Abstract
Cellular senescence is a complex stress response that permanently arrests the proliferation of cells at risk for oncogenic transformation. However, senescent cells can also drive phenotypes associated with aging. Although the senescence-associated growth arrest prevents the development of cancer and the metabolism of cancer cells has been studied in depth, the metabolic causes and consequences of cellular senescence were largely unexplored until recently. New findings reveal key roles for several aspects of cellular metabolism in the establishment and control of senescent phenotypes. These discoveries have important implications for both cancer and aging. In this review, we highlight some of the recent links between metabolism and phenotypes that are commonly associated with senescent cells.
Introduction
Cellular senescence is a multifaceted process that radically alters the phenotype of metazoan cells that have the ability to divide. Two hallmarks of cellular senescence are an essentially irreversible arrest of cell proliferation and the development of a multi-component senescence-associated secretory phenotype (SASP). Both features are induced in response to a variety of stresses and physiological signals, and can have beneficial or deleterious effects on the organism, depending on the context.
Many of the stresses that induce a senescence response cause genomic or epigenomic damage -- or metabolic deficits – all of which can put cells at risk for oncogenic transformation (Campisi and d’Adda di Fagagna, 2007). Thus, the senescence arrest protects organisms from developing cancer. In addition, the SASP can optimize certain physiological processes, such as wound healing and the formation of specific embryonic structures (Munoz-Espin and Serrano, 2014).
Senescent cells have been shown to accumulate with age in many tissues of vertebrate organisms (Adams, 2009; Campisi, 2013). In contrast to apoptotic and quiescent cells, senescent cells are highly metabolically active. Indeed, continued cell enlargement in the absence of cell division is a hallmark of senescent cells, suggesting an uncoupling of signals that link cell proliferation to cell size. The high metabolic activity of senescent cells may derive from the SASP, which entails the transcriptional activation and secretion of a myriad of factors that have potent biological activities on neighboring cells and the surrounding tissue. These activities include the promotion of inflammation, invasion, angiogenesis, and – ironically – cell proliferation. The SASP may explain how a relatively small number of senescent cells can have potent local and systemic effects that promote aging phenotypes and pathologies.
Senescent cells can contribute to a loss of tissue homeostasis by 1) limiting the regenerative capacity of a tissue due to the proliferative arrest, and 2) altering the functions of neighboring of cells by virtue of the SASP. Recent studies suggest that both the senescence arrest and the SASP are intimately linked to the metabolic state of the cell. Here, we summarize these studies and their relevance within the larger framework of cancer and aging.
General metabolic features of senescent cells
Decades ago, Otto Warburg observed that cancer cells generally differ from normal cells by the manner in which they metabolize glucose (Warburg, 1956). Cancer cells often elevate glycolysis over mitochondrial oxidative phosphorylation, even in the presence of high (atmospheric) oxygen. This “aerobic glycolysis” has been studied extensively for its role in tumor promotion in the decades since its discovery (Gatenby and Gillies, 2004; Kim and Dang, 2006; Vander Heiden et al., 2009). However, the study of glycolysis and other aspects of metabolism have only recently begun to be studied in the context of tumor suppression responses, such as cellular senescence. These new findings demonstrate how cancer, senescence, and aging can be linked in part through common metabolic processes.
It has been known for more than 30 years that as human fibroblast cultures accumulate senescent cells during serial passage, they gradually adopt a more glycolytic state (Bittles and Harper, 1984). Increased glycolysis has since been repeatedly associated with senescent cells in culture. Later, metabolic profiling similarly showed a significant shift to a more glycolytic state, but also a less energetic state, as cell cultures underwent replicative senecence (Zwerschke et al., 2003). Further, the senescent state was linked to increases in ADP and AMP relative to ATP (Figure 1), and addition of exogenous mononucleotides (e.g., AMP) to the medium was shown to result in a senescence arrest. Subsequent studies identified several markers of increased glycolysis in conditioned media from senescent fibroblasts (James et al., 2015), further linking these processes and suggesting that this metabolic shift is a potentially important attribute of senescent cells. Indeed, in a murine lymphoma model of chemotherapy-induced senescence, blockade of glucose utilization resulted in regression of the tumor mass and improved treatment outcomes (Dorr et al., 2013), presumably due to the elimination of senescent tumor cells and their SASP, which can promote inflammation and the proliferation of tumor cells that escape the chemotherapy-induced senescence. Together, these studies suggest a functional link between a more highly glycolytic state and senescent phenotypes.
Figure 1. Energy status regulates the senescence growth arrest.
Senescent cells have increased AMP:ATP and ADP:ATP ratios, which activate AMP-activated protein kinase (AMPK). AMP kinase in turn activates p53, which increases p21 transcription. AMPK also inactivates HuR, which increases the stability of the p21 and p16INK4a mRNAs. p21 and p16INK4a are cyclin-dependent kinase inhibitors that impose the senescence proliferative arrest by increasing activity of the pRB tumor suppressor. p53 and pRB might counterbalance each other in regulating glycolysis in senescent cells.
How might this metabolic shift drive senescence? AMP-activated protein kinase (AMPK) is a master regulator of cellular responses to energy stress. AMPK responds to increased AMP:ATP and ADP:ATP ratios (Figure 1) by activating a compensatory series of responses including fatty acid oxidation, inhibition of fatty acid synthesis, increased mitochondrial biogenesis, and stimulation of glucose uptake (Hardie et al., 2012). AMPK activation can induce cell cycle arrest and, ultimately, senescence through two distinct mechanisms (Figure 1). First, AMPK directly phosphorylates p53 on multiple residues, most notably on serine 15 (Jones et al., 2005). This p53 residue is also phosphorylated by ATM in response to genotoxic stress (Abraham, 2001) and is required for its activation and ability to arrest the cell cycle through transcriptional upregulation of the p21 cyclin-dependent kinase inhibitor. Indeed, AMPK itself is activated (phosphorylated) in an ATM-dependent manner in response to genotoxic stress (Luo et al., 2013). Second, AMPK also inhibits Hu antigen R (HuR)-dependent degradation of the mRNAs encoding the p21waf1 and p16INK4a cyclin-dependent kinase inhibitors, thereby resulting in a senescence arrest of cell proliferation (Figure 1) (Wang et al., 2003). Thus, AMPK acts as a bioenergetic sensor, mediating a metabolic checkpoint during energy stress that can ultimately result in senescence.
Beyond establishing the proliferative arrest of senescent cells, p53 also plays an important role in the regulation of glycolysis (Kruiswijk et al., 2015; Puzio-Kuter, 2011). First, p53 antagonizes the uptake of glucose by lowering expression of the glucose transporters GLUT1 and GLUT4 (Schwartzenberg-Bar-Yoseph et al., 2004). Second, p53 transcriptionally activates the expression of TP53-inducible glycolysis and apoptosis regulator (TIGAR), which dephosphorylates fructose-2,6-bisphosphate to antagonize the early stages of glycolysis (Bensaad et al., 2006). Third, p53 inhibits phosphoglycercate mutase (PGM), slowing the rate at which glucose is converted into pyruvate, thereby promoting senescence (Kondoh et al., 2005). Fourth, p53 binds glucose-6-phosphate dehydrogenase (G6PDH), inhibiting its activity and, consequently, the pentose phosphate pathway (PPP) (Jiang et al., 2011). Finally, p53 promotes mitochondrial oxidative phosphorylation by inducing expression of synthesis of cytochrome c oxidase 2 (SCO2), inhibiting pyruvate dehydrogenase kinase 2 (PDK2) through Parkin (PARK2) activation, and promoting fatty acid oxidation (Ide et al., 2009; Matoba et al., 2006; Zhang et al., 2011). Together, these activities of p53 antagonize glycolysis and promote mitochondrial respiration. Thus, p53 likely acts to limit the glycolytic activity of senescent cells (Figure 1). Notably, p53 also limits the extent of the SASP (Coppe et al., 2008), suggesting the SASP is at least partly regulated by glycolysis.
Pyruvate metabolism and senescence
Pyruvate is a key metabolite at the crossroads of glycolysis and mitochondrial respiration. As a net effect of glycolysis, 2 molecules of ATP and 2 molecules NAD+ are converted to NADH, and 2 molecules of pyruvate are produced (Lehninger et al., 2013). Pyruvate itself is active in multiple metabolic processes. As cells shift to a glycolytic phenotype, as described for senescent cells (James et al., 2015; Zwerschke et al., 2003), pyruvate and NADH become available substrates for lactate dehydrogenase, producing NAD+ and lactate. Lactate is then excreted. Because NADH is produced by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) early during glycolysis, excess NADH can inhibit GAPDH and therefore become rate limiting for glycolysis. Pyruvate also serves as the primary carbon source for acetyl-coenzyme A (acetyl-CoA, which ultimately enters the tricarboxylic acid (TCA) cycle (Lehninger et al., 2013). In this way, mitochondria generate much of their energy via pyruvate metabolism. Whether pyruvate becomes lactate or enters the TCA cycle is determined by phosphorylation of pyruvate dehydrogenase (PDH), which catalyzes the formation of acetyl-CoA from pyruvate; this decision determines whether a cell senesces in response to activated oncogenes (Kaplon et al., 2013). Thus, pyruvate metabolism is key to determining whether a cell can generate sufficient energy via glycolysis upon becoming senescent.
Excessive reactive oxygen species (ROS) can induce a senescence response (Ziegler et al., 2015), and pyruvate also helps scavenge ROS. Pyruvate interacts with hydrogen peroxide (H2O2) to form acetate, CO2 and H2O, thereby contributing to cellular antioxidant defenses (Desagher et al., 1997). It is unclear how important the antioxidant properties of pyruvate are in senescent cells. Ironically, superphysiological (millimolar) concentrations of pyruvate actually decrease the replicative lifespan of human fibroblasts, likely through increased mitochondrial ROS production (Xu and Finkel, 2002). These data suggest the antioxidant effects of pyruvate cannot overcome the increased ROS produced as a byproduct of its metabolism. Importantly, these experiments were performed in cells cultured in atmospheric oxygen, whereas most cells reside in tissues with significantly lower oxygen levels (Parrinello et al., 2003).
Malate and senescence
Malate is another key metabolite that regulates senescence. Malate is produced from fumarate by fumarase, and is decarboxylated to pyruvate and CO2, through reduction of either NAD+ to NADH (mitochondrial malic enzyme 2; ME2) or NADP+ to NADPH (cytosolic malic enzyme 1; ME1). Notably, ME1 and ME2 are reciprocally regulated by p53 in senescent cells – p53 suppresses their expression, whereas loss of either enzyme activates p53 and induces senescence (Jiang et al., 2013). ME1 and ME2 expression declines in senescent cells; overexpression of either enzyme extends the replicative life span of cultured fibroblasts. Mechanistically, loss of either malic enzyme decreases NADPH levels, thus reducing NADPH-dependent functions such as lipid synthesis, glucose consumption, glutaminolysis, and antioxidant defenses. Indeed, ME2 depletion increases ROS, which activates AMPK, causing AMPK-dependent phosphorylation (activation) of p53 and senescence (Jiang et al., 2013). Thus, malate metabolism can antagonize senescence by bolstering cellular antioxidant defenses.
An additional role for malate in preventing senescence derives from studies that entail an inhibition of malate dehydrogenase 1 (MDH1), a component of the malate-aspartate shuttle, which transfers reducing equivalents of NADH from the cytosol to the mitochondrial matrix (Lee et al., 2012). Like the malic enzymes, MDH1 levels and activity decline in senescent cells, and MDH1 depletion induces a senescence response. However, unlike ME2 depletion, which reduces mitochondrial NADPH, MDH1 depletion lowers the cytosolic NAD+/NADH ratio. Similarly, aminooxyacetate (AOA) inhibition of aspartate aminotransferase (GOT1), a malate-aspartate shuttle enzyme, decreases the cytosolic NAD+/NADH ratio and induces senescence, which is rescued by exogenous pyruvate (Wiley et al., 2016). Furthermore, MDH1 and GOT1 are required for NAD+-dependent synthesis of aspartate in response to electron transport chain (ETC) inhibition, and loss of either enzyme prevents cell proliferation (Birsoy et al., 2015). Thus, malate is at the nexus of many redox reactions, loss of which induces cellular senescence (Figure 2).
Figure 2. The electron transport chain (ETC) and malate-aspartate shuttle antagonize senescence.
Inhibition of glutamic-qxaloacetic transaminase 1 (GOT1) by aminooxyacetate (AOA) or ETC deficits decrease the cytosolic NAD+/NADH ratio, leading to AMPK activation. AMPK in turn phosphorylates and activated p53, which promotes senescence but suppresses the IL1R signaling arm of the SASP. Malate dehydrogenase (MDH1) and GOT1 are required for the synthesis of aspartate following ETC inhibition, and loss of either enzyme prevents pyruvate from rescuing cellular proliferation.
Mitochondrial dysfunction-associated senescence (MiDAS)
Multiple mitochondrial manipulations - loss of the mitochondrial sirtuins SIRT3 or SIRT5 or the mitochondrial chaperone HSPA9, inhibition of the electron transport chain (ETC) by rotenone or antimycin, or depletion of mitochondrial DNA – induce a senescence response. Unlike genotoxic stress or oncogene activation, mitochondrial dysfunction induces a distinct SASP that lacks key proinflammatory factors (Wiley et al., 2016) that depend on interleukin 1 receptor (IL1R) signaling (Orjalo et al., 2009). Mitochondrial dysfunction-associated senescence (MiDAS) illustrates the plasticity of the SASP. Notably, pyruvate not only prevents the MiDAS growth arrest, but also restores the IL1R-dependent arm of the SASP. MiDAS cells have a decreased NAD+/NADH ratio; addition of pyruvate (Wiley et al., 2016) restores this ratio, decreases the ADP/ATP ratio, restores aspartate synthesis (Birsoy et al., 2015) and promotes cell proliferation (Figure 2).
Like the loss of malic enzymes (Jiang et al., 2013), MiDAS activates AMPK, resulting in p53 activation and senescence (Figure 1). This p53 activity in turn inhibits the transcription factor NF-κB (Wiley et al., 2016) – a major regulator of the SASP (Freund et al., 2011). Thus, metabolic regulation of p53 during mitochondrial dysfunction controls two aspects of the senescence response (growth arrest and the SASP). Senescent cells accumulate in at least three murine models of mitochondrial dysfunction and premature aging (Kang et al., 2013; Velarde et al., 2012; Wiley et al., 2016), implicating mitochondrial dysfunction as a driver of cellular senescence in vivo.
Mitochondrial mass and DNA (mtDNA) content increase 2- to 3-fold in senescent cells (Correia-Melo et al., 2016), and elimination of either mtDNA or mitochondria antagonizes the senescence arrest and SASP. Increased mitochondria, generally driven by mTOR (mechanistic target of rapamycin) signaling (discussed below), can increase ROS and DNA damage. Indeed, lowering mitochondrial mass in senescent cells lowers mitochondrial ROS, 53BP1 foci (a marker of DNA double-strand breaks (DSB) and DNA damage response (DDR) signaling) (Correia-Melo et al., 2016), which is required for the arrest and IL1R arm of the SASP (Rodier et al., 2009). Thus, expansion of mitochondria in senescent cells might reinforce senescent phenotypes by promoting DDR signaling.
Many questions remain unanswered. For example, although reducing mitochondria in senescent cells decreases both ROS and 53BP1 foci (Correia-Melo et al, 2009), increased ROS in some forms of mitochondrial dysfunction does not always increase 53BP1 foci or DDR signaling. Thus, ROS may not be an obligate driver or reinforcer of senescent phenotypes. Likewise, it is not clear which factors drive the MiDAS SASP, nor the extent to which MiDAS occurs during natural aging. By contrast, much more is known about the role of NAD metabolism in senescence and aging.
Role of NAD metabolism in senescent phenotypes
As described above, low NAD+/NADH ratios promote cellular senescence (Figure 2) at least in part by limiting glycolysis and ATP production. Likewise, NADP+/NADPH ratio controls cellular redox status, but NAD+ can regulate cellular homeostasis and senescence independently of NADH or NADPH. NAD+ is the primary ADP-ribose donor for poly-ADP ribose polymerase (PARP) - a major mediator of DNA repair in response to genotoxic stress (Kim et al., 2005). PARP inhibition promotes DNA damage in irradiated tumor cells, sensitizing them to senescence (Efimova et al., 2010). In addition, PARP-1 is required for NF-κB activation and the proinflammatory SASP in melanoma cells (Ohanna et al., 2011). Thus, low NAD+ promotes senescence. Indeed, nicotinamide mononucleotide phosphoribosytransferase (NAMPT) inhibition induces senescence (van der Veer et al., 2007) without the IL1R arm of the SASP (Wiley et al., 2016), whereas NAMPT overexpression suppresses smooth muscle cell senescence (van der Veer et al., 2007). Notably, NAMPT expression is lost in several tissues during aging (Gomes et al., 2013; Stein and Imai, 2014; Yoshino et al., 2011).
NAD+ is a cofactor for sirtuins, proteins that remove acetyl, succinyl and similar groups from proteins, some of which promote longevity. Low NAD+ levels decrease sirtuin activity (Haigis and Sinclair, 2010). As noted above, SIRT3 or SIRT5 loss induces senescence in human cells and mice (Wiley et al., 2016), as does SIRT6 deficiency (Cardus et al., 2013; Michishita et al., 2008). SIRT1 deacetylates several senescence-associated substrates, including p53, Ku70, and NF-κB (Brooks and Gu, 2009; Kong et al., 2012; Saunders and Verdin, 2007; Yi and Luo, 2010). SIRT2 deacetylates BUBR1 (mitotic checkpoint kinase budding uninhibited by benzimidazole-related 1) in a NAD+-dependent manner (North et al., 2014). BUBR1 activity antagonizes senescence (Baker et al., 2004), and NAD+ or SIRT2 depletion destabilizes BUBR1 (North et al., 2014). Thus NAD+ regulates senescent phenotypes through several mechanisms.
Together, these studies show that both senescence and the SASP are subject to metabolic control. On one hand, senescence appears to be a stress checkpoint that guards against metabolic crisis and potential oncogenesis. On the other, senescence due to metabolic stress may be accompanied by its own set of secreted molecules (as in MiDAS), and thus have unique effects on tissue microenvironments. Understanding these pathways can provide new therapeutic targets for the treatment of maladies associated with senescence, ranging from cancer to aging and age-related diseases.
Metabolic features of oncogene-induced senescence
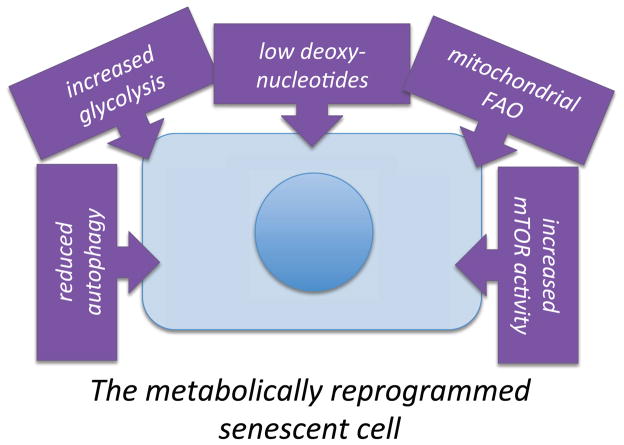
Activation of several proto-oncogenes induce a senescence response as a mechanism to protect against cancer (Courtois-Cox et al., 2008). Metabolic changes have been studied in the context of oncogenesis for some time, but only recently has metabolism been studied in the context of oncogene-induced senescence (OIS) and other forms of senescence (Figure 3).
Figure 3. The senescence response entails a complex series of metabolic changes (metabolic reprogramming).
See text for details.
Skewed balance between glycolysis and oxidative phosphorylation
OIS entails a key shift in the metabolism of pyruvate (Kaplon et al., 2013). Human fibroblasts transformed with BRAFV600E, a common oncogenic mutation and inducer of senescence, undergo several metabolic changes, including increased oxygen consumption, increased release of glutamate, and decreased pyruvate secretion. These changes are associated with decreased phosphorylation of pyruvate dehydrogenase (PDH), which catalyzes the conversion of pyruvate into acetyl-coenzyme A, allowing it to enter the TCA cycle. The decline in PDH phosphorylation results from increased expression of pyruvate dehydrogenase phosphatase (PDP2) and decreased levels of pyruvate dehydrogenase kinase (PDK1). Phosphorylation of PDH inhibits its activity, so the net effect of these alterations is increased TCA cycle activity and mitochondrial respiration. Further, PDP2 depletion or PDK1 overexpression allows BRAFV600E expressing cells to bypass senescence. PDK1 overexpression also potentiates malignant transformation of BRAFV600E-expressing melanocytes, while PDK1 depletion suppresses transformation. Since the ETC is a major source of ROS, mitochondria-derived oxidative stress might drive OIS. Alternatively, since anaerobic fermentation of pyruvate into lactate is required to maintain NAD+/NADH ratios under high glycolytic activity, reduced deoxynucleotides (discussed below) may partially explain how shunting pyruvate to mitochondria drives OIS in melanocytes, and how a metabolic switch to a more glycolytic phenotype allows escape from senescence (Aird et al., 2013). Thus, a tradeoff between glycolysis and mitochondrial metabolism can control senescence in response to oncogene activation.
Deoxynucleotide synthesis
Recent studies implicate the loss of deoxynucleotide synthesis in RAS-induced senescence (Aird et al., 2013). Oncogenic RAS lowers the mRNA and protein levels of ribonucleotide reductase (RRM2), which catalyzes the formation of deoxyribonucleotide diphosphates from ribonucleotide diphosphates. The depletion of deoxynucleotides causes replication forks to stall and eventually collapse, which is exacerbated by the stimulation of cell proliferation caused by oncogenic RAS. The resulting DNA DSBs elicit a senescence response. RRM2 overexpression can rescue OIS, whereas RRM2 depletion induces senescence (Aird et al., 2013). Further, RRM2 is downregulated in human benign nevi, which are often composed of senescent melanocytes (Salama et al., 2014), and is often overexpressed in human melanoma. Thus, nucleotide metabolism also plays a key role in senescent phenotypes, at least in the context of oncogene activation. Of additional interest, ATM deficiency allows RRM2-depleted cells to proliferate, reduces the incidence of stalled replication forks and DNA damage markers, and rescues the depletion of deoxynucleotides (Aird et al., 2015). ATM deficiency also increases glycolysis and – more specifically – the pentose phosphate pathway, which provides nucleotide precursors.
Retinoblastoma protein and regulation of glycolysis
The tumor suppressor and transcriptional regulator retinoblastoma protein (RB) mediates a second major pathway that promotes the senescence arrest, and also mediates metabolic changes in OIS. While p53 antagonizes glycolysis in favor of mitochondrial oxidation of pyruvate and fatty acids, RB stimulates glycolysis in OIS (Takebayashi et al., 2015). RB also promotes mitochondrial oxidative phosphorylation, either by regulating or complementing the activity of PDH1, which promotes mitochondrial catabolism of pyruvate produced by elevated glycolysis. RB-mediated mediated metabolic changes might regulate the senescence growth arrest by counterbalancing the inhibition of glycolysis by p53 (Figure 1). By contrast, RB activation by its upstream positive regulator p16INK4a is not sufficient for induction of a SASP (Coppe et al., 2011).
Role of fatty acid metabolism
OIS is also linked to increased fatty acid oxidation (beta-oxidation) (Quijano et al., 2012). Upon induction of senescence by oncogenic RAS, human fibroblasts increase oxygen consumption, while steady state levels of many fatty acids significantly decline. Inhibition of carnitine palmitoyltransferase (CPT1A) prevents the transfer of fatty acids into the mitochondria, which in turn prevents the increases in oxygen consumption. In accordance with this decrease in mitochondrial activity, much of the proinflammatory SASP is reduced. Thus, much like the case of MiDAS (Wiley et al., 2016), the bioenergetic state of the senescent cell strongly influences the proinflammatory arm of the SASP.
RAS-induced senescence is also linked to increased activity of arachidonate 5-lipoxygenase (ALOX5), which catalyzes the first step in the synthesis of several signaling lipids, including resolvins and leukotrienes (Catalano et al., 2005). In turn, overexpression of ALOX5 induces p53-dependent senescence. This senescence is not a consequence of leukotriene synthesis per se; rather, the elevated ROS produced by ALOX5 activity appears to be causal (Catalano et al., 2005). Little is known about the role of ALOX5 in the SASP, but leukotrienes are known to mediate inflammation and thus may themselves be SASP components.
Why does oncogene activation reprogram cellular metabolism? One possibility is that evolution selected for metabolic shifts that promote senescence in order to prevent tumorigenesis. Alternatively, since oncogene activation is often mitogenic, it may be that cells must reprogram metabolism in order to meet the energetic requirements of the mitogenic stimulus. Regardless of the reason, oncogene activation drives clear changes in cellular metabolism. These changes might offer opportunities to shift the metabolic balance in tumor cells from tumorigenesis to senescence. Indeed, inhibition of glycogen phosphorylase (PYGL) can suppress tumor growth by inducing senescence (Favaro et al., 2012).
Proteostasis in senescence
Senescent cells also display alterations in protein metabolism that can influence both the growth arrest and SASPs. There are two major ways in which cellular proteostasis is altered in senescent cells. First, accelerated degradation lowers the levels of proteins that antagonize senescent phenotype. Second, alterations in translational efficiency change levels of senescence-associated proteins.
Autophagy and protein damage
Multiple lines of evidence indicate a senescence-associated increase in autophagy, especially macroautophagy – the process by which protein complexes and organelles associate with membrane-bound autophagosomes that eventually fuse with lysosomes (Capparelli et al., 2012; Gamerdinger et al., 2009; Singh et al., 2012). In endothelial cells, activation of autophagy determines whether glycated collagen-induced stress results in senescence or cell death, although this determination may be due to autophagy of damaged lipids, rather than proteins (Patschan et al., 2008). Conversely, inhibition of autophagy predisposes cells to senescence (Kang et al., 2011; Wang et al., 2012). Recent evidence also links senescence-inducing genotoxic stress and ATM/ATR activation to the SASP through autophagy (Kang et al., 2015). In this case, loss of selective autophagy by p62/SQSTM1 stabilizes GATA4, which then induces transcription of a subset of SASP factors, including IL1A. Thus, although macroautophagy is elevated during senescence, loss of specific autophagic programs is essential for development of the SASP.
Autophagic degradation of damaged organelles and proteins occurs in the lysosome, a membrane-bound organelle that serves as a recycling center for cells. In agreement with activation of autophagy, there is also evidence for altered lysosomal activity in senescent cells. Indeed, the senescence-associated beta-galactosidase, a canonical marker of senescent cells, is a lysosomal protein (Lee et al., 2006). In addition, following induction of senescence, chromatin fragments bud from the nucleus, enter the cytoplasm, and are processed by lysosomes, resulting in a gradual depletion of total histone content (Ivanov et al., 2013). In addition, associations between the autophagy-regulating kinase mTOR and the lysosome occur at senescence, and disruption of this association prevents the synthesis of certain SASP factors (Narita et al., 2011). Lysosomes therefore play an important role in the senescent phenotype.
A second protein degradation complex – the proteasome – also changes in senescence. Senescent cells elevate selective proteasome activity; this senescence-associated protein degradation (SAPD) reduces the levels of selected proteins required for cell cycle progression, mitochondrial function, and RNA processing (Deschenes-Simard et al., 2013). Inactivation of individual SAPD targets is sufficient to induce senescence, confirming a role for the proteasome in senescence.
Proteins are targeted to the proteasome via the activity of ubiquitin-conjugating enzymes, many of which play key roles in senescence. For example, mouse double minute 2 (MDM2), a ubiquitin-conjugating enzyme, targets p53 for degradation, and its inhibition can drive senescence (Efeyan et al., 2007). MDM2 also antagonizes glycolysis and promotes senescence by targeting PGM for degradation (Mikawa et al., 2014). Protein ubiquitination states change notably with senescence, most notably in proteins responsible for regulating translation, including several eukaryotic initiation factors (eIFs), elongation factors, ribosomal subunits, and mTOR signaling complexes (Bengsch et al., 2015). The mitochondrial ubiquitin ligase MARCH5 antagonizes senescence (presumably MiDAS) by promoting degradation of mitofusin 1 and dynamin-regulated protein 1 (DRP1) (Park et al., 2010). Additionally, ubiquitin-conjugating enzymes can be targeted to proteins in a senescence-specific manner. For example, speckle-type POZ protein (SPOP) is an adapter protein that targets SENP7 (a desumoylase) for degradation in response to oncogene activation. Loss of SENP7 results in epigenetic changes that promote senescence (Zhu et al., 2015). SPOP is elevated during OIS, and overexpression of SPOP drives senescence, whereas knockdown of SPOP allows cells to escape OIS. Together, these studies confirm that protein degradation, whether from autophagy or the proteasome, plays an important role in senescent phenotypes.
Protein translation, rapamycin, and senescence
A growing body of literature now links cellular senescence to drugs that extend lifespan in multiple species. One such drug is rapamycin, an inhibitor of the mTORC1 complex of the mTOR kinase and an inducer of autophagy. Rapamycin reduces the IL1R-driven arm of the SASP (Herranz et al., 2015; Laberge et al., 2015) by inhibiting the translation of IL-1α and MAPKAPK2, key mediators of the proinflammatory elements of the SASP. Rapamycin also prevents the increase in mitochondrial DNA, biomass, and ROS associated with genotoxic stress-induced senescence (Correia-Melo et al., 2016). Rapamycin or mTOR inhibition prevents 4EBP1 phosphorylation through a pathway that is also inhibited by metabolic stresses such as calorie restriction (Syntichaki et al., 2007; Zid et al., 2009). Since rapamycin and calorie restriction extend lifespan in many species (Bove et al., 2011; Harrison et al., 2009; Heilbronn and Ravussin, 2003), these findings suggest senescence as a potential link between the overlapping effects of rapamycin and caloric restriction.
Thus, proteostasis is linked to senescence through both autophagy and translation. A key point of these studies is that these mechanisms are relatively easy to target. The links between life span extension through either calorie restriction or rapamycin and cellular senescence suggest mechanisms by which senescent phenotypes might be targeted for therapeutic intervention.
Conclusions
Recent advances have expanded our understanding of the interface between metabolism, cell signaling, and cell fates, particularly cellular senescence, which can drive a variety of age-related pathologies, including cancer. Our understanding of the metabolic reprogramming that occurs during cellular senescence (Figure 3) is still in infancy, so it is likely many new findings will emerge in the near future.
Links between senescence and NAD metabolism have been suspected for some time, but now it is clear that both NAD+ levels and NAD+/NADH ratios control aspects of senescent phenotypes through different pathways. Recently, compounds that elevate NAD+, including nicotinamide mononucleotide (NMN), nicotinamide riboside (NR) and P7C3 have been proposed as possible therapeutics for preventing several age-related pathologies (Gomes et al., 2013; Stein and Imai, 2014; Yoshino et al., 2011). Since elevating NAD+ can also increase the NAD+/NADH ratio, these compounds might prevent both MiDAS and other forms of senescence. A concern, however, is that restoring NAD+/NADH ratios in the context of mitochondrial dysfunction might also result in proliferation of cells with compromised mitochondria, as well as proinflammatory secretion that ultimately promotes tissue degeneration.
While the relationship between senescence and glycolysis has been confirmed in many studies, it less clear precisely why senescent cells become more glycolytic. Since senescent cells increase in size after arresting proliferation and the SASP demands considerable macromolecular biosynthesis, it is possible that senescent cells, like many cancer cells (Vander Heiden et al., 2009), favor glycolysis for its ability to provide precursors for a high demand for protein, lipid and other cellular components.
What therapeutic outcomes might result from these discoveries? Given the complexities of metabolism in general, and the role of metabolism in regulating both the causes and consequences of the senescence response, the field is ripe for applications of systems biology approaches. Indeed, this approach was recently taken to not only understand the dynamics of the senescence response but also to identify therapeutic targets for ameliorating its deleterious effects (Dalle Pezze et al., 2014).
With the wealth of data now emerging, targeting the metabolic differences between non-senescent and senescent cells could become an important strategy for eliminating the deleterious effects of senescence. Indeed, metabolic targeting was a strategy used to eliminate senescent tumor cells following chemotherapy (Dorr et al., 2013). Similarly, metabolism-targeting drugs such as rapamycin or etomoxir can suppress the proinflammatory SASP (Laberge et al., 2015; Quijano et al., 2012), preventing one of the most detrimental aspects of senescence. Metabolism-targeted approaches therefore hold great promise for dealing with the complications that arise as a result of senescence, the most global of which is aging and its associated pathologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christopher D. Wiley, Email: cwiley@buckinstitute.org.
Judith Campisi, Email: jcampisi@buckinstitute.org, jcampisi@lbl.gov.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Adams PD. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell. 2009;36:2–14. doi: 10.1016/j.molcel.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Aird KM, Worth AJ, Snyder NW, Lee JV, Sivanand S, Liu Q, Blair IA, Wellen KE, Zhang R. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Rep. 2015;11:893–901. doi: 10.1016/j.celrep.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Bengsch F, Tu Z, Tang HY, Zhu H, Speicher DW, Zhang R. Comprehensive analysis of the ubiquitinome during oncogene-induced senescence in human fibroblasts. Cell Cycle. 2015;14:1540–1547. doi: 10.1080/15384101.2015.1026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Harper N. Increased glycolysis in ageing cultured human diploid fibroblasts. Biosci Rep. 1984;4:751–756. doi: 10.1007/BF01128816. [DOI] [PubMed] [Google Scholar]
- Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12:437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Capparelli C, Chiavarina B, Whitaker-Menezes D, Pestell TG, Pestell RG, Hulit J, Ando S, Howell A, Martinez-Outschoorn UE, Sotgia F, et al. CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, “fueling” tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle. 2012;11:3599–3610. doi: 10.4161/cc.21884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardus A, Uryga AK, Walters G, Erusalimsky JD. SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovasc Res. 2013;97:571–579. doi: 10.1093/cvr/cvs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano A, Rodilossi S, Caprari P, Coppola V, Procopio A. 5-Lipoxygenase regulates senescence-like growth arrest by promoting ROS-dependent p53 activation. EMBO J. 2005;24:170–179. doi: 10.1038/sj.emboj.7600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Rodier F, Patil CK, Freund A, Desprez PY, Campisi J. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem. 2011;286:36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia-Melo C, Marques FD, Anderson R, Hewitt G, Hewitt R, Cole J, Carroll BM, Miwa S, Birch J, Merz A, et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016 doi: 10.15252/embj.201592862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- Dalle Pezze P, Nelson G, Otten EG, Korolchuk VI, Kirkwood TB, von Zglinicki T, Shanley DP. Dynamic modelling of pathways to cellular senescence reveals strategies for targeted interventions. PLoS Comput Biol. 2014;10:e1003728. doi: 10.1371/journal.pcbi.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desagher S, Glowinski J, Premont J. Pyruvate protects neurons against hydrogen peroxide-induced toxicity. J Neurosci. 1997;17:9060–9067. doi: 10.1523/JNEUROSCI.17-23-09060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschenes-Simard X, Gaumont-Leclerc MF, Bourdeau V, Lessard F, Moiseeva O, Forest V, Igelmann S, Mallette FA, Saba-El-Leil MK, Meloche S, et al. Tumor suppressor activity of the ERK/MAPK pathway by promoting selective protein degradation. Genes Dev. 2013;27:900–915. doi: 10.1101/gad.203984.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Dabritz JH, Lisec J, Lenze D, Gerhardt A, Schleicher K, et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501:421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Ortega-Molina A, Velasco-Miguel S, Herranz D, Vassilev LT, Serrano M. Induction of p53-dependent senescence by the MDM2 antagonist nutlin-3a in mouse cells of fibroblast origin. Cancer Res. 2007;67:7350–7357. doi: 10.1158/0008-5472.CAN-07-0200. [DOI] [PubMed] [Google Scholar]
- Efimova EV, Mauceri HJ, Golden DW, Labay E, Bindokas VP, Darga TE, Chakraborty C, Barreto-Andrade JC, Crawley C, Sutton HG, et al. Poly(ADP-ribose) polymerase inhibitor induces accelerated senescence in irradiated breast cancer cells and tumors. Cancer Res. 2010;70:6277–6282. doi: 10.1158/0008-5472.CAN-09-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Bensaad K, Chong MG, Tennant DA, Ferguson DJ, Snell C, Steers G, Turley H, Li JL, Gunther UL, et al. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 2012;16:751–764. doi: 10.1016/j.cmet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Freund A, Patil PK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30:1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronn LK, Ravussin E. Calorie restriction and aging: review of the literature and implications for studies in humans. Am J Clin Nutr. 2003;78:361–369. doi: 10.1093/ajcn/78.3.361. [DOI] [PubMed] [Google Scholar]
- Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ, Raguz S, Acosta JC, Innes AJ, Banito A, et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17:1205–1217. doi: 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Brown-Endres L, Chu K, Ongusaha PP, Ohtsuka T, El-Deiry WS, Aaronson SA, Lee SW. GAMT, a p53-inducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol Cell. 2009;36:379–392. doi: 10.1016/j.molcel.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. Lysosome-mediated processing of chromatin in senescence. J Cell Biol. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EL, Michalek RD, Pitiyage GN, de Castro AM, Vignola KS, Jones J, Mohney RP, Karoly ED, Prime SS, Parkinson EK. Senescent human fibroblasts show increased glycolysis and redox homeostasis with extracellular metabolomes that overlap with those of irreparable DNA damage, aging, and disease. J Proteome Res. 2015;14:1854–1871. doi: 10.1021/pr501221g. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, Lu T, Yankner BA, Campisi J, Elledge SJ. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HT, Lee KB, Kim SY, Choi HR, Park SC. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS One. 2011;6:e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Louboutin JP, Datta P, Landel CP, Martinez D, Zervos AS, Strayer DS, Fernandes-Alnemri T, Alnemri ES. Loss of HtrA2/Omi activity in non-neuronal tissues of adult mice causes premature aging. Cell Death Differ. 2013;20:259–269. doi: 10.1038/cdd.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaa EME, Cascante M, Shlomi T, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- Kong S, McBurney MW, Fang D. Sirtuin 1 in immune regulation and autoimmunity. Immunol Cell Biol. 2012;90:6–13. doi: 10.1038/icb.2011.102. [DOI] [PubMed] [Google Scholar]
- Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, Curran SC, Davalos AR, Wilson-Edell KA, Liu S, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17:1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- Lee SM, Dho SH, Ju SK, Maeng JS, Kim JY, Kwon KS. Cytosolic malate dehydrogenase regulates senescence in human fibroblasts. Biogerontology. 2012;13:525–536. doi: 10.1007/s10522-012-9397-0. [DOI] [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Lehninger principles of biochemistry. New York: W.H. Freeman; 2013. [Google Scholar]
- Luo L, Huang W, Tao R, Hu N, Xiao ZX, Luo Z. ATM and LKB1 dependent activation of AMPK sensitizes cancer cells to etoposide-induced apoptosis. Cancer Lett. 2013;328:114–119. doi: 10.1016/j.canlet.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Maruyama T, Okamoto K, Nakagama H, Lleonart ME, Tsusaka T, Hori K, Murakami I, Izumi T, Takaori-Kondo A, et al. Senescence-inducing stress promotes proteolysis of phosphoglycerate mutase via ubiquitin ligase Mdm2. J Cell Biol. 2014;204:729–745. doi: 10.1083/jcb.201306149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Rosenberg MA, Jeganathan KB, Hafner AV, Michan S, Dai J, Baker DJ, Cen Y, Wu LE, Sauve AA, et al. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 2014;33:1438–1453. doi: 10.15252/embj.201386907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, Bille K, Robert C, Bressac-de Paillerets B, Hofman P, et al. Senescent cells develop a PARP-1 and nuclear factor-{kappa}B-associated secretome (PNAS) Genes Dev. 2011;25:1245–1261. doi: 10.1101/gad.625811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci U S A. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YY, Lee S, Karbowski M, Neutzner A, Youle RJ, Cho H. Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J Cell Sci. 2010;123:619–626. doi: 10.1242/jcs.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patschan S, Chen J, Polotskaia A, Mendelev N, Cheng J, Patschan D, Goligorsky MS. Lipid mediators of autophagy in stress-induced premature senescence of endothelial cells. Am J Physiol Heart Circ Physiol. 2008;294:H1119–1129. doi: 10.1152/ajpheart.00713.2007. [DOI] [PubMed] [Google Scholar]
- Puzio-Kuter AM. The Role of p53 in Metabolic Regulation. Genes Cancer. 2011;2:385–391. doi: 10.1177/1947601911409738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano C, Cao L, Fergusson MM, Romero H, Liu J, Gutkind S, Rovira II, Mohney RP, Karoly ED, Finkel T. Oncogene-induced senescence results in marked metabolic and bioenergetic alterations. Cell Cycle. 2012;11:1383–1392. doi: 10.4161/cc.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- Singh K, Matsuyama S, Drazba JA, Almasan A. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy. 2012;8:236–251. doi: 10.4161/auto.8.2.18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LR, Imai SI. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 2014 doi: 10.1002/embj.201386917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Tanaka H, Hino S, Nakatsu Y, Igata T, Sakamoto A, Narita M, Nakao M. Retinoblastoma protein promotes oxidative phosphorylation through upregulation of glycolytic genes in oncogene-induced senescent cells. Aging Cell. 2015;14:689–697. doi: 10.1111/acel.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veer E, Ho C, O’Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde MC, Flynn JM, Day NU, Melov S, Campisi J. Mitochondrial oxidative stress caused by Sod2 deficiency promotes cellular senescence and aging phenotypes in the skin. Aging. 2012;4:3–12. doi: 10.18632/aging.100423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Lopez de Silanes I, Carling D, Gorospe M. Increased AMP:ATP ratio and AMP-activated protein kinase activity during cellular senescence linked to reduced HuR function. J Biol Chem. 2003;278:27016–27023. doi: 10.1074/jbc.M300318200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang XD, Lapi E, Sullivan A, Jia W, He YW, Ratnayaka I, Zhong S, Goldin RD, Goemans CG, et al. Autophagic activity dictates the cellular response to oncogenic RAS. Proc Natl Acad Sci U S A. 2012;109:13325–13330. doi: 10.1073/pnas.1120193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, Shirakawa K, Lim HW, Davis SS, Ramanathan A, et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016;23:303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Finkel T. A role for mitochondria as potential regulators of cellular life span. Biochem Biophys Res Commun. 2002;294:245–248. doi: 10.1016/S0006-291X(02)00464-3. [DOI] [PubMed] [Google Scholar]
- Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim Biophys Acta. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Ren S, Bitler BG, Aird KM, Tu Z, Skordalakes E, Zhu Y, Yan J, Sun Y, Zhang R. SPOP E3 Ubiquitin Ligase Adaptor Promotes Cellular Senescence by Degrading the SENP7 deSUMOylase. Cell Rep. 2015;13:1183–1193. doi: 10.1016/j.celrep.2015.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DV, Wiley CD, Velarde MC. Mitochondrial effectors of cellular senescence: beyond the free radical theory of aging. Aging Cell. 2015;14:1–7. doi: 10.1111/acel.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W, Mazurek S, Stockl P, Hutter E, Eigenbrodt E, Jansen-Durr P. Metabolic analysis of senescent human fibroblasts reveals a role for AMP in cellular senescence. Biochem J. 2003;376:403–411. doi: 10.1042/BJ20030816. [DOI] [PMC free article] [PubMed] [Google Scholar]