Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Clonal and subclonal mutations of NOTCH1 and TP53, clonal mutations of SF3B1, and ATM mutations in CLL have an impact on clinical outcome.
Clonal evolution in longitudinal samples occurs before and after treatment and may have an unfavorable impact on overall survival.
Abstract
Genomic studies have revealed the complex clonal heterogeneity of chronic lymphocytic leukemia (CLL). The acquisition and selection of genomic aberrations may be critical to understanding the progression of this disease. In this study, we have extensively characterized the mutational status of TP53, SF3B1, BIRC3, NOTCH1, and ATM in 406 untreated CLL cases by ultra-deep next-generation sequencing, which detected subclonal mutations down to 0.3% allele frequency. Clonal dynamics were examined in longitudinal samples of 48 CLL patients. We identified a high proportion of subclonal mutations, isolated or associated with clonal aberrations. TP53 mutations were present in 10.6% of patients (6.4% clonal, 4.2% subclonal), ATM mutations in 11.1% (7.8% clonal, 1.3% subclonal, 2% germ line mutations considered pathogenic), SF3B1 mutations in 12.6% (7.4% clonal, 5.2% subclonal), NOTCH1 mutations in 21.8% (14.2% clonal, 7.6% subclonal), and BIRC3 mutations in 4.2% (2% clonal, 2.2% subclonal). ATM mutations, clonal SF3B1, and both clonal and subclonal NOTCH1 mutations predicted for shorter time to first treatment irrespective of the immunoglobulin heavy-chain variable-region gene (IGHV) mutational status. Clonal and subclonal TP53 and clonal NOTCH1 mutations predicted for shorter overall survival together with the IGHV mutational status. Clonal evolution in longitudinal samples mainly occurred in cases with mutations in the initial samples and was observed not only after chemotherapy but also in untreated patients. These findings suggest that the characterization of the subclonal architecture and its dynamics in the evolution of the disease may be relevant for the management of CLL patients.
Introduction
The clinical course of patients with chronic lymphocytic leukemia (CLL) is highly heterogeneous.1,2 The mutational status of the immunoglobulin heavy-chain variable-region genes (IGHV) and deletions/mutations of 11q/ATM/BIRC3 and 17p/TP53 are important determinants of the clinical outcome of the disease.1 In recent years, next-generation sequencing (NGS) studies have provided a complete profile of somatic mutations in CLL.3-9 Few genes have mutations with mid/low frequencies around 11% to 15%, whereas a larger group of genes are mutated at much lower frequencies (2%-5%), highlighting a striking interpatient heterogeneity.10 The most commonly altered genes cluster in a limited number of pathways, including DNA damage response and cell cycle control, the nuclear factor–κB signaling pathway, messenger RNA processing, and NOTCH signaling among others.8,9,11 Multiple studies on population-based or clinical trial cohorts have demonstrated the adverse prognostic value of TP53, ATM, SF3B1, NOTCH1, and BIRC3 mutations.6,12-14
Combined copy number analysis13,15-20 and NGS11,12,21 have shown that CLL cases may be composed of heterogeneous tumor cell populations with subclonal mutations that may evolve over the course of the disease and influence its biological behavior. The acquisition and selection of genomic aberrations over the disease course may be critical to understanding the progression and resistance to treatment.22 In addition, the presence of subclonal driver mutations may influence a more aggressive evolution of the disease.14 The high sensitivity of ultra-deep NGS allows for the study of the clonal heterogeneity of tumors and the detection of very small mutated subclones.23,24 Recent studies have shown the clinical relevance of the detection of TP53 mutation at very low allele frequency.12,24 However, the presence and prognostic impact of minor mutated subclones of other genes with prognostic impact in CLL and their clonal dynamics in the evolution of the disease are not well known. The goals of this study were to explore the presence of clonal and subclonal mutations of TP53, ATM, SF3B1, NOTCH1, and BIRC3 in CLL using an ultra-deep NGS strategy, define the evolution of these subclones at different time points of the disease, and determine their influence in the outcome of patients.
Materials and methods
Patients and samples
Samples from 406 untreated CLL patients were included in this study (Table 1). In 48 patients, longitudinal samples obtained at different time points of the disease, including stable phase, progression before treatment, or relapse, were also examined. Tumor cells were purified from fresh or cryopreserved mononuclear cells using a cocktail of magnetically labeled antibodies as described (AutoMACS; Miltenyi Biotec).10 The median final fraction of tumor cells determined by flow cytometry was 98% with 85% samples having a tumor purity >90%. The frequency of mutant alleles detected by ultra-deep NGS was corrected for the specific tumor cell content of each tumor. DNA from purified normal blood cells from the same patients was also obtained. The study was approved by the Hospital Clínic ethics committee. All patients gave informed consent according to the International Cancer Genome Consortium guidelines.25
Table 1.
Patients’ baseline characteristics at the time of sampling
| Parameter | Category | CLL, n = 406 |
|---|---|---|
| Sex | % Male/Female | 57/43 |
| Age, y | Median (range) | 66 (19-94) |
| Time from diagnosis to sampling, mo | <12 | 206 |
| >12 | 200 | |
| Binet stage | A | 313 |
| B | 52 | |
| C | 15 | |
| Unknown | 26 | |
| Rai stage | 0 | 253 |
| I-II | 109 | |
| III-IV | 17 | |
| Unknown | 27 | |
| CNAs | Trisomy 12 | 52/376 (13.8%) |
| Del13q | 163/376 (43.4%) | |
| Del17p | 19/398 (4.7%) | |
| Del11q | 36/398 (9.0%) | |
| IGHV mutational status | Mutated | 218/382 (57.1%) |
| Patients treated during follow-up | n (%) | 208/406 (51.2%) |
| Follow-up from sampling, mo | Median (range) | 35 (6-224) |
Del, deletion; IGHV unmutated, ≥98% identity with germ line.
Molecular and genetic characterization
Copy number alterations (CNAs) were investigated as described (supplemental Methods, available on the Blood Web site).9 Targeted ultra-deep NGS of TP53 (exons 4-10), ATM (exons 2-63), BIRC3 (exons 2-9), SF3B1 (exons 14-16 and 18), and NOTCH1 (exons 26, 27, 34, and 3′ untranslated region [UTR]) was performed. Specific primers for TP53, ATM, BIRC3, and SF3B1 were designed with the D3 Assay Design web-based tool (https://www.fluidigm.com/assays) (supplemental Table 1). Amplicon libraries were generated using the Access-Array system (Fluidigm), pooled, and paired-end sequenced in MiSeq equipment (Illumina). A mean coverage >3000x was obtained for each gene. Across the whole target region, a coverage >1000x was obtained in >85% of the sequence in 95% of the samples (supplemental Figure 1). NOTCH1-specific primers were designed using the Primer3 program (supplemental Table 2).26,27 Long polymerase chain reaction (PCR) amplifications were performed using the KAPA HiFi DNA Polymerase HotStart ReadyMix (Kapa Biosystems) and normalized with the SequalPrep Normalization Plate kit (Invitrogen).28 Libraries were generated with the Nextera XT DNA Library Preparation kit (Illumina) and sequenced in a MiSeq. The average sequencing coverage was 2310x; a coverage >250x and >500x was obtained in >85% of the target region in 87% and 71% of the samples, respectively (supplemental Figure 1).
Bioinformatic workflow
Sequencing reads were mapped to the human reference genome (GRCh37) using the Burrows-Wheeler Aligner–MEM algorithm (version 0.7.10).29 Coverage along the targeted regions was analyzed using SAMtools (version 1.1)30 and custom scripts. Variant calling was performed using the VarScan2 (version 2.3.6).31 Moreover, the entire pipeline established on the MiSeq Reporter software (MSR; version 2.4.60) was run in parallel. All variants detected by any of these 2 algorithms were combined and annotated using ANNOVAR (version 2014Jul14)32 as well as custom scripts. Two additional callers, UnifiedGenotyper and HaplotypeCaller (Genome Analysis Toolkit [GATK], version 3.3-0),33 were tested but no additional variants were detected by these 2 algorithms. Variant calling was executed after performing the indel realignment and the base quality score recalibration steps defined in the GATK Best Practice recommendations.34,35 All programs were executed following the authors’ recommendations. The complete bioinformatic pipeline is shown in supplemental Figure 2, and a comparison of variant callers in supplemental Figure 3. Synonymous variants and polymorphisms described in the Single Nucleotide Polymorphism Database (dbSNP138) with a European population frequency higher than 1% (1000 Genomes Project database) were removed. TP53 and SF3B1 variants were considered as somatic mutations when, in addition to fulfilling the previous criteria, they were truncating, affected splicing sites, or were identified as somatic mutations in COSMIC (http://cancer.sanger.ac.uk/cosmic), the International Agency for Research on Cancer TP53 database (http://p53.iarc.fr/), or in our CLL-genome project database.9 NOTCH1 truncating mutations were considered somatic whereas all nontruncating variants were confirmed to be in the germ line by sequencing the respective normal DNA sample. All ATM and BIRC3 variants were investigated in the germ line DNA of the patient. All truncating BIRC3 mutations were identified as somatic whereas the missense variants were present in the germ line and not considered for further studies. ATM mutations identified in the germ line were classified as rare polymorphisms, mutations of unknown significance, rare missense mutations, and likely/definitively pathogenic according to previous criteria (supplemental Table 3).36-39 To assess the sensitivity of this methodology to detect low-frequency mutations, we performed 4 independent dilution experiments using DNA from 4 cases with TP53, ATM, and NOTCH1 mutations in >90% of the cells. Our approaches were able to call these mutations down to a variant allele frequency (VAF) <1% (supplemental Figure 4).
Verification of clonal and subclonal mutations
Sanger sequencing was used to verify a selected number of mutations. As in previous studies, our VAF threshold to detect mutant alleles by Sanger was 12% (supplemental Figure 5).24 Therefore, mutations were considered subclonal (ie, low-allele frequency) when VAF was <12% and clonal (ie, high-allele frequency) when VAF was ≥12%. To verify clonal mutations, Sanger sequencing was performed on 69 high-frequency mutations and in 233 unmutated regions/genes. All results obtained by our pipeline were confirmed by Sanger (supplemental Methods; supplemental Table 4). In addition, 43 cases carrying high frequency mutations were subjected to a second round of NGS showing concordant results in all cases. All subclonal mutations (VAF <12%) were verified by a second independent NGS experiment and/or confirmed by allele-specific PCR, as described (supplemental Table 5; supplemental Figure 6).24,40 According to the verification step, the specificity of our analysis on calling low-frequency mutations was 73%.
Statistical methods
Primary end points were overall survival (OS) and time to first treatment (TTT). OS was calculated from the date of sampling to the date of death or last follow-up. TTT was calculated from the date of sampling to the date of first treatment or last follow-up, considering disease-unrelated deaths as competing events. The log-rank test was used to compare Kaplan-Meier curves of OS; the Gray test was used to compare cumulative incidence curves of TTT. Multivariate analyses of prognostic factors were modeled using Cox and Fine-Gray regression models as previously described.41 Thresholds for VAF that offered the best prediction in terms of TTT or OS were calculated for every gene using maximally selected rank statistics and receiver operating characteristic curves (supplemental Results). All calculations were performed using R, version 3.2.2. Double-sided P values < .05 were considered significant. A detailed explanation of the statistical methods is available in supplemental Methods.
Results
Clonal and subclonal mutations
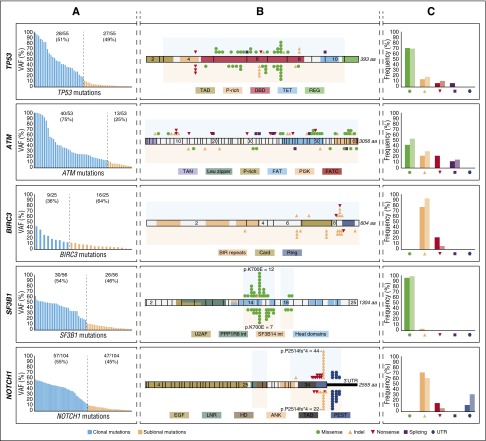
The minimal mutant allelic fractions observed in our cases were 0.3% for TP53 and NOTCH1, 0.5% for BIRC3, 1% for SF3B1, and 2% for ATM. Missense mutations were the most frequent aberration in SF3B1 and TP53, whereas all BIRC3 and NOTCH1 mutations were truncating. ATM-truncating variants accounted for half of the variants detected. Clonal and subclonal mutations had similar molecular features and gene distribution (Figure 1). Convergent evolution (acquisition of independent genetic mutations in the same gene) was observed in 30 cases (NOTCH1, 13; ATM, 12; TP53, 9; BIRC3, 5; SF3B1, 4).
Figure 1.
Molecular profile and schematic diagram of clonal and subclonal TP53, ATM, BIRC3, SF3B1, and NOTCH1 mutations. (A) VAF of the mutations identified by NGS in each of the studied genes. Blue bars correspond to clonal mutations (VAF ≥12%) whereas orange bars to the subclonal mutations (VAF <12%). (B) Schematic diagram of TP53, ATM, BIRC3, SF3B1, and NOTCH1. Exons are represented by boxes and the main protein domains are colored. Color-coded shapes indicate the position and type of the mutation. Variants represented on the top of the protein correspond to high-frequency mutations (clonal) whereas variants represented under the diagram correspond to low-frequency mutations (subclonal). Shaded area corresponds to the region sequenced. (C) Comparison of the molecular profile of the identified clonal and subclonal mutations. Each pair of bars represent clonal (dark) and subclonal (light) mutations. No statistical differences were observed by the Fisher exact test.
TP53.
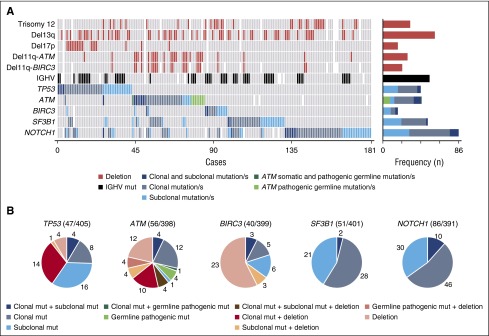
A total of 55 TP53 mutations were found in 43 of the 405 patients (10.6%) studied: 28 clonal (51%) and 27 subclonal (49%) (Figure 1A; supplemental Table 6). Mutations were mainly located at the DNA-binding domain of the protein (Figure 1B), and around 70% were missense (Figure 1C). Subclonal mutations were the only TP53 aberration in 16 of 405 patients (4%) and co-occurred with other abnormalities in 5 of 405 patients (1%): 4 TP53 clonal mutations and 1 17p deletion (Figure 2). In contrast, 14 of 22 patients (64%) with TP53 clonal mutations also had a 17p deletion. Isolated 17p deletions were only observed in 4 of 405 patients (1%) (Figure 2B).
Figure 2.
Graphical representation of gene aberrations observed in the entire cohort. (A) CNA, IGHV status, and mutational status of the studied genes are represented. Each column represents an untreated CLL case carrying at least 1 mutation in any of the studied genes. Bar plot on the right represents the number of times at which each CNA and IGHV status was observed in all mutated cases. Blue bar plots refers to the number of cases carrying isolated subclonal mutations, only clonal mutations, or both regarding the mutational status of the studied genes. Cases carrying ATM definitely/likely pathogenic germ line variants are also shown. (B) Incidence of TP53, ATM, BIRC3, SF3B1, and NOTCH1 alterations classified regarding its clonal representation in the study cohort. Del, deletion; mut, mutation/s.
ATM.
ATM had 126 variants in 95 patients. To determine whether these variants were somatic, we sequenced the germ line DNA of all mutated patients. Fifty-three mutations were classified as somatic (supplemental Table 7) and 73 as germ line variants. The latter were 67 missense (92%) and 6 truncating (8%) mutations. These germ line variants were classified as definitely (n = 8) or likely (n = 2) pathogenic (2 of these 10 were present in the same patient), rare missense (n = 33), variants of unknown significance (n = 12), or polymorphisms (n = 18) (supplemental Table 8). Interestingly, 4 of 9 cases (44%) with germ line pathogenic but only 3 of the 53 (6%) with nonpathogenic variants acquired 11q deletions (P < .01), suggesting a possible role of these germ line variants in the progression of the disease through deletion of the remaining allele (supplemental Figure 8).37 On the other hand, most somatic mutations (40 of 53, 75%) were clonal and 27% (13 of 53) subclonal (Figure 1A). Mutations were detected in 32 of 63 exons, with no hotspot regions (Figure 1B).
Combining both ATM somatic mutations and pathogenic germ line variants, 63 mutations were identified in 44 of 398 patients (11%). All but 1 subclonal alterations were detected in cases that already carried a clonal mutation (4 patients), 11q deletion (4 patients), or clonal mutation, and 11q deletion (4 patients) (Figure 2). Among the remaining 31 patients, 17 carried isolated clonal ATM mutations whereas in the other 14 cases, clonal ATM mutations coexisted with 11q deletions. Isolated 11q deletion without mutations was observed in 12 patients (Figure 2B; supplemental Figure 7). Both ATM mutations and 11q deletions mainly occur in IGHV-unmutated CLL (87%). Coexistence of TP53 and ATM mutations was only observed in 2 of 87 cases (2%).
BIRC3.
We found 25 BIRC3-truncating mutations in 17 of 399 patients (4%), 9 clonal (36%), and 16 subclonal (64%) (Figure 1A; supplemental Table 9). All but 1 mutation were located in exons 6 to 9 (Figure 1B). Nine of the 17 patients (53%) had subclonal mutations, 3 of which were associated with 11q deletions. Eight patients (47%) had clonal BIRC3 mutations, 3 coexisting with a subclonal mutation (Figure 2).
SF3B1.
We detected 56 SF3B1 mutations in 51 of 401 patients (13%), 30 clonal (54%), and 26 subclonal (46%) (Figure 1A; supplemental Table 10). Mutations were found in the 4 evaluated exons, although the hotspot p.K700E mutation was the most prevalent (19 of 56, 34%) (Figure 1B). All but 1 were missense mutations (Figure 1C). Clonal SF3B1 mutations were seen in 28 of 51 patients (55%), mutated subclones in 21 of 51 (41%), and only 2 of 51 cases (4%) had both clonal and subclonal mutations (Figure 2). SF3B1 mutations were found together with TP53 or ATM mutations in 5 and 13 CLL cases, respectively, and none with BIRC3.
NOTCH1.
We found 104 NOTCH1 mutations in 86 of 391 patients (22%), 57 clonal (55%), and 47 subclonal (45%) (Figure 1A; supplemental Table 11). All mutations were truncating and detected in exon 34 (82, 79%) or the 3′UTR region (22, 21%) (Figure 1B-C). p.P2514fs*4 (n = 66) and 3′UTR9 (n = 22) mutations accounted for 85% of all NOTCH1 mutations (Figure 1B). Interestingly, only subclonal NOTCH1 mutations were observed in 30 of 86 (35%) of the mutated cases, only clonal mutations in 46 cases (53%), and the remaining 10 cases (12%) carried both clonal and subclonal mutations (Figure 2). NOTCH1 mutations mostly occur in IGHV-unmutated CLL (82%), with no difference between clonal and subclonal alterations.
Clinical impact
Time to first treatment.
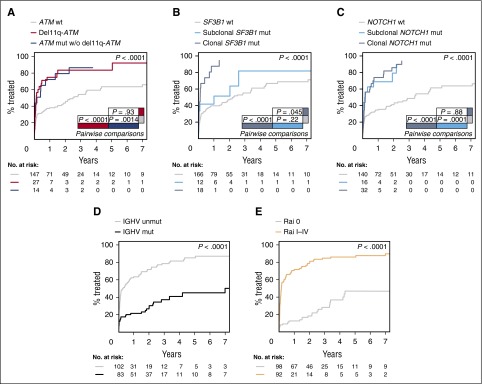
The impact of clonal and subclonal mutations was initially evaluated in the 206 patients in whom the sample was obtained within 1 year of diagnosis. ATM mutations had a significant effect on TTT independent of the presence of 11q deletions. Seventy-one percent of patients with ATM mutations and no 11q deletions had received therapy within 1 year of sampling compared with only 37% of unmutated patients (P = .0014). Patients with 11q deletions also had a significantly shorter TTT compared with patients without ATM disruption (74% vs 37%, P < .0001). There was no significant difference between patients with ATM mutations without 11q deletion and patients with 11q deletion (P = .93) (Figure 3A). SF3B1 clonal (P < .0001), but not subclonal (P = .22), mutations had a significant impact on TTT. At 1 year from sampling, 87% of patients with clonal mutations in SF3B1 had required therapy compared with 51% with subclonal mutations and 40% with wild-type (WT) SF3B1 (Figure 3B). In contrast, both clonal (P < .0001) and subclonal (P = .0001) NOTCH1 mutations predicted for a shorter TTT compared with patients with a WT NOTCH1 sequence (Figure 3C). Indeed, 74% of patients with clonal and 69% of patients with subclonal NOTCH1 mutations had received first-line therapy within 1 year of sampling compared with 34% of WT patients. Other covariates with a significant impact on TTT were IGHV mutational status (P < .0001) (Figure 3D) and Rai stage (P < .0001) (Figure 3E). All 5 covariates were independently associated with TTT according to the Fine and Gray regression model: IGHV mutational status (hazard ratio [HR] = 1.78; 95% confidence interval [CI], 1.06-2.98; P = .028), SF3B1 mutations (HR = 1.73; 95% CI, 1.05-2.86; P = .031), ATM aberrations (mutations/deletions) (HR = 1.36; 95% CI, 1.05-1.76; P = .021), NOTCH1 mutations (HR = 1.39; 95% CI, 1.11-1.76; P = .0049), and Rai stage (HR = 4.34; 95% CI, 2.71-6.96; P < .0001). On the other hand, the presence of TP53 or BIRC3 mutations, either clonal or subclonal, was not significantly associated with TTT (P = .63 and P = .97, respectively) (supplemental Figure 9). The TTT impact of clonal and subclonal mutations was also evaluated in the entire cohort of 406 patients with similar results for all 5 genes (supplemental Results; supplemental Figure 10).
Figure 3.
TTT according to gene aberrations. (A) Comparison of TTT among patients carrying ATM mutations without 11q deletion (blue line), 11q deletion (red line), and cases carrying a WT ATM gene (gray line) (P = .0014 for ATM mutations vs WT; P < .0001 for 11q deletion vs WT; P = .93 for ATM mutations vs 11q deletion). (B) Comparison of TTT among cases carrying isolated subclonal SF3B1 mutations (light blue line), clonal SF3B1 mutations (dark blue line), and cases carrying a WT SF3B1 gene (gray line) (P < .0001 for clonal mutations vs WT; P=.22 for subclonal mutations vs WT; P = .045 for clonal vs subclonal mutations). (C) Comparison of TTT among patients carrying subclonal NOTCH1 mutations (light blue line), clonal NOTCH1 mutations (dark blue line), or WT NOTCH1 gene sequence (gray line) (P < .0001 for clonal mutations vs WT; P=.0001 for subclonal mutations vs WT; P = .88 for clonal vs subclonal mutations). (D) Comparison of TTT among patients carrying the mutated (black line) or unmutated IGHV gene sequence (gray line) (P < .0001). (E) Comparison of TTT among patients diagnosed with Rai I-IV (orange line) or Rai 0 disease (gray line) (P < .0001). P, P values by Gray test.
Overall survival.
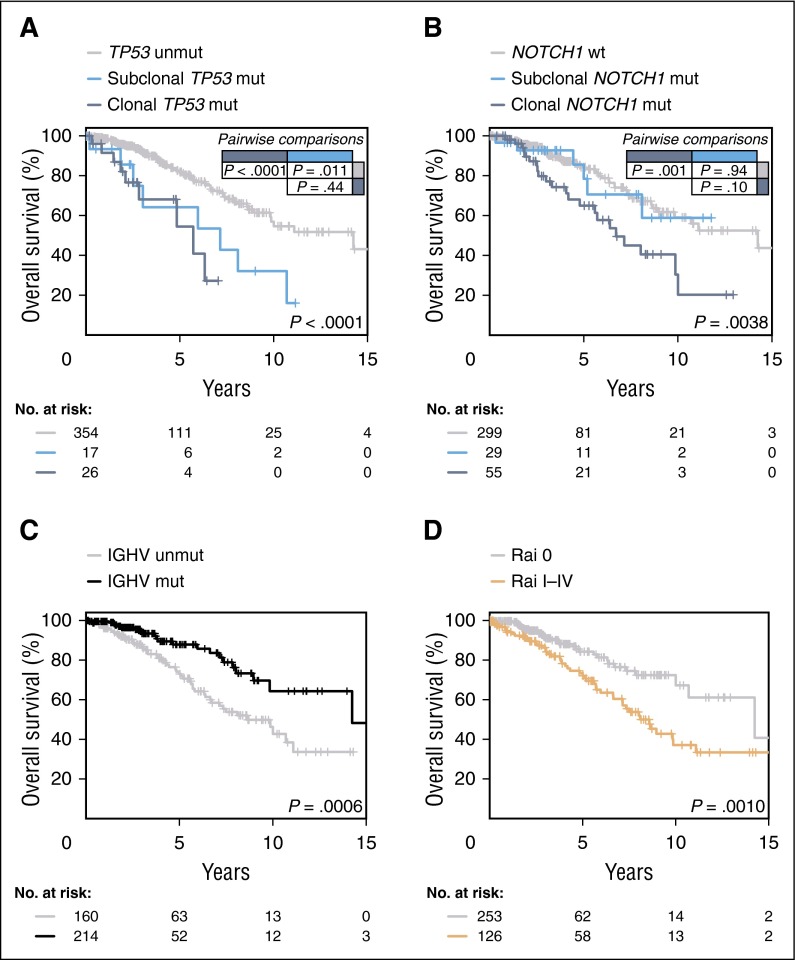
The 5-year OS of patients harboring TP53 mutations was significantly shorter for patients with both clonal (54%) and subclonal (64%) mutations compared with those with WT TP53 (82%) (P < .0001 and P = .011, respectively), with no significant difference between clonal and subclonal mutations (P=.44) (Figure 4A). Given the frequent co-occurrence of TP53 mutations with 17p deletions, we also evaluated the impact of isolated mutations vs 17p deletions. All 3 subgroups (17p deletions, TP53 clonal mutations without deletions and TP53 subclonal mutations without deletions) had prognostic impact compared with the WT sequence (P < .0001, P = .037, and P = .037, respectively [supplemental Figure 11]). Patients harboring clonal, but not subclonal, NOTCH1 mutations had a significantly shorter OS compared with those having a WT sequence (P = .001 and P = .94, respectively) (Figure 4B), whereas clonal BIRC3 (P = .049) or SF3B1 (P = .097) mutations had a trend toward a shorter OS compared with the WT cases (supplemental Figure 12). Finally, other covariates with a significant impact on OS by univariate analysis were IGHV mutational status (P = .0006) and Rai staging (P=.001) (Figure 4C-D). In contrast, neither 11q deletions or ATM mutations nor biallelic ATM inactivation had a significant impact on OS (P = .69 and P = .91, respectively) (supplemental Figure 13). A multivariate analysis revealed that CLL patients harboring TP53 aberrations (clonal and subclonal mutations/deletions) had a 1.71-fold increased risk of death (95% CI, 1.28-2.26; P = .0001), and also patients with clonal NOTCH1 mutations (HR, 1.5; 95% CI, 1.13-1.99; P = .0049). Other factors independently associated with a shorter OS were unmutated IGHV (HR = 1.84; 95% CI, 1.05-3.21; P = .032) and Rai stage I-IV (HR = 2.33; 95% CI, 1.41-3.85; P = .0009). The internal validity of the model was evaluated using bootstrapping, and the 4 covariates were selected for the model in 69% of 1000 replications.
Figure 4.
OS according to gene aberrations. (A) Comparison of OS among patients carrying subclonal TP53 mutations (light blue line), clonal TP53 mutations (dark blue line), and cases harboring an unmutated TP53 gene (gray line) (P < .0001 for clonal mutations vs WT; P = .011 for subclonal mutations vs WT; P = .44 for clonal vs subclonal mutations). (B) Comparison of OS from date of sampling between CLL patients carrying subclonal NOTCH1 mutations, clonal NOTCH1 mutations, and WT NOTCH1 gene (light blue, dark blue, and gray lines, respectively) (P = .001 for clonal mutations vs WT; P = .94 for subclonal mutations vs WT; P = .10 for clonal vs subclonal mutations). (C) Comparison of OS among patients carrying mutated (black line), and unmutated IGHV genes (gray line) (P = .0006). (D) Comparison of OS among patients diagnosed with Rai I-IV (orange line), or Rai 0 disease (gray line) (P = .001). P, P values by log-rank test.
Clonal evolution
We also performed a longitudinal analysis in 48 patients with sequential samples available (median time between samples [months]: 38, range: 1-198) (supplemental Table 12-13; supplemental Figure 14). Twenty of them lacked mutations in any of the 5 genes analyzed at sampling. Only 4 of the 17 cases (24%) in which the second sample was examined before treatment acquired mutations: 1 in TP53, 2 in NOTCH1, and the other acquired 2 mutations in BIRC3 (Figure 5 case 84). This contrasts with patients evaluated after therapy, in which all 3 (100%) acquired mutations in TP53 (2) or BIRC3 (1). Eight of the 28 cases (29%) with mutated genes at sampling expanded the mutated clone in the subsequent study: 5 at the moment of progression (3 TP53, 1 SF3B1, and 1 TP53 + SF3B1) and 3 at relapse posttreatment (1 TP53, 1 SF3B1, and 1 NOTCH1). Two of the latter 3 cases also acquired additional mutations after treatment (TP53 and SF3B1). Four mutations observed before treatment in 3 patients (1 TP53, 1 SF3B1, and 2 BIRC3 in cases 7, 75, and 84, respectively) were not detected in the relapsed sample after treatment. The negative detection was confirmed by allele-specific PCR and/or a second NGS round. More complex patterns of evolution involving several mutated subclones in the same case were also observed in 5 cases: 4 before treatment and 1 at relapse. No evolution was seen before treatment in 4 cases with small mutated subclones (1 ATM + SF3B1, 1 NOTCH1 + ATM + TP53, 1 ATM, and 1 SF3B1 + NOTCH1), and in 9 cases (4 before treatment, 5 after treatment) in which virtually all cells carried a driver mutated gene (with a VAF around 50% or 100%).
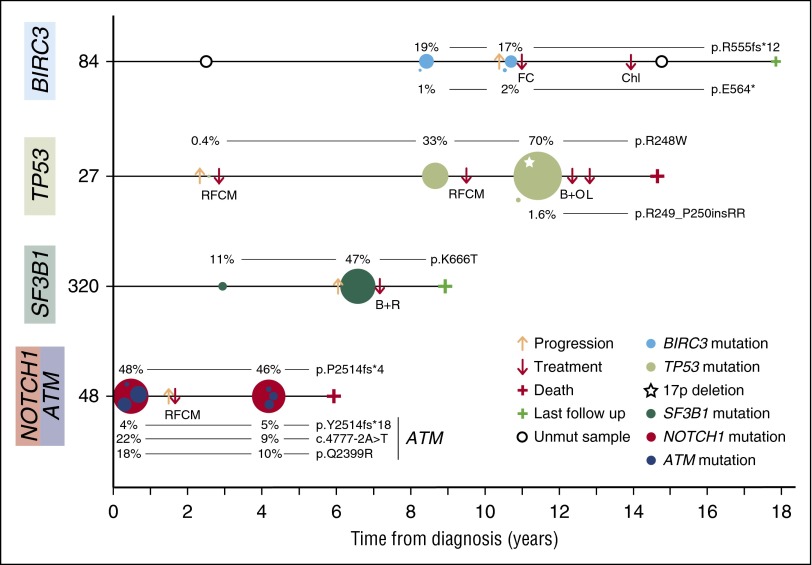
Figure 5.
Representative examples of clonal evolution observed in a 48-sample longitudinal analysis. Illustration of 4 representative CLL cases of clonal evolution showing the decrease or expansion of the BIRC3-, TP53-, SF3B1-, ATM-, or NOTCH1-mutated clone. Time 0 corresponds to the diagnosis time point. Each circle represents a unique mutation and its size is proportional to the VAF of the mutation corrected by the sample’s tumor purity. Each mutation is represented at the time point at which a tumor sample was collected. B+O, bendamustine, ofatumumab; B+R, bendamustine, rituximab; Chl, chlorambucil; FC, fludarabine, cyclophosphamide; L, lenalidomide; RFCM, rituximab, fludarabine, cyclophosphamide, mitoxantrone.
The allele frequency of TP53 mutations expanded before treatment in 5 cases. Three samples examined at relapse showed an expansion (Figure 5 case 27), persistence, or disappearance of the initial TP53-mutated clone. We also observed that SF3B1-mutated subclones also expanded before any treatment in 4 of the 6 cases (Figure 5 case 320). NOTCH1 mutations appeared before treatment in 3 patients, expanded in one after treatment and remained stable in the others with and without treatment (Figure 5 case 48). On the contrary, no evolution was observed in cases with clonal or subclonal ATM mutations (Figure 5 case 48), although the mean time between samples was relatively short in these cases.
We then evaluated the impact of “clonal evolution” on the OS of these 48 patients with sequential samples. Clonal evolution was observed in 21 patients, 13 of 34 untreated (38%) and 8 of 14 after treatment (57%). These patients had a significant shorter OS than those with no evidence of clonal evolution (HR = 2.95; 95% CI = 1.16-7.5; P = .023).
Discussion
Genomic studies in CLL have recently emphasized the complex heterogeneity of the disease.11,21 The characterization of the clonal architecture at early and subsequent phases of the disease may provide relevant information to orient management strategies more related to the biology of the tumor.22 Whole-genome and exome-sequencing studies have revealed a large number of driver genes and the influence of their subclonal heterogeneity in the outcome of the patients.9,10,14,42,43 However, these comprehensive approaches have limited power to detect very small subclones of mutated driver genes that can expand over time and influence the evolution of the disease.14 Recent studies using ultra-deep NGS have confirmed the clinical relevance of low-frequency TP53-mutated subclones on the outcome of CLL patients but whether this phenomenon occurs for other drivers is not well known.12,24
Using a highly sensitive NGS strategy, we have detected small subclones (down to 0.3% allele frequency) of 5 major CLL drivers (TP53, SF3B1, BIRC3, NOTCH1, and ATM) in a relative high proportion of patients (93 of 406, 23%). These subclonal mutations have similar molecular characteristics as their respective high-allele frequency mutations supporting a comparable pathogenic effect.7,24,42,44 In this sense, we have confirmed the unfavorable impact on OS of TP53 subclonal mutations, which was analogous to that of clonal alterations, even in the absence of deletions of the other allele.12,24 We have also observed that both clonal and subclonal NOTCH1 mutations and clonal, but not subclonal, SF3B1 mutations have a significant impact on TTT, independent of IGHV mutations. The unfavorable prognosis of clonal SF3B1 and NOTCH1 mutations has been confirmed in several studies but the impact of subclonal mutations had not been investigated.7,9,45-48 The prognostic value of the NOTCH1 subclonal mutations shown here is relevant because deep-sequencing approaches might be able to identify these high-risk patients that were undetected by classical techniques.
The mutational study of ATM is challenging due to its large size and the need to distinguish potential pathogenic mutations already present in the germ line from polymorphisms or nonpathogenic variants. In this study, we have completely characterized the mutational status of ATM in a large series of patients. Of note, isolated subclonal ATM mutations were uncommon (only 1 case of 44). We found that ATM mutations had a significant impact on TTT even in the absence of 11q deletions, suggesting that fluorescence in situ hybridization or CNA are not sufficient for a complete ATM characterization. No effect of ATM mutations or 11q deletions on OS was observed, as previously described.38,49,50
ATM germ line variants previously described as definitively/likely pathogenic were frequently associated with 11q deletions, confirming the hypothesis that these germ line variants may influence disease progression through loss of the other allele.37 Germ line variants considered as nonpathogenic had no impact on outcome and were rarely associated with 11q deletions. The advent of NGS platforms will certainly help to better characterize both somatic and germ line ATM mutations. The requirement of germ line DNA may be also relevant for BIRC3 and NOTCH1 because missense variants detected in the tumor sample were already present in the germ line. On the contrary, all TP53 and SF3B1 mutations detected had been previously confirmed already as somatic or pathogenic, suggesting that germ line DNA may be dispensable in these studies.
Our longitudinal study reveals the complex clonal evolution of this disease. We confirmed the expansion of most TP53-mutated clones after therapy observed also in other studies.12,20,24 However, TP53, SF3B1, and NOTCH1 mutations appeared de novo or expanded before any therapy in some patients, indicating that progressive dynamics of these clones are not only dependent on therapy selection. On the contrary, small ATM-mutated clones seem to be more stable, although the time between samples in our study was relatively short. We have also observed 2 subclonal (TP53, SF3B1) and 1 clonal (BIRC3) mutations that apparently disappeared under the detection threshold after treatment, suggesting that in some cases therapy may control these small subclones. Although the number of cases is limited, we observed that clonal evolution in longitudinal samples had an unfavorable impact on OS, suggesting that, in addition to the subclonal architecture of the tumor, the study of the clonal dynamics may provide relevant information to understanding the outcome of the patients.
In conclusion, this study shows the presence of a high number of clonal and subclonal mutations and convergent evolution of 5 driver genes in CLL and their impact on the outcome of the patients, as well as their possible patterns of clonal evolution. Particularly, clonal NOTCH1, SF3B1, and ATM mutations had an impact on shorter TTT, whereas clonal NOTCH1 and TP53 mutations conferred a shorter OS. Regarding the subclonal mutations detected in this study, only NOTCH1 subclonal mutations had an impact on TTT, whereas only subclonal TP53 mutations influenced OS. Therefore, once validated by prospective studies, targeted ultra-deep NGS may well become a common approach for the assessment of patients’ genomic alterations in daily practice and may be relevant for management strategies of CLL patients.
Acknowledgments
This work was mainly developed at the Centre Esther Koplowitz, Barcelona, Spain. The authors are indebted to the Genomics Core Facility of the Institut d’Investigacions Biomèdiques August Pi i Sunyer for the technical help. The authors are grateful to N. Villahoz and M. C. Muro for their excellent work in the coordination of the CLL Spanish Consortium and also thank S. Guijarro, C. Capdevila, L. Pla, and M. Sánchez for their excellent technical assistance. The authors are also very grateful to all patients with CLL who have participated in this study.
This work was supported by the Ministerio de Economía y Competitividad, grant no. SAF12-38432 (to E. Campo), Generalitat de Catalunya Suport Grups de Recerca AGAUR 2014-SGR-795 (to E. Campo), the Spanish Ministry of Science and Innovation through the Instituto de Salud Carlos III International Cancer Genome Consortium for Chronic Lymphocytic Leukemia (ICGC-CLL Genome Project), the Red Temática de Investigación Cooperativa en Cáncer grant RD12/0036/0036 (to E. Campo), RD12/0036/0023 (to A.L.-G.), RD12/0036/0069 (to M.G.), and the European Regional Development Fund “Una manera de fer Europa.”
E. Campo is an Academia Researcher of the “Institució Catalana de Recerca i Estudis Avançats” of the Generalitat de Catalunya.
Footnotes
The sequencing data reported in this article have been deposited in the European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena; accession number ERP013384).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.N., A.E., and E. Campo designed the study; F.N., M.P., P.J., A.E., X.S.P, C.L.-O., and E. Campo interpreted data; J.D. performed statistical analysis; F.N. performed the bioinformatic analysis; F.N., S.B., I.S., and D.M.-G. performed CNA analysis; F.N., C.R., A.N., and H.S.-C. performed and interpreted molecular studies; T.S., C.O., and E. Campo defined the classification of ATM germ line mutations; J.D., T.B., M. Aymerich, M.R., A.L.-G., N.V., D.C., M.G., M. Alcoceba, M.J.T., E. Colado, and E. Campo collected clinical and pathological data; and F.N., J.D., A.E., and E. Campo wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elías Campo, Unitat Hematopatologia, Hospital Clínic, Villarroel 170, 08036 Barcelona, Spain; e-mail: ecampo@clinic.ub.es.
References
- 1.Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 2.Hallek M. Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. Am J Hematol. 2013;88(9):803–816. doi: 10.1002/ajh.23491. [DOI] [PubMed] [Google Scholar]
- 3.Guièze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126(4):445–453. doi: 10.1182/blood-2015-02-585042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton L-A, Rosenquist R. Deciphering the molecular landscape in chronic lymphocytic leukemia: time frame of disease evolution. Haematologica. 2015;100(1):7–16. doi: 10.3324/haematol.2014.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villamor N, López-Guillermo A, López-Otín C, Campo E. Next-generation sequencing in chronic lymphocytic leukemia. Semin Hematol. 2013;50(4):286–295. doi: 10.1053/j.seminhematol.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Quesada V, Ramsay AJ, Rodríguez D, Puente XS, Campo E, López-Otín C. The genomic landscape of chronic lymphocytic leukemia: clinical implications. BMC Med. 2013;11:124. doi: 10.1186/1741-7015-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baliakas P, Hadzidimitriou A, Sutton L-A, et al. European Research Initiative on CLL (ERIC) Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329–336. doi: 10.1038/leu.2014.196. [DOI] [PubMed] [Google Scholar]
- 8.Strefford JC. The genomic landscape of chronic lymphocytic leukaemia: biological and clinical implications. Br J Haematol. 2015;169(1):14–31. doi: 10.1111/bjh.13254. [DOI] [PubMed] [Google Scholar]
- 9.Puente XS, Beà S, Valdés-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 10.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landau DA, Carter SL, Getz G, Wu CJ. Clonal evolution in hematological malignancies and therapeutic implications. Leukemia. 2014;28(1):34–43. doi: 10.1038/leu.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malcikova J, Stano-Kozubik K, Tichy B, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29(4):877–885. doi: 10.1038/leu.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stilgenbauer S, Sander S, Bullinger L, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92(9):1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 14.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brejcha M, Stoklasová M, Brychtová Y, et al. Clonal evolution in chronic lymphocytic leukemia detected by fluorescence in situ hybridization and conventional cytogenetics after stimulation with CpG oligonucleotides and interleukin-2: a prospective analysis. Leuk Res. 2014;38(2):170–175. doi: 10.1016/j.leukres.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Janssens A, Van Roy N, Poppe B, et al. High-risk clonal evolution in chronic B-lymphocytic leukemia: single-center interphase fluorescence in situ hybridization study and review of the literature. Eur J Haematol. 2012;89(1):72–80. doi: 10.1111/j.1600-0609.2012.01790.x. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer D, Pantic M, Skatulla I, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109(3):1202–1210. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 18.Ojha J, Ayres J, Secreto C, et al. Deep sequencing identifies genetic heterogeneity and recurrent convergent evolution in chronic lymphocytic leukemia. Blood. 2015;125(3):492–498. doi: 10.1182/blood-2014-06-580563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubor V, Krasnitz A, Troge JE, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA). Blood. 2009;113(6):1294–1303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- 20.Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN. Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19(11):2893–2904. doi: 10.1158/1078-0432.CCR-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuh A, Becq J, Humphray S, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120(20):4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- 22.Puente XS, López-Otín C. The evolutionary biography of chronic lymphocytic leukemia. Nat Genet. 2013;45(3):229–231. doi: 10.1038/ng.2556. [DOI] [PubMed] [Google Scholar]
- 23.Sutton L-A, Ljungström V, Mansouri L, et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach will facilitate implementation in a clinical setting. Haematologica. 2015;100(3):370–376. doi: 10.3324/haematol.2014.109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139–2147. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson TJ, Anderson W, Artez A, et al. International Cancer Genome Consortium. International network of cancer genome projects [published correction appears in Nature. 2010;465(7300):966]. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Untergasser A, Cutcutache I, Koressaar T, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 28.Harris JK, Sahl JW, Castoe TA, Wagner BD, Pollock DD, Spear JR. Comparison of normalization methods for construction of large, multiplex amplicon pools for next-generation sequencing. Appl Environ Microbiol. 2010;76(12):3863–3868. doi: 10.1128/AEM.02585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Auwera GA, Carneiro MO, Hartl C, et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;11(suppl 43):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavtigian SV, Oefner PJ, Babikyan D, et al. Australian Cancer Study; Breast Cancer Family Registries (BCFR); Kathleen Cuningham Foundation Consortium for Research into Familial Aspects of Breast Cancer (kConFab) Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 2009;85(4):427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skowronska A, Austen B, Powell JE, et al. ATM germline heterozygosity does not play a role in chronic lymphocytic leukemia initiation but influences rapid disease progression through loss of the remaining ATM allele. Haematologica. 2012;97(1):142–146. doi: 10.3324/haematol.2011.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowronska A, Parker A, Ahmed G, et al. Biallelic ATM inactivation significantly reduces survival in patients treated on the United Kingdom Leukemia Research Fund Chronic Lymphocytic Leukemia 4 trial. J Clin Oncol. 2012;30(36):4524–4532. doi: 10.1200/JCO.2011.41.0852. [DOI] [PubMed] [Google Scholar]
- 39.Austen B, Skowronska A, Baker C, et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J Clin Oncol. 2007;25(34):5448–5457. doi: 10.1200/JCO.2007.11.2649. [DOI] [PubMed] [Google Scholar]
- 40.Tiacci E, Schiavoni G, Forconi F, et al. Simple genetic diagnosis of hairy cell leukemia by sensitive detection of the BRAF-V600E mutation. Blood. 2012;119(1):192–195. doi: 10.1182/blood-2011-08-371179. [DOI] [PubMed] [Google Scholar]
- 41.Delgado J, Pereira A, Villamor N, López-Guillermo A, Rozman C. Survival analysis in hematologic malignancies: recommendations for clinicians. Haematologica. 2014;99(9):1410–1420. doi: 10.3324/haematol.2013.100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi D, Fangazio M, Rasi S, et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood. 2012;119(12):2854–2862. doi: 10.1182/blood-2011-12-395673. [DOI] [PubMed] [Google Scholar]
- 45.Oscier DG, Rose-Zerilli MJJ, Winkelmann N, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121(3):468–475. doi: 10.1182/blood-2012-05-429282. [DOI] [PubMed] [Google Scholar]
- 46.Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28(1):108–117. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 47.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247–3254. doi: 10.1182/blood-2014-01-546150. [DOI] [PubMed] [Google Scholar]
- 48.Villamor N, Conde L, Martínez-Trillos A, et al. NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia. 2013;27(5):1100–1106. doi: 10.1038/leu.2012.357. [DOI] [PubMed] [Google Scholar]
- 49.Tam CS, O’Brien S, Wierda W, et al. Long-term results of the fludarabine, cyclophosphamide, and rituximab regimen as initial therapy of chronic lymphocytic leukemia. Blood. 2008;112(4):975–980. doi: 10.1182/blood-2008-02-140582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hallek M, Fischer K, Fingerle-Rowson G, et al. International Group of Investigators; German Chronic Lymphocytic Leukaemia Study Group. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376(9747):1164–1174. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]