Abstract
Glioblastoma (GBM) is the most common adult primary brain tumor. Despite aggressive multimodal therapy, the survival of patients with GBM remains dismal. However, recent evidence has demonstrated the promise of bone marrow-derived mesenchymal stem cells (BM-hMSCs) as a therapeutic delivery vehicle for anti-glioma agents, due to their ability to migrate or home to human gliomas. While several studies have demonstrated the feasibility of harnessing the homing capacity of BM-hMSCs for targeted delivery of cancer therapeutics, it is now also evident, based on clinically relevant glioma stem cell (GSC) models of GBMs, that BM-hMSCs demonstrate variable tropism towards these tumors. In this study, we compared the lipid environment of GSC xenografts that attract BM-hMSCs (N=9) with those that do not attract (N=9), to identify lipid modalities that are conducive to homing of BM-hMSC to GBMs. We identified lipids directly from tissue by matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry (IMS) and electrospray ionization-tandem mass spectrometry (ESI-MS/MS) of lipid extracts. Several species of signaling lipids, including phosphatidic acid (PA 36:2, PA 40:5, PA 42:5, and PA 42:7) and diacylglycerol (DAG 34:0, DAG 34:1, DAG 36:1, DAG 38:4, DAG 38:6, and DAG 40:6) were lower in attracting xenografts. Molecular lipid images showed that PA (36:2), DAG (40:6), and docosahexaenoic acid (DHA) were decreased within tumor regions of attracting xenografts. Our results provide the first evidence for lipid signaling pathways and lipid-mediated tumor inflammatory responses in the homing of BM-hMSCs to GSC xenografts. Our studies provide new fundamental knowledge on the molecular correlates of the differential homing capacity of BM-hMSCs toward GSC xenografts.
Keywords: MALDI-Imaging Mass Spectrometry, glioblastoma, xenografts, lipidomics, phosphatidic acid, diacylglycerol, docosahexaenoic acid, bone marrow-derived mesenchymal stem cells
Graphical abstract
Introduction
Glioblastoma (GBM, World Health Organization (WHO) grade IV astrocytoma) is the most common (1-2) adult primary brain tumor, with a median survival rate of 14-15 months (2-4). Despite advances in treatment and an aggressive multimodal therapeutic approach including maximal surgical resection, followed by concurrent chemotherapy, radiotherapy, and adjuvant chemotherapy, the disease is nearly universally fatal. This poor outcome is largely due to poor penetration of therapeutic agents across the blood-tumor barrier (5-6) and resistance of glioma stem cells (GSCs) to standard treatments. We and others have shown that bone marrow-derived human mesenchymal stem cells (BM-hMSCs) show promise in addressing this deficiency because they selectively migrate or home to gliomas after systemic or intravascular delivery, making them attractive vehicles for targeted therapeutic delivery of anti-glioma agents (6-12). In fact, due to their intrinsic tropism towards gliomas, BM-hMSCs have been used to deliver immunotherapy (6-8), enzymes for chemotherapeutic pro-drugs (11), and oncolytic viruses (10) to human gliomas in animal models. However, despite the potential clinical application of BM-hMSCs in glioma therapy, the mechanism underlying the ability of BM-hMSCs to home to gliomas remains largely unknown. Deciphering this mechanism is not only of biological interest, but could be exploited also to identify the most appropriate patient groups for BM-hMSC-mediated delivery or to enhance the homing of BM-hMSCs to GBMs.
We have recently shown that in addition to the commonly used “professional” glioma cell lines, BM-hMSCs are also capable of homing to xenografts derived from human GSCs (13). Compared to commercially available rat and human glioma cell lines, which poorly mimic clinical tumors, GSCs faithfully recapitulate the genotype and phenotype of human GBMs in vivo (14) and therefore, offer a more clinically relevant model with high translational significance. Like cancer stem cells from many other tumors, GSCs, which are isolated directly from fresh surgical specimens of patient gliomas, represent a sub-population of cells in GBMs that have stem-like properties, including self-renewal (15-16). GSCs grow as spheroids in vitro and often express CD133 or CD15 on their surface (16-17). Most importantly, GSCs form tumors that mimic human GBMs when injected in small numbers (100-1000 cells) into the brains of athymic mice and are therefore thought to be the tumor-initiating cells that are often resistant to therapy and responsible for treatment failure (16-17).
Although xenografts formed from GSCs are capable of attracting BM-hMSCs, recent work from our group has indicated that not all GSC xenografts are capable of attracting BM-hMSCs equally (13). In our studies of a large group of GSCs we found that intracranial xenografts of some GSCs were capable of strongly attracting BM-hMSCs after intracarotid injection (GSC17, GSC274, GSC268), whereas others showed a more limited capacity to attract BM-hMSCs (GSC11, GSC229, GSC231). The identification of “attractor” GSCs and “non-attractor” GSCs is not only of therapeutic importance, but provides a unique model for understanding the mechanisms underlying the tropism of BM-hMSCs toward GSCs. Evidence suggests that intravascularly-delivered BM-hMSCs extravasate from the blood vessel endothelium via diapedesis in order to localize to the tumor mass (10). Extravasation is a complex multi-step process whereby cells within blood vessels migrate in response to soluble factors released from sites of inflammation and/or tissue damage. Lectins on the endothelial surface recognize and bind carbohydrates on glycolipids or glycoproteins on the opposing cell surface, inducing rolling adhesion of circulating cells. Complementary pairs of adhesion molecules on opposing cell surfaces mediate the strengthening of this interaction. Next, the cells begin the process of squeezing between the endothelial cells (diapedesis) that comprise the blood vasculature and migrate throughout the tumor parenchyma (10).
Several studies, focused on tumor-derived growth factors and cytokines, such as TGF-β (13), PDGF-BB (18), and SDF-1 (19), have provided clues about soluble factors that mediate BM-hMSC homing to gliomas. However, knowledge of lipids as molecular correlates of GSC xenograft differential homing of BM-hMSCs is lacking. Lipids are not only crucial to the maintenance of cellular structure, but are also important in signal transduction as second messenger molecules. The main structural components of the biological membranes, different classes of lipids and saturations can significantly modulate membrane fluidity, impacting membrane-dependent cellular functions (20-21). Lipids as signaling molecules can act independently (e.g. as receptor ligands) or in conjunction with proteins through structural-functional modulation (22-23). Alterations in lipids are strongly correlated with cancer and other human diseases (e.g. Alzheimer's disease) (24-25). Changes in lipid metabolism may influence biological processes, including growth, proliferation, invasion, and energy homeostasis through lipid signal transduction pathways (24). In glioma, this becomes especially relevant because the location of these tumors, the brain, contains the highest lipid content of any organ in the body. Lipids are critical in cell-to-cell communication in the brain and can act as inflammatory mediators. Given the diverse role of lipids in biological systems, we hypothesized that alterations of lipids between attractor and non-attractor GSCs may provide insight into the elements within the tumor environment that are favorable to or elicit cues for BM-hMSC homing. Consequently, we applied high throughput ESI-MS/MS and MALDI-IMS approaches to identify inherent underlying differences in lipid profiles between those tumors that attract BM-hMSCs (attractors) and those that do not (non-attractors).
We have previously performed lipidomic analysis of GSCs and GBM cells (26-27). For the first time, we have applied lipidomics and MALDI-IMS to GSC xenografts. We employed quantitative lipidomics and matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) from thin tissue sections. Lipidomic analysis by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) allows for the identification and quantification of lipid species (28-29), while MALDI-IMS permits localization and identification of lipid biomolecules within the GSC xenograft tissue. This technology allows direct detection of ionized analytes by rastering a laser across matrix-coated tissue in an ordered array. Each x-y coordinate contains a spectrum of lipid biomolecular ions. The combination of x-y coordinates with signal intensity produces an image representing an in situ molecular-histological map (30). The added advantage to this technology is the minimal sample processing, which preserves the integrity of the tissue and its compatibility with other imaging modalities (31).
Using this approach we now detected both global and tumor-specific differences in signaling lipids phosphatidic acid (PA 36:2, PA 40:5, PA 42:5, and PA 42:7) and diacylglycerol (DAG 34:0, DAG 34:1, DAG 36:1, DAG 38:4, DAG 38:6, and DAG 40:6) in attractor versus non-attractor GSC xenografts. Finally, our data reveals decreased levels of docosahexaenoic acid (DHA) in attractors. Taken together, these results implicate phospholipid signaling and lipid-mediated inflammation in BM-hMSCs homing.
Methods and Materials
Chemicals and reagents
LC-MS grade acetonitrile, methanol, and water were from J.T. Baker (Philipsburg, NJ). 2,5 dihydroxybenzoic acid (DHB) xylene, and ammonium acetate were obtained from Sigma-Aldrich (St. Louis, MO). Hematoxylin & eosin was purchased from Amresco (Solon, OH). Conductive indium tin oxide (ITO) coated glass slides were from Delta Technologies (Loveland, CO). Docosahexaenoic acid (22:6) lipid standard was purchased from Cayman Chemical Company.
Animals
Male athymic nude mice (nu/nu) were purchased from the Department of Experimental Radiation Oncology, The University of Texas M.D. Anderson Cancer Center (Houston, TX). Mice were housed n ≤ 5 per cage, on a 12 hr light-dark cycle with food and water ad libitum. Mice were allowed to feed until the time of surgery. All animal work was performed in accordance with institutional (MDACC) guidelines under Animal Care and Use Committee-approved protocols and in compliance with the USDA Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals.
Glioma Xenograft Model
GSCs (GSC17, GSC11, GSC229, GSC231, GSC268, GSC274) were established as previously described (15-16). Cells were harvested, counted, and resuspended in PBS for intracranial implantation. GSCs (5 × 106) were implanted in mice via the screw-guide method as previously described (32). Each cell line was injected into three mice for a total of eighteen animals. Based on previous experiment GSCs were classified as “attractors” (GSC17, GSC274, and GSC268) and “non-attractors” (GSC11, GSC229, GSC231) (13).
Tissue Dissection and Sectioning
Animals were anesthetized by intraperitoneal injection of ketamine/xylazine solution (200 mg ketamine and 20 mg xylazine in 17 ml of saline) at a dosage of 0.15 mg/10 g body weight and sacrificed by CO2 inhalation. Brains were removed immediately and flash frozen in liquid nitrogen vapor (33) and stored at -80°C. Brain tissue was sectioned at 10 μm along the coronal plane and thaw mounted on ITO glass slides in a block randomized fashion (34), ensuring each slide had at least two attractor (GSC xenografts GSC17, GSC274, and GSC268) and two non-attractor (GSC xenografts GSC11, GSC229, GSC231) sections derived from different cells and different biological replicates to minimize systemic bias during instrument run. Slides were stored at -80°C until further analysis by MALDI-IMS. Serial sections at 15 μm were taken at the same time for lipid extraction as previously described (28-29) and subsequent ESI-MS/MS analysis.
MALDI-IMS
Slides were dried in a vacuum desiccator for 1 hr, then washed with 50 mM ammonium acetate three times and allowed to dry for 3 hrs in vacuo. Matrix 2,5 dihydroxybenzoic acid was applied to slides through sublimation (35). Imaging experiments were acquired in a MALDI TOF/TOF mass spectrometer (Ultraflextreme, Bruker Daltonics, Bremen, Germany) with Flex Control 3.4 and FlexImaging 3.0 (Bruker Daltonics) software. For all analyses, data were collected at 100 μm lateral resolution with a 1 kHz UV laser (smartbeam II) in positive ion mode (500 shots/spot) and in negative ion mode (1,000 shots/spot) over m/z 200-1600. Imaging data were subsequently processed and analyzed by FlexImaging™ software. Ion mass-to-charge ratio (m/z) and lipid assignments were made through the LIPID Metabolites and Pathways Strategy Database (LipidMaps http://www.lipidmaps.org) (36-37). Images were normalized to the total ion current (TIC). Collision-induced dissociation (CID) fragmentation was to fragment selected lipids. The DHA lipid standard was spotted on a MALDI plate with DHB (50/50 v/v) and fragmented in negative ion mode using the same parameters described above (Supplemental Figure S1 and Supplemental Table 2).
Optical imaging
After MALDI-IMS acquisition and subsequent MS/MS fragmentation, slides were washed twice (15 sec each) with 100% ethanol to remove matrix. Samples were washed successively with 95% ethanol, 70% ethanol, and H2O (30 sec each) followed by hematoxylin staining for 3 min. Next, samples were washed with H2O, 70% then 95% ethanol (30 sec each) followed by eosin for 1 min. This was followed by 95% and 100% ethanol for 30 sec each and xylene for 2 min. Xenograft tissues were imaged with a Zeiss Stereomicroscope SteREO Discovery.V20 equipped with an AxioCam MRc5 (Carl Zeiss Microscopy, LLC, Thornwood, NY).
Lipid Extraction and Mass Spectrometric Analysis
Tissue samples were extracted with tert-butyl methyl ether and methanol (28-29) to which [2H8]arachidonic acid, [2H3]phytanic acid, [2H4]hexacosanoic acid, [2H28]hexadecanedioic acid [13C16]palmitic acid, [2H7]cholesterol sulfate, [2H5]MAG 18:1, [2H3]Carnitine 18:0, [13C3]DAG 36:2, [2H5]TAG 48:0, [2H31]PtdEtn 34:1, [2H54]PtdEtn 28:0, [2H31]PtdCh 34:1, [2H54]PtdCh 28:0, [2H62]PtdCh 32:0, [2H31]SM 16:0, [2H31]PtdSer 36:1, [2H31]PA 34:1, [2H62]PG 32:0 were added as internal standards. Glyburide was added as a lock mass. Extracts were dried by centrifugal vacuum evaporation prior to resuspension in isopropanol:methanol:chloroform (4:2:1) containing 7.5 mM ammonium acetate. Lipid analyses were performed by direct infusion in an Orbitrap mass spectrometer (Thermo Q-Exactive), with successive switching between polarity modes utilizing high-resolution (140,000 at 200 amu) data acquisition, with < 1 ppm mass accuracy. Washes between samples with hexane/ethyl acetate (3:2) were used to minimize ghost effects. In negative and positive ion ESI, the anions and cations, respectively were quantified and lipid identities validated by MS/MS. Precursor ions were selected at unit mass resolution and product ions analyzed at high resolution (140,000) utilizing collision cell energies of 10, 25, and 35.
Lipidomic Data Processing and Analysis
Measured lipid abundances were taken as the ratio of lipid peak area to the peak area of an internal standard corrected for protein concentration. Grubb's test was used to identify and remove outliers in the dataset. Data presented as mean ± SEM with p ≤ 0.05 considered significant.
Results
General strategy for mass spectrometric analysis and imaging of lipids
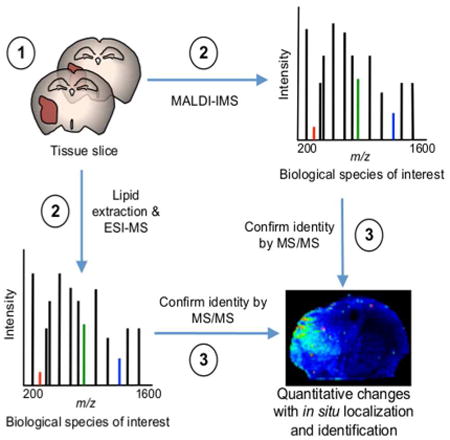
We employed the workflow outlined in Figure 1 to investigate lipid species, which were differentially expressed in xenograft tumors in the attractor or non-attractor groups, derived from intracranial implantation of human GSCs (13, 15-16, 32). Tissue slices were obtained from a total of eighteen xenografts. Slices (15 μm) were extracted and analyzed by ESI-MS/MS (28-29), or MALDI-IMS (10 μm) as described in Methods and Materials). We obtained quantitative measurements of lipid species with high resolution MS/MS confirmation of assignments (Supplemental Table 1). The MALDI-IMS technology allowed for in situ localization of lipid species from the lipidomics analysis as intact molecular ions with confirmation of selected m/z assignments by MS/MS.
Figure 1. Workflow outlining tissue sample preparation for combined lipidomics and MALDI-IMS.
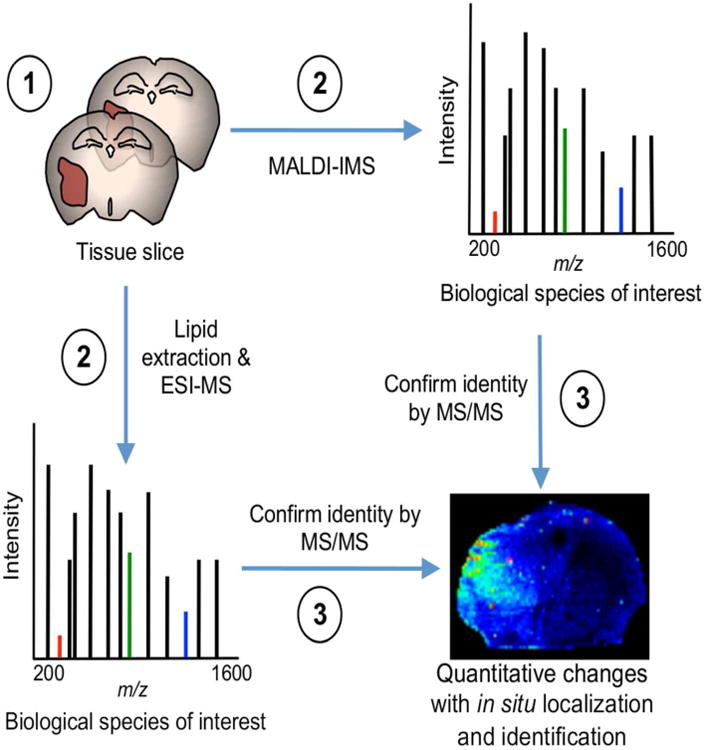
(1) Serial coronal sections from GSC xenografts at 10 and 15 μm. (2) MALDI-IMS (10 μm) and lipid extraction ESI-MS/MS (15 μm) experiments were performed in parallel. (3) The identity of biological species of interest was confirmed by MS/MS. Lipid extraction and ESI-MS/MS provide quantitative measures of lipid species at high-resolution with submillimass accuracy and MALDI-IMS provides lipid localization of identified lipids as intact molecular ions.
Identification of Phosphatidic Acid Alterations
Analysis of lipid extracts from tissue slices detected multiple species of phosphatidic acid (PA), and quantification of these detected species revealed a subset of PA species PA (36:2), PA (40:5), PA (42:5), and PA (42:7), that were significantly lower in tumors of the attractor phenotype compared with the non-attractor phenotype (Fig. 2A). This pattern held true even for the PA species that did not reach statistical significance (Supplemental Table 1). Intriguingly, species of lysophosphatidic acid (LPA), the common precursor of PA via the Kennedy Pathway (38-40), detected and validated by ESI-MS/MS (LPA 16:0, LPA 18:2, and LPA 18:3) were not statistically different between the two tumor phenotypes (Supplemental Table 1).
Figure 2. Negative ion MALDI-IMS of PA in GSC xenografts.
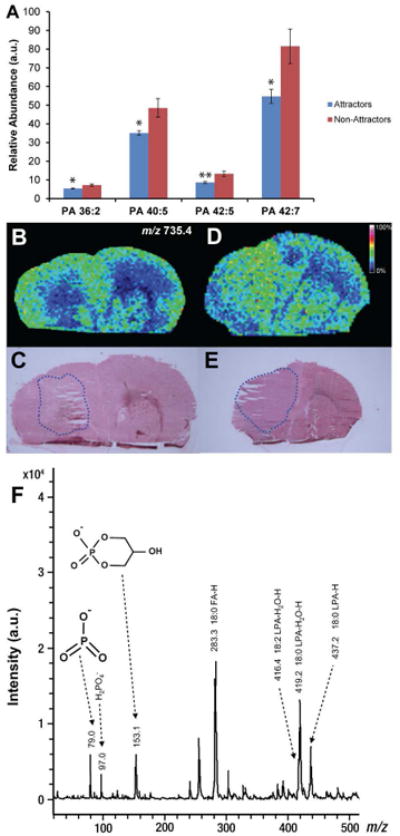
(A) Relative abundance of PA (36:2), PA (40:5), PA (42:5), and PA (42:7) as determined by ESI-MS/MS. Values are mean ± SEM (n = 9 for each phenotype) of the lipid abundance as the ratio of lipid peak area to the peak area of an internal standard corrected for protein concentration; *p < 0.05 and **p < 0.01 (Student's t-test). (B) Negative ion image derived from MALDI-IMS experiments of a representative attractor demonstrating the low signal intensity of PA (36:2) (m/z 735.4). Color intensity of the ion micrographs corresponds to signal strength. (C) H&E staining of corresponding tissue section used in MALDI-IMS experiment. Tumor area is outlined in blue. (D) Negative ion image derived from MALDI-IMS experiments of a representative non-attractor demonstrating the distribution and signal intensity of m/z 735.4. (E) H&E staining of corresponding tissue section used in MALDI-IMS experiment. Tumor area is outlined in blue. (F) Annotated MS/MS spectrum of PA (36:2) suggesting isoform PA (18:0/18:2). MS/MS product ions are shown in Table 1.
Next, we sought to localize the distribution of the significant PA species by MALDI-IMS in negative ion mode through inspection of the intact mass of anions. Of the significant PA species identified by ESI-MS/MS, PA (40:5), PA (42:5), and PA (42:7) did not specifically localize to the tumor region (data not shown), suggesting that the elevated levels quantified by ESI-MS/MS represent a global decrease of these species in the attracting xenografts. We did, however, find that PA (36:2) at m/z 735.4 [M+Cl]- was largely depleted in the tumor area of attractors (Fig. 2B), whereas it was consistently present in the tumor area of non-attractors (Fig. 2D). Corresponding tissue was stained for hematoxylin-eosin (H&E) to correlate lipid distribution with histological features (Fig. 2C, E, respectively). To confirm the PA (36:2) finding, we performed MS/MS analysis of m/z 735.4 in the MALDI-TOF/TOF directly on the tissue. The tissue-derived MALDI-MS/MS analysis of m/z 735.4 yielded product ions consistent with the structure of PA (18:0/18:2) (Fig. 2F and Table 1).
Table 1. Summary of results of negative and positive ion MS/MS by CID obtained directly from tumor sections oost-MALDI-IMS.
| Observed (m/z) | Product Ions (m/z) | Assignment |
|---|---|---|
| 735.4 [M+CI]- | 79.0, 97.0, 153.1, 283.3, 416.4, 419.2, 437.2 | PA (18:0/18:2) |
| 327.0 [M-H]- | 149.4, 178.5, 242.0, 283.2 | DHA (22:6) |
Identification of Diacylglycerol Alterations
ESI-MS/MS of GSC xenograft lipid extracts showed a number of DAG species (DAG 34:0, DAG 34:1, DAG 36:1, DAG 38:4, DAG 38:6, and DAG 40:6) that were significantly and differentially expressed between the two tumor phenotypes. All differentially expressed DAG species were significantly lower in the attractor GSCs compared with non-attractor GSCs (Fig. 3A). This trend was consistent even for detected DAG species that did not reach significance (Supplemental Table 1). We analyzed the distribution of the significant DAG species by positive ion mode MALDI-IMS. Of the significant DAG species (Fig. 3A), DAG (34:0), DAG (34:1), DAG (36:1), DAG (38:4), and DAG (38:6) did not consistently localize to any particular histological region (data not shown). This suggests that the lipid levels quantified by ESI-MS/MS represent a global decrease of these species in the attractor brains compared with the non-attractor brains. MALDI-IMS of DAG (40:6), on the other hand, was absent in the tumor area of attractor xenografts (Fig. 3B) but consistently present at m/z 707.1 [M+K]+ in the tumor area of non-attractors (Fig. 3D). Corresponding tissue was stained to correlate the MALDI-IMS lipid distribution with histopathological features in a representative attractor and non-attractor xenograft (Fig. 3C and E, respectively). Performing MALDI-MS/MS directly from the tissue on the precursor m/z 707.1, we were able to obtain some fragmentation (Supplemental Figure S2) though diacylglycerols are more amenable to ESI-MS/MS than MALDI-MS/MS. The MS/MS spectrum of m/z 707.1 [M+K]+ did reveal a product ion at m/z 323.6 which is consistent with the potassiated 18:0 FA at the C-1 (sn-1) position (41). This would leave the remaining fatty acyl to consist of C22:6 at either m/z 385 (22:6 FA cyclized at the glycerol backbone), an energetically favorable reaction, which forms a six-member ring (41), or m/z 367 [FA+H+K]+ likely at the C-2 (sn-2) position. However, these ions were not observed in our spectrum do to the difficulty of using MALDI to fragment DAG as well as the inherent difficulties of fragmenting DHA or DHA-containing lipids, which ionize better in negative ion mode.
Figure 3. Positive ion MALDI-IMS of DAG lipids in GSC xenografts.
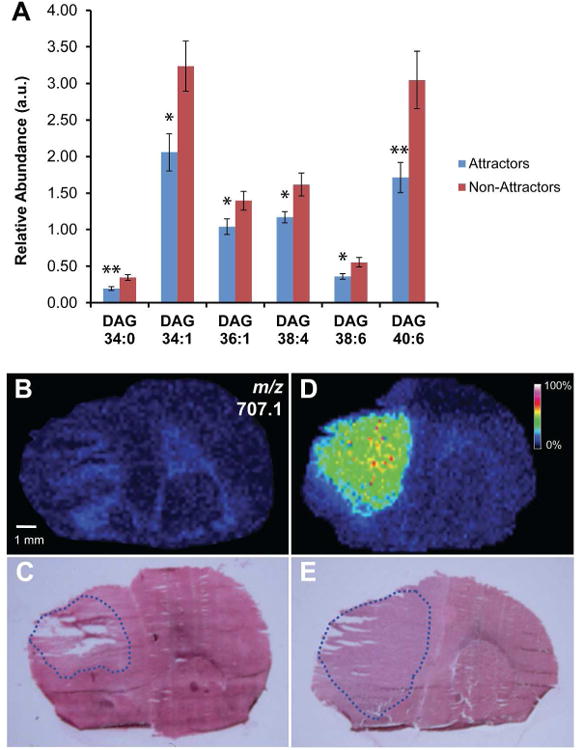
(A) Relative abundance of DAG (34:0), DAG (34:1), DAG (36:1), DAG (38:4), DAG (38:6), and DAG (40:6) as determined by ESI-MS/MS. Values are mean ± SEM (n = 9 for each phenotype) of the lipid abundance as the ratio of lipid peak area to the peak area of an internal standard corrected for protein concentration; *p < 0.05 and **p < 0.01 (Student's t-test). (B) Positive ion image from MALDI-IMS experiments of a representative attractor specimen, demonstrating the localization and signal intensity of DAG (40:6) (m/z 707.1). Image color intensity corresponds to signal strength. (C) H&E staining of corresponding tissue section analyzed by MALDI-IMS. The tumor area is outlined in blue. (D) Positive ion image of DAG (40:6), m/z 707.1 from MALDI-IMS experiments of a representative non-attractor specimen. (E) H&E staining of a corresponding tissue section analyzed by MALDI-IMS. The tumor area is outlined in blue.
Docosahexaenoic Acid
Docosahexaenoic Acid (DHA) is one of the most abundant polyunsaturated lipids in the CNS (42-43) and serves as a precursor for bioactive molecules (44-48). Lipid analysis by ESI-MS/MS data identified DHA as significantly decreased in tumors of the attractor phenotype (p = 0.046). By MALDI-IMS we found the intact mass of DHA anions, [M-H]- m/z 327.0, to be consistently decreased within the tumor area of attractors compared to non-attractors. Representative negative ion images of an attractor and non-attractor are shown in Figure 4 (A and C, respectively) with corresponding H&E staining (Fig. 4B and D, respectively) outlining the tumor area (blue). Quantified relative abundance of DHA from xenograft tissue using total lipid extract and ESI-MS/MS is shown in Figure 4E. MALDI-MS/MS fragmentation was performed on the parent anion. The spectrum of m/z 327.0 produced product ions that were consistent with its assignment of DHA (Table 1). The annotated MS/MS spectrum from the MALDI in negative ion mode confirming the identity of DHA can be seen in Figure 4F. The assignment was also confirmed with the MS/MS spectrum from a standard directly spotted on a MALDI plate with DHB (Supplemental Figure S1).
Figure 4. Tumor-specific localization of DHA.
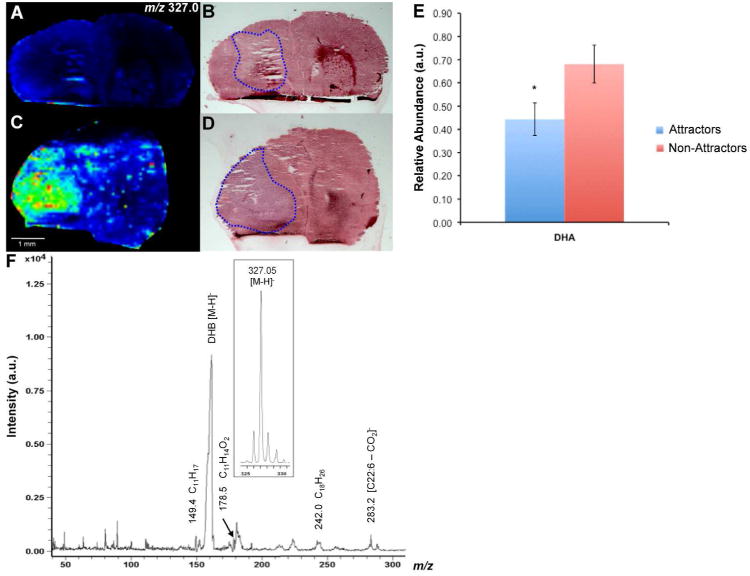
(A and C) Negative ion images derived from MALDI-IMS experiments showing the localization and signal intensity of DHA (22:6) m/z 327.0 in a representative non-attractor and attractor, respectively. (B and D) H&E stain of tissue sections used in MALDI-IMS experiments in a non-attractor and attractor, respectively. The tumor area is outlined in blue. (E) Relative abundance of DHA as determined by ESI-MS/MS (p = 0.046). (F) Annotated negative ion MS/MS spectrum of m/z 327.0 with precursor ion (inset). The most abundant peak, DHB [M-H]-, represents the matrix. MS/MS product ions are shown in Table 1.
Discussion
While several studies have demonstrated the feasibility of harnessing the homing capacity of BM-hMSCs for targeted delivery of cancer therapeutics (6-12), it is now evident, using a clinically relevant model of glioma, that BM-hMSCs demonstrate variable tropism towards these tumors (13). We demonstrate that specific lipid species including PA, DAG, and DHA species are differentially expressed in GSCs that support BM-hMSC homing, compared with those that do not. To the best of our knowledge, our study is the first to examine lipids in relation to BM-hMSC homing towards glioma. Our approach included ESI-MS/MS based quantitative lipidomics and MALDI-IMS to examine lipid profiles from GSC xenograft tissue. ESI-MS/MS provided high-resolution, high-mass accuracy quantitative data of lipid species (Supplemental Table 1). MALDI-IMS allowed visualization of the distribution of significant lipid species detected by ESI-MS/MS as molecular ions within the tissue; MALDI-MS/MS on tissue-derived molecular ions was provided.
We were intrigued by the results for phosphatidic acid (PA) and diacylglycerol (DAG) due to their capacity to act as signaling lipids and their interconnectivity through the Kennedy Pathway (40). Also of interest were docosahexaenoic acid (DHA), an abundant CNS lipid and precursor to many bioactive molecules (42-48). All were significantly decreased in the attractor phenotype (Fig. 2A and 3A, respectively), and this trend remained consistent for the species of PA and DAG that did not achieve significance in our lipid extract analysis (Supplemental Table 1). MALDI-IMS analysis demonstrated that of these lipids, PA (36:2) and DAG (40:6) had histopathological relevance, being absent in the tumor regions of attractor xenografts compared with the tumor region of non-attractors, where they were enriched (Fig. 2B-E and Fig. 3B-E, respectively). The DAG (40:6) potassiated molecular ion fragmentation spectrum suggests that the particular isoform is DHA containing DAG (22:6/18:0), in which the polyunsaturated fatty acid (PUFA) is located at the sn-1 position. Typically, but not invariably the sn-1 position will contain a saturated acyl chain and the sn-2 position will be occupied by an unsaturated acyl chain (e.g. PUFA) (49). Several lines of evidence indicate that the sn-1 position may be occupied by PUFAs (50) including DAG (22:6/18:0) (51). DHA (22:6) itself was lower in attractors vs non-attractors (Fig. 4E). Further investigation by MALDI-IMS demonstrated that, as with PA (36:2) and DAG (40:6), DHA expression levels correlated histopathologically with the tumor regions. DHA was largely depleted in the tumor region of attractor xenografts, but highly expressed in the tumors of non-attractors (Fig. 4A-D).
PA and DAG are multifunctional lipids that not only serve as a precursor to other lipids, but as biologically active signaling molecules (39, 52). Changes in these lipids may affect signaling pathways, nuclear signaling, and membrane ultrastructure in diverse and complex ways (38-39, 23; 53-54). In addition, DAGs are non-lamellar lipids that serve critical structural functions in the nuclear envelope, nucleoplasmic reticulum, membrane pore formation, membrane protein fusion, vesicle fusion, and membrane budding (53-54). All glycerophospholipids are derivatives of PA through DAG (40). Glycerophospholipids are important structural lipids as the main component of membranes (55-56). However, they may also affect signal transduction because they serve as reservoirs for signaling lipids via lipid remodeling at sn-2 (55-56). The overwhelming majority of glycerophospholipids detected in our dataset were decreased in attractors (Supplemental Table 1), which is consistent with our measured decreases in the precursors PA and DAG.
Intriguingly, while the detected PA species were consistently lower in the attractors, the lipid precursor to PA – LPA – was not significantly differentially expressed (Supplemental Table 1). The statistically significant difference in PA levels, but not LPA, suggests that there are underlying biochemical differences governing lipid metabolism between the tumor phenotypes. The acylation of LPA by LPAAT is the most common pathway of PA biosynthesis (39-40), though we do not exclude the possibility of alterations in PA levels due to differences in phospholipase D activity between the two tumor phenotypes (39). Likewise, PA may serve as a precursor to DAG within the Kennedy Pathway through enzymatic dephosphorylation (39-40, 52), though phospholipase C mediated cleavage of phosphatidylinositol 4,5-bisphosphate (PIP2) may also contribute to alterations in DAG biosynthesis (23).
While PA and DAG are effectors for a number of proteins, a point of convergence for both DAG and PA is the activation of protein kinase C (PKC) signaling. The decreased expression of both PA and DAG in attractors may point to a role for PKC signaling as a downstream effector in the BM-hMSC homing (39, 56). DAG governs the activation of both the classical and novel type of PKCs (59), whereas PA has been reported to activate select PKC isoforms representative of all three groups (classical, novel, and atypical) (39). The role of PKC in tumorigenesis is complex and in glioma poorly understood (56). This in part stems from past studies using non-selective pharmacological inhibitors of PKC, the relative contribution of each PKC isoform, and the variability of actions (56). Nonetheless, future studies are needed to dissect the role of PKC in the homing of BM-hMSCs to gliomas.
Lastly, DHA is the major PUFA in the CNS (42-44) and is a precursor for potent bioactive molecules (44). As with many other cancers, the balance of fatty acids – arachidonic acid (AA) and DHA – are dysregulated in glioma. A significant decrease in DHA and DHA-containing glycerophospholipids has been previously observed in malignant glioma compared to normal brain tissue (57-58). This decrease was accompanied by unchanged levels of AA (59-58). The mechanism of this disruption in the homeostatic DHA:AA ratio is unclear. However, one of most important functions of DHA is the resolution of inflammatory processes (44-46). The anti-inflammatory actions of DHA are the result of its metabolic products resolvins, neuroprotectins, and maresins (44-48). These molecules are transient lipids that are extremely difficult to measure. However, we can surmise that a decrease in DHA in attractor xenografts compared to non-attractors indicates relatively increased lipid-mediated inflammation.
Ultimately, the rationale to use BM-hMSCs for targeted delivery of cancer therapeutics is their unique ability to home and engraft into tissues that have undergone injury or severe stress (61-64). Solid tumors are considered “wounds that do not heal” (65) and gliomas are no exception. Gliomas not only possess an environment conducive to BM-hMSC engraftment (7, 66), but also elicit effective signals for BM-hMSC homing. In relation to the differential homing of BM-hMSCs in attractors vs non-attractors, the correlation between decreased DHA and the attractor phenotype may allow a permissive environment for BM-hMSC homing. The elevated levels of DHA in the non-attractor phenotype, on the other hand, may indicate relatively less inflammation correlating to less attraction to intravascularly delivered BM-hMSCs in these tumors. In this context, DHA may serve as a surrogate marker for relatively less inflammation. From a translational standpoint, our observation raises the possibility of DHA as a predictive measure of BM-hMSC homing efficacy (17). DHA radiotracers (11C) have been used recently to image DHA metabolism in the brain by positron emission tomography (PET) (67). More recently, longer-lived 18F-fluoro tracers have been developed to measure DHA content within the brain.
Presently, glioblastoma remains a nearly universally fatal disease despite aggressive treatment. Bone marrow-derived human mesenchymal stem cells show promise as cancer therapeutic delivery vehicles due to their intrinsic tropism for gliomas (6-12). In a clinically relevant orthotopic GSC xenograft model, these cells display variable tropism – the molecular basis of which is poorly understood. In the effort to understand BM-hMSC homing, the focus has been extensively confined to the evaluation of tumor-derived soluble factors (13, 18-19). However, lipids are not only critical to cell membrane properties, but also serve as important signaling molecules and have not been examined in previous homing studies. Our data are the first to suggest that PA and DAG, important lipid signaling hubs, may be involved in the homing process. Furthermore, the elevated levels of DHA in non-attractors compared to attractors may be indicator of relatively lower inflammatory response in these tumors, suggesting that differences in these processes between glioma tumors may regulate BM-hMSC homing efficacy (59-63). Taken together, our findings underscore the importance of lipid signaling in gliomas and provide new insights in advancing our understanding of underlying molecular differences that may mediate the differential homing capacity of BM-hMSCs to GSC xenografts and encourage further investigations.
Supplementary Material
Supplemental Figure S1. Annotated MS/MS spectrum of DHA (22:6) standard for comparison to tissue-acquired DHA in Figure 4F.
Supplemental Figure S2. Annotated MS/MS spectrum of DAG (40:6) suggesting DHA-containing DAG (22:6/18:0). Precursor ion of potassiated DAG (40:6) m/z 707.1 is shown (inset). Peak at m/z 191.9 is DHB matrix ion and the peak at 323.6 represents potassiated 18:0 FA [FA+K+]+
Acknowledgments
The authors gratefully acknowledge the financial support of the Cancer Prevention Research Institute of Texas (CPRIT) and The University of Texas Medical Branch to C.L.N. Grants from the National Cancer Institute CA115729 and 1P50 CA127001, The Broach Foundation for Brain Cancer Research, The Elias Family Fund, The National Brain Tumor Foundation, The Collaborative Ependymoma Research Network (CERN), The Gene Pennebaker Brain Cancer Fund, The Sorenson Foundation, and The Brian McCulloch Fund to F.F.L and Lincoln Memorial University (P.L.W.) are gratefully acknowledged.
Footnotes
Supporting Information: Supporting Information, this material is available free of charge via http://pubs.acs.org/
Author Contributions: N.C.W. conceived of the study, designed, and performed all MALDI-IMS experiments, data analysis and interpretation, and wrote the paper. P.L.W. contributed to the experimental design, performed all lipid extractions, and data acquisition on the Q-Exactive. J.G. grew stem cell lines and performed all animal work. C.F.L. contributed to the experimental design and interpretation of MALDI-TOF/TOF data. M.R.E. conceived the project and contributed critical review of MALDI-TOF/TOF data, technical support, and helpful discussions. C.L.N. and F.F.L conceived the project and supervised the work and critically revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest: The authors do not have any conflicts of interest to disclose.
References
- 1.Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neuro Oncol. 1999;1(1):44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berens ME, Giese A. …those left behind. Biology and oncology of invasive glioma cells. Neoplasia. 1999;1(3):208–19. doi: 10.1038/sj.neo.7900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO Groups, E. O. f. R. a. T. o. C. B. T. a. R.; Group, N. C. I. o. C. C. T. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Pardridge WM. Blood-brain barrier drug targeting: the future of brain drug development. Mol Interv. 2003;3(2):90–105. 51. doi: 10.1124/mi.3.2.90. [DOI] [PubMed] [Google Scholar]
- 6.Kosztowski T, Zaidi HA, Quiñones-Hinojosa A. Applications of neural and mesenchymal stem cells in the treatment of gliomas. Expert Rev Anticancer Ther. 2009;9(5):597–612. doi: 10.1586/era.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamizo A, Marini F, Amano T, Khan A, Studeny M, Gumin J, Chen J, Hentschel S, Vecil G, Dembinski J, Andreeff M, Lang FF. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65(8):3307–18. doi: 10.1158/0008-5472.CAN-04-1874. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura K, Ito Y, Kawano Y, Kurozumi K, Kobune M, Tsuda H, Bizen A, Honmou O, Niitsu Y, Hamada H. Antitumor effect of genetically engineered mesenchymal stem cells in a rat glioma model. Gene Ther. 2004;11(14):1155–64. doi: 10.1038/sj.gt.3302276. [DOI] [PubMed] [Google Scholar]
- 9.Studeny M, Marini FC, Dembinski JL, Zompetta C, Cabreira-Hansen M, Bekele BN, Champlin RE, Andreeff M. Mesenchymal stem cells: potential precursors for tumor stroma and targeted-delivery vehicles for anticancer agents. J Natl Cancer Inst. 2004;96(21):1593–603. doi: 10.1093/jnci/djh299. [DOI] [PubMed] [Google Scholar]
- 10.Yong RL, Shinojima N, Fueyo J, Gumin J, Vecil GG, Marini FC, Bogler O, Andreeff M, Lang FF. Human bone marrow-derived mesenchymal stem cells for intravascular delivery of oncolytic adenovirus Delta24-RGD to human gliomas. Cancer Res. 2009;69(23):8932–40. doi: 10.1158/0008-5472.CAN-08-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miletic H, Fischer Y, Litwak S, Giroglou T, Waerzeggers Y, Winkeler A, Li H, Himmelreich U, Lange C, Stenzel W, Deckert M, Neumann H, Jacobs AH, von Laer D. Bystander killing of malignant glioma by bone marrow-derived tumor-infiltrating progenitor cells expressing a suicide gene. Mol Ther. 2007;15(7):1373–81. doi: 10.1038/sj.mt.6300155. [DOI] [PubMed] [Google Scholar]
- 12.Doucette T, Rao G, Yang Y, Gumin J, Shinojima N, Bekele BN, Qiao W, Zhang W, Lang FF. Mesenchymal stem cells display tumor-specific tropism in an RCAS/Ntv-a glioma model. Neoplasia. 2011;13(8):716–25. doi: 10.1593/neo.101680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinojima N, Hossain A, Takezaki T, Fueyo J, Gumin J, Gao F, Nwajei F, Marini FC, Andreeff M, Kuratsu J, Lang FF. TGF-β mediates homing of bone marrow-derived human mesenchymal stem cells to glioma stem cells. Cancer Res. 2013;73(7):2333–44. doi: 10.1158/0008-5472.CAN-12-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–8. [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 18.Hata N, Shinojima N, Gumin J, Yong R, Marini F, Andreeff M, Lang FF. Platelet-derived growth factor BB mediates the tropism of human mesenchymal stem cells for malignant gliomas. Neurosurgery. 2010;66(1):144–56. doi: 10.1227/01.NEU.0000363149.58885.2E. discussion 156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son BR, Marquez-Curtis LA, Kucia M, Wysoczynski M, Turner AR, Ratajczak J, Ratajczak MZ, Janowska-Wieczorek A. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells. 2006;24(5):1254–64. doi: 10.1634/stemcells.2005-0271. [DOI] [PubMed] [Google Scholar]
- 20.Helmreich EJ. Environmental influences on signal transduction through membranes: a retrospective mini-review. Biophys Chem. 2003;100(1-3):519–34. doi: 10.1016/s0301-4622(02)00303-4. [DOI] [PubMed] [Google Scholar]
- 21.Aureli M, Grassi S, Prioni S, Sonnino S, Prinetti A. Lipid membrane domains in the brain. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbalip.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Fernandis AZ, Wenk MR. Membrane lipids as signaling molecules. Curr Opin Lipidol. 2007;18(2):121–8. doi: 10.1097/MOL.0b013e328082e4d5. [DOI] [PubMed] [Google Scholar]
- 23.Berridge MJ. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu Rev Biochem. 1987;56:159–93. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- 24.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–23. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 25.Wood PL. Lipidomics of Alzheimer's disease: current status. Alzheimers Res Ther. 2012;4(1):5. doi: 10.1186/alzrt103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He H, Nilsson CL, Emmett MR, Ji Y, Marshall AG, Kroes RA, Moskal JR, Colman H, Lang FF, Conrad CA. Polar lipid remodeling and increased sulfatide expression are associated with the glioma therapeutic candidates, wild type p53 elevation and the topoisomerase-1 inhibitor, irinotecan. Glycoconj J. 2010;27(1):27–38. doi: 10.1007/s10719-009-9249-6. [DOI] [PubMed] [Google Scholar]
- 27.He H, Conrad CA, Nilsson CL, Ji Y, Schaub TM, Marshall AG, Emmett MR. Method for lipidomic analysis: p53 expression modulation of sulfatide, ganglioside, and phospholipid composition of U87 MG glioblastoma cells. Anal Chem. 2007;79(22):8423–30. doi: 10.1021/ac071413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49(5):1137–46. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood PL. Lipidomics Analysis of Postmortem Interval: Preliminary Evaluation of Human Skeletal Muscle. In: Shirley NR, editor. Metabolomics. Vol. 3. 2013. p. 127. [Google Scholar]
- 30.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113(4):2309–42. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chughtai K, Jiang L, Greenwood TR, Glunde K, Heeren RM. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J Lipid Res. 2013;54(2):333–44. doi: 10.1194/jlr.M027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–33. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 33.Shavkunov AS, Wildburger NC, Nenov MN, James TF, Buzhdygan TP, Panova-Elektronova NI, Green TA, Veselenak RL, Bourne N, Laezza F. The fibroblast growth factor 14·voltage-gated sodium channel complex is a new target of glycogen synthase kinase 3 (GSK3) J Biol Chem. 2013;288(27):19370–85. doi: 10.1074/jbc.M112.445924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberg AL, Vitek O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J Proteome Res. 2009;8(5):2144–56. doi: 10.1021/pr8010099. [DOI] [PubMed] [Google Scholar]
- 35.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18(9):1646–52. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35(Web Server issue):W606–12. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;(50 Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45(3):250–78. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Bruntz RC, Lindsley CW, Brown HA. Phospholipase D Signaling Pathways and Phosphatidic Acid as Therapeutic Targets in Cancer. Pharmacol Rev. 2014;66(4):1033–1079. doi: 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy EP. Metabolism and function of membrane lipids. Klin Wochenschr. 1987;65(5):205–12. doi: 10.1007/BF01715846. [DOI] [PubMed] [Google Scholar]
- 41.Al-Saad KA, Siems WF, Hill HH, Zabrouskov V, Knowles NR. Structural analysis of phosphatidylcholines by post-source decay matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2003;14(4):373–82. doi: 10.1016/S1044-0305(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien JS, Sampson EL. Fatty acid and fatty aldehyde composition of the major brain lipids in normal human gray matter, white matter, and myelin. J Lipid Res. 1965;6(4):545–51. [PubMed] [Google Scholar]
- 43.Svennerholm L. Distribution and fatty acid composition of phosphoglycerides in normal human brain. J Lipid Res. 1968;9(5):570–9. [PubMed] [Google Scholar]
- 44.Hong S, Gronert K, Devchand PR, Moussignac RL, Serhan CN. Novel docosatrienes and 17S-resolvins generated from docosahexaenoic acid in murine brain, human blood, and glial cells. Autacoids in anti-inflammation. J Biol Chem. 2003;278(17):14677–87. doi: 10.1074/jbc.M300218200. [DOI] [PubMed] [Google Scholar]
- 45.Mukherjee PK, Marcheselli VL, Serhan CN, Bazan NG. Neuroprotectin D1: a docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc Natl Acad Sci U S A. 2004;101(22):8491–6. doi: 10.1073/pnas.0402531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcheselli VL, Hong S, Lukiw WJ, Tian XH, Gronert K, Musto A, Hardy M, Gimenez JM, Chiang N, Serhan CN, Bazan NG. Novel docosanoids inhibit brain ischemia-reperfusion-mediated leukocyte infiltration and pro-inflammatory gene expression. J Biol Chem. 2003;278(44):43807–17. doi: 10.1074/jbc.M305841200. [DOI] [PubMed] [Google Scholar]
- 47.Ariel A, Serhan CN. Resolvins and protectins in the termination program of acute inflammation. Trends Immunol. 2007;28(4):176–83. doi: 10.1016/j.it.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Schwab JM, Chiang N, Arita M, Serhan CN. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007;447(7146):869–74. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper RA. Lipids of human red cell membrane: normal composition and variability in disease. Semin Hematol. 1970;7(3):296–322. [PubMed] [Google Scholar]
- 50.Beermann C, Möbius M, Winterling N, Schmitt JJ, Boehm G. sn-position determination of phospholipid-linked fatty acids derived from erythrocytes by liquid chromatography electrospray ionization ion-trap mass spectrometry. Lipids. 2005;40(2):211–8. doi: 10.1007/s11745-005-1377-1. [DOI] [PubMed] [Google Scholar]
- 51.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shulga YV, Topham MK, Epand RM. Regulation and functions of diacylglycerol kinases. Chem Rev. 2011;111(10):6186–208. doi: 10.1021/cr1004106. [DOI] [PubMed] [Google Scholar]
- 53.Peddie CJ, Blight K, Wilson E, Melia C, Marrison J, Carzaniga R, Domart MC, O'Toole P, Larijani B, Collinson LM. Correlative and integrated light and electron microscopy of in-resin GFP fluorescence, used to localise diacylglycerol in mammalian cells. Ultramicroscopy. 2014;143:3–14. doi: 10.1016/j.ultramic.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Domart MC, Hobday TM, Peddie CJ, Chung GH, Wang A, Yeh K, Jethwa N, Zhang Q, Wakelam MJ, Woscholski R, Byrne RD, Collinson LM, Poccia DL, Larijani B. Acute manipulation of diacylglycerol reveals roles in nuclear envelope assembly & endoplasmic reticulum morphology. PLoS One. 2012;7(12):e51150. doi: 10.1371/journal.pone.0051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hermansson M, Hokynar K, Somerharju P. Mechanisms of glycerophospholipid homeostasis in mammalian cells. Prog Lipid Res. 2011;50(3):240–57. doi: 10.1016/j.plipres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 56.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7(4):281–94. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 58.Martin DD, Robbins ME, Spector AA, Wen BC, Hussey DH. The fatty acid composition of human gliomas differs from that found in nonmalignant brain tissue. Lipids. 1996;31(12):1283–8. doi: 10.1007/BF02587914. [DOI] [PubMed] [Google Scholar]
- 59.Albert DH, Anderson CE. Fatty acid composition at the 2-position of ether-linked and diacyl ethanolamine and choline phosphoglycerides of human brain tumors. Lipids. 1977;12(9):722–8. doi: 10.1007/BF02570902. [DOI] [PubMed] [Google Scholar]
- 60.Elsherbiny ME, Emara M, Godbout R. Interaction of brain fatty acid-binding protein with the polyunsaturated fatty acid environment as a potential determinant of poor prognosis in malignant glioma. Prog Lipid Res. 2013;52(4):562–70. doi: 10.1016/j.plipres.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahmood A, Lu D, Chopp M. Marrow stromal cell transplantation after traumatic brain injury promotes cellular proliferation within the brain. Neurosurgery. 2004;55(5):1185–93. doi: 10.1227/01.neu.0000141042.14476.3c. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Chopp M, Chen J, Wang L, Gautam SC, Xu YX, Zhang Z. Intrastriatal transplantation of bone marrow nonhematopoietic cells improves functional recovery after stroke in adult mice. J Cereb Blood Flow Metab. 2000;20(9):1311–9. doi: 10.1097/00004647-200009000-00006. [DOI] [PubMed] [Google Scholar]
- 63.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410(6829):701–5. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 65.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 66.Studeny M, Marini FC, Champlin RE, Zompetta C, Fidler IJ, Andreeff M. Bone marrow-derived mesenchymal stem cells as vehicles for interferon-beta delivery into tumors. Cancer Res. 2002;62(13):3603–8. [PubMed] [Google Scholar]
- 67.Basselin M, Ramadan E, Rapoport SI. Imaging brain signal transduction and metabolism via arachidonic and docosahexaenoic acid in animals and humans. Brain Res Bull. 2012;87(2-3):154–71. doi: 10.1016/j.brainresbull.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Annotated MS/MS spectrum of DHA (22:6) standard for comparison to tissue-acquired DHA in Figure 4F.
Supplemental Figure S2. Annotated MS/MS spectrum of DAG (40:6) suggesting DHA-containing DAG (22:6/18:0). Precursor ion of potassiated DAG (40:6) m/z 707.1 is shown (inset). Peak at m/z 191.9 is DHB matrix ion and the peak at 323.6 represents potassiated 18:0 FA [FA+K+]+


