Abstract
Background
Aspirin use is an effective strategy for the chemoprevention of colorectal cancer, even at low doses. However, in order to implement aspirin interventions, risk–benefit balances and biological mechanisms need to be better defined; to further this aim, we used a metabolomics approach.
Methods
We metabolically profiled 40 healthy, non-smoking men and women aged 20–45 years enrolled in a randomized, double-blind, crossover trial of 325 mg aspirin/d over 60 days. Gas and liquid chromatography–mass spectrometry were used to comprehensively profile participants’ plasma samples after aspirin and placebo interventions.
Results
A total of 363 metabolites, covering most human biochemical pathways, were measured. Compared to placebo-treated participants, plasma concentrations of the oncometabolite 2-hydroxyglutarate (R+S) decreased after aspirin treatment in both men and women (p = 0.005). This signal proved robust during 20-fold random splitting of the data using 80% of the samples in each split. We subsequently performed functional follow-up studies using targeted, enantiospecific detection in human colorectal cancer cell lines and observed an aspirin-induced reduction of (R)-2-hydroxyglutarate. We further showed that salicylate, the primary aspirin metabolite, inhibits the hydroxyacid–oxoacid transhydrogenase (HOT) mediated production of (R)-2-hydroxyglutarate, thereby providing mechanistic evidence for the clinically observed effects of aspirin on total-2-hydroxyglutarate.
Conclusion
Using a metabolomics approach with functional follow-up, we propose that a decrease in the oncometabolite (R)-2-hydroxyglutarate may identify an additional mechanism for aspirin or its metabolites in cancer prevention.
Impact
Reduction of the oncometabolite (R)-2-hydroxyglutarate identifies a novel, non-COX-inhibition-mediated mechanism of aspirin.
Keywords: aspirin, metabolomics, intervention, 2-hydroxyglutarate, oncometabolites
INTRODUCTION
Aspirin use is an effective strategy for the prevention of gastrointestinal tumors (1). Recent meta-analyses of randomized controlled trials with >14,000 individuals report a 24% reduced colorectal cancer (CRC) incidence by daily aspirin supplementation, as well as a 34% decrease in fatal CRC (2). These findings are strengthened by a systematic review including >100,000 participants enrolled in case-control, cohort and nested case-control studies (3). A meta-analysis of randomized trials of aspirin use for chemoprevention of metachronous colorectal adenomas showed a risk-reduction of subsequent lesions (4). Furthermore, aspirin treatment was observed to decrease overall cancer mortality by 15% (2), even at low doses (75–300 mg/day). However, the chemopreventive effects of aspirin must be balanced against its risks, including uncommon, but possibly severe, adverse effects such as gastrointestinal bleeding (5).
Inhibition of cyclooxygenases and reduced inflammation are known effects of aspirin (1), although mechanisms by which low-dose aspirin (75–300 mg) protects against gastrointestinal cancers remain incompletely understood. Metabolomics is perfectly suited to describe the phenotypic changes associated with pharmacologic challenges (pharmacometabolomics). Metabolites may be biomarkers or play a role themselves in driving the transformation of cells towards malignancy – such compounds are referred to as “oncometabolites” (6). Examples include the gain-of-function mutations in isocitrate dehydrogenases (IDH), which lead to the accumulation of (R)-2-hydroxyglutarate (7), a metabolite driving epigenetic pathogenesis.
We aimed to explore aspirin-mediated changes in the human plasma metabolome of individuals enrolled in a randomized, double-blind, crossover trial. This exploration may provide further insight into mechanisms associated with the protective effect of aspirin. Additionally, we tested signals from the clinical study in human colorectal cancer cell lines and in in vitro follow-up experiments.
MATERIALS AND METHODS
Study design
The Aspirin and Biology of the Colon (ABC) intervention study was a randomized, double-blind, placebo-controlled, crossover trial of 325 mg/day aspirin vs. placebo for 60 days. Participants were randomly assigned, blocked on sex and genotype, as to the order in which they would receive aspirin or placebo. The washout period between interventions was 3 months.
Twelve-hour fasting morning blood samples were drawn on days −5 and 55 of intervention period 1 as well as on days 1 and 55 of intervention period 2. Samples drawn before each intervention were used for clinical assessment. Samples drawn on day 55 (last clinic visit) of both intervention periods were used for metabolic profiling, enabling paired comparisons where each individual was treated both with placebo and aspirin (crossover design). Each individual served as his/her own control, thereby limiting the effects of intra-individual variability. Blood samples were collected in EDTA tubes, cooled to 4°C and, after centrifuging, the plasma was aliquoted and stored at −80°C until analysis.
Study population
Healthy men and women aged 20–45 years were recruited from among those who completed a cross-sectional study of diet and aspirin metabolism in the greater Seattle area between June 2003 and March 2007 (see Supplementary figure 1 and (8) for more detailed exclusion criteria and study design). Participants in this secondary study (ABC intervention) were selected on the basis of their UDP-glucuronosyltransferase 1A6 genotype, (UGT1A6*2 [T181A+R184S]) − 19 with UGT1A6*1/*1 and 21 with UGT1A6*2/*2. All participants with a *2/*2 genotype and sex-matched participants with a *1/*1 genotype were invited to consider participation in the trial. Given the aims of the parent study, individuals were selected for participation on the basis of the homozygous genotypes (*1/*1, *2/*2) to maximize the potential functional differences (see original study (8) for a more detailed description of the rationale, dropout characteristics and genotyping). Study participants’ characteristics are given in Table 1. No dropouts were attributed to the aspirin treatment.
Table 1.
Baseline characteristics of the participants in the ABC study; a randomized, double-blind, crossover trial of 325 mg aspirin/d over 60 days
| Characteristic | All participants (n ; Mean ± SD) |
|---|---|
| Age (years) | 31.0 ± 6.2 |
| Sex | |
| Women | 22 |
| Men | 18 |
| Ethnicity | |
| Asian | 5 |
| Black African American | 1 |
| Caucasian | 27 |
| Not answered | 2 |
| Other | 5 |
| Weight (kg) | 78.5 ± 16.8 |
| Height (cm) | 172.2 ± 9.4 |
| BMI (kg/m2) | 26.4 ± 4.8 |
Exclusion criteria included tobacco use, consumption of >2 alcoholic beverages/d (equivalent to 720 mL beer, 240 mL wine, or 90 mL hard liquor), regular use of prescription or over-the-counter medications, known aspirin intolerance, weight loss or gain of >4.5 kg in the past year, current or planned pregnancy, breastfeeding, chronic medical illness, history of gastrointestinal disorders (e.g., ulcerative colitis, Crohn’s disease, celiac sprue, hereditary non-polyposis colorectal cancer, familial adenomatous polyposis, pancreatic disease, previous gastrointestinal resection, radiation, or chemotherapy), and cancer (other than non-melanoma skin cancer).
To ensure that participants could take aspirin safely for 60 days, each participant underwent a clinical assessment that included a detailed medical history, measurement of blood pressure, complete blood count, liver and chemistry panel, blood urea nitrogen, serum creatinine, and urinalysis. Women also underwent a pregnancy test (8). Clinical laboratory assays were completed by a commercial lab (CLIA-licensed Quest Diagnostics, Seattle, WA). The study was approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center. All study participants gave written informed consent.
Metabolic profiling
Plasma samples were transferred blind to Metabolon (Durham, NC) for untargeted metabolic profiling using gas and liquid chromatography–mass spectrometry (GC–MS and LC–MS). Detailed sample preparation and data acquisition parameters can be found in the literature (9) and the Supplementary methods and materials.
Statistical analysis
Preprocessing
Possible batch effects were checked by principal component analysis (data not shown). Metabolites with more than 60% missing data were excluded. Remaining missing data were imputed as half the minimum value for each metabolite. After normalization (log-transformation, range, and median scaling), the data was checked for normality. As most of the metabolites diverged from a normal distribution, we used non-parametric tests in further analyses.
Fold changes
Fold changes (expressed as ×) were calculated from the raw, non-processed, peak abundances dividing plasma metabolite intensities of the aspirin by those of the placebo intervention.
All other statistical analyses were carried out using the uni- and multivariate tools available in the software toolbox Metaboanalyst 2.0 (10) on the pre-processed data matrices. Since untargeted metabolomics approaches are semi-quantitative, scaled intensities, rather than absolute concentrations, are reported.
Pairwise comparisons
We conducted pairwise comparisons for each metabolite between placebo and aspirin groups using paired Wilcoxon signed-rank tests. Metabolites that were considered statistically significant (p < 0.01) were further tested for robustness by 20× leave-n-out cross-validation, as previously published (11): data was split randomly using 80% of the study participants in each split and p-values were calculated for each of these 20 random tests. The false discovery rate (FDR) was estimated using the method of Benjamini and Hochberg (12). We further carried out an analysis stratified by sex, age, BMI, and dose group (high/low) based on a median split: using Wilcoxon tests, we tested if the change in the metabolite level between placebo and aspirin treatment was significantly different between these subgroups (i.e male vs. female or UGT1A6*1/*1 vs. UGT1A6*2/*2).
Multivariate analysis
A partial least squares-discriminant analysis (PLS-DA) was carried out on the full placebo vs. aspirin dataset stratified by sex to reduce dimensionality and for data visualization. We assessed the performance and validity of the model using 10× cross-validation and permutation testing within Metaboanalyst (10).
Pathway visualization
To visualize the effects of aspirin on a global metabolic scale, we used chemical similarity (Pubchem ID) and biochemical pathway mapping (KEGG ID) with the MetaMapR tool as previously described (13). We mapped metabolites as being decreased/increased after the aspirin intervention by a cutoff of a raw p-value of 0.1.
Targeted profiling of (R)- and (S)-2-hydroxyglutarate in cell lines
As part of the functional follow-up of our clinical findings, colon cancer cell lines HCT116, Caco2, KM12, and DLD1 were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum + 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. K073, SW48, and SW480 cells were cultured in Roswell Park Memorial Institute (RPMI) medium with the same supplements. Cell lines were regularly tested for contamination and authenticated (14). Cells (5 × 105/well) were incubated for 24 h with either 500 µM acetylsalicylic acid (aspirin, Sigma Aldrich, St Louis, USA) or vehicle control (0.1% dimethylsulfoxide) in triplicate with an independent replication. After incubation, cells were harvested and extracted using an optimized protocol for cell-culture metabolomics (15) (see also Supplementary methods and materials). To differentiate the two enantiomers of 2-hydroxyglutarate (2-hydroxyglutarate), derivatization following literature methodology (16) using a chiral auxiliary was applied. The resulting diastereomeric O-acetyl-di-(S)-2-butyl esters were quantified by GC–MS using stable isotope dilution analysis. Racemic 2-hydroxyglutarate-d4 (2-hydroxyglutarate-d4), synthesized herein, was used as an internal standard. Finally, to account for differences in the cellular mass extracted, the total amounts of S- and (R)-2-hydroxyglutarate were normalized to the sum of all metabolites present in the extract measured by 1H NMR as previously described (17). Free salicylate was quantified within the same chromatographic run using salicylic acid-d6 (internal standard, Sigma Aldrich, St Louis, USA) to ensure efficacy of aspirin treatment (see Supplementary figure 2). Detailed extraction procedures, synthesis of the internal standard, and chromatographic conditions can be found in the Supplementary methods and materials and Supplementary figure 3.
Inhibition studies of the hydroxyacid–oxoacid transhydrogenase (HOT) reaction
A rat liver mitochondrial/lysosomal fraction (10.03 mg protein/mL) was used as a source for the HOT enzyme (see Supplementary methods and materials for preparation). Inhibition studies with sodium salicylate were carried out using 0.6 mg total protein (60 µL) incubated with 50 µM substrate (2-oxoglutarate/4-hydroxybutyrate) for 1 h at 37°C in TRIS phosphate buffer (pH 7.8, total volume 100 µL). Final concentrations of sodium salicylate were 0, 5, 50, 250, and 500 µM. The reaction was stopped by the addition of 200 µL of MeOH (−80°C) including 0.1 nmol 2-hydroxyglutarate-d4 as internal standard (equivalent to 1 µM in 100 µL). After centrifugation at 12,000 g for 5 min, the supernatant was dried in vacuo and analyzed by GC–MS as previously described (17). 2-hydroxyglutarate was quantified using single ion monitoring: 129 m/z (quantifier for 2-hydroxyglutarate), 349 m/z (qualifier for 2-hydroxyglutarate), 132 m/z (quantifier for 2-hydroxyglutarate-d4), and 353 m/z (qualifier for 2-hydroxyglutarate-d4). Enzymatic activity was monitored using the formation of 2-hydroxyglutarate and expressed as relative HOT activity. An inhibition curve was fitted using the "log(inhibitor) vs. enzymatic activity" function in Graphpad 5.0 without constraints.
RESULTS
Effects of aspirin intervention on the plasma metabolome of study participants
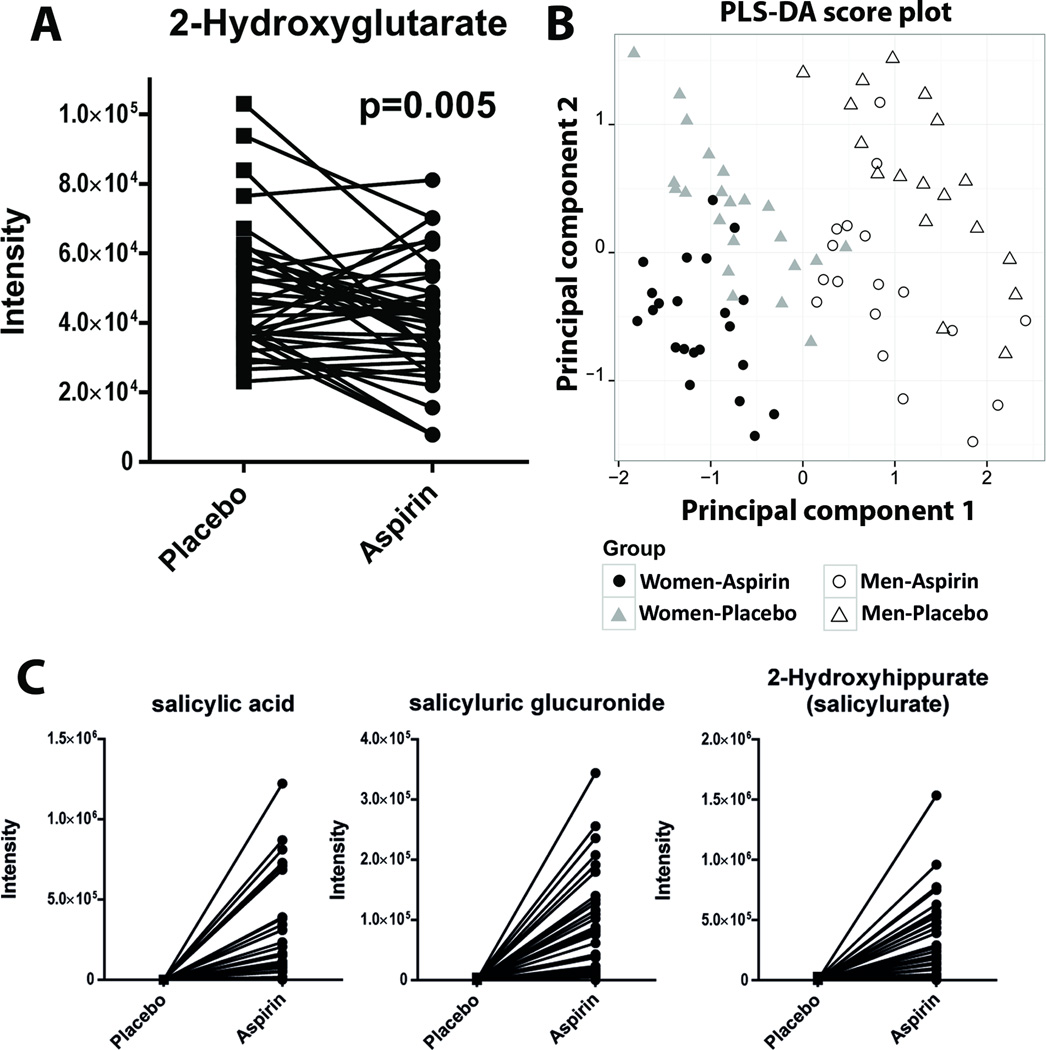
We profiled 387 metabolites in the plasma of study participants covering most major human biochemical pathways. After pre-processing, 363 metabolites were subject to further statistical analysis. Twenty metabolites differed (p < 0.05) in aspirin-treated samples compared to the control samples (see Table 2 and Supplementary Table 1). Five compounds were considered statistically significant applying a cutoff p-value of 0.01: the three aspirin metabolites (p < 0.001), as well as the oncometabolite 2-hydroxyglutarate (as the sum of R and S enantiomers; p = 0.005; see Fig. 1a) and dimethylarginine (as the sum of symmetric and asymmetric regioisomers; p = 0.002). The false discovery rate (FDR) was between 0.11 and 0.33 for dimethylarginine and 2-hydroxyglutarate. Thus, we evaluated the robustness of the above mentioned markers through a leave-n-out cross-validation approach as previously described (11). We cross-validated our dataset with 20× random splitting of the data, using 80% of the samples in each split. For all aspirin metabolites, p-values were <10−5 in all cross-validated test sets. For total-2-hydroxyglutarate, p-values varied between 0.0012–0.07 (mean: 0.021), and between 0.0009–0.087 (mean: 0.017) for dimethylarginine. In both cases, p-values were >0.05 only twice in 20 random splits, indicating statistically robust findings. For total-2-hydroxyglutarate, a reduction in the aspirin group was observed in every split (fold changes: 0.76–0.86), and higher levels of dimethylarginine in every split (fold changes: 1.07–1.17). 2-Hydroxyglutarate was reduced in all stratified subgroups and a very modest, but non-significant trend (p > 0.50) towards dose-dependence was observed when aspirin dose was expressed as mg/kg body weight (0.86× in the high-dose and 0.90× in the low-dose group; see also Table 2). The same was true for the difference by genotype (fold changes: 0.86× in 2*/2* and 0.90× in 1*/1*; see also Table 2), but this mild effect modification was also not statistically significant (p > 0.50). Dimethylarginine was 1.32× higher in men in response to the aspirin intervention (p < 0.001) but no difference was observed in women (p > 0.10).
Table 2.
Changes in metabolite concentrations after a 60 day aspirin (325 mg/d) intervention compared to placebo intervention in the ABC study; a randomized, double-blind, crossover trial. Metabolites with a paired Wilcoxon p < 0.10 are presented. Fold changes were calculated for the full dataset as well as stratified by sex, genotype, BMI, dose, and age group. Fold changes <1 indicate a reduction of the metabolite after the aspirin intervention compared to placebo; fold changes >1 indicate an increase of the metabolite after the aspirin intervention
| n (↑) | n (↓) | Fold change** |
p-value | Fold change** | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Age1 | BMI2 | Genotype | Dose3 | ||||||||||
| Not stratified | M | F | Young | Old | Low | High | 1*1 | 2*2 | Low | High | ||||
| Changes in metabolites (p < 0.01) | ||||||||||||||
| salicylate | 35 | 1 | 177 | <0.001 | 304 | 122 | 138 | 252 | 148 | 218 | 122 | 220 | 212 | 152 |
| salicyluric-glucuronide | 37 | 1 | 87 | <0.001 | 168 | 57 | 69 | 123 | 70 | 108 | 60 | 115 | 111 | 67 |
| salicylurate | 34 | 0 | 56 | <0.001 | 57 | 56 | 49 | 65 | 43 | 71 | 67 | 47 | 69 | 45 |
| dimethylarginine | 28 | 12 | 1.13 | 0.002 | 1.32* | 1.00* | 1.21 | 1.06 | 1.11 | 1.15 | 1.16 | 1.10 | 1.16 | 1.11 |
| 2-hydroxyglutarate | 14 | 26 | 0.88 | 0.005 | 0.91 | 0.86 | 0.85 | 0.90 | 0.90 | 0.86 | 0.90 | 0.86 | 0.90 | 0.86 |
Difference between the two groups significant at a level of p < 0.01
Fold changes for salicylate, salicylurate, and salicyluric-glucuronide were below the detection limit in 38, 32, and 44 samples, respectively. Instances occurred mainly in the placebo group. In order to calculate a fold change these missing values have been replaced by half the minimum observed value for the respective metabolite
Age group young: 21.3 – 31.1 y; old: 31.2 – 44.2 y.
BMI group low: 19.4 – 24.9 kg/m2; high: 25.0 – 44.7 kg/m2.
Dose group low: 2.2 – 4.2 mg/kg; high: 4.3 – 6.2 mg/kg.
Figure 1.
A) Paired intensity values of 2-hydroxyglutarate in placebo and aspirin treated individuals. One line corresponds to one individual. The p-value was based on paired Wilcoxon test; B) Score plot of a partial least squares-discriminant analysis (PLS-DA) on the metabolomics dataset comparing aspirin- and placebo-treated individuals by sex; C) Paired intensity values of the three main salicylic acid metabolites (salicylate, salicyluric-glucuronide, and salicyluric acid) in placebo and aspirin treated individuals.
Illustrating the effects of aspirin on a multivariate and global metabolite scale
We used a PLS-DA model to visualize our findings on a multivariate scale: branched chain amino acids as well as N-acetylcarnosine mainly contributed to principal component (PC) 1 of the model (separating men from women), whereas the three aspirin metabolites contributed to PC2 (intervention; see Fig. 1b). Overall, high discrimination accuracy was achieved in a model consisting of two principal components with no signs of overfitting as indicated by a 10× cross-validation. We further visualized the effects of aspirin on a global metabolic scale by mapping metabolites based on their biochemical (KEGG) and chemical (Pubchem ID) similarities (see Supplementary figure 4). Besides the impact on direct salicylate metabolites and downstream aromatic products, modest effects in nucleotide, steroid, and short-chain acyl-carnitine metabolism, as well as the urea cycle-associated metabolites, could be observed (see Supplementary figure 4). In addition to the prominent reduction of 2-hydroxyglutarate, we noted a decrease in malate, which was mapped in the close vicinity to 2-hydroxyglutarate due to its chemical similarity.
Salicylic acid metabolites as compliance markers
Salicylate, salicylurate (2-hydroxyhippurate), and salicyluric-glucuronide were all detected in the subjects’ plasma after aspirin treatment, whereas concentrations were below the detection limit following placebo treatment. However, abundances of these aspirin metabolites varied substantially across subjects (see Fig. 1c), possibly reflecting the variability in times between last dose and blood draw. Moreover, individual differences in the metabolism of aspirin likely played a role. For three participants levels of all aspirin metabolites were below the limit of detection. When the analysis was limited to only those participants who indeed had detectable aspirin metabolites in their plasma, the observed effect on 2-hydroxyglutarate was strengthened (FC: 0.82, p = 0.001, FDR = 0.08).
Effects of aspirin treatment on the concentration of 2-hydroxyglutarate enantiomers in human colorectal cancer cell lines
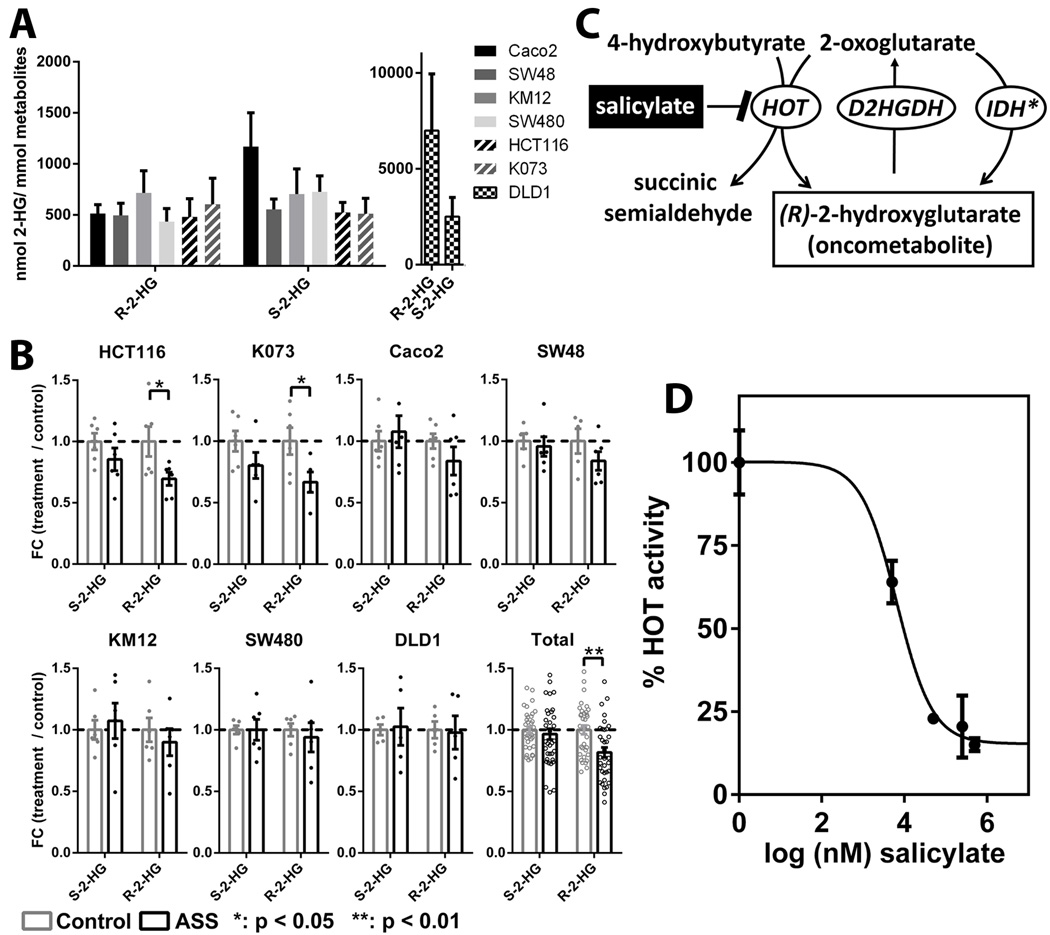
To further evaluate the findings of aspirin mediated effects on the oncometabolite 2-hydroxyglutarate, we tested the effects of aspirin treatment on intracellular concentrations of (R)- and (S)-2-hydroxyglutarate in colorectal cancer cell lines. In untreated (control) cell lines, a narrow intracellular concentration range of ~500 nmol per mmol total metabolites was observed for both enantiomers (Fig. 2a). DLD1 cells carry a IDH1 mutation (IHD1 G97D) (18) establishing a phenotype with elevated (R)-2-hydroxyglutarate concentrations (19), as evident from more than 10× higher concentrations of (R)-2-hydroxyglutarate compared to other cell lines in our dataset (Fig. 2a). This mimicked the situation of IDH-mutant tumors, where this oncometabolite has been identified (7, 20).
Figure 2.
A) Intracellular concentrations of both enantiomers of 2-hydroxyglutarate in different colorectal cancer cell lines (control treated), expressed as nmol/mmol total metabolites. Bars and error bars represent means ± SDs. B) Effects of 500 µM aspirin on (R)-2-hydroxyglutarate and (S)-2-hydroxyglutarate levels in colorectal cell lines. Triplicate values of biological replicates are expressed as relative fold change of treated cells vs. untreated controls. Bars and error bars indicate means ± SEMs. "Total" represents mean± SEM from all cell lines combined (IDH-mutant cell line DLD1 was excluded). Dots represent the actual datapoints. P-values are based on Wilcoxon signed rank test. C) Enzymatic production of (R)-2-hydroxyglutarate from 2-oxoglutarate via mutant IDH (IDH*; non-physiological) or via hydroxyacid–oxoacid transhydrogenase (HOT; physiological). (R)-2-hydroxyglutarate can be converted back to 2-oxoglutarate via D-2-hydroxyglutarate dehydrogenase (D2HGDH). D) Relative HOT activity (2-hydroxyglutarate formation) observed after incubations of a rat liver mitochondrial/lysosomal fraction with 50 µM substrates (2-oxoglutarate/4-hydroxybutyrate) in the presence of varying concentrations of the inhibitor sodium salicylate. Data represent means ± SDs of triplicate measurements.
Intracellular concentrations of 2-hydroxyglutarate showed stereospecific responses to aspirin treatment (Fig. 2b). Concentrations of the known oncometabolite (R)-2-hydroxyglutarate were reduced up to 34% in colorectal cancer cell lines (p < 0.05 for HCT116 and K073). Because the response was near uniform, we also evaluated the combined data of all non-IDH mutant cell lines; a 24% reduction of (R)-2-hydroxyglutarate (p < 0.01) was observed. No effects of aspirin on (R)-2-hydroxyglutarate concentrations were observed in the IDH-mutant DLD1 cells. (S)-2-hydroxyglutarate was not, or only marginally so, affected by aspirin treatment for all cell lines tested.
Inhibition of the hydroxyacid–oxoacid transhydrogenase (HOT) reaction by sodium salicylate
Under physiological (non-IDH mutant) conditions, (R)-2-hydroxyglutarate is produced by hydroxyacid–oxoacid transhydrogenase (HOT), (Fig. 2c). We therefore tested whether inhibition of the HOT-mediated production of (R)-2-hydroxyglutarate provided a novel mode of action for aspirin. We studied the inhibitory effects of salicylate, the primary aspirin metabolite, on HOT product formation and observed inhibitory effects at concentrations within the 5–500 µM range (Fig. 2d), which corresponds to physiologically achievable plasma concentrations in aspirin-treated individuals (21).
DISCUSSION
In this study, we used a metabolomics approach to test the effects of a 60-day, randomized, double-blind, placebo-controlled, crossover aspirin intervention trial on the plasma metabolomes of healthy humans. We discovered a novel, robust signal of reduction of the oncometabolite 2-hydroxyglutarate (sum of enantiomers). Subsequently, we confirmed these results in functional follow-up studies and showed that salicylate inhibits the HOT reaction, which is the physiological route for (R)-2-hydroxyglutarate production (see Fig. 2c).
Effects of aspirin on 2-hydroxyglutarate in human plasma and human cell lines
The intriguing result from the crossover study was a statistically significant reduction of the oncometabolite 2-hydroxyglutarate in both sexes. A slightly larger effect was observed for participants receiving a higher dose, based on their weight (mg/kg bodyweight) and participants with the 2*/2* genotype. However, these trends were non-significant, but would both support the concept of dose-dependence and an increased chemopreventive efficacy of aspirin in individuals with the UGT1A6 2*/2* genotype, consistent with the epidemiological data (22). The oncometabolic activities of 2-hydroxyglutarate depend on its stereochemistry. (R)-2-hydroxyglutarate has been described as being derived from the activity of mutated IDH1/IDH2 in glioma and leukemia (7, 20). It can promote epigenetically-driven carcinogenesis by inhibiting histone lysine demethylases and 5-methylcytosine hydroxylases (23). By contrast, (S)-2-hydroxyglutarate has only recently been reported to have possible oncometabolic activities in renal cancer (24). Differences between renal tumors with the highest and lowest (S)-2-hydroxyglutarate levels were mild compared to the differences observed for IDH-mutations ((R)-2-hydroxyglutarate), but still reflective of reduced 5-hydroxymethylcytosine in DNA (24).
We confirmed the effects of aspirin on both enantiomers of 2-hydroxyglutarate in targeted enantiospecific analyses of cell culture experiments. A ~30% reduction of the known oncometabolite (R)-2-hydroxyglutarate in HCT116 and K073 cells was observed following aspirin treatment. Although this did not meet statistical significance, (R)-2-hydroxyglutarate concentrations were lower after aspirin treatment in 2 additional colorectal cell lines (Caco2, SW48). Concentrations of (R)-2-hydroxyglutarate were high and unchanged by aspirin in DLD1 cells indicating that (R)-2-hydroxyglutarate concentrations in cells with IDH1 mutations are primarily affected by the altered enzyme activity. This finding further indicated that aspirin interacts with enzymes involved in the physiological metabolism and not mutant IDH1. We therefore hypothesized that either aspirin or one of its metabolites act stereospecifically and might be involved in affecting one of the enzymes having (R)-2-hydroxyglutarate as a substrate or product, such as HOT. In vitro enzyme inhibition assays showed that, at physiologically achievable concentrations, salicylate, the primary aspirin metabolite, inhibits the (R)-2-hydroxyglutarate formation carried out by HOT, providing mechanistic evidence for the observed clinically lowered 2-hydroxyglutarate levels. The effects of aspirin on 2-hydroxyglutarate were mild, both clinically as well as in cell culture models. However, considering long-term aspirin use, chronically lowered 2-hydroxyglutarate levels might be associated with slower rates of malignant transformation and progression in target tissues and reduce the oncometabolic activity in target cells. Additional results of our group further support a potential role of 2-hydroxyglutarate in CRC: we observed that total 2-hydroxyglutarate excretion in urine is reduced after CRC-tumor removal (17).
We also observed elevated levels of dimethylarginine in men (measured as sum of both symmetric and asymmetric regioisomers; SDMA + ADMA). Increased levels of ADMA are associated with a variety of chronic conditions, especially risk of cardiovascular diseases (25). Effects of aspirin on ADMA are well documented in the literature (26–28). In contrast to our results, the group of Hennekens (26, 27) reported a decrease in ADMA concentrations and a resulting increase of NO formation, after aspirin administration. It has to be noted that they did not report on fasting status, which could have affected ADMA levels. It is also possible, however, that the symmetric regioisomer (SDMA) may have masked the true and biologically important ADMA values in our study.
Overall effects of aspirin on the plasma metabolome
Overall, metabolic signatures in response to the aspirin intervention were modest with only a limited number of metabolites altered. Levels of direct aspirin metabolites were increased and served as compliance markers, with only 3 individuals exhibiting no aspirin metabolites in their plasma. A sub-analysis with only those participants with detectable aspirin in their plasma strengthened the overall effects observed on 2-hydroxyglutarate. However, based on the study protocol and the pharmacokinetics of aspirin it cannot definitely be established whether those individuals were non-compliant or whether their dosing timepoint was >>5 elimination half-lives prior to blood draw.
Strengths and limitations
The strengths of this study are its randomized, controlled, double-blind, crossover design, providing strong evidence for causality, while controlling for inter-individual differences. However, it has to be noted that the false positive rate for the observed effects regarding 2-hydroxyglutarate might be elevated, due to the number of comparisons. We have addressed this through a 20× random split cross-validation which showed strong consistency of the results. Moreover, our data from cell culture assays indicate an aspirin-induced reduction of (R)-2-hydroxyglutarate in colon cancer cell lines. Further in vitro experiments provide mechanistic evidence for salicylate being an inhibitor of the HOT reaction producing (R)-2-hydroxyglutarate. A limitation is that the metabolic profiling platform used in the human study was not able to differentiate between the two 2-hydroxyglutarate enantiomers in vivo. Thus, only our targeted functional studies were able to test for these effects. Furthermore, sampling of biospecimens only at the end of the intervention period cannot account for the full dynamics of the metabolome and the exact timepoint at which a first metabolic response to aspirin occurs. Finally, future studies sampling colon tissue specimen are required to assess if the observed systemic changes in plasma are reflected in individuals' target tissues or may originate from systemic metabolism, i.e. liver metabolism.
Conclusion
We have metabolically profiled the plasma of healthy individuals enrolled in a randomized, double-blind, crossover, placebo-controlled aspirin intervention. The oncometabolite 2-hydroxyglutarate was dose-dependently reduced in both men and women. We further observed an aspirin-induced reduction of (R)-2-hydroxyglutarate in colorectal cancer cell lines. Subsequently, we showed that salicylate, the primary aspirin metabolite, inhibits the HOT-mediated production of (R)-2-hydroxyglutarate in vitro, thereby providing mechanistic evidence for the clinically observed effects. Taken together, we propose that a decrease in the oncometabolite 2-hydroxyglutarate may identify an additional mechanism for aspirin or its metabolites in cancer prevention.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Ross Prentice for statistical advice, Dr. Bert Vogelstein for providing colorectal cancer cell lines, Andrea Breuer for providing rat liver mitochondrial/lysosomal fractions, and we would also like to thank the ABC study participants for their time and contribution.
Financial support: This study was supported by the US National Institutes of Health (R01 CA094954; J.W. Lampe, R01 CA112516; C.M. Ulrich, J.D. Potter), the German Consortium for Translation Cancer Research (DKTK; C.M. Ulrich), and institutional funding of the Division of Preventive Oncology (C.M. Ulrich), National Center for Tumor Diseases, Heidelberg. D.B. Liesenfeld and C. Weigel both received fellowships from the Helmholtz International Graduate School for Cancer Research.
Footnotes
Conflicts of interest: Dr. Ulrich has participated in an aspirin workshop organized by Bayer AG in 2013. There are no further conflicts of interest.
REFERENCES
- 1.Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat Rev Cancer. 2006;6:130–140. doi: 10.1038/nrc1801. [DOI] [PubMed] [Google Scholar]
- 2.Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- 3.Algra AM, Rothwell PM. Effects of regular aspirin on long-term cancer incidence and metastasis: a systematic comparison of evidence from observational studies versus randomised trials. The Lancet Oncology. 2012;13:518–527. doi: 10.1016/S1470-2045(12)70112-2. [DOI] [PubMed] [Google Scholar]
- 4.Cole BF, Logan RF, Halabi S, Benamouzig R, Sandler RS, Grainge MJ, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. Journal of the National Cancer Institute. 2009;101:256–266. doi: 10.1093/jnci/djn485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potter JD. Aspirin and cancer prevention and treatment: are we there yet? Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:1439–1440. doi: 10.1158/1055-9965.EPI-12-0837. [DOI] [PubMed] [Google Scholar]
- 6.Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. The Journal of Clinical Investigation. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas SS, Makar KW, Li L, Zheng Y, Yang P, Levy L, et al. Tissue-specific patterns of gene expression in the epithelium and stroma of normal colon in healthy individuals in an aspirin intervention trial. BMC medical genetics. 2015;16:18. doi: 10.1186/s12881-015-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical chemistry. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 10.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:2. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao J. Linear Model Selection by Cross-validation. Journal of the American Statistical Association. 1993;88:486–494. [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 13.Grapov D, Wanichthanarak K, Fiehn O. MetaMapR: pathway independent metabolomic network analysis incorporating unknowns. Bioinformatics (Oxford, England) 2015 doi: 10.1093/bioinformatics/btv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro F, Dirks WG, Fähnrich S, Hotz-Wagenblatt A, Pawlita M, Schmitt M. High-throughput SNP-based authentication of human cell lines. International Journal of Cancer. 2013;132:308–314. doi: 10.1002/ijc.27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng Q, Huang W, Collette T, Ekman D, Tan C. A direct cell quenching method for cell-culture based metabolomics. Metabolomics. 2009;5:199–208. [Google Scholar]
- 16.Engqvist MK, Kuhn A, Wienstroer J, Weber K, Jansen EE, Jakobs C, et al. Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. The Journal of biological chemistry. 2011;286:11382–11390. doi: 10.1074/jbc.M110.194175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liesenfeld D, Habermann N, Toth R, Owen R, Frei E, Böhm J, et al. Changes in urinary metabolic profiles of colorectal cancer patients enrolled in a prospective cohort study (ColoCare) Metabolomics. 2014:1–15. doi: 10.1007/s11306-014-0758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Human mutation. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 19.Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, et al. Identification of additional IDH mutations associated with oncometabolite R(−)-2-hydroxyglutarate production. Oncogene. 2012;31:2491–2498. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science (New York, NY) 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfonso L, Ai G, Spitale RC, Bhat GJ. Molecular targets of aspirin and cancer prevention. British journal of cancer. 2014;111:61–67. doi: 10.1038/bjc.2014.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigler J, Whitton J, Lampe JW, Fosdick L, Bostick RM, Potter JD. CYP2C9 and UGT1A6 genotypes modulate the protective effect of aspirin on colon adenoma risk. Cancer research. 2001;61:3566–3569. [PubMed] [Google Scholar]
- 23.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim EH, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, et al. l-2-Hydroxyglutarate: An Epigenetic Modifier and Putative Oncometabolite in Renal Cancer. Cancer discovery. 2014;4:1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooke JP. Asymmetrical dimethylarginine: the Uber marker? Circulation. 2004;109:1813–1818. doi: 10.1161/01.CIR.0000126823.07732.D5. [DOI] [PubMed] [Google Scholar]
- 26.Hennekens CH, Schneider WR, Pokov A, Hetzel S, Demets D, Serebruany V, et al. A randomized trial of aspirin at clinically relevant doses and nitric oxide formation in humans. Journal of cardiovascular pharmacology and therapeutics. 2010;15:344–348. doi: 10.1177/1074248410375091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hetzel S, DeMets D, Schneider R, Borzak S, Schneider W, Serebruany V, et al. Aspirin increases nitric oxide formation in chronic stable coronary disease. Journal of cardiovascular pharmacology and therapeutics. 2013;18:217–221. doi: 10.1177/1074248413482753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroecksnadel K, Weiss G, Stanger O, Teerlink T, Fuchs D. Increased asymmetric dimethylarginine concentrations in stimulated peripheral blood mononuclear cells. Scandinavian journal of immunology. 2007;65:525–529. doi: 10.1111/j.1365-3083.2007.01935.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.