Abstract
FACT (facilitates chromatin transcription) consists of two essential subunits, Spt16 and Pob3, and functions as a histone chaperone. Mutation of spt16 results in a global loss of nucleosomes as well as aberrant transcription. Here, we show that the majority of nucleosome changes upon Spt16 depletion are alterations in nucleosome fuzziness and position shift. Most nucleosomal changes are suppressed by the inhibition of RNA polymerase II (Pol II) activity. Surprisingly, a small subgroup of nucleosome changes is resistant to transcriptional inhibition. Notably, Spt16 and distinct histone modifications are enriched at this subgroup of nucleosomes. We also report 1,037 Spt16-suppressed noncoding transcripts (SNTs) and found that the SNT start sites are enriched with the subgroup of nucleosomes resistant to Pol II inhibition. Finally, the nucleosomes at genes overlapping SNTs are more susceptible to changes upon Spt16 depletion than those without SNTs. Taken together, our results support a model in which Spt16 has a role in maintaining local nucleosome stability to inhibit initiation of SNT transcription, which once initiated drives additional nucleosome loss upon Spt16 depletion.
INTRODUCTION
Chromatin, which is an organized complex of DNA, RNA, and associated proteins, encodes epigenetic information and maintains the stability of the genome (1, 2). Over the years, many chromatin regulators, including histone-modifying enzymes, chromatin-remodeling complexes, and histone chaperones, have been identified. It has become increasingly clear that many of these chromatin regulators modulate transcription and chromatin stability (3). However, the dynamic interplay between transcription, including both coding and noncoding transcription, and chromatin stability is largely underexplored.
The basic repeating unit of chromatin is the nucleosome, consisting of 146 bp of DNA wrapped around one histone octamer (one histone H3-H4 tetramer and two H2A-H2B dimers) (4). Nucleosomes are assembled via two major nucleosome assembly pathways: DNA replication-coupled (RC) nucleosome assembly and DNA replication-independent (RI) nucleosome assembly (2, 5–7). During DNA replication, the parental and newly synthesized H3-H4 tetramers are assembled into different nucleosomes via an RC nucleosome assembly process to maintain chromatin state and gene expression (8). RI nucleosome assembly functions to replace nucleosome H3-H4 with free H3-H4 tetramers in a process called H3-H4 turnover or exchange (7). H3-H4 turnover can occur in both a transcription-dependent and -independent manner (9). Genome-wide studies show that histone exchange occurs at both active and inactive gene promoters, and H3-H4 turnover is likely locus specific. For instance, in Saccharomyces cerevisiae and Drosophila melanogaster, rapid H3-H4 turnover is observed at nucleosomes near the promoter and at gene regulatory elements (10, 11). In contrast, the H3-H4 molecules in nucleosomes in the middle of open reading frames (ORFs) are relatively stable (12, 13). Therefore, the stability of nucleosomes within the gene body may help suppress intragenic cryptic transcription.
Nucleosome mapping through micrococcal nuclease (MNase) digestion followed by deep sequencing reveals that there are approximately 60,000 nucleosomes in the budding yeast genome (14). The nucleosomes in a given cell population are characterized by nucleosome position, the fuzziness of the nucleosome, and nucleosome occupancy (15). In a cell population, each nucleosome unit has a preferred position, which is referred to as the nucleosome position. A shift from this preferred position in the cell population is defined as a position shift. Nucleosome fuzziness refers to a deviation of a nucleosome unit from the preferred position while remaining centered in position. Nucleosome occupancy refers to the frequency at which a nucleosome unit occupies its preferred position. While the underlying DNA sequence can dictate nucleosome positioning in vitro (16), nucleosome positioning in cells is also regulated by chromatin regulators (17). Furthermore, transcription-generated torsion stress can destabilize nucleosomes (18). Therefore, there is an intimate connection between transcription and nucleosome stability in general.
FACT (facilitates chromatin transcription) consists of two essential subunits, Spt16 and SSRP1 (Pob3 subunit of yeast FACT) (19). FACT is a histone chaperone for both H3-H4 and H2A-H2B and has the ability to alter the structure of nucleosomes in vitro, without ATP hydrolysis (20, 21). In yeast cells, mutations in Spt16 or Pob3 result in many defects that are attributed to gene transcription and/or DNA replication. For instance, it has been shown that mutations in SPT16 or POB3 (SSRP1 homolog) exhibit the Spt− phenotype, an indication of altered stringency in the regulation of transcription initiation (22). Spt16 mutations also lead to reduced levels of histone H3 on chromatin, and it has been proposed that Spt16 is important for the deposition of parental H3 following gene transcription (23). Third, mutations in Spt16 result in a global loss of nucleosome positioning and increased levels of DNA-RNA hybrids (24, 25). It has been proposed that FACT has the ability to resolve R-loops and maintain genome integrity following gene transcription. Finally, mutations in Spt16 lead to an increase in cryptic transcription at a subset of genes (26) and the accumulation of H2A.Z in gene bodies (27). However, it remains largely unknown whether these phenotypes observed in spt16 mutant cells, such as the loss of nucleosomes, increased cryptic transcription, and the formation of DNA-RNA hybrids, reflect a direct role for FACT in these processes.
Here, we report a remarkable interplay between nucleosome stability and noncoding transcription in spt16 mutant cells. We show that both the loss of nucleosomes within gene bodies and increased noncoding transcription upon Spt16 depletion are largely suppressed by the inactivation of Rbp1, a subunit of RNA polymerase II (Pol II). Furthermore, we provide evidence supporting the idea that cryptic noncoding transcription is associated with the majority of nucleosome changes observed in the spt16 mutant cells. Finally, we present results indicating that Spt16 maintains the stability of a subgroup of nucleosomes. Alternations at this subgroup of nucleosomes in spt16 mutant cells lead to initiation of cryptic noncoding transcription, which in turn drives additional loss of nucleosome stability in the spt16 mutant cells.
MATERIALS AND METHODS
Genome-wide data sets, yeast strains, and antibodies.
Information of correlation among repeats for micrococcal nuclease (MNase) digestion followed by deep sequencing (MNase-seq) and transcriptome sequencing (RNA-seq) is listed in Table S1 in the supplemental material. The information regarding sequencing depth, single-end versus paired-end sequencing and the number of repeats for chromatin immunoprecipitation-sequencing (ChIP-seq), MNase-Seq, and RNA-seq is listed in Table S2. Saccharomyces cerevisiae strains used in this study were derived from the parental W303 background (leu2-3,112 ura3-1 his3-11 trp1-1 ade2-1 can1-100). Standard yeast media and manipulations were used to generate all the yeast strains. All yeast strains are listed in Table S3.
ChIP assay.
Exponentially growing cells with tandem affinity purification (TAP)-tagged Spt16 (Spt16-TAP) were synchronized with 5 μg/ml α-factor for 3 h to G1 phase at 25°C. Spt16-TAP was fully functional (see Fig. S8 in the supplemental material). The resulting samples were used to perform ChIP assays as previously described (28, 29). The ChIP DNA was used to construct a library for deep sequencing using Illumina sequencing platforms. The wild-type (WT) w3031a strain without any tag was used as the negative control. In general, our Spt16 ChIP-seq results are consistent with the Spt16 ChIP-seq results published by Foltman et al. (30).
MNase-seq to map nucleosome positions.
Wild-type and the G-to-D change at position 132 encoded by spt16 [spt16(G132D)], rpb1-1, and spt16(G132D) rpb1-1 mutant cells were arrested in G1 phase using α-factor at 25°C. These cells were either kept at 25°C or shifted to the nonpermissive temperature (37°C) for 45 min. Cells were cross-linked by 0.1% paraformaldehyde for 15 min and then treated with 0.125 M glycine. The pellets were subjected to different amounts of MNase digestion as previously reported (9). Samples with similar digestion patterns were chosen, and DNA with the size of mononucleosomes was purified to construct libraries according to the manufacturer's guide (Illumina). The library was sequenced using the Illumina 1G genome analyzer or HiSeq 2000 sequencing system. The sequencing results were used to determine nucleosome positions genome-wide as previously described (14, 31, 32). Briefly, the reads were mapped back to the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/) reference genome. The reads mapping to the plus and minus strands of the reference genome were separated. Each read was shifted toward the 3′ direction for half of the estimated fragment size, which was estimated by the distribution of distances between reads on the plus and minus strands. In addition, the comprehensive bioinformatic pipeline, DANPOS (32), was also used to identify the nucleosome peaks and nucleosome fuzziness, positioning, and occupancy. Briefly, nucleosome peaks were first identified at single-nucleotide resolution. Five statistical tests were used to calculate differential signals at each nucleosome between samples and to estimate the degree to which each category a nucleosome belong based on the differential signals. Transcription start sites (TSS) are from the SGD database (33).
Transcriptome analysis of yeast genome by RNA-Seq.
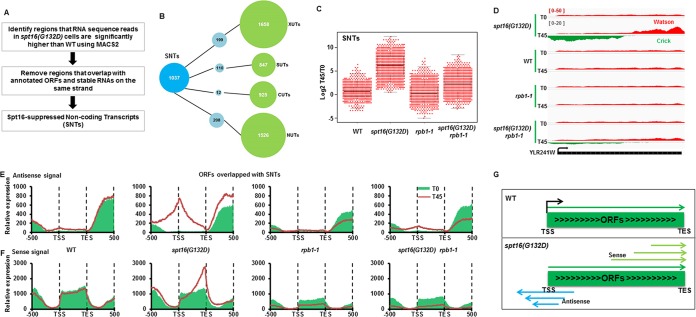
Wild-type and different mutant cells were grown to an optical density at 600 nm (OD600) of around 0.4 and then arrested to G1 phase with α-factor at 25°C. These cells were collected just before the temperature shift (T0) or 45 min after the shift to the nonpermissive temperature (37°C) (T45). Total RNAs were extracted as previously described by the phenol/freeze method (34). RNA qualities were checked with the Fragment Analyzer (Advanced Analytical Technologies, Inc. [AATI]). Total RNA after depletion of rRNA or poly(A) RNA purified by using a Dynabeads oligo(dT)25 kit according to the manufacturer's instructions (Invitrogen) were fragmented and used to construct a library for Illumina sequencing with the TruSeq stranded mRNA sample prep kit (Illumina). For Illumina sequencing, RNAs were subjected to sequencing on the HiSeq 2000 sequencing machine, and 100-nucleotide reads were generated. Similar results were obtained. RNA sequencing quality was checked by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and mapped to yeast genome and gene annotations from Refseq gene using TopHat (35). Cufflinks (36) was used to quantify FPKM (fragments per kilobase per million) values. The tag densities were normalized based on tRNAs present in WT and mutant strains. As shown in Fig. 3A, Spt16-suppressed noncoding transcripts (SNTs) were identified by comparing regions with an spt16(G132D)/WT ratio significantly larger than the background ratio by magnetically activated cell sorting (MACS) (37) after removal of the SNTs that overlapped with annotation ORFs and stable RNAs on the same strand. The SNT start sites in Fig. 5E were defined as the start position at the gene body where RNA-seq sequence reads of spt16(G132D) were significantly higher than WT. To compare SNTs with other noncoding RNAs (ncRNAs), cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs) from Xu et al. (38) and Xrn1-sensitive unstable transcripts (XUTs) from van Dijk et al. (39) were used.
FIG 3.
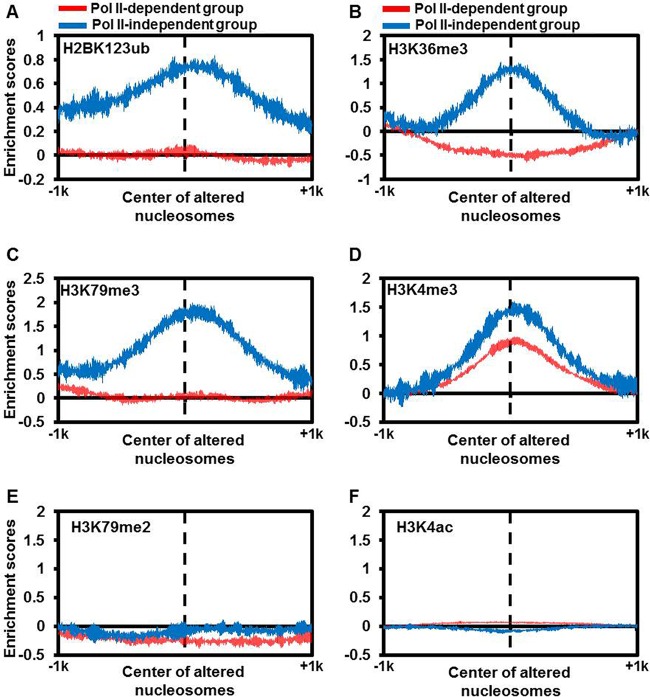
H2BK123ub, H3K36me3, and H3K79me3 are enriched for the “Pol II-independent” subgroup of nucleosomes. The enrichment score for each of the six histone modifications H2BK123ub (A), H3K36me3 (B), H3K79me3 (C), H3K4me3 (D), H3K79me2 (E), and H3K4ac (F) was calculated for two subgroups of nucleosomes, as defined in the legend to Fig. 2A. The histone modification ChIP-chip/Seq data sets from previous studies (42, 67, 68) were used to calculate read density from 1 kb downstream to 1 kb upstream of the center of each altered nucleosome defined in the legend to Fig. 2B.
FIG 5.
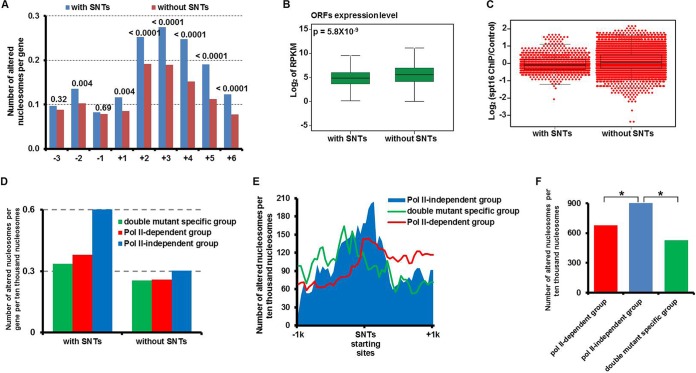
Correlations between the nucleosome changes and SNTs upon Spt16 depletion. (A) Nucleosomes at genes that overlap SNTs are significantly more susceptible to changes upon Spt16 depletion than those without SNTs. The ORFs were separated into two groups, ORFs that overlapped SNTs (with SNTs) and ORFs that did not overlap SNTs (without SNTs). The average number of altered nucleosomes from the −3 to +6 position at each group of ORFs in the spt16(G132D) cells at 37°C for 45 min was calculated. The value above each pair of bars in the bar graph is the P value calculated by Fisher's exact test. (B) The average gene expression level of the ORFs that overlapped the SNTs is lower than those without SNTs. The average expression of the ORFs in each subgroup was calculated. The P value was calculated by a two-tailed Student t test. (C) The Spt16 occupancy is lower at ORFs with SNTs. Spt16-TAP ChIP-seq was performed in G1 phase cells. The Spt16 occupancy levels (log2) (Spt16 ChIP read density/control ChIP read density) at individual ORFs with or without SNTs were calculated and plotted as a dot-box plot, with each dot representing one ORF. (D) ORFs with SNTs are enriched with “Pol II-independent” subgroup nucleosomes as defined in the legend to Fig. 2A. ORFs were separated into two groups depending on whether they overlapped the SNTs, and the number of altered nucleosomes in each subgroup defined in the legend to Fig. 2A at the +2 to +6 position of each ORF with or without SNTs was calculated. (E and F) The SNT start sites are significantly enriched with the “Pol II-independent” subgroup of nucleosomes. (E) A 40-bp sliding window was used to count the three subgroups of altered nucleosomes defined in the legend to Fig. 2A that spanned from kb −1 to +1 from the start site of each SNT. The total number of altered nucleosomes at all SNTs in each subgroup was plotted against each bin. (F) Total number of altered nucleosomes of each subgroup 100 bp surrounding the starting sites of SNTs was determined. The chi-square test was applied to calculate the P values for the Pol II-independent group versus other subgroups of nucleosomes. The values that were significantly different (P < 0.01) are indicated by a bar and an asterisk.
Microarray data accession number.
MNase-seq, ChIP-seq, and RNA-seq data sets have been deposited at Gene Expression Omnibus under accession number GSE66215.
RESULTS
Spt16 depletion results in a global change in chromatin structure.
Spt16, a subunit of the FACT complex, is essential for cell viability. The reduction in Spt16 levels using a temperature-sensitive (ts) mutant, such as spt16(G132D), results in nucleosome alterations, increased cryptic gene transcription, and the formation of R-loops (24, 25). However, the mechanism underlying these changes is not well understood. Therefore, we first determined to what extent the chromatin structure was altered in the mutant strain using MNase-seq. Briefly, bulk chromatin isolated from the wild-type (WT) and spt16(G132D) mutant cells at permissive and nonpermissive temperatures was digested with MNase, which preferentially cleaves the linker DNA between nucleosomes and is commonly used to probe chromatin structure. At the nonpermissive temperature (37°C), Spt16(G132D) mutant proteins were depleted upon raising the temperature to 37°C for 45 min compared to wild-type Spt16 (see Fig. S1A in the supplemental material). The MNase-digested fragments were subjected to next-generation sequencing. On average, we mapped the locations of approximately 66,800 nucleosomes of the wild-type and spt16(G132D) cells (data not shown). The average nucleosome position profile of all genes with transcription start sites (TSS) revealed a nucleosome-free region (NFR) and two well-positioned nucleosomes (+1 and −1 nucleosomes) surrounding the TSS (Fig. 1A; see Fig. S1B and D in the supplemental material), which is in agreement with many published results (14). In addition, well-positioned nucleosomes flanking the replication origins were also observed (31) (see Fig. S1C). At the permissive temperature (25°C), the average nucleosome position and occupancy at the TSS sites were similar between the wild-type and spt16(G132D) mutant cells (see Fig. S1B). At the nonpermissive temperature (T45), the nucleosome positions and occupancy in the wild-type strain were not noticeably changed (see Fig. S1B). In contrast, the nucleosomes at the gene bodies after the +1 nucleosome (Fig. 1A; see Fig. S1D) or at regions beyond the +1 nucleosomes of the replication origins (see Fig. S1C) were significantly altered in the spt16(G132D) mutant cells, with a slight alteration at +1 nucleosomes. These results were observed in three independent repeats (see Table S1 in the supplemental material) and are consistent with published results (data not shown) (24), indicating that inactivation of Spt16 leads to a global change in nucleosome organization.
FIG 1.
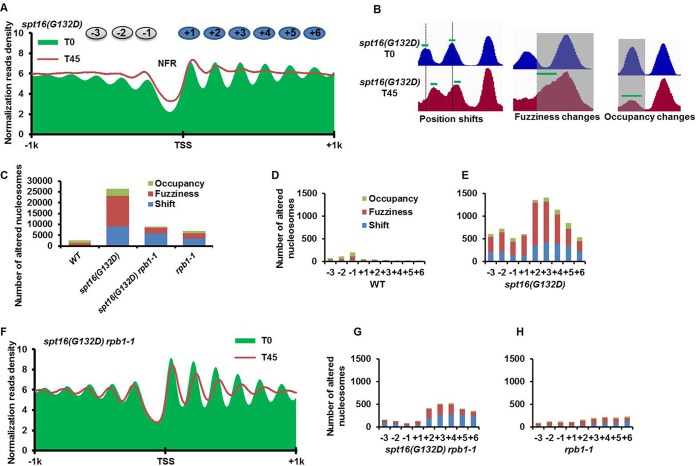
Spt16 depletion results in global changes in nucleosome organization, and most of these changes can be suppressed by RNA polymerase II (Pol II) inhibition. (A) The nucleosomes surrounding the transcription start sites (TSS) are altered in spt16(G132D) cells at the nonpermissive temperature. The average nucleosome profiles centered at the TSS in the spt16(G132D) mutant cells before and after the restrictive temperature treatment are plotted as green areas and red lines, respectively. Cells were arrested at G1 phase at 25°C (T0), shifted to 37°C for 45 min to deplete Spt16(G132D) mutant proteins (T45) and were then used for nucleosome mapping by MNase-seq. (B) Snapshots of the three categories of nucleosome changes (position shift, fuzziness, and occupancy) in the spt16(G132D) mutant cells. The green bars indicate the centers of the nucleosomes in each category, as defined by the DANPOS algorithm, that are used for the calculations in Fig. 2C to G. (C to H) Inhibition of Pol II transcription suppresses most of the nucleosome changes observed in spt16(G132D) mutant cells. (C) The numbers of nucleosome changes in each category (position shift, fuzziness, and occupancy) in the four strains are indicated. (D to H) Comparison of the nucleosome changes in each category from the −3 to +6 nucleosomes surrounding each TSS in wild-type (D), spt16(G132D) (E), rpb1-1 mutant (H), and spt16(G132D) rpb1-1 double mutant cells (G). (F) The average nucleosome profile surrounding the TSS in the spt16(G132D) rpb1-1 cells at 25°C (T0) and 37°C for 45 min (T45). Please note that all nucleosomal changes were repeated three times and are reproducible.
Nucleosomes become fuzzy in spt16(G132D) cells at the nonpermissive temperature.
To gain further insight into the impact of Spt16 inactivation on nucleosome changes, we further grouped the nucleosome changes into position shift, fuzziness, and occupancy in the wild-type and spt16(G132D) cells using the DANPOS method (32) (Fig. 1B and C). A total of 28,035 nucleosomes were altered in the spt16(G132D) cells compared to 2,500 nucleosomes changed in wild-type cells at 37°C. Approximately 15,000 and 10,000 nucleosomal changes were classified as fuzziness and position shift, respectively, in spt16(G132D) cells (Fig. 1B and C). These results suggest that the nucleosomes within a gene body deviate from their preferred positions much more frequently in the spt16(G132D) mutant cells. To determine where these changes occurred in reference to the well-positioned nucleosomes surrounding the TSS, we determined the number of altered nucleosomes from the −3 to +6 positions surrounding each TSS (Fig. 1D and E). In wild-type cells, most of the altered nucleosomes were located from the −3 to −1 positions (Fig. 1D). In contrast, most of the altered nucleosomes in the spt16(G132D) mutant cells were located within the gene body, starting from the +2 nucleosome (Fig. 1E). Taken together, these results indicate that upon Spt16 inactivation/depletion, the nucleosomes become fuzzy and/or shift from their preferred positions, and most of these changes occur within the gene body after the +1 position from the TSS.
Inhibition of ongoing gene transcription restores most of the nucleosomes that were altered by Spt16 depletion.
To gauge the impact of gene transcription on the nucleosome changes, we first used thiolutin, an inhibitor of transcription, which has been used before to study FACT's role in nucleosome reassembly during gene transcription (23). We observed, however, that in contrast to the degradation of Spt16(G132D) proteins in cells treated with dimethyl sulfoxide (DMSO), Spt16(G132D) protein levels in cells treated with thiolutin did not change significantly upon shifting the temperature to 37°C (see Fig. S2A in the supplemental material), suggesting that thiolutin inhibits not only gene transcription but also the degradation of Spt16(G132D) proteins. Therefore, we employed a temperature-sensitive mutation in the RPB1 gene (rpb1-1), which encodes the catalytic subunit of RNA polymerase II and did not affect Spt16 depletion at 37°C (see Fig. S2B). In agreement with published results (40), low levels of nucleosome changes were detected in the rpb1-1 single mutant cells at the nonpermissive temperature of 37°C, with the nucleosome position shifted slightly toward the transcription termination site (TES) compared to the wild-type cells (see Fig. S3A to C). Remarkably, at 37°C, the nucleosomes in the spt16(G132D) rpb1-1 double mutant cells were largely restored compared to the spt16(G132D) mutant cells, based on analysis of the average nucleosome localization profiles starting at TSS using single-end sequencing (Fig. 1F). Similar results were obtained using MNase paired-end sequencing (see Fig. S3D), suggesting that these changes may not be due to the extent of MNase digestion in different strains/conditions, which is known to affect average nucleosome occupancy (40). Moreover, nucleosomal changes detected using the DANPOS method showed that nucleosome changes in occupancy and fuzziness in spt16(G132D) cells were suppressed to a large degree upon inhibiting Pol II, and the nucleosome position shift was reduced to a lesser extent (Fig. 1C; also compare Fig. 1G to E and H). Interestingly, after shifting the temperature to 37°C for 45 min, a significant higher percentage of spt16(G132D) rbp1-1 double mutant cells was viable than spt16(G132D) cells (see Fig. S3E), indicating that inhibition of Pol II transcription suppresses Spt16 depletion-caused cell death. Together, these results strongly suggest that ongoing transcription catalyzed by Pol II is responsible for most of the nucleosomal changes in the spt16(G132D) cells at the nonpermissive temperature.
Spt16 is enriched at the “Pol II-independent” subgroup of nucleosomes that are resistant to transcription inhibition.
While the majority of nucleosomal changes in spt16(G132D) cells were restored upon inactivation/depletion of Rpb1, a significant number of nucleosome changes were detected in the gene body region in the spt16(G132D) rpb1-1 double mutant cells, and most of the nucleosome changes are from the +2 to +6 nucleosomes (Fig. 1F and G). This result suggests that factors other than Pol II-mediated transcription may also contribute to nucleosome changes. We then separated total yeast genes into three groups based on their expression level (high, medium, and low expression) and quantified the nucleosomal changes at these three groups of genes. Surprisingly, the nucleosome changes at these three groups of genes were similar in the spt16(G132D) mutant cells, irrespective of gene expression status (see Fig. S4D to F in the supplemental material). Nucleosomes at each group of genes in WT cells were not altered (see Fig. S4A to C). Interestingly, the nucleosomes at the silent HML locus (see Fig. S4G) but not at the HMR locus (see Fig. S4H) were also altered in the spt16(G132D) cells at 37°C, despite the fact that changes in transcription at both the HML and HMR loci were not detected by RNA-seq (see Fig. S4I). These results support the notion that factors other than transcription also contribute to alterations of some nucleosomes in the spt16(G132D) mutant cells.
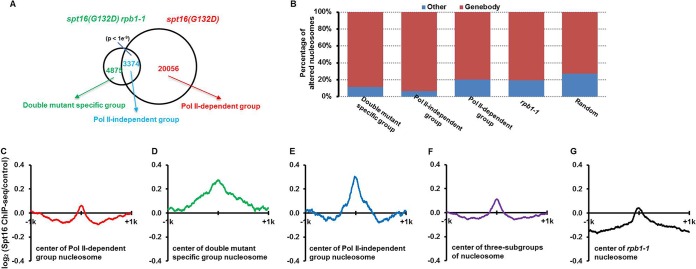
We grouped the nucleosomes altered in spt16(G132D) cells at 37°C into three categories based on whether they were sensitive to Rpb1 inactivation (Fig. 2A). The majority of altered nucleosomes (20,056) in the spt16(G132D) cells were not changed in the spt16(G132D) rpb1-1 double mutant cells (Fig. 2A), supporting the idea that the inhibition of ongoing transcription prevents changes in the majority of nucleosomes upon Spt16 depletion. We named this group the “Pol II-dependent” group. About 4,875 altered nucleosomes were detected only in the spt16(G132D) rpb1-1 double mutant cells. We named this group the “double mutant-specific” group. Notably, approximately 3,374 nucleosomes were altered in both the spt16(G132D) single mutant cells and spt16(G132D) rpb1-1 double mutant cells at the nonpermissive temperature. While it is possible that low levels of transcription after inactivation of Pol II still contribute to nucleosomal changes of this subgroup of nucleosomes, this subgroup of nucleosomes has distinct properties from those of “Pol II-dependent” nucleosomes (see below). For instance, the majority of this subgroup of nucleosomes was located within gene bodies compared to the other subgroups of nucleosomes, including those that were affected only in the spt16(G132D) cells (Fig. 2B). Therefore, we called this group the “Pol II-independent” group of altered nucleosomes.
FIG 2.
A subgroup of nucleosomes altered after Spt16 depletion is resistant to transcription inhibition. (A) The nucleosomes that are altered upon Spt16 depletion in the spt16(G132D) single mutant cells and spt16(G132D) rpb1-1 double mutant cells are separated into three subgroups: nucleosomes altered only in the spt16(G132D) rpb1-1 double mutant cells (double mutant-specific group [4,875]), nucleosomes altered in both the spt16(G132D) and spt16(G132D) rpb1-1 double mutant cells (Pol II-independent group [3,374]), and nucleosomes altered only in the spt16(G132D) cells (Pol II-dependent group [20,056]). A total of 28,305 random nucleosomes, which is equal to the total number of altered nucleosomes in the three groups, were chosen for analysis. (B) The percentage of nucleosomes from each subgroup in panel A at the gene body regions versus other regions was calculated. The nucleosomes altered in the rpb1-1 strain and a random set of nucleosomes were used as controls. (C to G) Spt16 is enriched at the “Pol II-independent” subgroup of nucleosomes. Spt16 ChIP was performed, and the Spt16 ChIP-seq normalized reads from 1 kb upstream to 1 kb downstream of centers of each subgroup of altered nucleosome defined in the legend to Fig. 1A and B were calculated. For comparisons, we also included 6,543 nucleosomes altered in rbp1-1 mutant cells.
To determine whether Spt16 has any roles in this subgroup of nucleosomes, we performed Spt16 ChIP-seq. Consistent with published results (30), Spt16 was enriched at the gene bodies of highly expressed genes (see Fig. S5 in the supplemental material), suggesting that our Spt16 ChIP-seq results are reliable. Next, we compared the levels of Spt16 at the center of the different subgroups of altered nucleosomes using ChIP-Seq (Fig. 2C to G). When the different subgroups of nucleosomes defined in the legend to Fig. 2A were compared, we found that Spt16 was enriched at the centers (defined as in the legend to Fig. 1B) of two subgroups of altered nucleosomes: the “Pol II-independent” subgroup and “double mutant-specific” subgroup (Fig. 2D and E). In contrast, Spt16 was not enriched at the “Pol II-dependent” subgroup (Fig. 2C) or nucleosomes were not altered in the rpb1-1 single mutant cells (Fig. 2F). Together, these results are consistent with the idea that the nucleosome changes in the “Pol II-independent” subgroup of nucleosomes are likely caused by Spt16 depletion itself and are independent of gene transcription.
The “Pol II-independent” subgroup of nucleosomes contains a unique histone modification pattern.
To gain insight into the nucleosome properties of the “Pol II-independent” subgroup, we compared the histone modification patterns at this subgroup of nucleosomes to those at the “Pol II-dependent” subgroup because histone modifications are associated with and/or impact nucleosome changes (40, 41). To do this, we calculated the chromatin immunoprecipitation with microarray technology (ChIP-chip) or ChIP-seq read densities of 18 histone modifications surrounding the center of each nucleosome in these two subgroups using published data sets (42, 43). Compared to the “Pol II-dependent” subgroup, the center of the “Pol II-independent” subgroup of nucleosomes was enriched with three histone modifications (H2BK123ub [monoubiquitination of histone 2B on lysine 123], H3K36me3 [trimethylation of histone H3 at lysine 36], and H3K79me3) but not in the 15 other histone modifications analyzed, including H3K79me2 (dimethylation of histone H3 at lysine 79) and H3K4ac (acetylation of histone H3 at lysine 4) (Fig. 3 and data not shown). We noticed that yeast cells lacking H2BK123ub also exhibit a global reduction in nucleosome occupancy (44). Moreover, in an in vitro transcription system, H2B monoubiquitylation is important for the function of FACT in gene transcription (45). Together, these results suggest that the “Pol II-independent” subgroup of nucleosomes, which is enriched with Spt16, is also associated with a specific pattern of histone modifications, some of which is linked to FACT's function.
Increased coding gene expression is not associated with nucleosome alterations in spt16 mutant cells.
There was an apparent discrepancy between coding gene transcription and nucleosome changes, where the nucleosome changes in the spt16(G132D) cells were independent of the level of coding gene expression (see Fig. S4D and E in the supplemental material), but the inactivation of Pol II transcription largely suppresses these nucleosome changes (Fig. 2A). Therefore, we analyzed how ongoing transcription is associated with the nucleosome changes at different subgroups of nucleosomes. To do this, we first examined the changes in both mRNA and noncoding RNA transcription in the spt16(G132D) cells via strand-specific RNA-seq. To compare the samples under different conditions, in particular inactivation of Pol II, we normalized the RNA-seq reads using the average expression of tRNA genes, which are transcribed by Pol III, in each sample. We noticed that unmodified tRNAs/misfolded tRNAs in yeast can be polyadenylated for degradation (46, 47). Moreover, another study used tRNAs as normalization to identify noncoding RNA (39). Finally, while FACT is required for Pol III function (48), total tRNA synthesis is not affected in cells lacking Nhp6 (49), a protein required for yeast FACT to bind nucleosomes (50). Therefore, we conclude that normalization of RNA-seq using tRNAs is our best option to compare changes in coding and noncoding transcription in spt16(G132D) and rpb1-1 single and double mutant cells.
Upon shifting to the nonpermissive temperature, the expression of 1,576 genes was increased in the spt16(G132D) mutant cells compared with wild-type cells (see Fig. S6A and B in the supplemental material), and most of these changes were suppressed in spt16(G132D) rbp1-1 double mutant cells (see Fig. S6C and D). Remarkably, the nucleosome changes at the genes with increased expression were similar to those without altered expression upon Spt16 depletion in the spt16(G132D) cells (see Fig. S6E), suggesting that increased coding transcription of these genes may not be the primary driver of the nucleosome changes in the spt16(G132D) cells.
Bidirectional cryptic transcription from the gene body is detected in the spt16(G132D) cells.
It is known that inactivation of Spt16 also results in increased cryptic transcription (26). Therefore, we first used an unbiased approach to identify the noncoding transcripts in the spt16(G132D) cells at the nonpermissive temperature and then determined whether noncoding transcription is associated with the nucleosome changes at different subgroups of nucleosomes. Briefly, we used the MACS program, which was developed to analyze broad ChIP-seq peaks by bins, to identify regions where the mRNA sequence reads in the spt16(G132D) cells were significantly higher than in the wild type at 37°C. The regions that overlapped with annotated genes and stable RNAs on the same strand were then removed (Fig. 4A). Using this method, we identified 1,037 noncoding RNAs, most of which were antisense to the ORFs. Moreover, half of the noncoding transcripts did not overlap with the noncoding RNAs identified in other studies, including NUTs, XUTs, and CUTs (51). Therefore, we called these 1,037 noncoding transcripts SNTs for Spt16-suppressed noncoding transcripts (Fig. 4A and B). To determine whether these SNTs were transcribed by Pol II, we compared the expression levels of each SNT in the WT, spt16(G132D), rbp1-1, and spt16(G132D) rbp1-1 mutant cells. SNT expression in the spt16(G132D) cells was significantly higher than that in wild-type cells, consistent with the idea that Spt16 depletion results in increased SNT expression (Fig. 4C). Moreover, the expression of SNTs in the spt16(G132D) rbp1-1 double mutant cells was significantly lower than that in the spt16(G132D) cells, suggesting that these SNTs are mainly transcribed by Pol II (Fig. 4C). We noted that it has been shown that cryptic transcription detected in spt6 mutant cells in budding yeast is also Pol II dependent (52). In short, we detected 1,037 Pol II-dependent noncoding RNAs in the spt16(G132D) cells at the nonpermissive temperature, approximately half of which overlapped with previously identified noncoding RNAs.
FIG 4.
Inactivation of Spt16 results in bidirectional cryptic transcription. (A) Schematic diagram for the identification of Spt16-suppressed noncoding transcripts (SNTs). (B) Overlap of SNTs with other previously annotated noncoding RNAs, cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs) from Xu et al. (38), Xrn1-sensitive unstable transcripts (XUTs) from van Dijk et al. (39), and Nrd1 unterminated transcripts (NUTs) from Schulz et al. (69). If an SNT shares more than 50% length with each known ncRNA, it would be counted as one that overlaps with the known ncRNAs. (C) SNTs are produced by RNA polymerase II. To compare the expression of each SNT in the WT, spt16(G132D), rpb1-1, and spt16(G132D) rpb1-1 mutant cells, the log2 ratio of the RPKM of the individual SNTs at 37°C for 45 min (T45) over 0 min (T0) [log2(T45/T0)] was calculated and plotted using a dot-box plot. Each dot represents one SNT. (D) An SNT overlapping with YLR241W shows increased SNT transcripts in the spt16(G132D) mutant cells at T45 compared to T0 in both the sense and antisense directions. RNA-seq reads for the Watson strand and Crick strand are shown. (E and F) Both antisense and sense noncoding transcripts increase at ORFs that overlapped SNTs. The yeast ORFs were separated into two groups based on whether they overlapped an SNT. Antisense (E) and sense (F) of RNA-Seq reads of ORFs that overlapped SNTs were calculated and plotted from 500 bp upstream TSS to 500 bp downstream TES. The results for ORFs without overlapped SNTs are presented in Fig. S7 in the supplemental material. Red (T0, 25°C) and green lines (T45, 37°C) represent treatments at two different temperatures. (G) A model for the generation of noncoding transcripts through bidirectional cryptic transcription in the spt16(G132D) mutant cells. It is likely that SNT initiates transcription at multiple start sites and thus generates the SNT transcripts shown in panels E and F.
An inspection of the individual SNTs located at each ORF revealed that both the antisense and sense transcripts were elevated (Fig. 4D). Therefore, we calculated the average levels of both the antisense and sense transcripts of each ORF that overlapped the SNTs. The SNTs started from the gene body and peaked close to the TSS, whereas the sense transcripts at these ORFs peaked close to the TES (Fig. 4E and F). Similar results were obtained using RNA-seq data sets obtained using total RNA depleted of rRNA (see Fig. S7A and B in the supplemental material). The same analysis was also applied to the ORFs that did not overlap with SNTs. Interestingly, the sense transcripts were not altered at the ORFs without detectable SNTs (see Fig. S7C and D), suggesting that the increased expression of ORFs in the sense direction is likely due to increased cryptic transcription. Finally, transcription inhibition using the rbp1-1 mutant suppressed both the sense and antisense transcripts (Fig. 4E and F). Together, these results strongly suggest that cryptic transcription initiates at multiple start sites of a gene body and then proceeds bidirectionally upon inactivation/depletion of Spt16 (Fig. 4G).
Noncoding transcription is associated with nucleosome changes in spt16(G132D) mutant cells.
Because increased coding transcription was not associated with the nucleosome changes observed in the spt16(G132D) cells, we asked whether cryptic noncoding transcription had any relationship with the nucleosome changes observed in the spt16(G132D) cells. To answer this question, we first compared the nucleosome changes at the ORFs overlapping with SNTs to those without SNTs in the spt16(G132D) cells. Significantly more nucleosomes were altered at genes with SNTs than those without SNTs (Fig. 5A). This result suggests that cryptic noncoding transcription likely drives the nucleosome changes at the gene body in the spt16(G132D) mutant cells. Consistent with this interpretation, the expression of ORFs with SNTs in the wild-type cells was significantly lower than those without SNTs (Fig. 5B), which again argues against the idea that mRNA transcription is the primary force that drives the nucleosome alterations at these genes that overlapped the SNTs in the spt16(G132D) cells. Moreover, the Spt16 levels at ORFs with SNTs were lower than those without SNTs (Fig. 5C), providing an explanation for why this group of genes was more susceptible to Spt16 depletion.
To gain further insight into the relationship between the nucleosome changes and SNTs, we next asked which subgroup of nucleosomes, which was altered in the spt16(G132D) cells and defined in the legend to Fig. 2A, was enriched at the ORFs with SNTs. To do this, we separated the ORFs into two groups, ORFs with SNTs and ORFs without SNTs, calculated the number of altered nucleosomes from +2 to +6 at each ORF, and normalized the values to the total number of nucleosomes of each group. We found that nucleosomes altered at the ORFs with SNTs were enriched with the “Pol II-independent” subgroup of nucleosomes (Fig. 5D). In contrast, nucleosomes altered at the ORFs without an overlapping SNT did not exhibit any enrichment. These results suggest that SNT production in the spt16(G132D) cells is associated with a nucleosome change that is insensitive to Pol II inhibition.
To further explore this relationship, we asked where the “Pol II-independent” subgroup of nucleosomes was localized at the SNTs by calculating the number of altered nucleosomes from kb −1 to kb +1 surrounding the start site of the SNTs. We observed that the “Pol II-independent” subgroup of nucleosomes was localized preferentially at the midpoint of the SNT start sites compared to two other subgroups of nucleosome changes in the spt16(G132D) cells (Fig. 5E). To analyze whether the difference between “Pol II-dependent” and “Pol II-independent” subgroup of nucleosomes at the starting sites of SNTs was statistically significant, we determined the number of altered nucleosomes 100 bp surrounding the starting sites of SNTs and determined the P value. We found that the “Pol II-independent” subgroup of nucleosomes was significantly more enriched at the starting sites of SNTs than the “Pol II-dependent” subgroup of nucleosomes (Fig. 5F). These results raise the possibility that these nucleosome changes in the “Pol II-independent” subgroup facilitate the initiation of SNT cryptic transcription.
DISCUSSION
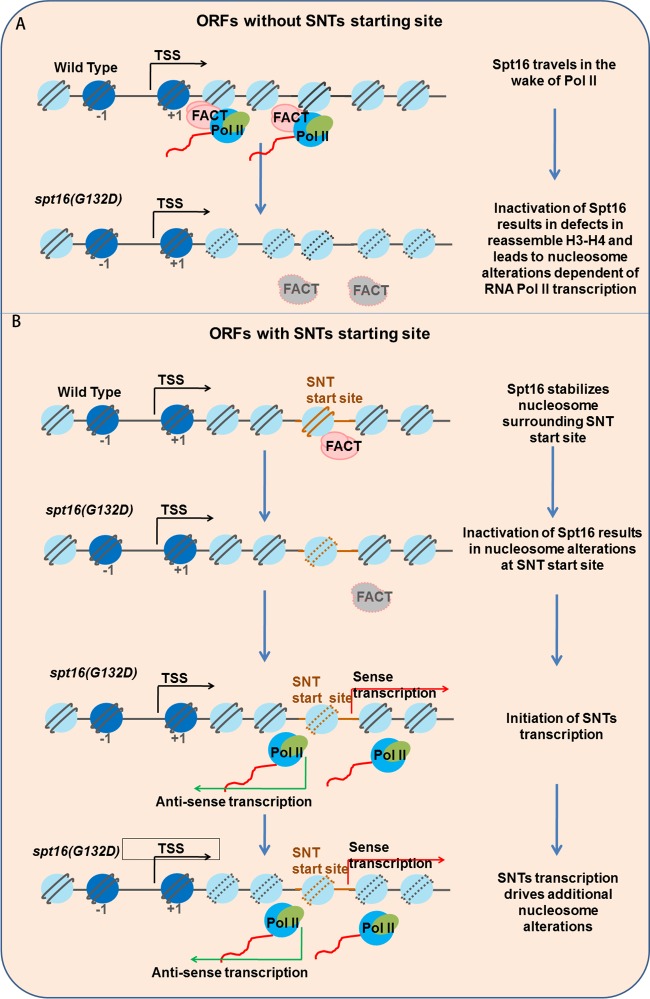
It is known that Spt16 inactivation results in global changes to nucleosomes, altered coding and cryptic noncoding transcription. However, the relationships among these changes upon Spt16 inactivation are not clear. Here, we show that Spt16 depletion results in global changes in nucleosome fuzziness and position shifts and that the majority of these changes are suppressed by inhibiting RNA polymerase II. Notably, a subgroup of nucleosome changes are resistant to Pol II inhibition, and this “Pol II-independent” subgroup of nucleosomes has properties that are distinct from the nucleosome changes that are sensitive to Pol II inhibition, including the enrichment of Spt16 as well as distinct histone modifications. In addition, we show that Spt16 depletion also leads to bidirectional cryptic gene transcription from regions at the gene body and that the nucleosomes at the centers of these sites are enriched with the “Pol II-independent” subgroup of nucleosomes. These results support the model that FACT likely has multiple functions to maintain genomic nucleosome integrity (Fig. 6). At highly expressed ORFs where Spt16 is enriched, Spt16 travels with RNA Pol II and restores nucleosome organization during transcription (Fig. 6A). However, at the ORFs with SNTs where transcription is generally low, Spt16 may also have a role in directly protecting nucleosome stability surrounding the SNT start sites. Upon Spt16 depletion, the nucleosomes within this subgroup become fuzzy or shift from their original positions, leading to the initiation of cryptic transcription. The ongoing cryptic transcription, in turn, promotes additional changes to the nucleosomes within gene bodies (Fig. 6B). Together, these studies reveal a role for FACT in maintaining local nucleosome stability and a novel interplay between noncoding transcription and the maintenance of nucleosome stability.
FIG 6.
A model for Spt16 to maintain nucleosome stability through different mechanisms. (A) At about 678 highly expressed genes without SNT start sites, Spt16 travels with RNA polymerase II (Pol II) and is involved in restoring nucleosome organization during transcription. In the spt16(G132D) mutant cells, the inactivation/depletion of Spt16 results in dramatic changes to the nucleosome positions that are dependent on RNA Pol II-mediated transcription. (B) At 931 genes whose coding regions overlap SNTs, Spt16 maintains nucleosome stability at nucleosomes surrounding the SNT start sites and prevents SNT initiation. Upon depletion of Spt16, the nucleosomes at the SNT start sites become altered and noncoding transcription initiates. Bidirectional noncoding transcription triggers additional nucleosome changes.
Noncoding transcripts have been detected at promoters and enhancers. For instance, divergent transcription has been detected at the promoters of many genes from yeast to human cells (53–55). In human cells, bidirectional transcripts from enhancers (eRNAs) have also been detected in mammalian cells (56, 57). Both enhancers and promoters are located in nucleosome-free regions. The functions of noncoding transcripts at the promoters and enhancers are not well understood, and one study indicates that eRNAs can promote cell-type-specific chromatin accessibility (58). Therefore, our observations on SNTs that can drive nucleosomal changes are consistent with noting that noncoding transcription at promoters and enhancers may help to create and/or maintain the nucleosome-free regions at these sites and thereby facilitate the association of transcription factors with these DNA regulatory elements.
Dramatic changes in nucleosomes have been observed in several other studies. For instance, loss of chromatin remodeling complex Isw2 leads to global nucleosome changes and increased noncoding transcription (59). In both mammalian and yeast cells lacking HMGB1 protein and Nhp6, a global reduction of histone protein levels and nucleosome occupancy is detected. The reduction in nucleosome occupancy is nonuniform and is likely due to different sites competing for available histones (60). In addition, depletion of histone H3 proteins in S. cerevisiae also results in alterations in nucleosome occupancy and positions at a defined subset of nucleosomes, and interestingly, nucleosomes altered by H3 depletion do not overlap with those altered in cells lacking Nhp6a/Nhp6b (61). Schizosaccharomyces pombe cells lacking chromatin remodeler Chd1 (Hrp3 in S. pombe) exhibit global changes in nucleosomes and production of antisense transcripts (62–64). Finally, S. pombe yeast cells lacking histone chaperone Spt6 also exhibit dramatic changes in nucleosome positioning and occupancy and increased cryptic transcription (65).
The results of our present study appear to have some discrepancies with some of the general conclusions of these studies, but in fact, this study provides more evidence to deepen the understanding of current models. First, most of these studies proposed that nucleosomal changes are the causes of antisense transcription but not vice versa. Our studies are consistent with this idea because we show that nucleosomes at SNT starting sites are more susceptible to changes upon Spt16 inactivation and propose that these nucleosomal changes drive the initiation of antisense transcription (Fig. 6B). Our studies extend these studies by showing that antisense transcription, once initiated, can further promote the loss of nucleosomes in spt16(G132D) cells, thereby revealing an intimate connection among nucleosomal changes and antisense transcription.
Second, it has been shown that depletion of Nhp6a/Nhp6b leads to changes in nucleosome occupancy and/or positioning. While Nhp6 is not part of the stable Pob3-Spt16 complex, Nhp6 can bind nucleosomes and recruit yeast FACT to these nucleosomes (50). In contrast, we observed that in spt16(G132D) cells, most changed nucleosomes are due to fuzziness changes based on analysis using the DANPOS method. It is possible that the dramatic nucleosomal changes upon Spt16 reduction make it difficult for the DNAPOS program to differentiate occupancy changes from fuzziness changes. Nonetheless, the fact that far more nucleosomes became fuzzy upon Spt16 depletion suggests that Spt16 and Nhp6 may also have distinct functions in chromatin dynamics.
Finally, FACT is proposed to reassemble H3-H4 in the wake of gene transcription (23). One line of evidence supporting this conclusion is that treating cells with thiolutin prevents loss of H3 from chromatin after Spt16 depletion. We observed that thiolutin inhibits the degradation of Spt16(G132D) mutant proteins at nonpermissive temperature, indicating that the inhibition of histone H3 loss by thiolutin in spt16(G132D) cells at the nonpermissive temperature may be due to inhibition of degradation of the Spt16(G132D) mutant proteins. Despite this, our conclusion that noncoding transcription also promotes nucleosomal loss in spt16(G132D) cells does not contradict the conclusion per se. In fact, we observed that nucleosomes were also lost at gene bodies where SNTs were not detected. In general, the expression level of this group of coding genes is higher than those with SNTs (Fig. 5B). While it is possible that high transcript levels prevent us from identifying SNTs at these genes, we notice that the ability to identify cryptic transcription in set2Δ mutant cells is independent of transcription frequency (66). Nonetheless, it is equally likely that the nucleosome loss is due to a compromised role of FACT in nucleosome assembly during gene transcription (Fig. 6A).
Supplementary Material
ACKNOWLEDGMENTS
We thank Tim Formosa for helpful discussions. We thank Alain Verreault, Tim Formosa, and Michael Hampsey for plasmids and yeast strains. We thank Yun Zhang and Wenping Ma of the NGS group at Peking University for assistance with library preparation and Illumina sequencing.
This work was supported by grants from the NIH (GM81838) to Z.Z., NIH (GM104097) to D.M.M., National Science Foundation for Excellent Young Scholars of China Grant (31322017) and National Natural Science Foundation of China Grant (31370767) to Q.L. Q.L. is also a scholar from the Chinese 1000 Plan-The Young Talents Group.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00152-16.
REFERENCES
- 1.Haushalter KA, Kadonaga JT. 2003. Chromatin assembly by DNA-translocating motors. Nat Rev Mol Cell Biol 4:613–620. doi: 10.1038/nrm1177. [DOI] [PubMed] [Google Scholar]
- 2.Ransom M, Dennehey BK, Tyler JK. 2010. Chaperoning histones during DNA replication and repair. Cell 140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatesh S, Workman JL. 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 4.Luger K. 2006. Dynamic nucleosomes. Chromosome Res 14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 5.Groth A, Rocha W, Verreault A, Almouzni G. 2007. Chromatin challenges during DNA replication and repair. Cell 128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Burgess RJ, Zhang Z. 2013. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szenker E, Ray-Gallet D, Almouzni G. 2011. The double face of the histone variant H3.3. Cell Res 21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. 2010. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet 4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamai A, Imoberdorf RM, Strubin M. 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell 25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 13.Mito Y, Henikoff JG, Henikoff S. 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science 315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 14.Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pugh BF. 2010. A preoccupied position on nucleosomes. Nat Struct Mol Biol 17:923. doi: 10.1038/nsmb0810-923. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teves SS, Henikoff S. 2014. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol 21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinberg D, Sims RJ III. 2006. de FACTo nucleosome dynamics. J Biol Chem 281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 20.Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. 2009. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell 35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler DD, Luger K. 2011. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malone EA, Clark CD, Chiang A, Winston F. 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol 11:5710–5717. doi: 10.1128/MCB.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamai A, Puglisi A, Strubin M. 2009. Histone chaperone Spt16 promotes redeposition of the original H3-H4 histones evicted by elongating RNA polymerase. Mol Cell 35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 24.van Bakel H, Tsui K, Gebbia M, Mnaimneh S, Hughes TR, Nislow C. 2013. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet 9:e1003479. doi: 10.1371/journal.pgen.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. 2014. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev 28:735–748. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeronimo C, Watanabe S, Kaplan CD, Peterson CL, Robert F. 2015. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol Cell 58:1113–1123. doi: 10.1016/j.molcel.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Li Q, McCullough L, Kettelkamp C, Formosa T, Zhang Z. 2010. Ubiquitylation of FACT by the Cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev 24:1485–1490. doi: 10.1101/gad.1887310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foltman M, Evrin C, De Piccoli G, Jones RC, Edmondson RD, Katou Y, Nakato R, Shirahige K, Labib K. 2013. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 3:892–904. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 31.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W. 2013. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23:341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt ME, Brown TA, Trumpower BL. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, Nicolas A, Thermes C, Morillon A. 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 40.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zentner GE, Tsukiyama T, Henikoff S. 2013. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet 9:e1003317. doi: 10.1371/journal.pgen.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. 2011. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev 25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, Bonneil E, Thibault P, Verreault A, Festenstein RJ. 2011. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet 7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. 2011. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 46.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 47.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, Russell J, Lee SC, Zomerdijk JC. 2009. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J 28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kruppa M, Moir RD, Kolodrubetz D, Willis IM. 2001. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol Cell 7:309–318. doi: 10.1016/S1097-2765(01)00179-4. [DOI] [PubMed] [Google Scholar]
- 50.Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smolle M, Workman JL. 2013. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta 1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 53.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 54.Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lam MT, Li W, Rosenfeld MG, Glass CK. 2014. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Natoli G, Andrau JC. 2012. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 58.Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. 2013. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 60.Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, Grianti P, Pagani M, Bonaldi T, Ragoussis J, Friedman N, Camilloni G, Bianchi ME, Agresti A. 2011. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol 9:e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gossett AJ, Lieb JD. 2012. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet 8:e1002771. doi: 10.1371/journal.pgen.1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shim YS, Choi Y, Kang K, Cho K, Oh S, Lee J, Grewal SI, Lee D. 2012. Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin. EMBO J 31:4375–4387. doi: 10.1038/emboj.2012.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hennig BP, Bendrin K, Zhou Y, Fischer T. 2012. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep 13:997–1003. doi: 10.1038/embor.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pointner J, Persson J, Prasad P, Norman-Axelsson U, Stralfors A, Khorosjutina O, Krietenstein N, Svensson JP, Ekwall K, Korber P. 2012. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J 31:4388–4403. doi: 10.1038/emboj.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bahler J, Park PJ, Winston F. 2013. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol 33:4779–4792. doi: 10.1128/MCB.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. 2009. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulz D, Schwalb B, Kiesel A, Baejen C, Torkler P, Gagneur J, Soeding J, Cramer P. 2013. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155:1075–1087. doi: 10.1016/j.cell.2013.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.