VEGF and FGF receptor inhibitor) on ERK activation by conditioned medium (CM). Interestingly, ERK activation was inhibited by dovitinib in both sunitinib-sensitive and sunitinib-resistant cultures (Fig. 7B). To further evaluate this finding, similar experiments were performed with a second VEGFR/FGFR inhibitor, PD173074. Like dovitinib, PD173074 uniformly inhibited ERK in EC (Fig. 7B).
We sought to obtain more-direct evidence implicating FGFRs in the activation of ERK by tumor cell CM. However, we could not detect phosphorylation of FGFR or FRS2, an adaptor protein that mediates FGFR signaling (60), in EC (data not shown). Furthermore, even with the addition of recombinant FGF2, no phospho-FGFR or FRS2 could be detected in EC (data not shown). This was surprising, as FGF2 was able to induce EC proliferation (Fig. 8B) and FGF2 effects were dovitinib sensitive (Fig. 9A). One explanation is that our assays lacked the sensitivity needed for the detection of FGFR activation by CM in EC.
FIG 9.
FGFR activation by RCC. (A) Proliferation of HUVEC and HDMEC treated with Ve, Su (100 nM), or Do (500 nM) in the absence or presence of VEGF (100 ng/ml) or FGF2 (50 ng/ml) or both. Data are means ± SEM. n = 4. (B) Western blot analysis of MEFs pretreated with Ve, Su, Do, or PD and stimulated with VEGF (50 ng/ml), FGF2 (25 ng/ml), or insulin (300 nM). (C) Western blot analysis of MEFs pretreated with Ve, Su, Do, or PD and stimulated with conditioned media (CM) obtained from the indicated RCC cells. Su was used at 200 nM, Do at 1 μM, and PD at 100 nM for MEFs. (S), sunitinib sensitive; (R), sunitinib resistant; −, cells in the same base medium that was used to collect RCC-conditioned media.
To study the potential of CM from RCC cells to activate FGFRs, we turned to fibroblasts (mouse embryonic fibroblasts [MEFs]). MEFs express FGFRs and were used to elucidate the functional relationship of FGFR and FRS2 (61, 62). FRS2 is an accepted readout for FGFR signaling, and in our system FRS2 was activated by FGF2 but not by other growth factors such as VEGF or insulin (Fig. 9B). We observed that, as for EC, the addition of CM to MEFs resulted in ERK activation (Fig. 9C). Interestingly, in every instance examined, RCC-CM induced FRS2 phosphorylation in MEFs (Fig. 9C). Pretreatment with either dovitinib or PD compound blocked the activation of FRS2 by CM (Fig. 9C). Overall, these data suggest that RCCs secrete ligands (most likely FGFs) that are able to induce FRS2 activation in other cell types.
The observation that CM from sunitinib-sensitive RCC lines activated FGFR was unexpected. FGFR activation would have been predicted to render these cell lines sunitinib resistant, at least on the basis of results of experiments showing that FGF2 is sufficient to overcome sunitinib effects. In some instances, it appeared that the magnitude of FRS2 phosphorylation by CM from sunitinib-sensitive lines was lower than that with sunitinib-resistant lines, which could provide a potential explanation (Fig. 9C). Alternatively, other signals may be required to cooperate with FGFs, which may be secreted at low levels by RCC cells in vivo. Independently of these considerations, our data also show that RCC cells signal not only to neighboring EC but also to other stromal cells, such as fibroblasts, which could also be implicated in regulating angiogenesis.
Mapping the expression of FGFs in sunitinib-sensitive and -resistant primary RCC cells.
Because RCC cells uniformly activated FGFRs but no significant differences between sensitive and resistant RCC cells in the levels of FGF2 were observed, we evaluated other FGFs. In humans, there are 22 known FGFs, which we evaluated by qRT-PCR. In addition, we also measured the expression of two other angiogenic factors, hepatocyte growth factor (HGF) and interleukin-8 (IL-8). The expression levels of these growth factors are shown in Fig. 10. While there is no common FGF that is uniformly upregulated in sunitinib-resistant lines, we found different sets of FGFs upregulated in different sunitinib-resistant RCC lines (Table 5). However, some of these FGFs were also upregulated in some of the sensitive lines. In addition, not all FGFs bind FGFR1 and induce angiogenesis. Nevertheless, the upregulation of different FGFs in different RCC lines does suggest that FGFR signaling may play a role in RCC pathogenesis.
TABLE 5.
FGF expression in sunitinib-resistant RCC primary culturesa
| GF or cytokine | RCC line |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TG26 |
TG127 |
PF22 |
T258 |
T241 |
PF114 |
T121 |
||||||||
| R/S ratio | P value | R/S | P value | R/S | P value | R/S | P value | R/S | P value | R/S | P value | R/S | P value | |
| FGF1 | 6.26 | 2.4 × 10−6 | 1.94 | 0.013 | 38.2 | 2.00 × 10−5 | 11.6 | 0.028 | ||||||
| FGF2 | 3.03 | 0.010 | 3.48 | 0.0035 | 6.45 | 3.6 × 10−5 | 3.40 | 0.015 | ||||||
| FGF5 | 9.89 | 0.0021 | 6.68 | 7.4 × 10−5 | ||||||||||
| FGF7 | 44.8 | 0.0028 | ||||||||||||
| FGF10 | 2.34 | 0.040 | ||||||||||||
| FGF11 | 5.26 | 0.031 | ||||||||||||
| FGF13 | 28.4 | 0.0041 | 2.14 | 0.0044 | 4.79 | 0.0067 | 15.5 | 2.3 × 10−7 | ||||||
| FGF18 | 4.8 | 0.036 | ||||||||||||
| FGF20 | 3.12 | 0.026 | ||||||||||||
| FGF22 | 2.57 | 0.0082 | ||||||||||||
| HGF | 8.14 | 0.0082 | 16.7 | 2.9 × 10−5 | 4.16 | 0.02 | ||||||||
R/S ratio, expression value of individual resistant line divided by the average of the expression values of all sensitive lines. There was no expression detected from FGF3, FGF4, FGF8, FGF14, FGF16, FGF17, FGF19, FGF21, and FGF23. There was no observable increase for FGF6, FGF9, FGF12, or IL-8. GF, growth factor.
Dovitinib suppresses the effects of sunitinib-resistant RCC cell lines on EC survival.
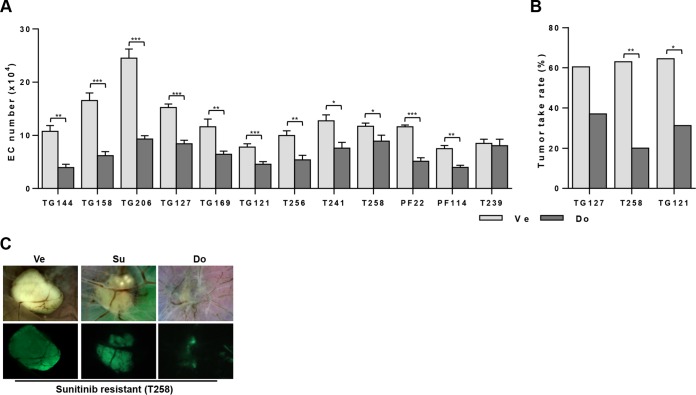
Given that ERK activation in EC was uniformly inhibited by both dovitinib and PD173074 (dovitinib/PD173074), we hypothesized that tumor-induced EC survival should be blocked by treatment with these inhibitors. We performed coculture experiments with dovitinib. Dovitinib was used at concentrations that did not affect RCC proliferation (the IC50 of dovitinib for RCC cells is in the micromolar range [2.5 to 9 μM] [data not shown]). Provocatively, dovitinib inhibited RCC-induced EC survival in the majority of tumors (Fig. 11A and Table 6). All sunitinib-sensitive lines were sensitive to dovitinib, and only one of the resistant lines examined was resistant to it (Fig. 11A and Table 6).
FIG 11.
Suppression of RCC-mediated EC survival and tumor formation by dovinitib treatment. (A) Coculture assay testing the ability of dovitinib to suppress EC survival by the different RCC lines. Data are means ± SEM. n = 4 for TG144, n = 4 for TG158, n = 7 for TG206, n = 4 for TG127, n = 6 for TG169, n = 12 for TG121, n = 4 for T256, n = 3 for T241, n = 4 for T258, n = 4 for PF22, n = 4 for PF114, and n = 2 for T239. (B) Tumor take rates of sunitinib-resistant tumors upon dovitinib treatment. See Table 7 for more details. (C) Bright-field and fluorescence images of a sunitinib-resistant RCC line (T258) after treatment with Ve, Su, or Do. *, P < 0.05; **, P < 0.01, ***, P < 0.001.
TABLE 6.
Sensitivity to dovitinib in coculturesa
| RCC cell line according to sunitinib sensitivity or resistance | Do/Ve ratio | P value |
|---|---|---|
| Sensitive | ||
| TG144 (n = 4) | 0.37 | 0.0015 |
| TG158 (n = 4) | 0.37 | 0.0007 |
| TG206 (n = 7) | 0.38 | <0.0001 |
| T264 (n = 4) | 0.42 | 0.0008 |
| TG142 (n = 4) | 0.45 | 0.0009 |
| TG191 (n = 8) | 0.52 | 0.0026 |
| Resistant | ||
| PF22 (n = 4) | 0.44 | 0.0005 |
| PF114 (n = 4) | 0.53 | 0.0023 |
| T256 (n = 4) | 0.54 | 0.0095 |
| T249 (n = 4) | 0.55 | 0.016 |
| TG127 (n = 4) | 0.55 | 0.0003 |
| TG169 (n = 12) | 0.56 | 0.0053 |
| TG121 (n = 12) | 0.59 | 0.0006 |
| T241 (n = 3) | 0.60 | 0.030 |
| T258 (n = 4) | 0.76 | 0.044 |
| T239 (n = 2) | 0.95 | 0.79 |
List of cell lines evaluated for dovitinib sensitivity in coculture assays with EC. Cell lines are ranked based on Do/Ve ratios (ratio of EC numbers in RCC cocultures in the presence of dovitinib or vehicle). The single cell line resistant to dovitinib is indicated in boldface characters.
Next, we tested the effects of dovitinib on 3 sunitinib-resistant RCC lines using the CAM assay. Dovitinib administration reduced tumor engraftment rates (Fig. 11B and Table 7) and inhibited the growth of all tumors (Fig. 11C). Overall, these data show that dovitinib can block angiogenesis and EC proliferation by sunitinib-resistant RCC cells.
TABLE 7.
Dovitinib sensitivity in CAM assaysa
| RCC cell line | Treatment | Tumor engraftment rate (%) (no. of samples with tumors/total no. of samples) | Do/Ve | P value |
|---|---|---|---|---|
| TG127 | Vehicle (n = 43) | 60.5 (26/43) | 0.61 | 0.085 |
| Dovitinib (n = 27) | 37.0 (10/27) | |||
| T258 | Vehicle (n = 27) | 63.0 (17/27) | 0.32 | 0.002 |
| Dovitinib (n = 25) | 20.0 (5/25) | |||
| TG121 | Vehicle(n = 31) | 64.5 (20/31) | 0.49 | 0.012 |
| Dovitinib (n = 32) | 31.2 (10/32) |
List of sunitinib-resistant cell lines evaluated for dovitinib sensitivity in CAM assays.
FGFR-independent paracrine stimulation of fibroblasts by RCC.
Next, we examined the effects of dovitinib/PD173074 on fibroblasts. Whereas dovitinib/PD173074 fairly consistently inhibited CM-induced FRS2 phosphorylation in fibroblasts, they failed to uniformly downregulate ERK phosphorylation (Fig. 9C). MEFs cultured with CM from several lines (i.e., T285, T258, PF114, and PF22) failed to inhibit ERK phosphorylation in response to dovitinib/PD173074. Notably, with one exception (T285), all these lines were sunitinib resistant. In contrast, dovitinib/PD173074 effectively blocked ERK activation mediated by CM from sunitinib-sensitive lines (T279 and TG143). In these lines, ERK activation was coupled to the activation of FGFR (or of another dovitinib/PD173074-sensitive receptor). In the second group, where ERK was uncoupled, there was still phosphorylation of FRS2 (indicating that FGFR were being activated), but FGFR inhibition was not sufficient to suppress ERK activation. The simplest explanation is that these tumors secrete additional growth factors that are sufficient to support fibroblast proliferation. Inasmuch as there appeared to be a correlation between tumors which showed sustained ERK activation despite dovitinib/PD173074 and sunitinib resistance, these paracrine mechanisms may confer resistance to all of these drugs (dovitinib/PD173074 and sunitinib). This notion may explain why dovitinib was not superior in a phase 3 trial to sorafenib (another VEGFR inhibitor, which, like sunitinib, is approved for advanced RCC) (63).
Interestingly, in contrast to fibroblasts, where ERK inhibition by dovitinib/PD173074 was variable, ERK phosphorylation was consistently inhibited in EC regardless of the CM (Fig. 7B). These data suggest that mitogenic signaling to EC by RCC involves, exclusively, dovitinib/PD173074-sensitive receptors such as VEGF and FGF receptors. In contrast, fibroblasts could be activated by some tumors independently of dovitinib/PD173074, and these pathways may contribute to resistance.
DISCUSSION
Here, we evaluate an important clinical problem: resistance to drugs targeting angiogenesis. Using approaches that combine primary tumor cultures with target cells (EC and fibroblasts) or a CAM assay, we provide insight into RCC paracrine signaling and potential mechanisms of resistance.
Sunitinib was originally designed to inhibit cells expressing platelet-derived growth factor receptors (PDGFRs) and VEGFRs (32); however, data concerning the cell types inhibited by sunitinib in kidney cancer remain controversial (35–37). This controversy results from a lack of appropriate experimental systems. Ideally, the effects of sunitinib on different cell types would be analyzed in tumor biopsy specimens from treated patients. However, this is challenging. To study how sunitinib acts, we developed several experimental systems, including in vitro stimulation experiments using tumor cell conditioned medium, coculture assays, ex vivo CAM assays, and tumor grafts. Our data strongly suggest that sunitinib does not directly act on tumor cells. Even at concentrations 10 times higher than the Cmax in patients, sunitinib did not affect mitogenic signaling or the proliferation of RCC tumor cell lines. In addition, studies of the effect of sunitinib administration on human RCC orthotopically implanted in mice showed that at concentrations matching human exposures (25), sunitinib does not inhibit tumor cells. We believe that inhibition of mitogenic signaling in tumor cells over prolonged treatment courses represents an indirect effect from reduced angiogenesis.
We show that primary RCC cultures from patient tumors support the survival of EC in vitro. The ability of RCC to support EC survival in vitro was inhibited by sunitinib in approximately 50% of tumors. We observed a correlation between sunitinib sensitivity and ERK inhibition in EC. Sunitinib-sensitive tumors were unable to sustain ERK phosphorylation in EC in the presence of the drug. In contrast, sunitinib-resistant tumors uniformly sustained ERK activation in EC despite the presence of sunitinib. While such tests would not be easy to incorporate into clinical practice, our data suggest that ERK activation in EC cocultured with tumor samples could serve as an indicator of sunitinib sensitivity. Importantly, the sunitinib sensitivity seen in cocultures in vitro was reproduced by an independent CAM assay in vivo, which highlights its reliability.
We also found a relationship between VEGF secretion and sunitinib sensitivity. There is controversy in the literature as to the role of VEGF in predicting tumor sensitivity to sunitinib. Plasma VEGF levels do not appear to correlate with sunitinib sensitivity in patients (64, 65). However, VEGF levels in plasma may not signify VEGF production by the tumor, and other cells produce VEGF. On the other hand, VEGF mRNA levels in tumors may correlate with sunitinib responsiveness (66). While this approach focused on mRNA rather than protein measurements, we show that the tumors that produce largest amounts of VEGF are the most sensitive to sunitinib in our assays. Our data suggest that tumor VEGF levels could serve as a surrogate biomarker of sunitinib sensitivity in RCC cells.
We show that RCC cells activate FGFR in both EC and fibroblasts. FGF2 was previously proposed as a mediator of anti-VEGF therapy resistance (23, 24). FGF2 emerged from studies of resistance in a pancreatic β-cell transgenic tumor model expressing a simian viral oncoprotein (24). A second study (23) showed that FGF2 is sufficient to overcome sunitinib effects on EC. However, neither study evaluated the role of this pathway in renal cancer. We show that RCCs express a variety of FGFs and uniformly activate FGFR (as determined by the use of phospho-FRS2). Furthermore, FGFR activation likely contributes to the activation of ERK in EC in a subset of tumors.
Interestingly, RCC signaled not only to EC but also to fibroblasts. In fact, RCC signaled to fibroblasts through dovitinib-sensitive and -insensitive pathways. While RCC-induced ERK activation was uniformly inhibited by dovitinib/PD173074 in EC, this was not the case in fibroblasts. These data show that ERK activation by some RCC in fibroblasts can occur independently of PDGF and FGF receptors, which are inhibited by dovitinib and PD173074.
Our data suggest that RCCs may be broadly classified into 3 subtypes. One subtype (sunitinib sensitive) is characterized by high VEGF production and VEGF-dependent (sunitinib and dovitinib sensitive) activation of EC. A second subtype (sunitinib resistant and dovitinib sensitive) is characterized by lower production of VEGF but higher production of FGFs, resulting in sunitinib-resistant/dovitinib-sensitive activation of EC and fibroblasts. A third subtype (resistant to both sunitinib and dovitinib) produces at least one other growth factor able to activate fibroblasts (or other cell types) independently of dovitinib.
We previously showed that dovitinib had significantly higher activity than sunitinib against human RCC in tumor graft-bearing mice. These results are in contrast to the results of a phase III trial of dovitinib in advanced ccRCC, where dovitinib was found to have activity comparable to that of sorafenib (a VEGR inhibitor like sunitinib) (63). The greater sensitivity to dovitinib of RCC in mice could have several explanations. For example, some tumors may be resistant to dovitinib in humans but not in mice, which could result from the activation of stromal cells (or other cell types) by RCC in humans but not in mice. In fact, there is a precedent for this (i.e., human HGF fails to activate the cognate receptor in mouse cells) (67–69). In addition, tumors in humans may be able to recruit angiogenic cell types (70) whose levels may be diminished in immunocompromised NOD/SCID mice. Notwithstanding the results of experiments performed with tumor grafts, another potential explanation for the results of the phase III trial is a paucity of tumors in the second clade—that is, of tumors that sustain EC and fibroblast activation in a sunitinib-resistant but dovitinib-sensitive manner.
In summary, our results provide a foundation for understanding molecular mechanisms of resistance to antiangiogenic agents and an experimental framework to further dissect the pathways.
ACKNOWLEDGMENTS
We thank patients and their families for graciously donating samples for this study. We thank members of the Brugarolas laboratory for helpful discussions and reading of the manuscript, Payal Kapur for taking IHC pictures, William G. Kaelin for RCC3 and WT8 cell lines, and Osamu Ogawa for Caki-1 cells. We also thank Shane Alexander for generating 786-O cells with EV or VHL.
This work was supported by a T32CA124334 and subsequently a F32CA1360872 fellowship to T.A.T. and by the following grants to J.B.: RO1CA129387, RSG115739, RP160440, and RP101075. Tumor samples were collected through the UTSW Tissue resource, supported in part through grant 1P30CA142543. J.B. is a Virginia Murchison Linthicum Scholar in Medical Research at UT Southwestern.
The content is solely the responsibility of the authors and does not represent official views from any of the granting agencies.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Mukeria A, Holcatova I, Schmidt LS, Toro JR, Karami S, Hung R, Gerard GF, Linehan WM, Merino M, Zbar B, Boffetta P, Brennan P, Rothman N, Chow WH, Waldman FM, Moore LE. 2008. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res 14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linehan WM, Walther MM, Zbar B. 2003. The genetic basis of cancer of the kidney. J Urol 170:2163–2172. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- 3.Duan DR, Pause A, Burgess WH, Aso T, Chen DY, Garrett KP, Conaway RC, Conaway JW, Linehan WM, Klausner RD. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 4.Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. 2000. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci U S A 97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science 269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 6.Kishida T, Stackhouse TM, Chen F, Lerman MI, Zbar B. 1995. Cellular proteins that bind the von Hippel-Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res 55:4544–4548. [PubMed] [Google Scholar]
- 7.Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, Kaelin WG Jr. 1998. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol 18:732–741. doi: 10.1128/MCB.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pause A, Lee S, Worrell RA, Chen DY, Burgess WH, Linehan WM, Klausner RD. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci U S A 94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 10.Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, Maher ER, Pugh CW, Ratcliffe PJ, Maxwell PH. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem 275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 11.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 12.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol 2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 13.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E. 1998. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 14.Ferrara N, Hillan KJ, Gerber HP, Novotny W. 2004. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov 3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 15.Escudier B, Bellmunt J, Negrier S, Bajetta E, Melichar B, Bracarda S, Ravaud A, Golding S, Jethwa S, Sneller V. 2010. Phase III trial of bevacizumab plus interferon alfa-2a in patients with metastatic renal cell carcinoma (AVOREN): final analysis of overall survival. J Clin Oncol 28:2144–2150. doi: 10.1200/JCO.2009.26.7849. [DOI] [PubMed] [Google Scholar]
- 16.Rini BI, Halabi S, Rosenberg JE, Stadler WM, Vaena DA, Archer L, Atkins JN, Picus J, Czaykowski P, Dutcher J, Small EJ. 2010. Phase III trial of bevacizumab plus interferon alfa versus interferon alfa monotherapy in patients with metastatic renal cell carcinoma: final results of CALGB 90206. J Clin Oncol 28:2137–2143. doi: 10.1200/JCO.2009.26.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, Negrier S, Szczylik C, Pili R, Bjarnason GA, Garcia-del-Muro X, Sosman JA, Solska E, Wilding G, Thompson JA, Kim ST, Chen I, Huang X, Figlin RA. 2009. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol 27:3584–3590. doi: 10.1200/JCO.2008.20.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, Chen I, Bycott PW, Baum CM, Figlin RA. 2007. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 19.Engelman JA, Janne PA. 2008. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 20.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 21.Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, Takigawa N, Takata M, Kiura K, Tanimoto M. 2007. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res 67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 22.Yamasaki F, Johansen MJ, Zhang D, Krishnamurthy S, Felix E, Bartholomeusz C, Aguilar RJ, Kurisu K, Mills GB, Hortobagyi GN, Ueno NT. 2007. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res 67:5779–5788. doi: 10.1158/0008-5472.CAN-06-3020. [DOI] [PubMed] [Google Scholar]
- 23.Welti JC, Gourlaouen M, Powles T, Kudahetti SC, Wilson P, Berney DM, Reynolds AR. 2011. Fibroblast growth factor 2 regulates endothelial cell sensitivity to sunitinib. Oncogene 30:1183–1193. doi: 10.1038/onc.2010.503. [DOI] [PubMed] [Google Scholar]
- 24.Casanovas O, Hicklin DJ, Bergers G, Hanahan D. 2005. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell 8:299–309. doi: 10.1016/j.ccr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Sivanand S, Pena-Llopis S, Zhao H, Kucejova B, Spence P, Pavia-Jimenez A, Yamasaki T, McBride DJ, Gillen J, Wolff NC, Morlock L, Lotan Y, Raj GV, Sagalowsky A, Margulis V, Cadeddu JA, Ross MT, Bentley DR, Kabbani W, Xie XJ, Kapur P, Williams NS, Brugarolas J. 2012. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med 4:137ra175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucejova B, Pena-Llopis S, Yamasaki T, Sivanand S, Tran TA, Alexander S, Wolff NC, Lotan Y, Xie XJ, Kabbani W, Kapur P, Brugarolas J. 2011. Interplay between pVHL and mTORC1 pathways in clear-cell renal cell carcinoma. Mol Cancer Res 9:1255–1265. doi: 10.1158/1541-7786.MCR-11-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vega-Rubin-de-Celis S, Abdallah Z, Kinch L, Grishin NV, Brugarolas J, Zhang X. 2010. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry 49:2491–2501. doi: 10.1021/bi902135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peña-Llopis S, Brugarolas J. 2013. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat Protoc 8:2240–2255. doi: 10.1038/nprot.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. 2010. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nat Protoc 5:1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong HS, Chambers AF, Lewis JD. 2012. Assessing cancer cell migration and metastatic growth in vivo in the chick embryo using fluorescence intravital imaging. Methods Mol Biol 872:1–14. doi: 10.1007/978-1-61779-797-2_1. [DOI] [PubMed] [Google Scholar]
- 31.Cho CF, Ablack A, Leong HS, Zijlstra A, Lewis J. 21 June 2011. Evaluation of nanoparticle uptake in tumors in real time using intravital imaging. J Vis Exp doi: 10.3791/2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Liang C, Shirazian S, Zhou Y, Miller T, Cui J, Fukuda JY, Chu JY, Nematalla A, Wang XY, Chen H, Sistla A, Luu TC, Tang F, Wei J, Tang C. 2003. Discovery of 5-5-fluoro-2-oxo-1,2-dihydroindol-(3Z)-ylidenemethyl-2,4-dimethyl-1H-p yrrole-3-carboxylic acid (2-diethylaminoethyl)amide, a novel tyrosine kinase inhibitor targeting vascular endothelial and platelet-derived growth factor receptor tyrosine kinase. J Med Chem 46:1116–1119. doi: 10.1021/jm0204183. [DOI] [PubMed] [Google Scholar]
- 33.Faivre S, Demetri G, Sargent W, Raymond E. 2007. Molecular basis for sunitinib efficacy and future clinical development. Nat Rev Drug Discov 6:734–745. doi: 10.1038/nrd2380. [DOI] [PubMed] [Google Scholar]
- 34.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bender C, Ullrich A. 28 February 2012. PRKX, TTBK2 and RSK4 expression causes sunitinib resistance in kidney carcinoma- and melanoma-cell lines. Int J Cancer doi: 10.1002/ijc.26486. [DOI] [PubMed] [Google Scholar]
- 36.Gotink KJ, Broxterman HJ, Labots M, de Haas RR, Dekker H, Honeywell RJ, Rudek MA, Beerepoot LV, Musters RJ, Jansen G, Griffioen AW, Assaraf YG, Pili R, Peters GJ, Verheul HM. 2011. Lysosomal sequestration of sunitinib: a novel mechanism of drug resistance. Clin Cancer Res 17:7337–7346. doi: 10.1158/1078-0432.CCR-11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang D, Ding Y, Li Y, Luo WM, Zhang ZF, Snider J, Vandenbeldt K, Qian CN, Teh BT. 2010. Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res 70:1053–1062. doi: 10.1158/0008-5472.CAN-09-3722. [DOI] [PubMed] [Google Scholar]
- 38.Faivre S, Delbaldo C, Vera K, Robert C, Lozahic S, Lassau N, Bello C, Deprimo S, Brega N, Massimini G, Armand JP, Scigalla P, Raymond E. 2006. Safety, pharmacokinetic, and antitumor activity of SU11248, a novel oral multitarget tyrosine kinase inhibitor, in patients with cancer. J Clin Oncol 24:25–35. doi: 10.1200/JCO.2005.02.2194. [DOI] [PubMed] [Google Scholar]
- 39.Atkins MB, Hidalgo M, Stadler WM, Logan TF, Dutcher JP, Hudes GR, Park Y, Liou SH, Marshall B, Boni JP, Dukart G, Sherman ML. 2004. Randomized phase II study of multiple dose levels of CCI-779, a novel mammalian target of rapamycin kinase inhibitor, in patients with advanced refractory renal cell carcinoma. J Clin Oncol 22:909–918. doi: 10.1200/JCO.2004.08.185. [DOI] [PubMed] [Google Scholar]
- 40.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG Jr, Signoretti S. 2009. Patterns of gene expression and copy-number alterations in von Hippel-Lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res 69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaelin WG. 2012. Molecular biology of clear cell renal cell carcinoma, p 283–303. In Figlin RA, Rathmell WK, Rini BI (ed), Renal cell carcinoma: translational biology, personalized medicine, and novel therapeutic targets. Springer, New York, NY. [Google Scholar]
- 42.Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V, Ferrara N. 1998. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. J Biol Chem 273:30336–30343. [DOI] [PubMed] [Google Scholar]
- 43.Gospodarowicz D, Moran J, Braun D, Birdwell C. 1976. Clonal growth of bovine vascular endothelial cells: fibroblast growth factor as a survival agent. Proc Natl Acad Sci U S A 73:4120–4124. doi: 10.1073/pnas.73.11.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cenni E, Perut F, Granchi D, Avnet S, Amato I, Brandi ML, Giunti A, Baldini N. 2007. Inhibition of angiogenesis via FGF-2 blockage in primitive and bone metastatic renal cell carcinoma. Anticancer Res 27:315–319. [PubMed] [Google Scholar]
- 45.Nakagawa M, Emoto A, Hanada T, Nasu N, Nomura Y. 1997. Tubulogenesis by microvascular endothelial cells is mediated by vascular endothelial growth factor (VEGF) in renal cell carcinoma. Br J Urol 79:681–687. doi: 10.1046/j.1464-410X.1997.00140.x. [DOI] [PubMed] [Google Scholar]
- 46.Shi W, Siemann DW. 2002. Inhibition of renal cell carcinoma angiogenesis and growth by antisense oligonucleotides targeting vascular endothelial growth factor. Br J Cancer 87:119–126. doi: 10.1038/sj.bjc.6600416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, Lubensky I, Duan DR, Florence C, Pozzatti R, Walther MM, Bander NH, Grossman HB, Brauch H, Pomer S, Brooks JD, Isaacs WB, Lerman MI, Zbar B, Linehan WM. 1994. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 48.Iliopoulos O, Kibel A, Gray S, Kaelin WG Jr. 1995. Tumour suppression by the human von Hippel-Lindau gene product. Nat Med 1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 49.Potapova O, Laird AD, Nannini MA, Barone A, Li G, Moss KG, Cherrington JM, Mendel DB. 2006. Contribution of individual targets to the antitumor efficacy of the multitargeted receptor tyrosine kinase inhibitor SU11248. Mol Cancer Ther 5:1280–1289. doi: 10.1158/1535-7163.MCT-03-0156. [DOI] [PubMed] [Google Scholar]
- 50.Ribatti D, Nico B, Vacca A, Presta M. 2006. The gelatin sponge-chorioallantoic membrane assay. Nat Protoc 1:85–91. doi: 10.1038/nprot.2006.13. [DOI] [PubMed] [Google Scholar]
- 51.Ribatti D, Nico B, Vacca A, Roncali L, Burri PH, Djonov V. 2001. Chorioallantoic membrane capillary bed: a useful target for studying angiogenesis and anti-angiogenesis in vivo. Anat Rec 264:317–324. doi: 10.1002/ar.10021. [DOI] [PubMed] [Google Scholar]
- 52.Ausprunk DH, Knighton DR, Folkman J. 1975. Vascularization of normal and neoplastic tissues grafted to the chick chorioallantois. Role of host and preexisting graft blood vessels. Am J Pathol 79:597–618. [PMC free article] [PubMed] [Google Scholar]
- 53.Ribatti D. 2008. The chick embryo chorioallantoic membrane in the study of tumor angiogenesis. Rom J Morphol Embryol 49:131–135. [PubMed] [Google Scholar]
- 54.Pan CC, Chen PC, Ho DM. 2004. The diagnostic utility of MOC31, BerEP4, RCC marker and CD10 in the classification of renal cell carcinoma and renal oncocytoma: an immunohistochemical analysis of 328 cases. Histopathology 45:452–459. doi: 10.1111/j.1365-2559.2004.01962.x. [DOI] [PubMed] [Google Scholar]
- 55.Shen SS, Truong LD, Scarpelli M, Lopez-Beltran A. 2012. Role of immunohistochemistry in diagnosing renal neoplasms: when is it really useful? Arch Pathol Lab Med 136:410–417. doi: 10.5858/arpa.2011-0472-RA. [DOI] [PubMed] [Google Scholar]
- 56.Bikfalvi A, Klein S, Pintucci G, Rifkin DB. 1997. Biological roles of fibroblast growth factor-2. Endocr Rev 18:26–45. [DOI] [PubMed] [Google Scholar]
- 57.Chow LQ, Eckhardt SG. 2007. Sunitinib: from rational design to clinical efficacy. J Clin Oncol 25:884–896. doi: 10.1200/JCO.2006.06.3602. [DOI] [PubMed] [Google Scholar]
- 58.Mendel DB, Laird AD, Xin X, Louie SG, Christensen JG, Li G, Schreck RE, Abrams TJ, Ngai TJ, Lee LB, Murray LJ, Carver J, Chan E, Moss KG, Haznedar JO, Sukbuntherng J, Blake RA, Sun L, Tang C, Miller T, Shirazian S, McMahon G, Cherrington JM. 2003. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res 9:327–337. [PubMed] [Google Scholar]
- 59.Basilico C, Moscatelli D. 1992. The FGF family of growth factors and oncogenes. Adv Cancer Res 59:115–165. doi: 10.1016/S0065-230X(08)60305-X. [DOI] [PubMed] [Google Scholar]
- 60.Turner N, Grose R. 2010. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 61.Hadari YR, Gotoh N, Kouhara H, Lax I, Schlessinger J. 2001. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc Natl Acad Sci U S A 98:8578–8583. doi: 10.1073/pnas.161259898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lax I, Wong A, Lamothe B, Lee A, Frost A, Hawes J, Schlessinger J. 2002. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol Cell 10:709–719. doi: 10.1016/S1097-2765(02)00689-5. [DOI] [PubMed] [Google Scholar]
- 63.Motzer RJ, Porta C, Vogelzang NJ, Sternberg CN, Szczylik C, Zolnierek J, Kollmannsberger C, Rha SY, Bjarnason GA, Melichar B, De Giorgi U, Grunwald V, Davis ID, Lee JL, Esteban E, Urbanowitz G, Cai C, Squires M, Marker M, Shi MM, Escudier B. 2014. Dovitinib versus sorafenib for third-line targeted treatment of patients with metastatic renal cell carcinoma: an open-label, randomised phase 3 trial. Lancet Oncol 15:286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rini BI, Michaelson MD, Rosenberg JE, Bukowski RM, Sosman JA, Stadler WM, Hutson TE, Margolin K, Harmon CS, DePrimo SE, Kim ST, Chen I, George DJ. 2008. Antitumor activity and biomarker analysis of sunitinib in patients with bevacizumab-refractory metastatic renal cell carcinoma. J Clin Oncol 26:3743–3748. doi: 10.1200/JCO.2007.15.5416. [DOI] [PubMed] [Google Scholar]
- 65.Tran HT, Liu Y, Zurita AJ, Lin Y, Baker-Neblett KL, Martin AM, Figlin RA, Hutson TE, Sternberg CN, Amado RG, Pandite LN, Heymach JV. 2 July 2012. Prognostic or predictive plasma cytokines and angiogenic factors for patients treated with pazopanib for metastatic renal-cell cancer: a retrospective analysis of phase 2 and phase 3 trials. Lancet Oncol doi: 10.1016/S1470-2045(12)70241-3. [DOI] [PubMed] [Google Scholar]
- 66.Paule B, Bastien L, Deslandes E, Cussenot O, Podgorniak MP, Allory Y, Naimi B, Porcher R, de La Taille A, Menashi S, Calvo F, Mourah S. 2010. Soluble isoforms of vascular endothelial growth factor are predictors of response to sunitinib in metastatic renal cell carcinomas. PLoS One 5:e10715. doi: 10.1371/journal.pone.0010715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhargava M, Joseph A, Knesel J, Halaban R, Li Y, Pang S, Goldberg I, Setter E, Donovan MA, Zarnegar R, Michalopoulos GA, Nakamura T, Faletto D, Rosen EM. 1992. Scatter factor and hepatocyte growth factor: activities, properties, and mechanism. Cell Growth Differ 3:11–20. [PubMed] [Google Scholar]
- 68.Rong S, Bodescot M, Blair D, Dunn J, Nakamura T, Mizuno K, Park M, Chan A, Aaronson S, Vande Woude GF. 1992. Tumorigenicity of the met proto-oncogene and the gene for hepatocyte growth factor. Mol Cell Biol 12:5152–5158. doi: 10.1128/MCB.12.11.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang YW, Su Y, Lanning N, Gustafson M, Shinomiya N, Zhao P, Cao B, Tsarfaty G, Wang LM, Hay R, Vande Woude GF. 2005. Enhanced growth of human met-expressing xenografts in a new strain of immunocompromised mice transgenic for human hepatocyte growth factor/scatter factor. Oncogene 24:101–106. doi: 10.1038/sj.onc.1208181. [DOI] [PubMed] [Google Scholar]
- 70.Shojaei F, Wu X, Malik AK, Zhong C, Baldwin ME, Schanz S, Fuh G, Gerber HP, Ferrara N. 2007. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+ Gr1+ myeloid cells. Nat Biotechnol 25:911–920. doi: 10.1038/nbt1323. [DOI] [PubMed] [Google Scholar]