In this study, Nemajerova et al. identify TP73, a p53 homolog, as a key regulator of airway multiciliogenesis. They show that mice with TP73 deficiency suffer from chronic respiratory tract infections due to profound defects in ciliogenesis and complete loss of mucociliary clearance and identify the downstream components of TP73 during multiciliogenesis.
Keywords: TAp73, p73, TP73, motile multiciliogenesis, central transcriptional regulator, airways
Abstract
Motile multiciliated cells (MCCs) have critical roles in respiratory health and disease and are essential for cleaning inhaled pollutants and pathogens from airways. Despite their significance for human disease, the transcriptional control that governs multiciliogenesis remains poorly understood. Here we identify TP73, a p53 homolog, as governing the program for airway multiciliogenesis. Mice with TP73 deficiency suffer from chronic respiratory tract infections due to profound defects in ciliogenesis and complete loss of mucociliary clearance. Organotypic airway cultures pinpoint TAp73 as necessary and sufficient for basal body docking, axonemal extension, and motility during the differentiation of MCC progenitors. Mechanistically, cross-species genomic analyses and complete ciliary rescue of knockout MCCs identify TAp73 as the conserved central transcriptional integrator of multiciliogenesis. TAp73 directly activates the key regulators FoxJ1, Rfx2, Rfx3, and miR34bc plus nearly 50 structural and functional ciliary genes, some of which are associated with human ciliopathies. Our results position TAp73 as a novel central regulator of MCC differentiation.
Cilia are highly conserved, microtubule-based surface organelles with essential functions in animal cells. Most vertebrate cells form a single, immotile cilium that senses mechanical or chemical stimuli. However, distinct epithelia—those lining airways, ependyma, and the oviduct—undergo multiciliogenesis by amplifying their centrioles to nucleate hundreds of motile cilia per cell that beat vigorously to generate directional fluid flow across tissue surfaces. Each motile cilium, composed of an axonemal 9+2 microtubular shaft with dynein arms and nucleated from a basal body (BB), is a complex nanomachine with >600 proteins required for assembly, structure, and function (Nigg and Raff 2009; Choksi et al. 2014). Thus, multiciliated cells (MCCs) are key components in respiration, neurogenesis, and fertility, and defective MCCs underlie a diverse group of poorly understood human diseases.
Essential for respiratory health, MCCs clean inhaled pollutants and pathogens from the airways (mucociliary clearance). Defective MCCs, either acquired or due to genetic ciliopathies like primary ciliary dyskinesia (PCD) (Horani et al. 2016), impair mucociliary transport, resulting in chronic airway inflammation and secondary alveolar destruction (Fliegauf et al. 2007).
Conducting airways are lined by polarized pseudostratified epithelium composed of ciliated, secretory, goblet, and basal cells (Hogan et al. 2014). Previous studies have indicated that sustained Notch activation inhibits ciliated cell fate and enables differentiation into secretory cells (Tsao et al. 2009; Morimoto et al. 2010). Conversely, microRNAs of the miR-34/449 family initiate MCC differentiation by repressing cell cycle regulatory genes and the Notch pathway (Marcet et al. 2011). Beyond this initial step, miR-34/449 is also required for BB maturation and docking to the apical plasma membrane (Song et al. 2014; Chevalier et al. 2015).
The motile ciliogenesis differentiation cascade is under the control of the coiled-coil protein Mcidas (also known as Multicilin) (Stubbs et al. 2012), a transcriptional coactivator required for centriole biogenesis (Tan et al. 2013; Ma et al. 2014). In frog skin MCCs, a model for mammalian airway MCCs, Mcidas forms a complex with E2F4/E2F5 to activate centriole amplification genes (Ma et al. 2014). Recessive MCIDAS mutations in humans result in severe mucociliary clearance disorders (Boon et al. 2014). Myb and FoxJ1 function downstream from Mcidas (Brooks and Wallingford 2014). Myb is transiently expressed in MCC fated progenitors for mouse airways and ependyma, promoting centriole amplification and FoxJ1 induction, but switches off during MCC maturation (Tan et al. 2013; Brooks and Wallingford 2014).
FoxJ1 is currently the most upstream key transcription factor (TF) known that is essential for all motile cilia, a role conserved across eukaryotes and required for apical BB docking, axonemal extension, and ciliary motility through broad gene network control (Brooks and Wallingford 2014; Choksi et al. 2014). FoxJ1 knockout mice lack cilia in the airways, choroid plexus, and oviduct and lack motile primary cilia in embryonic nodal cells, causing body axis asymmetry defects (Chen et al. 1998; Brody et al. 2000; You et al. 2004; Choksi et al. 2014).
Evolutionarily conserved Rfx2–4 TFs partner with FoxJ1 in ciliogenesis (Choksi et al. 2014). They are broadly required for expressing core components of immotile and motile cilia (Kistler et al. 2015). Rfx2 knockdown causes ciliogenic defects in frog skin MCCs and organs of laterality (Chung et al. 2014). Rfx3 is required for multiciliogenesis in mouse ependyma. Rfx3 loss causes hydrocephalus, a hallmark of ciliary defects (Baas et al. 2006).
TP73 belongs to the TP53/TP63 TF family (Kaghad et al. 1997). The TP73 locus encodes two classes of isoforms, each undergoing C-terminal exon splicing. The P1 promoter yields TAp73 isoforms containing the N-terminal transactivation domain. The P2 promoter yields N-terminally truncated isoforms (ΔNp73) that are dominant-negative inhibitors of p53/TAp63/TAp73. TAp73 is a tumor suppressor (Flores et al. 2005; Nemajerova et al. 2010; Alexandrova and Moll 2012). Notably, aging TAp73-specific knockout mice (TAp73 knockout) develop lung cancer (Tomasini et al. 2008). In human small cell lung cancer, TP73 undergoes recurrent oncogenic rearrangements (George et al. 2015).
In addition, TP73 has crucial developmental roles reflected in the diverse phenotype of global TP73 knockout mice (p73 knockout, missing both isoform classes) with brain defects, sterility, and rhinitis/otitis (Yang et al. 2000). The latter is thought to be caused by an unspecified immune defect (Yang et al. 2000). Interestingly, multiciliated ependymal cells that line ventricles and generate cerebrospinal fluid flow express p73, and p73 knockout ependyma displays impaired ciliogenesis (Medina-Bolivar et al. 2014; Gonzalez-Cano et al. 2015). While p73 knockout mice have severe hydrocephalus, isoform-specific knockout brains show milder discrete cortical phenotypes (Yang et al. 2000; Tomasini et al. 2008; Tissir et al. 2009). Female TAp73 knockout mice are infertile, reportedly from poor oocyte quality and ovarian egg release defects (Tomasini et al. 2008). In testis, TAp73 regulates germ cell maturation, and male TAp73 knockouts are strongly subfertile (Holembowski et al. 2014).
Given the strong resemblance between p73 knockout phenotypes (hydrocephalus, infertility, and rhinitis/otitis) and mouse models of PCD ciliopathies, we hypothesized that p73 regulates motile multiciliogenesis. Here, we identify TAp73 as directing a broad ciliogenic network topology, acting upstream of known central nodes like FoxJ1, Rfx2, Rfx3, and miR34bc.
Results
TAp73 deficiency causes profound ciliogenesis defects in airways
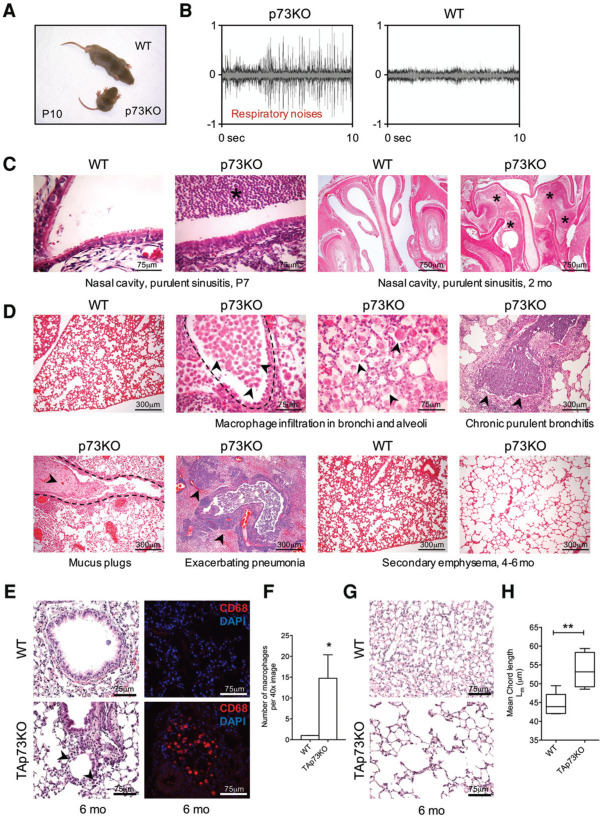
We compared global p73-deficient mice (p73 knockout) with wild-type littermates to determine the effects of p73 (TP73 gene) loss of function on airway ciliogenesis (Fig. 1A). By 3 wk of age, all p73 knockout mice showed severe respiratory distress characterized by constant coughing and sneezing (Fig. 1B; Supplemental Fig. S1A; Supplemental Movie S1). This distinct auditory phenotype predicted the knockout genotype with 100% accuracy (n=>120 mice). The underlying cause was chronic upper and lower respiratory tract infections, including purulent sinusitis/rhinitis, chronic bronchitis, exacerbating pneumonia, and inflammation-mediated alveolar destruction accompanied by secondary emphysema (Fig. 1C,D). Isoform-specific TAp73 knockout mice developed similar impaired pulmonary function with chronic inflammation, macrophage infiltration, and emphysema (Fig. 1E–H; Supplemental Fig. S1B–F).
Figure 1.
TP73 deficiency causes chronic upper and lower respiratory tract infections eventually leading to secondary emphysema. (A) Postnatal day 10 (P10) wild-type (WT) and p73 knockout (p73KO) littermates. While 20% of p73 knockout mice die young from severe hydrocephalus, 80% survive long-term when pups are separated from littermate competition (Nemajerova et al. 2010). (B) Coughing/sneezing phenotype of p73 knockout mice. Audiograms of knockout and wild-type mice at P42. Wild-type mice are silent (minor amplitudes in the wild-type track are due to rustling sounds from bedding material). See also Supplemental Figure S1A and Supplemental Movie S1. (C,D) p73 knockout mice suffer from chronic upper and lower respiratory tract infections since a young age. This includes purulent sinusitis, macrophage infiltration of bronchi and lung parenchyma, chronic bronchitis with mucus plugs, and exacerbating pneumonia. Over time, this leads to inflammation-mediated alveolar destruction with secondary emphysema. An asterisk denotes purulent exudate. Two images from overlapping areas of the same WT lung are shown. The WT image in the bottom row emphasizes the difference from the emphysematous p73KO lung. (E–H) TAp73 knockout (TAp73KO) mice have a similar phenotype. (E) Macrophage invasion in TAp73 knockout lungs stained with H&E, CD68 (red), and DAPI (blue). (F ) Quantification of CD68 immunostaining. n=3 mice per genotype at 6 mo; 12 images per mouse. (G,H) Emphysema with quantification of mean chord length at 13 mo. n=4 per genotype.
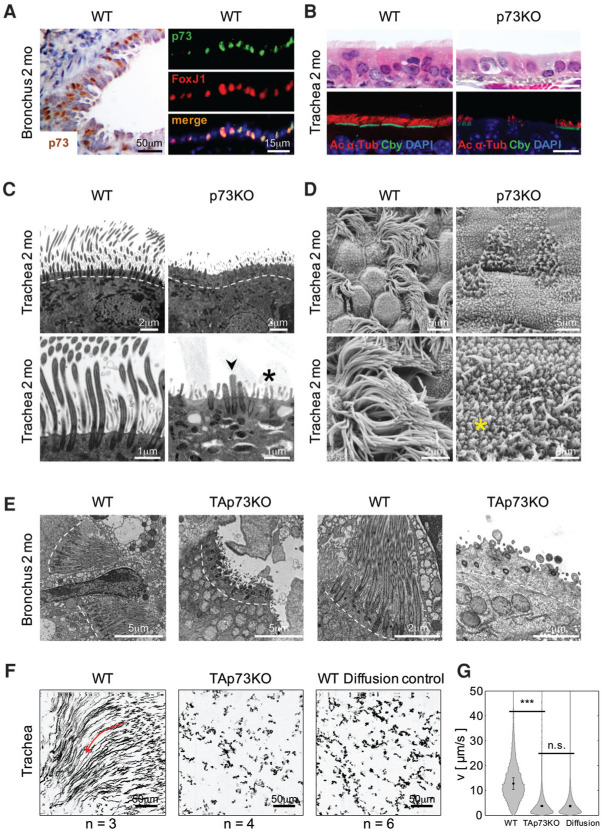
Since p73 knockout mice have normal lymphoid (Nemajerova et al. 2010) and granulocytic blood counts (Yang et al. 2000), this phenotype suggested defective mucociliary clearance rather than immune defects. Indeed, wild-type airway epithelium showed strong nuclear p73 staining in suprabasal cells harboring MCCs, among other cell types (Fig. 2A left). Colocalization studies with FoxJ1 identified these p73-positive cells as MCCs (Fig. 2A, right). p73 knockout and TAp73 knockout MCCs exhibited marked reduction in cilia number and length. The defect encompassed upper and lower airways and consisted of MCCs with far fewer and shorter cilia, indicated by markedly decreased levels of axonemal marker Ac α-Tub (Fig. 2B; Supplemental Fig. S2A–D). Ultrastructural analyses further highlighted the severe ciliary defects in p73 knockout and TAp73 knockout airways. In contrast to the abundant long wild-type cilia, knockout MCCs produced far fewer or almost no cilia. The cilia that did form were short and stubby (Fig. 2C–E; Supplemental Fig. S2E,F). The functional consequence was complete loss of mucociliary transport activity in TAp73 knockout and p73 knockout airway mucosa (Fig. 2F,G; Supplemental Fig. S2G,H; Supplemental Movie S2).
Figure 2.
TAp73 deficiency causes profound ciliogenesis defects in airway ciliated cells in vivo. (A, left) The pseudostratified airway epithelium of wild-type (WT) bronchus shows nuclear p73 immunostaining in many suprabasal cells where MCCs reside. The tracheal and bronchial epithelium from p73 knockout mice was devoid of staining, indicating antibody specificity (data not shown). (Right) The suprabasal p73-positive cells coexpress FoxJ1, a marker of MCC cells. Immunofluorescence. Lungs were from 2-mo-old mice. (B–E) Airway epithelia of p73 knockout (p73KO) and TAp73 knockout (TAp73KO) mice exhibit severe defects in ciliogenesis. (B) The p73 knockout trachea exhibits marked loss of cilia. H&E and immunofluorescence staining for axonemal marker Ac α-Tub and BB marker Chibby (Cby). DAPI counterstain. Bar, 10 µm. (C–E) Photomicrographs of transmission electron microscopy (TEM) (C,E) and scanning electron microscopy (SEM) (D) of the trachea (C,D) and bronchus (E) from 8-wk-old p73 knockout and TAp73 knockout mice and their wild-type littermates. In contrast to the abundant, long broom-like cilia of wild-type cells, knockout MCCs have far fewer and shorter cilia (ciliary stumps; arrowhead). Interspersed microvilli are preserved (asterisk). (F,G) TAp73 depletion causes severe mucociliary clearance defects. Fluorescent microspheres were applied to tracheal explants and tracked by high-speed confocal microscopy. (F ) Bead trajectories aggregated from 2000 images over 32 sec. Particles in wild-type tracheae have long, directed trajectories along a flow field (arrow). In contrast, directional movement in the knockout trachea is strongly reduced, similar to the dead wild-type diffusion control. (G) Particle velocity histograms from all measurements. n=13,683 tracks for wild-type; n=15,951 tracks for knockout; n=19,096, tracks for control. See also Supplemental Movie S2 and Supplemental Figure S2, G and H.
p73 is required for BB docking and axonemal extension
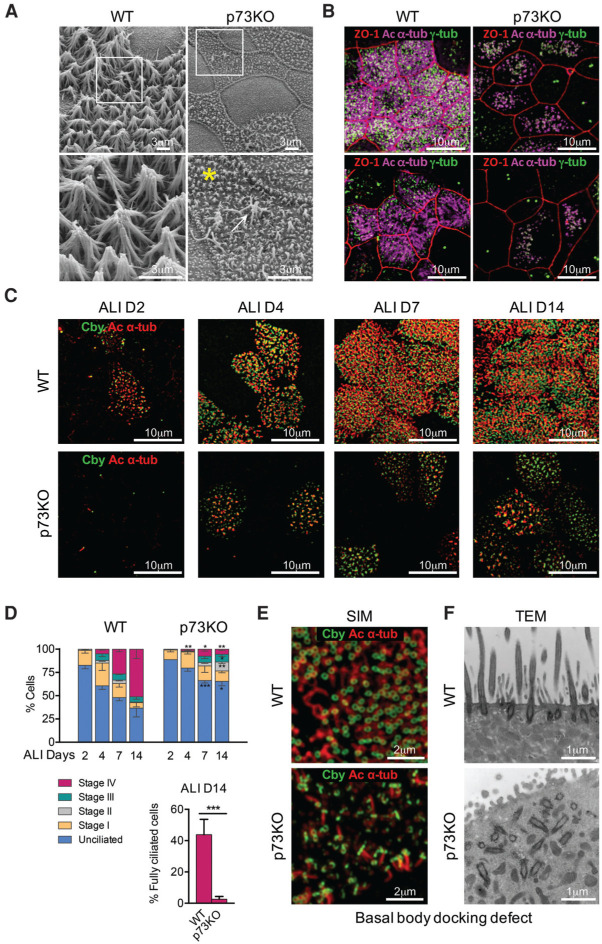
To exclude the possibility that the defective ciliary phenotype was secondary to inflammation, we triggered mucociliary differentiation in isolated adult tracheal progenitor cells through air–liquid interface (ALI) culture conditions (You et al. 2002) that recapitulate the physiological process in morphology and transcriptome (Dvorak et al. 2011). At day 14 (D14) after ALI, mouse tracheal epithelial cells (MTECs) consist of ∼50% MCCs plus all assorted nonciliated cells (You et al. 2002). Indeed, p73 knockout MTECs displayed similar defects in cilia numbers and length as previously observed in vivo. Specifically, the abundant long cilia at the apical surface of wild-type MCCs were almost absent in knockout cells (Fig. 3A–C; Supplemental Fig. S3B–D). In contrast, the interspersed short thin microvilli were preserved (Fig. 3A; Supplemental Fig. S3D). Thus, p73 is essential for ciliation.
Figure 3.
TP73 deficiency in primary organotypic airway cultures recapitulates the profound ciliogenesis defects present in vivo. (A) SEM photomicrographs of MTECs from wild-type (WT) and p73 knockout (p73KO) tracheae at ALI D14. The abundant, long cilia of wild-type MCCs are severely decreased in number and length in p73 knockout MCCs. Only rare normal-length cilia are formed (arrow). Boxed areas are shown as magnifications. Microvilli are preserved (asterisk). (B) Superresolution structured illumination microscopy (SIM). Surface view SIM photomicrographs of wild-type and p73 knockout MTECs at D14, immunostained for ZO-1 marking cell borders (red), axonemal marker Ac α-tub (magenta), and BB marker γ-tub (green). Two different representative MTEC cultures derived from independent animals are shown. (C,D) Quantitation of ciliogenesis stages. MTECs from wild-type and p73 knockout trachea at ALI D2, D4, D7, and D14. The percentages of unciliated cells, cells partially ciliated in stages I–IV of ciliogenesis, and fully ciliated cells were quantified based on immunostained confocal images for axonemal marker Ac α-tub (red) and BB marker Cby (green). Stages are defined as unciliated; stage I, appearance of centrosomal protein foci; stage II, centriole replication; stage III, centriole dispersion/migration; and stage IV, cilium elongation (Tan et al. 2013). (C ) Representative surface view. SIM images. (D) Quantitation of stages. “Fully,” as defined in this assay, is at least 20 cilia per cell. Notably, even fully differentiated knockout cells had far fewer cilia per cell than wild type (which typically have >100 cilia), and any cilia still formed were much shorter. Over 250 cells per genotype per ALI day were counted from three independent MTEC preparations (>750 total cells for each condition). Error bars represent standard error of the mean. P-values show a comparison of wild type versus knockout for each differentiation stage. (∗) P<0.05; (∗∗) P<0.02; (∗∗∗) P<0.001. (E,F ) TP73 deficiency causes docking defects at the apical membrane of MCCs. (E) p73 knockout MCCs show perturbed polarity of the ring-shaped Cby-positive distal appendage structures that facilitate BB docking to the apical membrane via ciliary vesicle formation (Burke et al. 2014). Wild-type and p73 knockout MTECs at D14 and SIM images immunostained for Cby (green) and Ac α-tub (red). (F ) Many BBs fail to properly dock along the apical surface and hence do not give rise to axonemal outgrowth. TEM images of wild-type and p73 knockout MTECs at D14. For TAp73 knockout bronchus, see also Figure 2E and Supplemental Fig. S2E. A, B, C, E, and F are representative images of at least five independent biological replicas of MTEC cultures.
We next determined the timing of p73 action during ciliogenesis. Both mouse and analogous human primary ALI cultures induced TAp73 expression early after ciliogenesis initiation (Supplemental Fig. S3A). We then quantified MCCs at sequential differentiation stages through Ac α-Tub (axonemal marker) and Chibby (Cby; BB marker) immunostaining (Fig. 3C,D; Tan et al. 2013). The early ciliary stages I and II in p73 knockout MCCs appeared relatively intact. Instead, marked defects became apparent by D4 and progressively worsened during stages III and IV until D14 (Fig. 3C,D). Stage IV cells were strongly reduced, with a reciprocal increase in cells that were either unciliated or early stage differentiation-arrested, implying that multiciliogenesis-arrested cells are either dying, dedifferentiating to an unciliated state, or being outcompeted by unciliated cells in the culture (Fig. 3C,D). The largest reduction occurred in “fully” ciliated cells, but even these cells had fewer and shorter cilia than wild-type controls (Fig. 3D).
Consistent with largely unperturbed early stages I and II, p73 deficiency did not cause major reductions in BB numbers, which maintained Cby expression in lateral views of airway mucosa (Fig. 2B; Supplemental Fig. S2A). However, although BB numbers were similar, top-down cell surface views revealed a marked loss of BB signal, indicating a severe docking defect at the apical membrane (Fig. 3B,C; Supplemental Fig. S3C). Moreover, p73 knockout BBs showed perturbed polarity (Fig. 3E). Transmission electron microscopy (TEM) analysis of p73 knockout MTECs confirmed that BBs were not properly aligned along the apical surface and had no extended axonemes (Fig. 3F; Supplemental Fig. S3E). Similarly, BB docking defects were apparent in p73 knockout and TAp73 knockout tracheal as well as bronchial MCCs in vivo (Fig. 2C,E; Supplemental Fig. S2E). Together, these results suggest that p73 acts after MCC fate specification and centriole multiplication as a major upstream ciliogenesis regulator, required for BB docking and axonemal extension.
TP73 deficiency broadly attenuates cilium gene expression
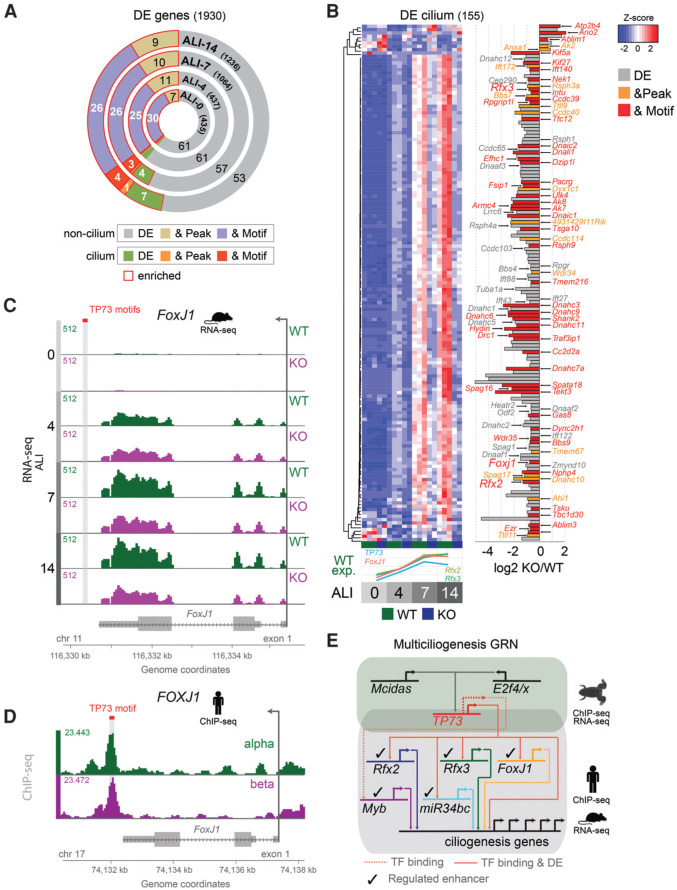
To determine the mechanistic role of p73 in multiciliogenesis, we compared RNA and small RNA (sRNA) expression profiles of wild-type and p73 knockout MTECs at ALI D0, D4, D7, and D14 of differentiation. Raw data showed good clustering by genotype and differentiation stage (Supplemental Fig. S4A,B; Supplemental Table S1). We detected 435 differentially expressed genes (DEGs) at ALI D0, 437 DEGs at D4, 1064 DEGs at D7, and 1236 DEGs at D14 by RNA sequencing (RNA-seq) analysis and no DEGs at ALI D0, one DEG at D4, three DEGs at D7, and 43 DEGs at D14 by sRNA-seq analysis (Fig. 4A; Supplemental Fig. S4C,D; Supplemental Table S2).
Figure 4.
TAp73 is a central regulator of ciliogenesis. RNA-seq gene expression of differentiating wild-type versus p73 knockout MTECs (at ALI D0, D4, D7, and D14) was compared with human TAp73 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data (peaks and TP73 DNA-binding motifs). (A) Overview of DEGs, DEGs that contain TP73-binding sites (& Peak), and DEGs that are bound and contain a TP73 motif (& Motif). DEGs are further split into the categories “cilium” for genes with known ciliogenesis function and “noncilium” for all others. Significant enrichment of overlap with gene ontology (GO) terms was tested using a two-sided Fisher's exact test (red outline, enriched). Ciliogenesis genes show significant enrichment starting at ALI D7. At D14, 12% of all 1236 DEGs are ciliogenic, many of which are bound by TP73 (5%) and contain a motif (4%). The total number of DEGs for each differentiation stage is shown in parentheses. (B, left panel) Hierarchical clustering of the 155 ciliogenesis DEG s during differentiation (ALI D0, D4, D7, and D14) with individual samples ([green] wild-type [WT]; [blue] p73 knockout [KO]). Expression differences are shown as color-coded Z-scores (blue to red). (Bottom plot) Z-score-normalized expression values of TP73 (blue), FoxJ1 (red), Rfx2 (olive), and Rfx3 (cyan) in wild-type MTECs during differentiation stages at ALI D0, D4, D7, and D14. (Right panel) Bar plot displaying the maximum log2 expression differences (estimated across all time points) between wild-type and p73 knockout samples. DEGs are shown in gray (DE) (e.g., Cep290, Odf2, Bbs4, Ift27, Ift43, Ift88, Ift122, Dnahc1, Dnahc2, and Dnahc12). DEGs that contain a TP73 peak are shown in yellow (& Peak), and DEGs that contain a TP73 peak plus a TP73 consensus motif are shown in red (& Motif). All 51 “red” genes and all 16 “yellow” genes as well as select “gray” genes are listed. This list also comprises eight human PCD-associated genes (HYDIN, DNAIC1, DRC1, ARMC4, DNAHC11, DNAIC2, CCDC39, and RSPH9 in red). Another 17 human PCD-associated DEGs contain TP73 peaks but no motif, suggesting indirect DNA binding (CCDC40, CCDC114, RSPH3a, and DYX1C1, in yellow), or are “simple” DEGs (RSPH1, RSPH4a, DNAHC5, DNAAF1, DNAAF2, DNAAF3, LRRC6, CCDC65, CCDC103, ZMYND10, SPAG1, RPGR, and HEATR2 in gray) (Supplemental Table S2). (C ) RNA-seq tracks of average FoxJ1 expression during MTEC differentiation (ALI D0, D4, D7, and D14) for wild-type (green) and p73 knockout (purple) samples. Of note, the predicted TP73-binding motifs in the murine FoxJ1 gene (shown in red) lie in a region similar to that in the humangene downstream from the 3′ end of FOXJ1, suggesting conserved TP73-dependent gene regulation between humans and mice. (D) TAp73 ChIP-seq genome tracks of the human FOXJ1 locus (GSE15780). Merged ChIP-seq tracks of TAp73α (green) and TAp73β (purple) are shown. (C,D) The transcriptional start site is marked with an arrow. (E) TP73-centric gene regulatory network of multiciliogenesis. Solid lines show direct binding of TFs to target genes (based on ChIP-seq) with differential target gene expression (based on RNA-seq). Dotted lines signify binding without evidence for differential target gene expression. A checkmark indicates that LUC reporter assays validated the TP73-mediated increase in gene expression for a given binding site and gene. The green background denotes ChIP-seq and RNA-seq data from experiments in Xenopus laevis, and the gray background indicates human ChIP-seq and murine RNA-seq data. See also Supplemental Figures S4–S6.
The RNA-seq DEGs showed mostly increased expression during early differentiation and mostly decreased expression during later differentiation (Fig. 4B; Supplemental Fig. S4C). Marked differential expression at D7 and D14 coincided with TP73 induction in wild-type MTECs (Fig. 4B, bottom; Supplemental Table S2) and was enriched for genes involved in ciliogenesis (Fig. 4A; Supplemental Fig. S5A,B). Most differentially expressed sRNAs appeared at later stages (Supplemental Fig. S4D). Thus, TP73 mainly activates transcription either directly or indirectly during later differentiation (D7 and D14).
To determine how TP73 activates ciliogenesis, we compiled ciliogenesis-related genes from SysCilia (van Dam et al. 2013), gene ontology (GO), and several publications (Supplemental Table S2). Of the 620 annotated cilium genes, 155 were differentially expressed (eight up-regulated and 147 down-regulated), encompassing various structural and functional components of the ciliary machinery (Fig. 4B; Supplemental Table S2). Whereas, at D0, five of eight cilium genes in p73 knockout MTECs were up-regulated, at D14, 143 of 147 cilium genes (97%) were down-regulated, indicating a transcriptional activating role for TP73 (Fig. 4B). Importantly, down-regulated cilium genes included key ciliogenesis regulators FoxJ1, Rfx2, Rfx3, and miR34bc (Fig. 4B,C; Supplemental Figs. S4C,D, S6A,B; Supplemental Table S2). In addition to the known ciliogenic roles of Rfx2 in embryonic frog skin and Rfx3 in mouse ependyma, our data now also implicate Rfx2 and Rfx3 in airway multiciliogenesis.
TAp73 is a central transcriptional activator of airway MCC differentiation
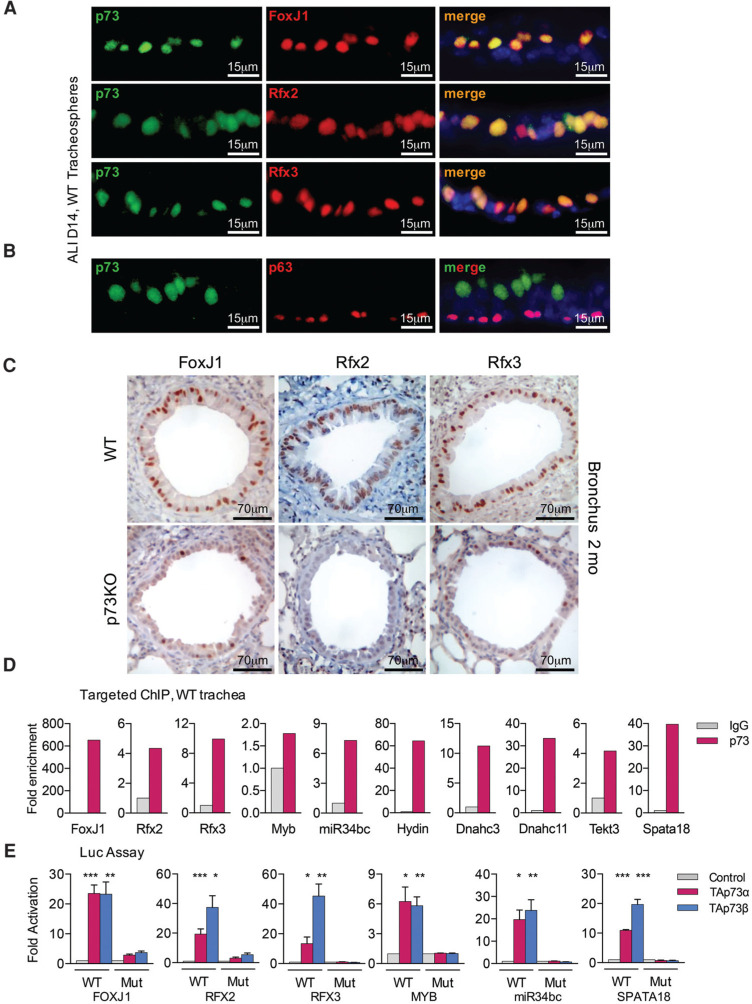
Analysis of wild-type MTECs confirmed that p73 is already coexpressed in suprabasal (p63-negative) FoxJ1-positive cells at D2 and maintained in FoxJ1/Rfx2/Rfx3-positive cells until D14 (Fig. 5A,B; Supplemental Fig. S7A,B). Moreover, p73/TAp73 knockout airways showed significantly reduced levels of FoxJ1, Rfx2, Rfx3, and Dnali1 proteins (Fig. 5C; Supplemental Fig. S7C,E,F). In confirmation, TAp73 knockout lungs showed reduced mRNA expression of FoxJ1 and Rfx2 as well as ciliary structural and motility genes (Supplemental Fig. S7D). Hence, the decrease in cilliogenesis regulators and effectors mirrors the cellular phenotype, suggesting causality.
Figure 5.
Validation of direct TAp73 target genes. (A,B) p73 is coexpressed in FoxJ1-positive, Rfx2-positive, and Rfx3-positive (A) as well as p63-negative (B) suprabasal cells. Confocal immunofluorescence images of wild-type (WT) MTECs at D14 grown as three-dimensional (3D) tracheospheres in Matrigel. Lateral views. (C ) p73-deficient airways show marked loss of FoxJ1, Rfx2, and Rfx3 protein expression. Representative bronchi from wild-type versus p73 knockout (p73KO) mice analyzed by immunohistochemistry. n=3 each. See also Supplemental Figure S7. (D) Endogenous p73 directly binds to regulatory regions of core ciliogenesis genes. Targeted ChIP assays from 60 mouse tracheae. Immunoprecipitations with pan-p73-specifc antibody versus IgG control. (E) TAp73-dependent direct responsiveness of human ciliogenesis genes. Luciferase reporter assays in Saos2 cells transfected with empty plasmid, TAp73α, or TAp73β. Wild-type reporters contain the putative TP73-binding motifs within the cognate DNA regions. Mutant (Mut) control reporters contain the same cognate DNA regions but lack the TP73-binding motifs. Data derived from at least three independent experiments per gene. Error bars represent standard error of the mean. See also Supplemental Table S5.
We next determined whether the cellular phenotype and RNA expression changes resulted from direct TAp73 binding to DEG promoter or enhancer regions. To this end, we analyzed our previously published TAp73α and TAp73β ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data from human Saos cells (Koeppel et al. 2011). Of note, TAp73α is by far the most abundant splice variant in mice and humans, while TAp73β is the most active splice variant in humans, although it is not detectable in many mouse tissues, including the lung (Grespi et al. 2012). Isoform-specific peaks were annotated to their potential target mouse genes, which resulted in 5017 TAp73α, 6100 TAp73β, and 6899 merged TP73 murine peak-containing genes (mPCGs) (Supplemental Tables S2, S3). We used de novo motif analysis to differentiate between motif-containing and motif-noncontaining peaks, since peaks with consensus motifs likely bind directly to TAp73's DNA-binding domain (Supplemental Table S3). We observed that 65% of merged peaks contained a highly conserved TP73 consensus motif that resembles TP53 and TP63 motifs (Supplemental Fig. S5C,D; Supplemental Table S3). We next overlapped TP73 mPCGs with our MTEC DEG list. This revealed that 40% (775 of 1930) of all DEGs are TP73-bound, and 69% of those also contain a consensus motif (Supplemental Fig. S4C; Supplemental Tables S2, S3). However, of all mouse genes, only 18% have TP73-bound peaks, indicating that DEGs are enriched for peaks (Supplemental Tables S2, S3). GO analysis of peak-containing DEGs showed cilium term enrichment at later stages of differentiation (Supplemental Fig. S5E). Forty-three percent of cilium-annotated DEGs contained TP73 peaks. Seventy-six percent of those also contained TP73 consensus motifs (Fig. 4B; Supplemental Table S3).
The 51 cilium DEGs that are bound and contain consensus motifs include key transcriptional regulators FoxJ1, Rfx2, and Rfx3 (Fig. 4B–D; Supplemental Figs. S4D, S6A, B). Both TAp73α and TAp73β directly bind to a putative FOXJ1 enhancer element immediately downstream from the coding region (Fig. 4D) and to intron 1 of human RFX2 and RFX3 (Supplemental Fig. S6A,B). The binding of TAp73 to FOXJ1, RFX2, and RFX3 enhancers/introns seems conserved between humans and mice, given TP73-binding motifs in corresponding mouse genes (Fig. 4C,D; Supplemental Fig. S6A,B). Of the 45 sRNA DEGs, three are TP73-bound and contain a motif, including MCC regulatory miR34bc (Supplemental Fig. S4D; Song et al. 2014). Key structural and functional ciliary genes associated with intraflagellar transport, transition fibers, BBs, planar cell polarity, and motility are differentially expressed and bound and contain a TP73 motif, including eight human PCD-associated genes (Fig. 4B, “red” genes). Another 17 human PCD-associated DEGs contain only TP73 peaks, suggesting indirect DNA binding (Fig. 4B “yellow” genes) or represent “simple” DEGs (Supplemental Table S2). Cilium DEGs are mostly down-regulated at D7 and D14, coinciding with TAp73 induction (Fig. 4B, bottom; Supplemental Fig. S3A). These results imply that TAp73 acts as central transcriptional ciliogenesis activator through both direct and indirect binding to regulatory elements of cilium genes (Fig. 4E; Supplemental Table S4).
These data were corroborated by targeted ChIP of endogenous mouse p73 protein from wild-type mouse tracheae using a pan-p73-specific antibody that we validated. It confirmed the enrichment of p73 binding to regulatory regions of the gamut of ciliogenic target genes identified above (Fig. 5D; Supplemental Fig. S7G,H). Notably, particularly strong binding was seen at the FoxJ1 gene (Fig. 5D). Moreover, we performed targeted ChIP of human TAp73α and TAp73β in Saos2 cells using a TAp73 isoform-specific antibody, which confirmed that the TAp73 isoforms indeed bind to these genes (Supplemental Fig. S7I). Luciferase reporter assays confirmed direct TAp73 responsiveness of cognate DNA regions containing TP73-binding motifs in FOXJ1, RFX2, RFX3, MYB, miR34bc, and SPATA18 gene loci. The corresponding controls lacking the TP73-binding motif showed no reporter gene induction (Fig. 5E; Supplemental Table S5).
We also asked how TAp73 itself might be regulated during multiciliogenesis. Previous work identified Multicilin (also known as Mcidas), a small coiled-coil transcriptional coregulator, as necessary in early stage MCC differentiation. Multicilin binds to E2F4/5 to promote transcription in centriole biogenesis (Stubbs et al. 2012; Ma et al. 2014). We reanalyzed published ChIP-seq/RNA-seq data of the ectoderm (animal caps) of Xenopus laevis embryos expressing transgenic Multicilin in the presence or absence of E2F4 (Ma et al. 2014). Of 92 X. laevis DEGs bound by the Multicilin/E2F4 complex, seven are TFs, and one is the X. laevis TP73 homolog (Supplemental Fig. S6C). In our wild-type differentiation system, Mcidas expression increased by D4 and peaked by D7, mirroring TP73, FoxJ1, Rfx2, and Rfx3 induction (Supplemental Fig. S4C, bottom). Importantly, ectopic expression of Mcidas in wild-type MTECs (ALI D0) induced TAp73 expression (Supplemental Fig. S6). Moreover, luciferase assays confirmed direct MCIDAS/E2F4 responsiveness of a p73 promoter region (Supplemental Fig. S6E).
Together, our results place TP73 at the center of a gene regulatory network controlling multiciliogenesis (Fig. 4E). Our data further show that Mcidas directly regulates TP73. This network is conserved between mice and humans, given the concerted temporal expression patterns between our MTECs and human airway epithelium (Supplemental Fig. S6F,G; Supplemental Table S2; Marcet et al. 2011).
TAp73 is necessary and sufficient to drive multiciliogenesis
Finally, since TAp73 contains the transactivation domain and binds to cilium genes (Figs. 4D, 5D,E; Supplemental Figs. S5C,D, S6A,B), we wished to independently confirm that the TP73 isoform class TAp73 controls these cellular and molecular phenotypes. Further implicating TAp73, knockout airways were severely ciliogenesis-defective and showed reduced mRNA of FoxJ1, Rfx2, and motility genes and reduced FoxJ1 and Dnali1 protein (Supplemental Fig. S7C,D,F).
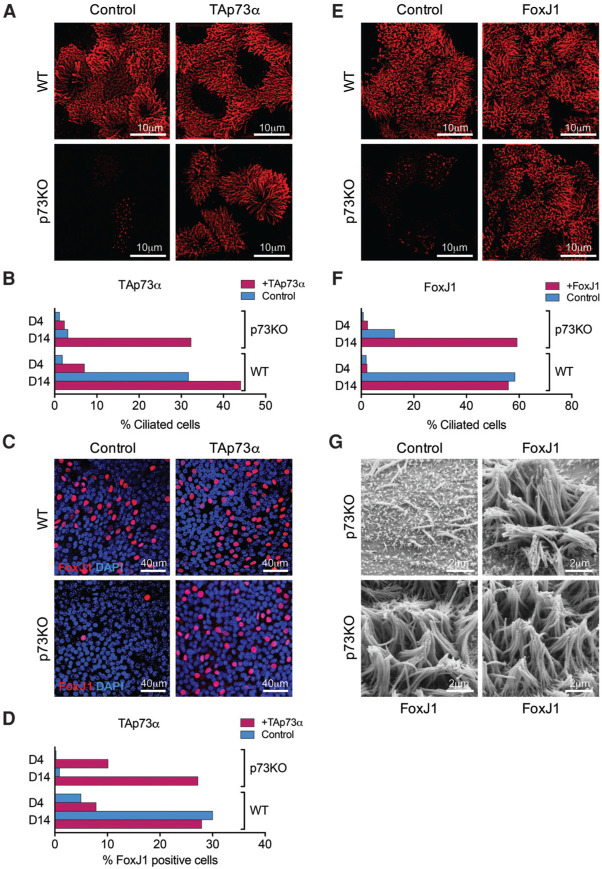
To this end, we performed rescue experiments in global p73 knockout MTECs (missing both isoform classes). Indeed, using lentiviral mouse TAp73α restricted to MCC fated cells, we were able to fully rescue cilia biogenesis. TAp73α reconstituted p73 knockout MCCs produced the full set of long motile cilia indistinguishable from wild-type cilia (Fig. 6A,B). Thus, TAp73 is necessary and sufficient to engage the full ciliogenic program. Moreover, when we compared p73 knockout control versus p73 knockout rescued by TAp73α, we saw the percentage of FoxJ1-positive cells rise to essentially wild-type levels (Fig. 6C,D). Furthermore, up to 88% of the TAp73 reconstituted p73 knockout MCCs now turn on FoxJ1 (Supplemental Fig. S8A,B). In addition, TAp73α is capable of inducing mRNA of FoxJ1, Rfx2, and other ciliary genes (Supplemental Fig. S8C). These data confirm that TAp73 acts through FoxJ1.
Figure 6.
TAp73 is necessary and sufficient to drive motile multiciliogenesis and functions upstream of FoxJ1 in the transcriptional network. Functional rescue. (A) MCC-specific TAp73α is necessary and sufficient to fully rescue ciliogenesis in p73 knockout airways. TAp73α reconstituted p73 knockout (p73KO) MCCs produced large numbers of long motile cilia indistinguishable from wild-type (WT) cilia. Wild-type and p73 knockout MTECs uninfected (Control) or infected at seeding with lentivirus targeting mouse TAp73α expression to MCC fated cells. Representative SIM images on ALI D14 stained for Ac α-tub. (B) Quantitation of phenotypic rescue in various percentages of fully ciliated cells at ALI D4 and D14. Data were derived from five wild-type and five p73 knockout mice. (C,D) Reintroduction of TAp73α induces expression of FoxJ1 in p73 knockout MTEC cultures to wild-type levels. Wild-type and p73 knockout MTECs uninfected (Control) or infected at seeding with lentivirus targeting mouse TAp73α expression to MCC fated cells. (C ) Representative confocal images on ALI D14 stained for FoxJ1. (D) Quantitation of FoxJ1-positive cells at ALI D4 and D14. Data were derived from five wild-type and five p73 knockout mice. (E–G) Epistasis analysis. p73 knockout MCCs are 100% rescued by FoxJ1. Wild-type and p73 knockout MTECs uninfected (Control) or infected at seeding with lentivirus targeting mouse FoxJ1 expression to MCC fated cells. (E) Representative SIM images on ALI D14 stained for Ac α-tub. (F ) Quantitation of rescue in various percentages of fully ciliated cells at ALI D4 and D14. (G) Representative SEM images of p73 knockout MTECs on ALI D14 uninfected or infected with FoxJ1. See also Supplemental Figure S8D for representative confocal images used for F.
Next, we performed epistasis analyses to establish that TAp73 lies upstream of motile multiciliogenesis transcriptional activators. We selected FoxJ1, given its crucial role in the ciliogenesis axis. Indeed, the severe ciliogenesis defect of p73 knockout MTECs, which express reduced levels of endogenous FoxJ1 (Fig. 5C; Supplemental Fig. S7E), was completely rescued by lentiviral mouse FoxJ1 restricted to MCCs (Fig. 6E–G). Phenotypic rescue of p73 knockout MCCs at D14 was 100% compared with control and FoxJ1-infected wild-type MCCs. Rescued p73 knockout MCCs exhibit long and motile cilia indistinguishable from control wild-type MCCs, confirming that TAp73 lies upstream of FoxJ1 in the transcriptional hierarchy. These data suggest that suppressed residual FoxJ1 levels present in p73 knockout MCCs (Fig. 5C; Supplemental Fig. S7C,E) are insufficient and require induction by TAp73 to drive productive ciliogenesis. We saw no rescue of p73 knockout ciliogenesis in MCCs targeted with lentiviral Rfx2 or Rfx3, indicating that ectopic Rfx2 or Rfx3 overexpression alone cannot compensate for TAp73 loss to activate or bypass FoxJ1. Taken together, these data identify TAp73 as a central transcriptional regulator of multiciliogenesis upstream of FoxJ1, Rfx2, Rfx3, miR34bc, Myb, and other structural ciliary genes and downstream from Mcidas (Fig. 7).
Figure 7.
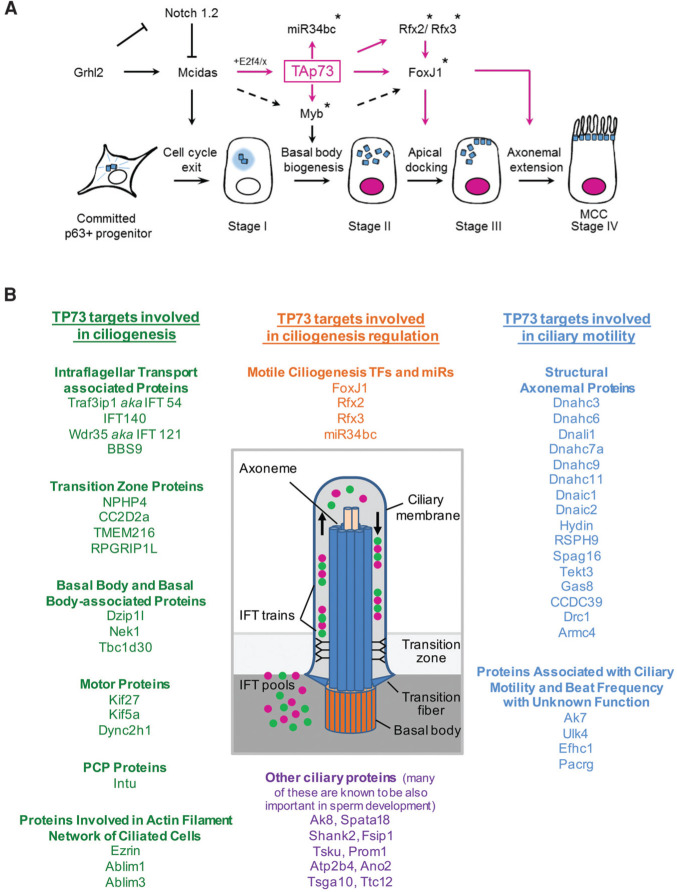
Summary of results and a proposed model of TAp73 as a central regulator of motile multiciliogenesis. (A) Regulatory transcriptional cascade of airway multiciliogenesis based on data generated here, mapping TAp73 as the proximal node (magenta). Asterisks indicates direct targets of TAp73. Grhl2 was recently mapped (Gao et al. 2015). (B) Direct target genes of TAp73 involved in the regulation, formation, and function of cilia (also indicated in red text in Fig. 4B and Supplemental Fig. S4D).
Discussion
Here we established TAp73, a homolog of the p53 tumor suppressor, as a key transcriptional regulator of airway multiciliogenesis. We propose that TAp73 acts as the central integrator of the complex MCC differentiation program, positioned downstream from Mcidas and upstream of FoxJ1, Rfx2, Rfx3, miR34bc, and Myb (Fig. 7A). Phenotypic characterization in two different TP73-deficient mouse strains and organotypic cultures, coupled with cross-species RNA-seq/ChIP-seq analyses and complete rescue of ciliogenesis by TAp73 reconstitution in p73 knockout cells, reveal that TAp73 is essential for BB docking, axonemal extension, and motility.
TP73's function in airway epithelium was unknown. In contrast, its homolog, TP63, is essential for tracheobronchial epithelium development and homeostasis through maintaining a self-renewing stem cell pool (Hogan et al. 2014). Lung regeneration in response to viral and chemical damage depends on pre-existing, intrinsically committed p63+Krt5+ distal airway stem cells that undergo proliferative expansion and assemble into nascent alveoli to replace damaged parenchyma (Kumar et al. 2011; Zuo et al. 2015).
TAp73 governs multiciliogenesis by controlling key ciliogenic TFs (notably, FoxJ1, Rfx2, and Rfx3) and by post-transcriptional processes via a key regulatory microRNA, miR34bc. Moreover, nearly 50 additional genes involved in ciliary assembly, structure, and motility are direct TAp73 targets (Fig. 7B). Eight of these genes, when mutated, are associated with human ciliopathies (HYDIN, DNAIC1, DRC1, ARMC4, DNAHC11, DNAIC2, CCDC39, and RSPH9). These data explain why TP73 deficiency in mice causes a PCD-like airway phenotype of chronic inflammation and impaired pulmonary function. By governing mucociliary clearance, TAp73 has a major role in protecting lung health.
One key TAp73 target is FoxJ1. FoxJ1 is considered a central ciliogenesis regulator, since FoxJ1-deficient mice do not develop any motile cilia (Brody et al. 2000). Notably, TAp73 depletion phenocopies FoxJ1 depletion, except for left–right asymmetry defects, implicating TAp73 functions in motile multiciliogenesis rather than nodal ciliogenesis. We found no overt defects in primary immotile and nodal cilia in p73 knockout cells. Since cilia production is completely rescued by FoxJ1 in p73 knockout MCCs, we propose that TAp73 regulates multiciliogenesis largely through FoxJ1.
Rfx2 and Rfx3 are also TAp73 targets. In motile cilia, FoxJ1 and Rfx3 mutually cross-regulate their expression and regulate an overlapping subset of genes involved in ciliary motility (Yu et al. 2008; El Zein et al. 2009; Alten et al. 2012; Didon et al. 2013; Choksi et al. 2014). Interestingly, we failed to observe a rescue of ciliogenesis in p73 knockout cells with MCC-directed reconstitution of Rfx2 or Rfx3, suggesting that ectopic expression of Rfx2 and Rfx3 alone is unable to compensate for TAp73 loss or reduced FoxJ1 levels. This implies that Rfx2 and Rfx3 require TAp73 and/or FoxJ1 for multiciliogenesis program activation despite regulating common targets.
We also found direct TAp73α and TAp73β binding to the MYB gene. Its regulatory region contains a TP73-binding motif and exhibits TAp73α/β-dependent responsiveness in luciferase assays (Fig. 5D,E). However, Myb was only marginally down-regulated in p73 knockout MTECs and thus is not classified as a DEG, likely due to its direct responsiveness to Mcidas/E2F4, which remained unaltered in knockouts (Tan et al. 2013). This suggests that Myb regulation involves several control mechanisms, one being TAp73.
To identify upstream regulators of TAp73, we performed a bioinformatics analysis of existing ChIP-seq/RNA-seq data sets from Xenopus embryos that probed the role of transcriptional coactivator Mcidas in MCCs (Ma et al. 2014). It predicted direct transcriptional control of the TAp73 gene by the Mcidas/E2F4 complex, which we confirmed in murine MTECs and human reporter assays. Mcidas functions in motile multicilia but not motile monocilia or kinocilia formation (Choksi et al. 2014). Previous studies suggest that the Mcidas/E2F4/E2F5 complex acts in a temporal window in early ciliation to drive centriole biogenesis (Stubbs et al. 2012; Ma et al. 2014). However, it remains unclear which upstream signal induces the switch for BB docking and axonemal extension during later stages. We propose TAp73 as this switch and speculate that Mcidas might function throughout all stages by first inducing TAp73 and then switching DNA-binding partners from E2F4/E2F5 to TAp73 to form a functional transcriptional complex that in turn activates Foxj1 and other target genes to complete ciliogenesis. This is worth exploring in future experiments.
In summary, the discovery of “nuts and bolts” structure/function components of the complex motile ciliary apparatus has progressed rapidly and now encompasses an extensive list. However, the transcriptional program that controls these ciliary effectors remains largely undetermined. Here, we show that TAp73 acts upstream to govern known central transcriptional nodes—notably, FoxJ1, Rfx2/3, miR34bc, and Myb (Fig. 7A)—and also controls a plethora of multiciliogenic effectors (Fig. 7B). Our findings could have profound implications for understanding the underlying mechanism disrupted in patients with ciliopathy-associated chronic respiratory diseases.
Materials and methods
Mice
p73 knockout mice with a deletion of Ex5/6 were a gift from Dr. Frank McKeon (Yang et al. 2000). TAp73 knockout mice with a deletion of Ex2/3 of the TP73 gene were a gift from Dr. Tak Mak (Tomasini et al. 2008). All animal experiments were performed with approval of the animal care and use committees at the University of Stony Brook and the University of Göttingen.
Mucociliary transport
The flow of fluorescent beads across the mucosal surface of tracheae was recorded by high-speed video camera on a confocal microscope at 61 Hz over 2000 frames. For details, see the Supplemental Material.
Histology, immunostaining, superresolution structured illumination microscopy (SIM), TEM, and scanning electron microscopy (SEM)
For histology, immunostaining, superresolution SIM, TEM, and SEM, see the Supplemental Material.
MTEC cultures and lentiviral infection
Primary MTECs were established as described (You et al. 2002). Upon confluence, the ALI condition was created by removing the apical chamber medium and switching the bottom chamber to differentiation medium. For viral infection of MTECs, isolated epithelial cells were mixed with lentiviral supernatants from HEK293T cells at the time of seeding, repeated once 24 h later. For quantitation of ciliary stages, the percentages of unciliated MCCs, partly ciliated MCCs (stages I–IV), and fully ciliated MCCs were determined from MTECs of wild-type and p73 knockout trachea at ALI D2, D4, D7, and D14 immunostained for Ac α-Tub and Cby. Confocal images of >250 cells per genotype per ALI day from each of three independent MTEC preparations were counted. For quantitating functional rescue, wild-type and p73 knockout MTECs infected with lentiviral FoxJ1-driven mouse TAp73α or -FoxJ1 at seeding or uninfected were stained for Ac α-Tub and scored at ALI D4 and D14. Scored cells (number of fully differentiated cells; i.e., at least 20 fully extended Ac α-tub-positive cilia per cell) were determined as the percentage of total cells. At least 750 cells per genotype per ALI day were counted.
TAp73 ChIP-seq and targeted ChIP
For ChIP-seq, we used our published genome-wide TAp73-binding sites (Gene Expression Omnibus [GEO] data set GSE15780) (Koeppel et al. 2011). The TAp73 ChIP-seq samples were aligned to the Homo sapiens hg19 genome using Bowtie 2 (version 2.0.2). For targeted ChIP on mouse tissue, freshly harvested tracheae were surgically removed. The respiratory epithelium was cross-linked by 1% formaldehyde for 15 min and scraped into PBS buffer with protease inhibitors (Roche). Two independent experiments were done—one from 30 tracheae and another from 60 tracheae of wild-type mice. Cells were sonicated, and chromatin was harvested, split into two equal aliquots, and immunoprecipitated with an antibody specific to p73 (Abcam, EP436Y) or IgG control. For additional targeted ChIP, chromatin was harvested from Saos2 cells expressing TAp73α, TAp73β, or empty vector; cross-linked; and incubated overnight with protein A/G plus agarose beads and 2 µg of TAp73 antibody or IgG (Abcam, ab14430 or ab2410). ChIP samples were extensively washed and decrosslinked, and purified DNA was analyzed by quantitative RT– PCR. Myoglobin D promoter served as a negative control. Fold enrichment was calculated over the recovery from IgG-ChIP, and significance was determined by unpaired one-tailed Student's t-test.
RNA-seq and sRNA-seq
Gene expression for wild-type and p73 knockout MTECs at ALI D0, D4, D7, and D14 was measured. RNA was isolated with RNeasy kit (Qiagen). RNA-seq and sRNA-seq libraries were prepared by TruSeq kits (Illumina). Library quality and sample concentration were checked before sequencing (50-base-pair single end) on a HiSeq 2000 (Illumina) using TruSeq SR cluster kit version 3-cBot-HS and TruSeq SBS kit version 3-HS. All experiments ran in triplicate, sequencing three biological replicates per condition. Murine RNA-seq data were aligned to the Mus musculus mm10 genome using STAR aligner (version 2.3.0e_r291) with default options. Read counts for all genes and all exons (Ensembl annotation version 72) were obtained using FeaturesCount (version 1.4.6) (Liao et al. 2014). To identify enriched GO categories, the Web service WebGestalt was used. GO category enrichment was assessed by calculating the fold change between observed and expected numbers of genes of a given GO category, where terms were scored enriched if they had an adjusted P-value of <0.1 (GEO no. GSE75717).
Acknowledgments
We thank Steve Brody for FoxJ1 antibody, and S. Van Horn from the Central Microscopy Imaging Center and J. Quinn for technical assistance with TEM and SEM, respectively. M.L. is supported by Deutsche Forschungsgemeinschaft (LI2405/2-1 and BA1641/12) and a Dorothea Schlözer Fellowship. C.G.S. is supported by a FAZIT Scholarship. K.G. and M. Wienken are supported by Scholarships of the German National Academic Foundation.M.A. and D.S. are supported by the Cluster of Excellence and Deutsche Forschungsgemeinschaft Research Center Nanoscale Microscopy and Molecular Physiology of the Brain. U.M.M. is supported by the National Institutes of Health/National Heart, Lung, and Blood Institute (R01HL129223), Deutsche Forschungs-gemeinschaft (MO 1998/2-1), and the Stony Brook Foundation. K.-I.T. is supported by the National Institutes of Health/National Heart, Lung, and Blood Institute (R01HL107493).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.279836.116.
Note added in proof
During the revision process of this work, a similar study by Marshall et al. (2016) was published.
References
- Alexandrova EM, Moll UM. 2012. Role of p53 family members p73 and p63 in human hematological malignancies. Leuk Lymphoma 53: 2116–2129. [DOI] [PubMed] [Google Scholar]
- Alten L, Schuster-Gossler K, Beckers A, Groos S, Ulmer B, Hegermann J, Ochs M, Gossler A. 2012. Differential regulation of node formation, nodal ciliogenesis and cilia positioning by Noto and Foxj1. Development 139: 1276–1284. [DOI] [PubMed] [Google Scholar]
- Baas D, Meiniel A, Benadiba C, Bonnafe E, Meiniel O, Reith W, Durand B. 2006. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur J Neurosci 24: 1020–1030. [DOI] [PubMed] [Google Scholar]
- Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, Dougherty GW, Raidt J, Werner C, Amirav I, et al. 2014. MCI-DAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun 5: 4418. [DOI] [PubMed] [Google Scholar]
- Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. 2000. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Respir Cell Mol Biol 23: 45–51. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. 2014. Multiciliated Cells. Curr Biol 24: R973–R982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MC, Li FQ, Cyge B, Arashiro T, Brechbuhl HM, Chen X, Siller SS, Weiss MA, O'Connell CB, Love D, et al. 2014. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J Cell Biol 207: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Knowles HJ, Hebert JL, Hackett BP. 1998. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest 102: 1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B, Adamiok A, Mercey O, Revinski DR, Zaragosi LE, Pasini A, Kodjabachian L, Barbry P, Marcet B. 2015. miR-34/449 control apical actin network formation during multicilio-genesis through small GTPase pathways. Nat Commun 6: 8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Lauter G, Swoboda P, Roy S. 2014. Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141: 1427–1441. [DOI] [PubMed] [Google Scholar]
- Chung MI, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, Wallingford JB. 2014. Coordinated genomic control of ciliogenesis and cell movement by RFX2. Elife 3: e01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didon L, Zwick RK, Chao IW, Walters MS, Wang R, Hackett NR, Crystal RG. 2013. RFX3 modulation of FOXJ1 regulation of cilia genes in the human airway epithelium. Respir Res 14: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG. 2011. Do airway epithelium air–liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol 44: 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zein L, Ait-Lounis A, Morle L, Thomas J, Chhin B, Spassky N, Reith W, Durand B. 2009. RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J Cell Sci 122: 3180–3189. [DOI] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893. [DOI] [PubMed] [Google Scholar]
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. 2005. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7: 363–373. [DOI] [PubMed] [Google Scholar]
- Gao X, Bali AS, Randell SH, Hogan BL. 2015. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basalstem cells. J Cell Biol 211: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cano L, Fuertes-Alvarez S, Robledinos-Anton N, Bizy A, Villena-Cortes A, Farinas I, Marques MM, Marin MC. 2015. p73 is required for ependymal cell maturation and neurogenic SVZ cytoarchitecture. Dev Neurobiol 10.1002/dneu.22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grespi F, Amelio I, Tucci P, Annicchiarico-Petruzzelli M, Melino G. 2012. Tissue-specific expression of p73 C-terminal isoforms in mice. Cell Cycle 11: 4474–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. 2014. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15: 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holembowski L, Kramer D, Riedel D, Sordella R, Nemajerova A, Dobbelstein M, Moll UM. 2014. TAp73 is essential for germ cell adhesion and maturation in testis. J Cell Biol 204: 1173–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Ferkol TW, Dutcher SK, Brody SL. 2016. Genetics and biology of primary ciliary dyskinesia. Paediatr Respir Rev 18: 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90: 809–819. [DOI] [PubMed] [Google Scholar]
- Kistler WS, Baas D, Lemeille S, Paschaki M, Seguin-Estevez Q, Barras E, Ma W, Duteyrat JL, Morle L, Durand B, et al. 2015. RFX2 is a major transcriptional regulator of spermiogenesis. PLoS Genet 11: e1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel M, van Heeringen SJ, Kramer D, Smeenk L, Janssen-Megens E, Hartmann M, Stunnenberg HG, Lohrum M. 2011. Crosstalk between c-Jun and TAp73α/β contributes to the apoptosis-survival balance. Nucleic Acids Res 39: 6069–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, et al. 2011. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147: 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Ma L, Quigley I, Omran H, Kintner C. 2014. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev 28: 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi LE, Cibois M, Robbe-Sermesant K, Jolly T, Cardinaud B, Moreilhon C, et al. 2011. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol 13: 693–699. [DOI] [PubMed] [Google Scholar]
- Marshall CB, Mays DJ, Beeler JS, Rosenbluth JM, Boyd KL, Santos Guasch GL, Shaver TM, Tang LJ, Liu Q, Shyr Y, et al. 2016. p73 is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Rep 14: 2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Bolivar C, Gonzalez-Arnay E, Talos F, Gonzalez-Gomez M, Moll UM, Meyer G. 2014. Cortical hypoplasia and ventriculomegaly of pp73–deficient mice: developmental and adult analysis. J Comp Neurol 522: 2663–2679. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Liu Z, Cheng HT, Winters N, Bader D, Kopan R. 2010. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci 123: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemajerova A, Petrenko O, Trumper L, Palacios G, Moll UM. 2010. Loss of p73 promotes dissemination of Myc-induced B cell lymphomas in mice. J Clin Invest 120: 2070–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678. [DOI] [PubMed] [Google Scholar]
- Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, et al. 2014. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510: 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C. 2012. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FE, Vladar EK, Ma L, Fuentealba LC, Hoh R, Espinoza FH, Axelrod JD, Alvarez-Buylla A, Stearns T, Kintner C, et al. 2013. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development 140: 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet AM. 2009. ΔNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci 106: 16871–16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. 2008. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 22: 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. 2009. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136: 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJ, Wheway G, Slaats GG, Group SS, Huynen MA, Giles RH. 2013. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404: 99–103. [DOI] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, Brody SL. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 283: L1315–L1321. [DOI] [PubMed] [Google Scholar]
- You Y, Huang T, Richer EJ, Schmidt JE, Zabner J, Borok Z, Brody SL. 2004. Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 286: L650–L657. [DOI] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, Roy S. 2008. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 40: 1445–1453. [DOI] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. 2015. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature 517: 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]