SUMMARY
Since the first H7N9 human case in Shanghai, February 19, 2013, the emerging avian-origin H7N9 influenza A virus has become an epizootic virus in China, posing a potential pandemic threat to public health. From April 2 to April 28, 2013, 422 oral-pharyngeal and cloacal swabs were collected from birds and environmental surfaces at five live poultry markets (LPMs) and 13 backyard poultry farms (BPFs) across three cities, Wuxi, Suzhou, and Nanjing, in the Yangtze Delta Region. A total of 22 isolates were recovered, and 6 were subtyped as H7N9, 9 as H9N2, 4 as H7N9/H9N2, and 3 un-subtyped influenza A viruses. Genomic sequences showed that the HA and NA genes of the H7N9 viruses were similar to those of the H7N9 human isolates as well as other avian origin H7N9 isolates in the region but the PB1, PA, NP, and MP genes of the sequenced viruses were, however, more diverse. Among the four H7N9/H9N2 mixed infections, three were from LPM whereas the other one from the ducks at one BPF, which were H7N9 negative in serological analyses. A survey of the bird trading records of the LPMs and BPFs indicates that trading was a likely route for virus transmission across these regions. Our results suggested that a better biosecurity and more effective vaccination should be implemented in backyard farms besides biosecurity management in LPMs.
Keywords: H7N9, Influenza A virus, live poultry market, backyard poultry farm, avian influenza virus, wild bird, domestic poultry
INTRODUCTION
Influenza A virus is a segmented, negative strand of RNA virus. Migratory waterfowl, especially Anseriformes and Charadriiformes, are proposed as the reservoirs for influenza A viruses (20). They maintain a large influenza virus genetic pool, which contributes to the appearance of new strains in humans, lower mammals, and other birds. The backyard poultry has been shown to be one of the sites for transmission of virus from wild birds to domestic poultry, and even to humans (1, 10, 22).
On February 19, 2013, a novel H7N9 low pathogenic avian influenza virus was reported to cause a fatal human case in Shanghai (3, 21). Since then, this virus has caused at least 588 laboratory confirmed human cases (http://www.who.int/influenza/human_animal_interface/influenza_h7n9/en/). This virus has been widely distributed in domestic poultry, especially the birds in the live poultry markets. A modeling study suggested that the density of live poultry market is an important predictor of H7N9 infection risk in markets where this virus is circulating (4). Genetic analyses showed that the HA and NA genes were derived from viruses in migratory waterfowl and six other internal genes from the influenza viruses, most likely H9N2, in domestic poultry (3). These viruses have become epizootic in the eastern and southeastern China and evolved into multiple regionally distinct lineages with different reassortant genotypes (12, 13). It is still unclear whether these diverse H7N9 genes were multiple introductions from wild birds or these reassortments were generated in LPMs or domestic poultry farms. Nevertheless, the details on emergence of H7N9 virus were not clear, especially during earlier stage of H7N9 outbreaks.
The goal of this study is to assess the role of backyard birds in emergence and spread of H7N9 influenza virus in early stage of H7N9 outbreaks. This study was carried out in three cities Wuxi, Suzhou, and Nanjing of Jiangsu Province, in the Yangtze Delta Region, that contained the first H7N9 human case and the major human cases (107 out of 137) during the first wave of H7N9 outbreaks.
Materials and Methods
Sample collection and storage
From April 2 to April 28 of 2013, a total 422 swabs, including 356 pairs of oral-pharyngeal and cloacal swabs and 66 environmental samples were collected (e.g. droppings, cage, and drainage water). Each swab was placed in an individual tube with cold sterile PBS containing 2,000 U/mL penicillin and 1,000 ug/mL streptomycin. These tubes were kept in an ice box before and during shipping to the laboratory, and stored at -80 degree C. All manipulations of these samples were conducted under Biosafety Level 2 containment facilities.
RNA extraction and reverse transcription
Solid debris of the samples was pelleted by centrifugation at 4,000 × g for 5 min, and the supernatants were collected. For rapid determination of the presence of any H7 and H9 viruses, primers specific for these subtypes were designed, based on the alignment of H7 and H9 gene sequences. Total viral RNA was extracted using the AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Axygen Inc., Hangzhou, China) from the supernatants of the samples according to the manufacturer’s instruction. Complementary DNAs were synthesized with primer Uni12 (AGCAAAAGCAGG) using the AMV reverse transcriptase (TaKaRa Biotechnology, Dalian, China) for 1 h at 42 °C. PCR amplification was performed with specific primers for H7 and H9 using Ex Taq DNA Polymerase according to the manufacturer’s protocol (TaKaRa Biotechnology, Dalian, China). Primers were H7-1120F (5’-AATGCACARGGAGGAGGAACT -3’), H7-1620R (5’-TGAYGCCCCGAAGCTAAACCA -3’), H9-184F (5’-CTYCACACAGAGCACAATGG -3’) and H9-691R (5’-GYCACACTTGTTGTTGTRTC -3’).
All samples positive for H7 or H9 influenza A viruses were selected for genome sequencing. PCR was performed with influenza A virus specific primers for eight genes (8) using Ex Taq DNA Polymerase according to the manufacturer’s protocol ( TaKaRa Biotechnology, Dalian, China) to sequence full length genome. PCR products were purified using an AxyPrep DNA Gel Extraction Kit (Axygen Inc., Hangzhou, China) in accordance with manufacturer’s recommendations. The products were cloned into the pMD-18T vector (TaKaRa Biotechnology, Dalian, China) and sequenced by Genscript Company, Nanjing, China. Sequence data were compiled and edited using the Lasergene sequence analysis software package (DNAStar, Inc., Madison, WI). The accession numbers of these genes were KT779566-KT779633.
Serological Assays
The birds from BPFs were vaccinated through intramuscular injection with inactivated H5N1 and H9N2 vaccines, which were supplied by Chinese government. The serological responses were measured using hemagglutinination inhibition assay. The hemagglutinination inhibition assays were performed according to OIE guidelines using 1% chicken red blood cells (2).
Phylogenetic analysis and molecular characterization
The phylogenetic analyses were performed using maximum likelihood by GARLI version (24) and bootstrap resampling analyses using PAUP* 4.0 Beta (18) with a neighborhood joining method, as described earlier (19). A total of 1,410 HA sequences of the H7 subtypes of influenza A viruses were downloaded from Influenza Research Database (IRD; http://www.fludb.org), and the phylogenetic analysis was used to identify those HA genes belonging to the Eurasian lineage for further analysis; 46 H7 sequences were included in the final uses in the phylogenetic trees.
Poultry trading routing survey
Along with influenza swab sample collection, a poultry trading survey was performed, including poultry source, climate, bird species, worker information, and farm or LPM size.
RESULTS AND DISCUSSION
The onset of the first case of H7N9 human infection in Wuxi city, Jiangsu province, was March 8, 2013. From April 2 to April 28, we sampled five LPMs and 13 BPFs; the number of birds in each sampling location ranged from about ten to 5,000. The sampled LPMs and BPFs were located across three cities, Wuxi, Suzhou, and Nanjing, in the Yangtze Delta Region (Figure 1). A total of 422 samples were collected, including oral-pharyngeal and cloacal swabs from broiler chicken, hen, duck, goose, pigeon, and quail, and environmental surfaces (swabs from droppings, cages, and water drainage). Twenty-two of the samples were influenza A virus positive as determined by M-gene based quantitative RT-PCR (Table 1). Genomic sequencing of these clinical samples showed that 6 were H7N9, 9 were H9N2, 4 were co-infected with both H7N9 and H9N2, and three un-subtyped influenza A viruses. Three samples with both H7N9 and H9N2 were from LPMs and the other one sample co-infected with H7N9 and H9N2 virus was from one BPF (Table 2).
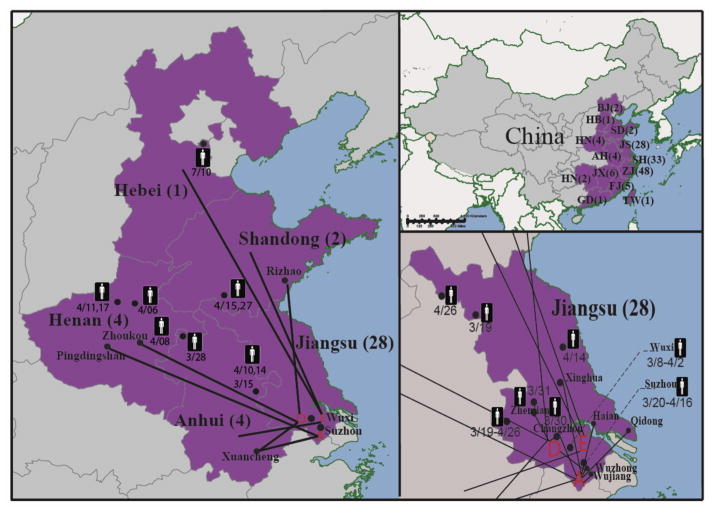
Figure 1.
The H7N9 positive LPMs and the poultry trades associated with these LPMs. (A) Geographical distributions of reported human case during early stage of H7N9 outbreaks. The number of the human cases is indicated in the parenthesis. Abbreviations: AH, Anhui; BJ, Beijing; FJ, Fujian; GD, Guangdong; HB, Hebei; HN, Henan; HN, Hunan; JX, Jiangxi; SD, Shandong; SH, Shanghai; TW, Taiwan; ZJ, Zhejiang. (B) Poultry trade between H7N9 positive LPMs in Jiangsu and nearby provinces. Our surveillance recovered H7N9 viruses from LPM A (Suzhou, 4/2/2013), D (Wuxi, 4/5/2013), and E (Wuxi, 4/5/2013). The birds from these markets were trading with at least 8 regions from Anhui, Henan, Hebei, and Shandong. These four regions were later reported with H7N9 human cases. (C) Poultry trade between H7N9 positive LPMs and nearby cities in Jiangsu province. The disease onset dates of the H7N9 cases are notified.
TABLE 1.
Results for influenza virological surveillance from Jiangsu Province in April of 2013
| Categories | Bird species | Total number of samples | IAVs | Number of positive samples
|
other IAVs | ||
|---|---|---|---|---|---|---|---|
| H7N9 | H9N2 | H7N9+H9N2 | |||||
| Backyard (13) | Chicken | 20 | 0 | 0 | 0 | 0 | 0 |
| Duck | 48 | 1 | 0 | 0 | 1 | 0 | |
| Goose | 20 | 0 | 0 | 0 | 0 | 0 | |
| Pigeon | 50 | 0 | 0 | 0 | 0 | 0 | |
|
| |||||||
| LPM (5) | Chicken | 116 | 12 | 3 | 7 | 2 | 0 |
| Duck | 48 | 6 | 2 | 0 | 1 | 3 | |
| Goose | 9 | 0 | 0 | 0 | 0 | 0 | |
| Pigeon | 33 | 1 | 1 | 0 | 0 | 0 | |
| Quail | 12 | 0 | 0 | 0 | 0 | 0 | |
| water (2)a | 31 | 0 | 0 | 0 | 0 | 0 | |
| Wetland Environment (3) | Bird dropping | 35 | 2 | 0 | 2 | 0 | 0 |
| Summary | 422 | 22 | 6 | 9 | 4 | 3 | |
The market was cleaned before sampling.
TABLE 2.
Co-infections of H7N9 influenza A virus with other subtypes of influenza A viruses
| Sample Index | Date | Location | Species | H7N9 | H9N2 | Other subtypes |
|---|---|---|---|---|---|---|
| 040201H02 | 04/02/2013 | LBM-A | Hen | + | − | − |
|
| ||||||
| 040201H04 | 04/02/2013 | LBM-A | Hen | + | − | − |
|
| ||||||
| 040205H | 04/02/2013 | LBM-A | Duck | − | − | + |
|
|
||||||
| 040207GH01 | 04/02/2013 | LBM-A | Sanhuang Broiler | − | + | − |
|
| ||||||
| 040207GH02 | 04/02/2013 | LBM-A | Sanhuang Broiler | − | + | − |
|
| ||||||
| 040207GH03 | 04/02/2013 | LBM-A | Sanhuang Broiler | − | + | − |
|
| ||||||
| 040207GH04 | 04/02/2013 | LBM-A | Sanhuang Broiler | − | + | − |
|
| ||||||
| 040207GH05 | 04/02/2013 | LBM-A | Sanhuang Broiler | + | + | − |
|
| ||||||
| 040207GH06 | 04/02/2013 | LBM-A | Sanhuang Broiler | − | + | − |
|
| ||||||
| 040208GH | 04/02/2013 | LBM-A | Duck | − | − | + |
|
| ||||||
| 04030201Ga | 04/03/2013 | LBM-B | Chicken | + | − | − |
| 04030201H | 04/03/2013 | LBM-B | Chicken | − | + | − |
|
| ||||||
| 04030202H | 04/03/2013 | LBM-B | Chicken* | − | + | − |
|
| ||||||
| 04030203GH | 04/03/2013 | LBM-C | Chicken | − | + | − |
|
| ||||||
| 04030204GH | 04/03/2013 | LBM-C | Wild Duck | + | + | − |
|
| ||||||
| 0405001G | 04/05/2013 | LBM-D | Duck | + | − | − |
|
| ||||||
| 0405005G | 04/05/2013 | LBM-D | Chicken | + | − | − |
| 0405006G | 04/05/2013 | LBM-D | Duck | + | − | − |
| 0405007G | 04/05/2013 | LBM-E | Pigeon | + | − | − |
| 0405015G04 | 04/05/2013 | LBM-E | Duck | − | − | + |
| 04060308 | 04/06/2013 | Lakebank | Bird Droppings | − | + | − |
|
| ||||||
| 04060311 | 04/06/2013 | Lakebank | Bird Droppings | − | + | − |
| 040802G | 04/08/2013 | DPF | Duck | + | + | − |
This chicken was slaughtered and de-feathered.
04030201G and 04030201H were sampled from the same bird.
In order to identify the potential sources and transmission routes of the H7N9 outbreaks, we conducted a survey of the bird trading records of the LPMs and BPFs targeted in this study. The results of the survey showed that in addition to nearby cities in Jiangsu province, active trading occurred between three of the LPMs and at least eight locations in Anhui, Hebei, Henan, and Shandong (Figure S1). Four of these areas, including three in Jiangsu and one in Henan, subsequently reported H7N9 human cases (Figure 1). These results indicate that trading was a likely route for virus transmission across these regions.
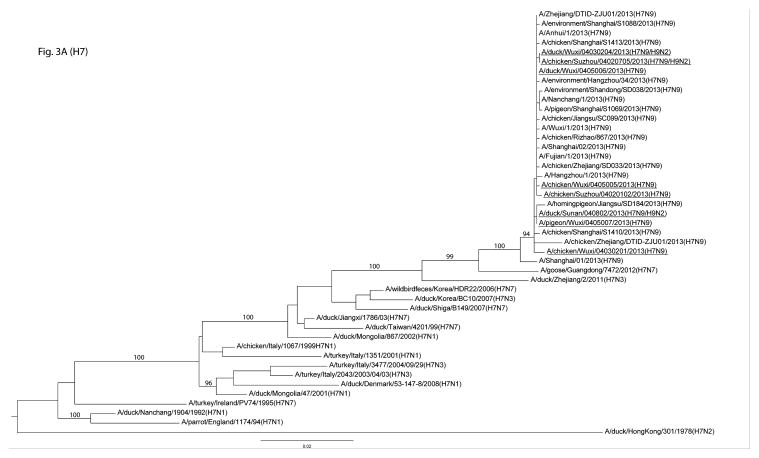
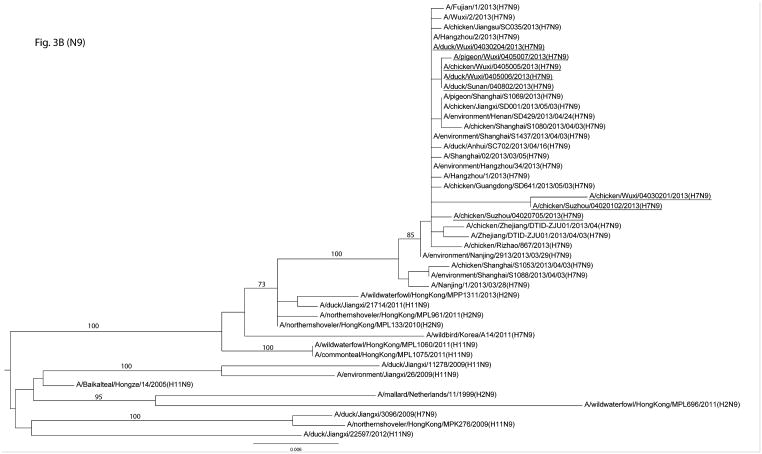
The genomes of 19 H7 and/or H9 were determined from positive samples. The HA and NA genes of the H7N9 viruses were similar to those of the H7N9 human isolates as well as other avian isolates in the region (3, 15) (Figure 3A&B). The internal genes, such as the PB1, PA, NP, and MP, of the sequenced viruses were, however, more diverse. For example, the PB1, PA, and NP gene segments belonged to two genetic clusters and the MP gene segment to three clusters (Figure S2 A–F). Our genetic analysis showed that the viruses isolated from humans were phylogenetically close to the viruses we identified in the LPMs (Figure 3A&B and Figure S2 A–F). In addition, the genes of the H9 viruses were genetically diverse, belonging to BJ94-like and G1-like genetic lineages (Figure S3 A&B). Molecular characterization showed that key residues in the receptor binding sites of these H7N9 viruses were 213Q and 217L or 217G (213 and 217 correspond to 222 and 226 in H3 numbering, respectively) (Table 3).
Figure 3.
Phylogenetic analyses of HA and NA genes of H7N9 low pathogenic avian influenza viruses. The genes from this surveillance are underlined, and bootstrap values for representative lineages are marked.
TABLE 3.
Molecular characterization of mutations at the receptor binding sites across H7N9 isolates
| Isolate | Location | 213 (222)* | 217 (226)* | 219 (228)* |
|---|---|---|---|---|
| A/chicken/040201/2013(H7N9) | LBM-A | G | L | G |
|
| ||||
| A/chicken/040207/2013(H7N9) | LBM-A | G | L | G |
|
| ||||
| A/chicken/04030201/2013(H7N9) | LBM-B | G | G | G |
|
| ||||
| A/duck/04030204/2013(H7N9) | LBM-C | G | L | G |
|
| ||||
| A/chicken/0405005/2013(H7N9) | LBM-D | G | L | G |
| A/duck/0405006/2013(H7N9) | LBM-D | G | L | G |
| A/pigeon/0405007/2013(H7N9) | LBM-E | G | G | G |
| A/duck/040802/2013(H7N9) | DPF | G | G | G |
a position corresponding to that in H3 influenza A virus.
The BPF with H7N9 and H9N2 positive samples had about 100 free-ranging ducks and 80 free-ranging hens; all positives were found in the ducks. The ducks hunted food along the river and returned to the farm in the evening, giving them a higher chance of exposure to migratory birds (although it is unclear whether H7N9 viruses were pre-existing in the migratory birds). The hens were vaccinated with H5N1 and H9N2 inactivated vaccines according to the avian influenza vaccination policies implemented by the Chinese Ministry of Agriculture. The 180 birds in this farm were culled following confirmation of the positive H7N9 results. Serologic evaluation through hemagglutinination inhibition assays showed that the duck population was H7N9 negative, demonstrating that the birds were probably infected with H7N9 not long before our sample collection. It is unclear whether the lack of H7N9 influenza A viruses among hens was due to vaccination, clearance of infection, resistance to infection, or exposure to low levels of viral loads in the environments or through shedding from other birds.
In this study, 4 out of 22 isolates were mixed infections with H7N9 and H9N2 viruses; one mixed infection was from BPF, and the other three from LPMs. The results suggested the co-circulating of multiple subtypes of influenza A viruses in both BPF and LPM where we performed influenza surveillance. Of the note, 9 of 15 subtyped single infection isolates were H9N2 virus, indicating the high prevalence of H9N2 virus in the region. The H9N2 virus has been endemic among domestic poultry in China since it was first isolated from chickens in China in 1994 (7, 14). The internal genes of contemporary H9N2 viruses contributed to the emergence and genomic diversity of highly pathogenic avian influenza H5N1 viruses (6, 23). Similarly, the mixed infection of H7N9 and H9N2 has contributed to the emergence of this novel H7N9 virus (3, 21) and genomic diversity of this virus (11), which could facilitate rapid evolution of this new virus.
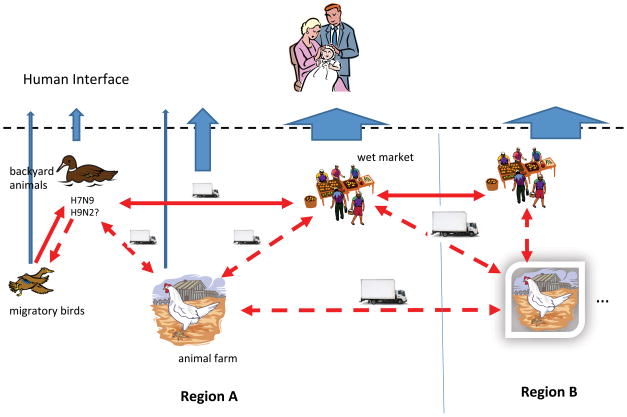
Introduction of influenza A virus from wild bird to domestic poultry, especially domestic duck, were frequently detected in China. Domestic duck facilitates the interaction between genetic pools in wild bird and domestic poultry, maintained the wild bird-origin virus and spread it to chicken (9). More recently, the emergence of H10N8 viruses in Southern China highlights the threat brought by the current influenza ecosystem (16). Based on the finding in this study and early research, we propose a simple model to describe the emergence and spread of the novel H7N9 virus (Figure 2). Although LPM could be one of the potential sources for emergence of H7N9 virus, we hypothesize that it would also be possible that the H7N9 viruses emerged in backyard birds through interactions with migratory waterfowl. It is likely that the H7N9 viruses could emerge in the backyard birds through co-infections of two or more different viruses in the same birds, as being supported in this study by detecting at least one mixed infection of H7N9 and H9N2 in one of 13 BPFs we sampled. The backyard birds then disseminated the H7N9 viruses into the LPMs, especially wholesale LPMs, through poultry trade. The wholesale markets served as the local source of infection, spreading virus to individual local LPMs. Due to less frequent cleaning, the H7N9 viruses circulated in the LPMs, potentially leading to the generation of multiple genotypes through reassortment with other LPM viruses (12). Birds and transportation vectors such as vehicles are the likely vectors that spread the viruses; a hypothesis supported by the temporal appearance and genetic similarity of H7N9 positives samples in our study and the reported human cases (Figure 1). Unfortunately, we were unable to collect avian samples in the regions supplying birds to the LPMs we sampled. Nevertheless, our model describes five major risk animal-influenza components, including LPM, transportation vectors, BPF, domestic poultry farms, and wild bird populations.
Figure 2.
An emergence and transmission model of H7N9 low pathogenic avian influenza viruses in Yangtze Delta Region. The H7N9 low pathogenic avian influenza viruses could emerge in backyard birds at BPFs through the interactions among migratory waterfowl and backyard birds. The birds from BPFs can carry H7N9 viruses into the LPMs, especially wholesale LPMs, where these viruses can quickly spread to other birds on the wholesale LPMs. The wholesale markets serve as the sources supplying these viruses into individual local LPMs. The model describes five major risk components in animal-influenza components, including LPM, transportation vectors, BPF, domestic poultry farm, and wild bird population. Among these risk components, LPMs are still primary sources for influenza exposures and infections for man.
Among these risk components, LPMs are still the primary sources for human exposure and infection (19). Instead, wild bird populations could be the smallest risk component for public health due to the relatively fewest opportunities for human-wild bird interactions. On the other hand, wild bird populations component is not negligible because the potential bi-dimensional transmission of influenza A viruses between wild birds and domestic poultry. In this study, one H7N9 isolate was identified in wild duck at LPM-C (Table 2); however, when and where the viral infection in this wild duck was not known. Nevertheless, bi-dimensional transmission of influenza A viruses between wild birds and domestic poultry are supported by the spread of clade 2.3.4.4 Gs/Gd/96-like H5 highly pathogenic avian influenza viruses, which emerged perhaps in China (5), from Southeast or Eastern Asia to Europe and North America (17).
Every year China produces approximately 10.75 billion broilers, 3 billion waterfowl, and 1.25 billion hens. In Jiangsu province, over 1 billion domestic poultries were produced in 2012, including approximately 600 million broilers, 160 million hens, 240 million ducks, and 60 million geese. Farm sizes range from ten to over 50,000 birds and there are a large number of BPFs in this province. There are at least 1.99 million of BPFs with less than 500 birds, producing about 25 percent of the domestic poultry in Jiangsu. In Jiangsu’s Capital, Nanjing City, there are alone over 163,100 BPFs (Jiangsu Animal Disease Control Center, unpublished data). Thousands of LPMs are located across Jiangsu, including those in suburbs of large cities such as Shanghai, Nanjing, Suzhou, and Wuxi. The above model and prior experience suggest that two concurrent procedures in BPFs and LPMs could be implemented in order to minimize influenza emergence and transmission. In BPFs, better biosecurity and more effective vaccination procedures (e.g. H5N1 and H9N2), especially in waterfowl, should be implemented. In LPMs, the consistent implementation of cleaning procedures and maintenance of a clean market without live birds (e.g. 1 day/week) should be used to reduce viral loads in LPMs (11). In order to reduce transmission of influenza A viruses from BPF-origin waterfowl to other bird species, these waterfowl should be separated from land-based bird species in both BPF and LPMs. Longitudinal influenza surveillance should also be implemented primarily in BPFs and LPMs to monitor influenza infection dynamics, and separate abnormal influenza situations from normal situations, providing a risk assessment and early warning platform.
Supplementary Material
Figure S1. Temporal and geospatial association between the live poultry markets we sampled and the areas with trades to these markets.
Figure S2. Phylogenetic analyses of PB2, PB1, PA, NP, MP, and NS genes of H7N9 low pathogenic avian influenza viruses. The genes from this surveillance are underlined, and bootstrap values for representative lineages are marked.
Figure S3. Phylogenetic analyses of HA and NA genes of H9N2 low pathogenic avian influenza viruses. The genes from this surveillance are underlined, and bootstrap values for representative lineages are marked.
Acknowledgments
We are grateful for the assistance from our colleagues at the Centers for Animal Disease Control, Jiangsu Province. XFW, LPL, and YX were partially supported by R01AI116744 and P20GM103646 from NIH.
Abbreviations
- LPM
live poultry markets
- BPF
backyard poultry farms
References
- 1.Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology. 2004;323:24–36. doi: 10.1016/j.virol.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Edwards S. OIE laboratory standards for avian influenza. Developments in biologicals. 2006;124:159–162. [PubMed] [Google Scholar]
- 3.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert M, Golding N, Zhou H, Wint GR, Robinson TP, Tatem AJ, Lai S, Zhou S, Jiang H, Guo D, Huang Z, Messina JP, Xiao X, Linard C, Van Boeckel TP, Martin V, Bhatt S, Gething PW, Farrar JJ, Hay SI, Yu H. Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia. Nat Commun. 2014;5:4116. doi: 10.1038/ncomms5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu M, Zhao G, Zhao K, Zhong L, Huang J, Wan H, Wang X, Liu W, Liu H, Peng D, Liu X. Novel variants of clade 2.3.4 highly pathogenic avian influenza A(H5N1) viruses, China. Emerg Infect Dis. 2013;19:2021–2024. doi: 10.3201/eid1912.130340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Y, Shortridge K, Krauss S, Chin P, Dyrting K, Ellis T, Webster R, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. Journal of virology. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo X, Liao M, Xin C. Sequence of HA gene of avian influenza A/Chicken/Guangdong/SS/1994 (H9N2) virus. Avian diseases. 2003;47:1118–1121. doi: 10.1637/0005-2086-47.s3.1118. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 9.Huang K, Bahl J, Fan X, Vijaykrishna D, Cheung C, Webby R, Webster R, Chen H, Smith GJ, Peiris J. Establishment of an H6N2 influenza virus lineage in domestic ducks in southern China. Journal of virology. 2010;84:6978–6986. doi: 10.1128/JVI.00256-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman JA. China's heath care system and avian influenza preparedness. J Infect Dis. 2008;197(Suppl 1):S7–13. doi: 10.1086/524990. [DOI] [PubMed] [Google Scholar]
- 11.Kung NY, Guan Y, Perkins NR, Bissett L, Ellis T, Sims L, Morris RS, Shortridge KF, Peiris JS. The impact of a monthly rest day on avian influenza virus isolation rates in retail live poultry markets in Hong Kong. Avian Dis. 2003;47:1037–1041. doi: 10.1637/0005-2086-47.s3.1037. [DOI] [PubMed] [Google Scholar]
- 12.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, Duan L, Chen P, Jiang J, Zhang Y, Li L, Poon LL, Webby RJ, Smith DK, Leung GM, Peiris JS, Holmes EC, Guan Y, Zhu H. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 16.Ma C, Lam TTY, Chai Y, Wang J, Fan X, Hong W, Zhang Y, Li L, Liu Y, Smith DK. Emergence and evolution of H10 subtype influenza viruses in poultry in China. Journal of virology. 2015;89:3534–3541. doi: 10.1128/JVI.03167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasick J, Berhane Y, Joseph T, Bowes V, Hisanaga T, Handel K, Alexandersen S. Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada, 2014. Scientific reports. 2015;5:9484. doi: 10.1038/srep09484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swofford DL. PAUP*: Phylogenic analysis using Parsimony. Sinauer; Sunderland, Massachusetts: 1998. [Google Scholar]
- 19.Wan XF, Dong L, Lan Y, Long LP, Xu C, Zou S, Li Z, Wen L, Cai Z, Wang W, Li X, Yuan F, Sui H, Zhang Y, Dong J, Sun S, Gao Y, Wang M, Bai T, Yang L, Li D, Yang W, Yu H, Wang S, Feng Z, Wang Y, Guo Y, Webby RJ, Shu Y. Indications that Live Poultry Markets Are a Major Source of Human H5N1 Influenza Virus Infection in China. J Virol. 2011;85:13432–13438. doi: 10.1128/JVI.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. Overview of the emergence and characteristics of the avian influenza A (H7N9) virus. 2013 [Google Scholar]
- 22.Woolcock PR, Suarez DL, Kuney D. Low-pathogenicity avian influenza virus (H6N2) in chickens in California, 2000–02. Avian Dis. 2003;47:872–881. doi: 10.1637/0005-2086-47.s3.872. [DOI] [PubMed] [Google Scholar]
- 23.Zhao ZM, Shortridge KF, Garcia M, Guan Y, Wan XF. Genotypic diversity of H5N1 highly pathogenic avian influenza viruses. J Gen Virol. 2008;89:2182–2193. doi: 10.1099/vir.0.2008/001875-0. [DOI] [PubMed] [Google Scholar]
- 24.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas; Austin: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Temporal and geospatial association between the live poultry markets we sampled and the areas with trades to these markets.
Figure S2. Phylogenetic analyses of PB2, PB1, PA, NP, MP, and NS genes of H7N9 low pathogenic avian influenza viruses. The genes from this surveillance are underlined, and bootstrap values for representative lineages are marked.
Figure S3. Phylogenetic analyses of HA and NA genes of H9N2 low pathogenic avian influenza viruses. The genes from this surveillance are underlined, and bootstrap values for representative lineages are marked.