Abstract
Relatively little is known regarding mitochondrial metabolism in neuronal differentiation of embryonic stem (ES) cells. By using a small molecule, present research has investigated the pattern of cellular energy metabolism in neural progenitor cells derived from mouse ES cells. Flavonoid compound 4a faithfully facilitated ES cells to differentiate into neurons morphologically and functionally. The expression and localization of peroxisome proliferator-activated receptors (PPARs) were examined in neural progenitor cells. PPAR-β expression showed robust upregulation compared to solvent control. Treatment with PPAR-β agonist L165041 alone or together with compound 4a significantly promoted neuronal differentiation, while antagonist GSK0660 blocked the neurogenesis-promoting effect of compound 4a. Consistently, knockdown of PPAR-β in ES cells abolished compound 4a-induced neuronal differentiation. Interestingly, we found that mitochondrial fusion protein Mfn2 was also abolished by sh-PPAR-β, resulting in abnormal mitochondrial Ca2+ ([Ca2+]M) transients as well as impaired mitochondrial bioenergetics. In conclusion, we demonstrated that by modulating mitochondrial energy metabolism through Mfn2 and mitochondrial Ca2+, PPAR-β took an important role in neuronal differentiation induced by flavonoid compound 4a.
Introduction
Stem cell differentiation is associated with changes in metabolism and function. Understanding these changes during neuronal differentiation is important in the context of stem cell research, neurodegenerative diseases and regenerative medicine. While much has been learned about the molecular events involved in neuronal differentiation [1], relatively little is known regarding their bioenergetic demands and how closely their energy metabolism is governed by the genetic developmental programme [2].
Mitochondria are vital ATP-generating cellular organelles, and they are particularly essential in the nervous system [3]. The increased mitochondrial activity associated with differentiation provides ATP to support fundamental energy-demanding processes involved in neuronal differentiation [1]. Mitochondria dynamics are essential for neuronal functions since they regulate mitochondrial number, location, morphology and function [1]. Mitochondrial fission and fusion are regulated by two distinct protein complexes. Drp (dynamin-related protein) and Fis1, mediate mitochondrial fission, whereas mitofusins (Mfn1 and Mfn2) and OPA1 (Optic Atrophy-1) regulate mitochondrial fusion [1, 3, 4]. In mammals, mutations in Mfn2 cause Charcot-Marie-Tooth (CMT) type 2A, a peripheral neuropathy characterized by axonal degeneration [5]. Cerebellum-specific Mfn2-knockout mice revealed the importance of mitochondrial fusion in protecting cerebellar neurodegeneration [6]. These findings suggested a key role of Mfn2 in nervous system. Furthermore, recent reports demonstrated a new function of Mfn2, which was tethering the endoplasmic reticulum and mitochondria to control the efficiency of mitochondrial uptake of Ca2+ ions [7–9]. However, relatively little is known about the potential role of Mfn2 and its upstream determinants in cellular energy metabolism of neuronal differentiation.
Mitochondrial biogenesis and dynamics require coordinated changes in the metabolic enzymes of oxidative phosphorylation and fatty acid oxidation [10]. Peroxisome proliferator-activated receptors (PPARs) are lipid-activated transcription factors belonging to the nuclear receptor superfamily [10]. Three PPAR isotypes (α, β, and γ) have been distinguished by tissue- and developmental-specific expression patterns [11]. PPAR-α, which is mainly rich in tissues that have high-energy demands, was involved in cardiomyocyte differentiation of mouse ES cells in vitro [12]. PPAR-γ is found primarily in the adipose tissue and plays an important role in adipose differentiation [13]. PPAR-β is the most ubiquitously expressed with a controversial role [10, 11]. The important role of lipid molecules in brain development is well known [14]. All three PPAR isotypes are expressed in the brain, while PPAR-β is the most abundant subtype [15]. Recent findings demonstrated that modulation of PPAR-β expression might be an important part of brain pathology [16]. The presence and possible modulation of these receptors were also examined in embryonic rat cortical neurons during their in vitro maturation [14]. The results suggested a potential role of PPAR-β in neuronal maturation. In addition, a neuronal differentiating effect of PPAR-β was demonstrated in human neuroblastoma cell line SH-SY5Y [17, 18]. Moreover, it was reported that retinoic acid (RA) induced neurogenesis by activating both retinoic acid receptors (RARs) and PPAR-β in P19 mouse embryonal carcinoma cell line [19]. However, the PPAR isotype expressions and their downstream effects during neuronal differentiation of ES cells have not been investigated so far.
The role of small molecules in stem cell biology is emerging [20]. Such molecules will likely provide new insights into mitochondrial metabolism in neuronal differentiation of ES cells, and may ultimately contribute to effective medicine for tissue repair and regeneration [21]. Our previous work showed that some natural flavonoid compounds, icaritin (ICT) [22] and isobavachin (IBA) [23] had significant neurogenesis-inducing activities. In the present study, we used a newly-screened flavonoid compound 4a as a probe of underlying biology, and aimed to elucidate PPARs expressions and several elements of cellular energy metabolism in neuronal differentiation of mouse ES cells.
Results
Flavonoid compound 4a promoted neuronal differentiation of mouse ES cells
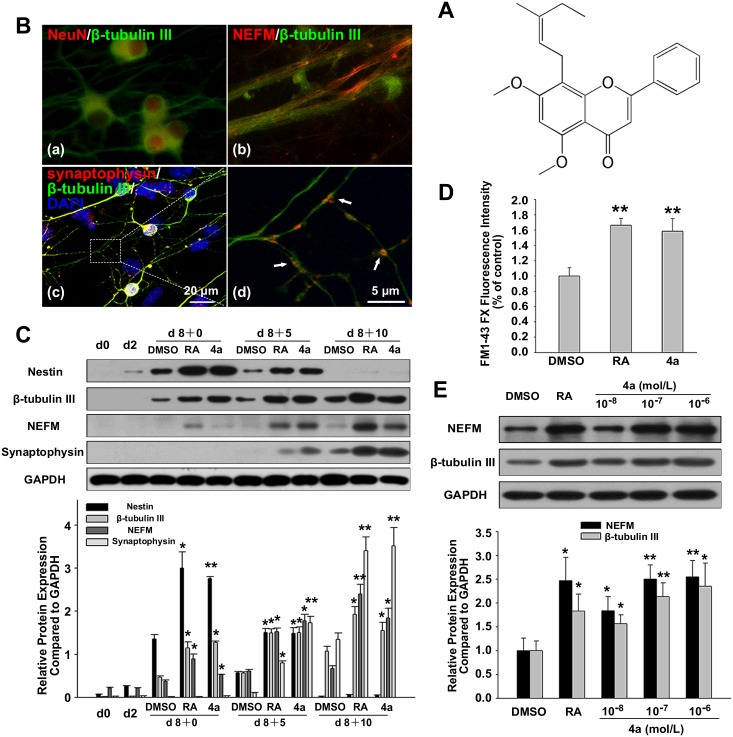
Compound 4a (5,7-dimethoxy-8-(3-methyl-pent-2-enyl)-2-phenyl-chromen-4-one) was offered in this case by Prof. Dr. Yong-ping Yu, which were synthesized by previous methods [24]. The structure of compound 4a was shown in Fig 1A. To induce neuronal differentiation, a typical 4−/4+ protocol was used (S1 Fig). After compound 4a treatment, the expression and localization of neuron-specific proteins were evaluated by immunocytochemistry. Among them, β-tubulin III and neuronal nuclei (NeuN) [25] were neuron cytoplasm and nucleus house-keeping marker, neurofilament 160 (NEFM) [26] was axons marker, and synaptophysin [27] was synaptic vesicles marker. The results in Fig 1B showed that compound 4a could induce neuron-specific proteins expression. In consistent with this, western blot analysis showed compound 4a could upregulate the neural specific proteins expression in a developmental way, providing the fundamentals for synaptic vesicle recycling (Fig 1C). Nestin is a neural progenitor marker, which expressed at early differentiation stage. Compound 4a induced Nestin expression robustly on day 8 of differentiation (Fig 1C), indicating that its neurogenesis-inducing effect appeared as early as neural progenitor cells formation period. The neuronal property of synaptic vesicle recycling was detected by FM 1-43FX. The dye can be internalized from the culture medium during synaptic vesicle recycling, in response to a high concentration of potassium ions in the medium [28]. As a result, cells that possess the neurogenic function display increased FM1-43FX fluorescence. The fluorescence intensity in ES-derived neurons induced by 4a was similar to that of cells treated with retinoic acid (RA) (Fig 1D). Since synaptic vesicle recycling is a neuron-specific function, we confirmed compound 4a could induce functional neuronal differentiation. Semiquantitative analysis indicated that the neurogenesis-inducing effect of compound 4a was in a dose-dependent manner at the terminal differentiation point (Fig 1E).
Fig 1. Flavonoid compound 4a promoted neuronal differentiation of mouse ES cells.
A: Structure of compound 4a. B: (a-c) Double Immunofluoresence staining for neural specific markers in ES-derived neurons induced by 4a on d 8+10. (d) The arrows indicated the areas of synaptophysin/β-tubulin III colocalization. Nuclei were stained with DAPI. Bar = 20 μm (a-c), and 5 μm (d). C: Developmental-dependent neural specific proteins expression induced by 4a (10−7 mol/L) during the differentiation course. D: Compound 4a promoted depolarization-induced synaptic vesicle recycling function of ES-derived neurons. The neuronal property of synaptic vesicle recycling was detected by FM 1-43FX. The data was obtained from the index of fluorescence intensity normalized to MTT absorbance. E: Dose-dependent neural specific proteins expression in ES-derived neurons induced by 4a on d 8+10. Data were represented as the mean ± S.D. of three independent experiments. Statistical significance was set as *P<0.05, **P<0.01 vs. DMSO control. Retinoic acid (RA) was used as a positive control.
PPAR-β was involved in compound 4a-induced neuronal differentiation of ES cells
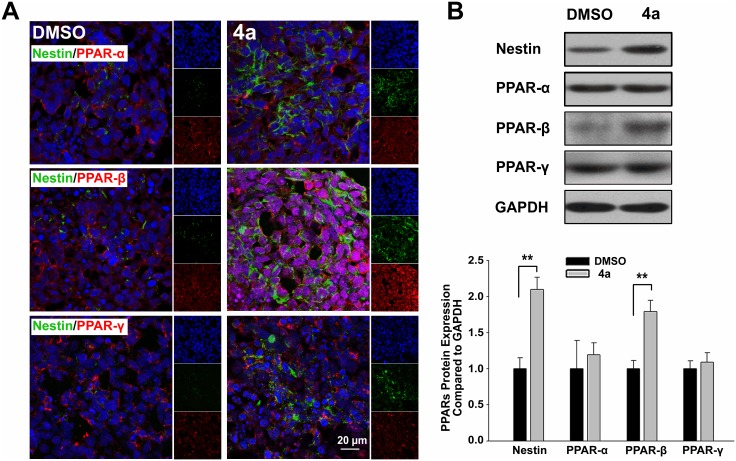
A developmental-dependent expression of PPAR isotypes were identified by western blot analysis (S2 Fig). Data showed that after compound 4a treatment, PPAR-α deceased during the differentiation, while PPAR-β gradually increased both in the early (d 8) and late (d 8+10) stage. PPAR-γ, however, did not change during neuronal differentiation. Therefore, we focused on the early stage of neuronal differentiation in the following research. Expression and distribution of PPARs were revealed by double staining PPAR isotypes with neural progenitor marker Nestin on d 8. The percentages of Nestin positive cells were counted in five different EBs frozen sections respectively, and the result was about 22.2±5.9% in DMSO group and 67.8±3.8% in 4a group. Fig 2A showed that all PPARs were expressed in ES-derived neural progenitors. Typically, PPAR-β was mainly found in cell nuclei with higher fluorescence intensity. The increased expression of PPAR-β was also identified by western blot analysis, while both PPAR-α and PPAR-γ stayed the same (Fig 2B), suggesting a key role of PPAR-β in early neuronal differentiation of ES cells induced by compound 4a.
Fig 2. PPAR-β was involved in neuronal differentiation of ES cells at early stage.
A: Double immunostaining for PPARs and Nestin in ES-derived neural progenitor cells. After treatment with compound 4a, EBs (d 8) was prepared for frozen section. The expression and localization of PPARs and Nestin were examined. Nuclei were stained with DAPI, and DMSO was set as solvent control. B: PPARs protein expression in ES-derived neural progenitor cells. Data were represented as the mean ± S.D. of three independent experiments. Statistical significance was set as **P<0.01 vs. DMSO control.
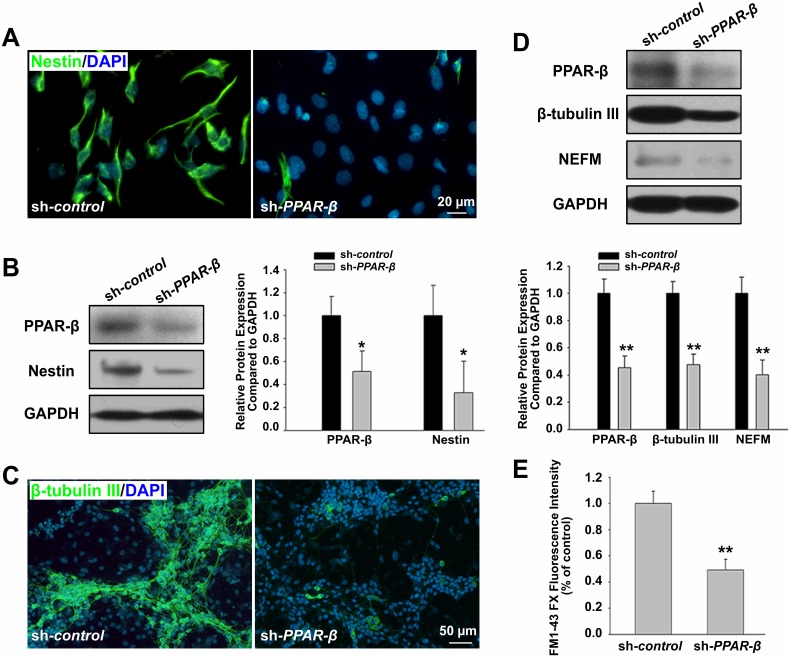
The Effects of PPAR-β agonist L165041 and antagonist GSK0660 on compound 4a-induced neuronal differentiation were then evaluated (S3 Fig). Treatment with PPAR-β agonist L165041 alone or together with compound 4a significantly promoted neuronal differentiation, while antagonist GSK0660 blocked the neurogenesis-promoting effect of compound 4a. We further investigated whether PPAR-β knockdown in ES cells could block compound 4a-induced neurogenesis of ES cells. Transfection and silencing efficiency of sh-PPAR-β plasmid was evaluated before the experiment (S4 Fig). Viability and pluripotency of ES cells were not affected by sh-PPAR-β. Consistently, knockdown of PPAR-β abolished compound 4a-induced neuronal differentiation. The results showed that Nestin was downregulated in neural progenitor cells (Fig 3A and 3B), followed by the decreasing of neuron-specific proteins β-tubulin III, NEFM expression (Fig 3C and 3D) and synaptic vesicle recycling as indicated by FM1-43FX fluorescence intensity (Fig 3E).
Fig 3. Effects of PPAR-β gene silencing on 4a-induced neuronal differentiation of ES cells.
ES cells were treated with sh-PPAR-β plasmid for 24 h, and then cultured according to 4-/4+ protocol. A-B: Nestin expression was detected by immunostaining and western blot assay on d 8. Nuclei were stained with DAPI. Bar = 20 μm. C: β-tubulin III expression was detected by immunostaining on d 8+10. Nuclei were stained with DAPI. Bar = 50 μm. D: β-tubulin III and NEFM expression was detected by western blot assay on d 8+10. The data was obtained from the index of each proteins compared with GAPDH. E: The neuronal property of synaptic vesicle recycling was detected by FM1-43FX. The data was obtained from the index of fluorescence intensity normalized to MTT absorbance. Statistical significance was set as *P<0.05, **P<0.01 vs. sh-control group. Data represent mean ± S.D. of three independent experiments.
Effects of PPAR-β knockdown on mitochondrial energy metabolism in compound 4a-induced neural progenitor cells
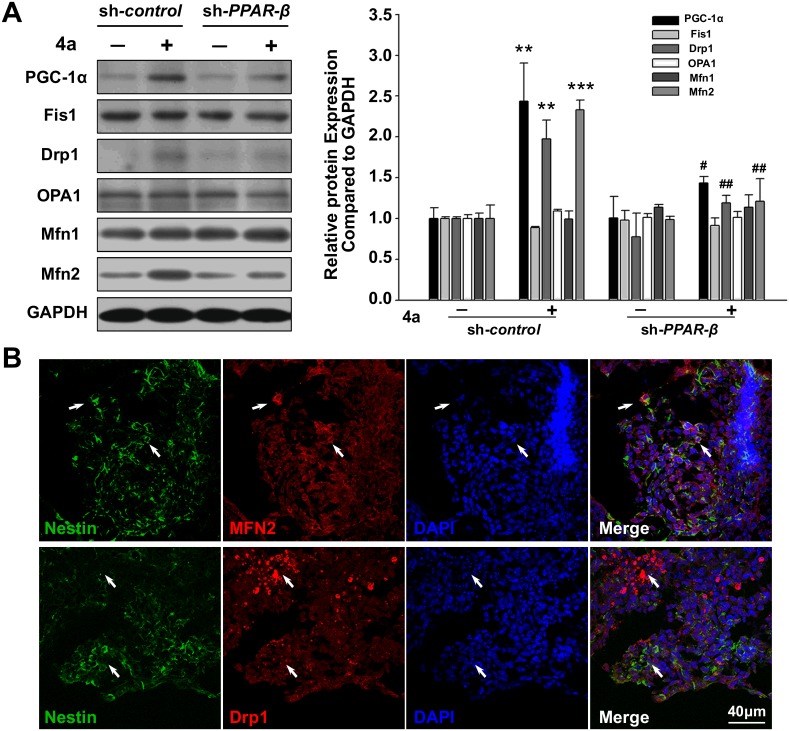
Compound 4a could increase the protein expressions of mitochondrial biogenesis regulator PGC-1α, mitochondrial fission protein Drp1 and mitochondrial fusion protein Mfn2, which were abolished by sh-PPAR-β (Fig 4A). Three regulators of mitochondrial biogenesis, PGC-1α, Nrf1 and TFAM mRNA expression were examined (S5 Fig). Consistently, compound 4a could upregulate PGC-1α, Nrf1 and TFAM mRNA expression, which were blocked by sh-PPAR-β, suggesting a key role of PPAR-β in mitochondrial biogenesis. We further investigated the relationship between the expressions of Drp1/Mfn2 and Nestin in neural progenitor cells. The results showed that Mfn2 was positively detected in the Nestin-positive cells. However, Drp1 was mostly detected in the Nestin-negetive cells with pyknotic nuclei (Fig 4B). The results indicated that a close relationship between PPAR-β and Mfn2 took an important role in early neuronal differentiation induced by compound 4a, whereas Drp1 might help to induce apoptosis [29] of non-neural progenitor cells.
Fig 4. Effects of PPAR-β gene silencing on mitochondrial system of 4a-induced neural progenitor cells.
A: Effects of PPAR-β gene silencing on mitochondrial fission and fusion function. PGC-1α is a master regulator of mitochondrial biogenesis. Fis1 and Drp1 mediate mitochondrial fission, whereas OPA1 and mitofusins (Mfn1 and Mfn2) mediate mitochondrial fusion. B: Double immunostaining for Drp1/Nestin and Mfn2/Nestin in 4a-induced neural progenitor cells. Bar = 40 μm. Statistical significance was set as **P<0.01 and ***P<0.001 vs. sh-control group, #P<0.05 and ##P<0.01 vs. 4a+sh-control group. Data represent mean ± S.D. of three independent experiments.
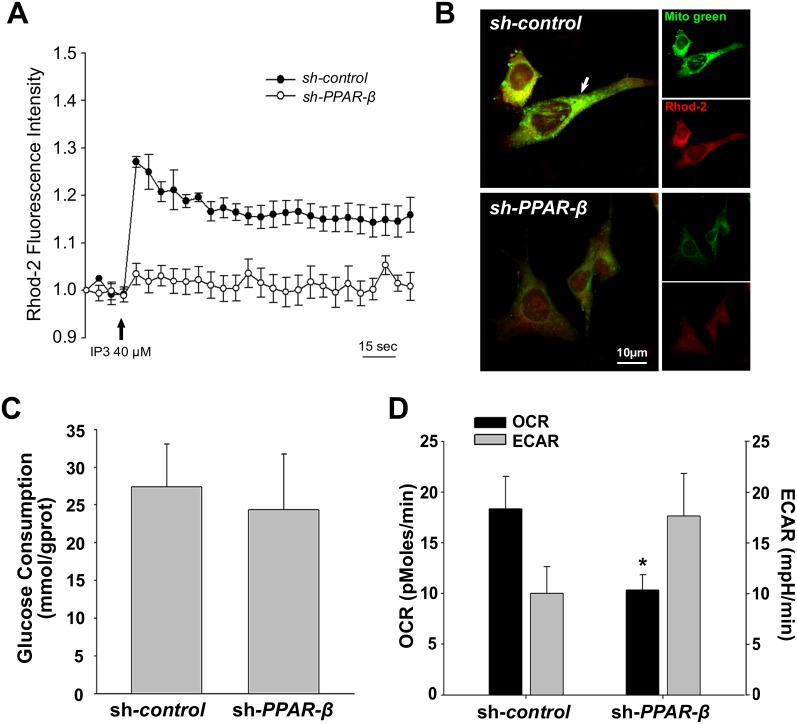
Mfn2 was reported to build a bridge between endoplasmic reticulum and mitochondria to control the efficiency of mitochondrial uptake of Ca2+ ions [7–9]. To investigate the effects of PPAR-β gene silencing on mitochondrial Ca2+ ([Ca2+]M), ES-derived neural progenitor cells were loaded with 4 μM Rhod-2AM prior to the experiment and then stimulated with 40 μM IP3 at the time as indicated. The results showed that PPAR-β knockdown affected [Ca2+]M transients in ES-derived neural progenitor cells (Fig 5A). IP3-evoked [Ca2+]M transients recorded in sh-control ES-derived neural progenitor cells appeared with a sharp peak, suggesting that the ability of the [Ca2+]M buffering still kept normal. In contrast, the same ability almost disappeared in sh-PPAR-β group. [Ca2+]M concentration was revealed by co-staining of Rhod-2AM with Mito green (Fig 5B). Compared to sh-control group, sh-PPAR-β decreased the fluorescence intensity of Mito green and Rhod-2, indicating that mitochondrial activity and [Ca2+]M concentration was downregulated by sh-PPAR-β. The results were consistent with the previous finding that mitochondrial biogenesis and Mfn2 were affected by sh-PPAR-β. [Ca2+]M could stimulate three rate-limiting enzymes in the Krebs cycle, resulting in accelerated ATP production [30]. Although glucose consumption in ES-derived neural progenitor cells was not affected by sh-PPAR-β (Fig 5C), energy metabolism pathway was shifted. The results in Fig 5D showed that basal oxygen consumption rate (OCR) was decreased and extracellular acidification rate (ECAR) was increased in sh-PPAR-β ES-derived neural progenitor cells.
Fig 5. Effects of PPAR-β gene silencing on mitochondrial energy metabolism of 4a-induced neural progenitor cells.
A: Mitochondrial Ca2+ ([Ca2+]M) transients was measured with Rhod-2AM. ES-derived neural progenitor cells were loaded with 4 μM Rhod-2AM prior to the experiment and then stimulated with 40 μM IP3 at the time as indicated. B: [Ca2+]M concentration was revealed by co-staining of Rhod-2AM with Mito green. Bar = 10 μm. C: Glucose consumption was detected by a commercial glucose monitor. D: Basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in ES-derived neural progenitor cells were measured by a XF96 Extracellular Flux Analyzer, and were normalized to cell numbers. Statistical significance was set as *P<0.05 vs. sh-control group.
Discussion
Recent studies link changes in energy metabolism with the fate of ES cells [31, 32]. Relative shifts in metabolism from ES cells to lineage-specific differentiation place variable demands on mitochondrial biogenesis and activity for cell types with distinct energetic and biosynthetic requirements [32]. Neurons are excitable cells that need large amounts of energy to support their survival and functions [33]. By generating energy and regulating subcellular Ca2+, mitochondria may play essential roles in controlling fundamental processes in neuroplasticity, including neural differentiation, neurite outgrowth, and synaptic vesicle recycling [1]. Here we used flavonoid compound 4a as a probe to dissect the involvement of mitochondrial energy metabolism in neuronal differentiation of ES cells. We further demonstrated the role of PPAR-β on the underlying mechanism, and other related molecular targets including Mfn2 and mitochondrial Ca2+.
It has been shown that flavonoids are key compounds for the development of a new generation of therapeutic agents that are clinically effective in treating neurodegenerative diseases [34]. Our previous work found that ICT [22] and IBA [23] had significant neurogenesis-inducing activities. Based on the core molecular scaffolds, a synthetic flavonoid library was designed and synthesized, with 2”, 3”-unsaturated alkyl groups at C-8 position as well as hydroxy or methoxy groups at different positions [24]. A cell-based phenotypic screen was undertaken to identify chemical inducers of neuronal differentiation of P19 embryonal carcinoma (EC) and mouse ES cells (data not shown). Here flavonoid compound 4a faithfully facilitated ES cells to differentiate into neurons morphologically and functionally. Neural specific proteins were upregulated in a developmental way, providing the fundamentals for neurite outgrowth and synaptic vesicle recycling. Importantly, Nestin expression was promoted at early differentiation stage, indicating that the neurogenesis-inducing effect of compound 4a appeared as early as neural progenitor cells formation. The FM 1-43FX fluorescence intensity in ES-derived neurons induced by 4a was similar to that of cells treated with RA. Since synaptic vesicle recycling is a neuron-specific function, we confirmed compound 4a could induce functional neuronal differentiation.
PPARs, which serve as lipid-activated transcription factors, play key roles in the regulation of differentiation and mitochondrial energy metabolism. It is therefore we examined the role of PPARs on the underlying mechanism of compound 4a-induced neuronal differentiation. Expression and distribution of PPARs were revealed by double staining PPAR isotypes with neural progenitor marker Nestin. The results showed that PPAR-β had very good colocalization with Nestin-positive cells. Compound 4a could promote PPAR-β protein expression. Treatment with PPAR-β antagonist GSK0660 abolished the neurogenesis stimulatory effect of compound 4a with downregulation of neural specific proteins. Consistently, knockdown of PPAR-β blocked compound 4a induced neurogenesis of ES cells, indicating the important role of PPAR-β in neuronal differentiation.
Mitochondria help neurons to meet their high energy demands of proper neuronal function [3]. Furthermore, mitochondrial biogenesis, together with mitochondrial fission and fusion, helps the transmission of energy across long distances, which is particularly essential in neurons [3]. We then analyzed the downstream events of PPAR-β involved in compound 4a-induced neural progenitor cells. Mitochondrial biogenesis was upregulated by compound 4a treatment, which was altered by PPAR-β knockdown, suggesting a key role of PPAR-β in mitochondrial biogenesis. Mfn2 expression is crucial in mitochondrial metabolism through the maintenance of the mitochondrial network architecture [35]. Furthermore, Mfn2 could tether endoplasmic reticulum to mitochondria, thereby increasing the efficiency of mitochondrial Ca2+ uptake and ATP production [7–9]. Recent research demonstrated that Mfn2 was a PPAR-β-selective target, which might play an important role in regulating myocardial energy homeostasis [36]. In accordance with these findings, present study showed that compound 4a increased the protein expression of mitochondrial fusion protein Mfn2, which were abolished by sh-PPAR-β. Moreover, Mfn2 was positively detected in the Nestin-positive cells, indicating that a close relationship between PPAR-β and Mfn2 took an important role in early neuronal differentiation induced by compound 4a.
Mitochondrial Ca2+ could stimulate three rate-limiting enzymes in the Krebs cycle, as a key regulator of mitochondrial ATP production in mammalian cells [30]. By determining the intermediates of energy metabolism, we found that PPAR-β knockdown affected mitochondrial Ca2+ buffering activity and intracellular Ca2+ homeostasis. Besides, PPAR-β knockdown reduced mitochondrial Ca2+ concentration. Consistently, energy metabolism pathway in ES-derived neural progenitor cells was shifted. OCR was decreased and ECAR was increased in sh-PPAR-β ES-derived neural progenitor cells, although glucose consumption was not affected. Because OCR and ECAR are indicators of mitochondrial respiration and anaerobic glycolysis pathway, the shift of mitochondrial respiration to anaerobic glycolysis might result in less ATP production and failing to fulfill the adequate level of bioenergetic capacity of neuronal differentiation.
Conclusion
In summary, our observation demonstrated that by modulating mitochondrial energy metabolism through Mfn2 and mitochondrial Ca2+, PPAR-β took an important role in neuronal differentiation induced by flavonoid compound 4a. This study provides novel insights for the role of mitochondria in the differentiation of neurons from ES cells. Further work is needed to focus on the action mechanism between compound 4a and PPAR-β. The effect of other flavonoids on PPAR-β expression should also be investigated.
Materials and Methods
Retinoic acid (RA), dimethylsulfoxide (DMSO), FM 1-43FX, MTT, β-mercaptoethanol (β-ME), 4,6-Diamidino-2-phenylindole (DAPI), inositol 1,4,5-triphosphate (IP3), L165041 and GSK0660 were purchased from Sigma-Aldrich (St. Louis, MO, USA). DMEM medium, fetal bovine serum (FBS), B27 supplement, neuralbasal medium was obtained from Gibco BRL (Burlington, Ontario, Canada). Non-essential amino acids (NEAA) stock solution was purchased from Hyclon (Logan, USA). Recombinant mouse leukemia inhibitory factor (LIF) was obtained from Millipore (CA, USA). Antibodies were used as follows: β-tubulin III, neuronal nuclei (NeuN), and neurofilament 160 (NEFM) were purchased from Sigma-Aldrich (St. Louis, MO, USA); synaptophysin and GAPDH were products of Cell Signaling; Nestin was from Millipore (CA, USA); PPAR-α, -β, -γ, Mfn1 and Mfn2 were ordered from Abcam; PGC-1α, Fis1, Drp1 and OPA1 were purchased from Santa Cruz Biotechnology. Secondary antibodies were purchased from Multisciences. Rhod-2AM was purchased from Dojindo Molecular Technologies. MitoTracker Green was a product of Life technologies.
Cell culture and differentiation scheme
Mouse embryonic fibroblasts (MEF) cells were prepared as previously described [37, 38]. ICR mice (22±3 g) were obtained from the Experimental Animal Center, Zhejiang University, Hangzhou, China (License No.: scxk-Zhejiang-2004-0014). Mice were housed under 12 h light/12 h dark and 21±1°C conditions. To obtain fetuses, mice (3 female and 1 male) were housed together at 17:00. The following morning, when a copulation plug was detected, was defined as d 0 of gestation. All efforts were made to minimize the number of animals used, and their suffering. Fetuses were obtained from the mice on d 13 of gestation for the preparation of MEF cells. The MEF cells of generation three to generation five were used as feeder cells, which were treated with 1 mg/L mitomycin C for 1 h and plated at an appropriate density.
Mouse ES cells (D3 line, ATCC, CRL-1934) were routinely cultured on MEF cells in DMEM, supplemented with 10% FBS, 0.1 mM β-ME, 1×NEAA and 1×106 U/L LIF. To induce neuronal differentiation, LIF was withdrawn from the medium and a typical 4−/4+ protocol was used as previously described [22, 39]. Briefly, drops (30 μL) containing about 900 ES cells were placed onto the lids of Petri dishes and the cells were cultured in hanging drops for 2 days. After EBs formed, they were transferred into agar coated Petri dishes and continued to be suspension cultured for another 2 days. On d 4, compound 4a (final, 10−7 mol/L) was added into the medium and the EBs were continually suspension cultured for another 4 days. On d 8, EBs was planted onto poly-D-lysine-coated culture plates to induce the differentiation in neurobasal medium supplemented with B27, with the treatment of each compound. The culture treated with 10−7 mol/L RA was considered as positive control and 0.1% DMSO was set as solvent control.
To investigate whether PPAR-β agonist or antagonist would affect the neurogenesis of ES cells, EBs were treated with either L165041 (10−5 mol/L) or GSK0660 (10−8 mol/L) as previously described [10].
Immunocytochemistry analysis
Immunostaining was performed on neuronal cultures (d 8+10) at the end of differentiation course and frozen EBs (d 8) section as previously described [22, 39]. Neuronal cultures seeded on coverslips were washed with PBS solution and then fixed for 10 min in ice-cold methanol, while frozen EBs sections were fixed in 4% paraformaldehyde for 2 h [40]. Non-specific binding sites were blocked with 10% FBS in PBS for 1 h. The cells were incubated with primary antibodies at appropriate dilutions in 0.5% triton X-100 in PBS overnight at 4°C, and then with the corresponding fluorescent secondary antibodies at a dilution of 1:200 for 2 h. Nuclear staining was performed with 2 μg/mL DAPI and the immunostaining results were visualized by microscopic examination using Leica DMI 3000B or on a confocal microscope (Fluoview FV1000, Olympus). For neuronal cultures, primary antibodies were used at the following dilutions: β-tubulin III mouse monoclonal (1:1000) and rabbit polyclonal (1:1000), NeuN rabbit polyclonal (1:1000), synaptophysin rabbit polyclonal (1:100), NEFM mouse monoclonal (1:100). While for frozen EBs section, primary antibodies were used at the following dilutions: Nestin mouse monoclonal (1:1000), PPAR-α rabbit polyclonal (1:1000), PPAR-β rabbit polyclonal (1:500), PPAR-γ rabbit polyclonal (1:500), Mfn2 rabbit polyclonal (1:500), Drp1 rabbit polyclonal (1:1000). Negative controls were performed by omitting the primary antibody. Experiments were repeated independently at least three times.
Western blot analysis
Western blotting was performed as previously described [22, 39]. The cells were collected in RIPA buffer and lysed 30 min on ice. The lysates were centrifuged at 14000 rpm for 30 min at 4°C. An aliquot of 40 μg of the supernatant protein from each sample separated electrophoretically on 8% SDS-PAGE. Subsequently, proteins were transferred onto PVDF membranes and blocked for 1 h in 5% nonfat milk in PBS, followed by an overnight incubation at 4°C with respective antibody. The specific dilutions of the primary antibodies were as follows: Nestin (1:1000), β-tubulin III (1:1000), NEFM (1:500), synaptophysin (1:1000), PPAR-α (1:1000), PPAR-β (1:1000), PPAR-γ rabbit polyclonal (1:1000), PGC-1α (1:1000), Fis (1:1000), Drp1 (1:1000), OPA1 (1:1000), Mfn1 (1:1000), Mfn2 (1:1000) and GAPDH (1:10000). Then the membranes were incubated with HRP-conjugated secondary antibody (1:400). The proteins were visualized with an enhanced chemiluminescent substrate. The density of the products was quantitated using Quantity One version 4.6.2 software (Bio-Rad).
FM 1-43FX fluorescent analysis
FM 1-43FX fluorescent analysis protocol was used as previously described [28, 41]. In brief, at the end of neuronal differentiation course, remove the culture medium from neuronal culture. Wash the cells for 30 s two times with 100 μl Krebs-Ringer buffer (115 mM NaCl, 5.9 mM KCl, 1.2 mM MgCl2, 1.2 mM NaH2PO4, 1.2 mM Na2SO4, 2.5 mM CaCl2, 25 mM NaHCO3 and 10 mM glucose) prewarmed to 37°C. Add 100 μl of prewarmed Krebs-Ringer buffer containing 100 mM potassium ions (20.9 mM NaCl, 100 mM KCl, 1.2 mM MgCl2, 1.2 mM NaH2PO4, 1.2 mM Na2SO4, 2.5 mM CaCl2, 25 mM NaHCO3 and 10 mM glucose) and 2 mM FM 1-43FX to the cells. Incubate the cells for 5 min at 37°C. Wash the cells for 30 s three times with 100 μl Krebs-Ringer buffer to remove excess FM 1-43FX. Add 100 μl Krebs-Ringer buffer to the cells. Measure the fluorescent intensity of the cells using DTX-880 Multimode Detector. The results across each well were then normalized by the MTT assay.
MTT assay
MTT labeling mixture (final, 0.5 mg/mL) was added to each well and incubated for additional 4 h. The formazan precipitate was dissolved in 200 μL DMSO and measured with a DTX-880 Multimode Detector at 570 nm. Assays were performed in triplicate in three independent experiments.
RNA silencing
Mouse PPAR-β shRNAs (sh-PPAR-β) with the sequences, 5’-GGAGCATCCTCACCGGC-AA-3’ and 5’-GCAGCTGGTCACTGAGCAT-3’[42] were constructed into pGPU6/GFP/Neo plasmid by GenePharm. In the PPAR-β knockdown experiments, these two plasmids were used at a 1:1 ratio. Scrambled shRNA (sh-control) with the sequence 5’-GTTCTCCGAA-CGTGTCACGT-3’ was used as negative control. ES cells were transfected with either sh-PPAR-β plasmid or sh-control plasmid (2 μg/mL) with lipofectamine 2000 according to the manufacturer protocol. GFP expression of ES cells was examined 48 h after transfection. To assess gene silencing, protein level of PPAR-β was determined by western blot 72 h after transfection. Viability and pluripotency of ES cells was also evaluated after transfection with sh-PPAR-β plasmid.
[Ca2+]M measurements with Rhod-2
ES-derived neural progenitor cells were loaded with 4 μmol/L Rhod-2AM [43] prior to the experiment and then stimulated with 40 μmol/L IP3 at the time as indicated. Single cell fluorescence was excited at 545nm and images of the emitted fluorescence obtained by a cooled CCD camera mounted on the microscope equipped with a polychromator (IX S1, Olympus). All analyses of [Ca2+]M transients were processed at a single-cell level and expressed as the relative fluorescence intensity. [Ca2+]M concentration was also revealed by co-staining of Rhod-2AM (4 μmol/L) with Mito green (200 nM) [44]. The images were visualized by confocal microscope (Fluoview FV1000, Olympus).
Glucose consumption
Cell culture supernatants of a 48 h culture of ES-derived neural progenitor cells were used to quantify glucose consumption using a commercial glucose monitor (Accu-Chek, Roche). Amounts of glucose consumption were normalized to the number of cells [45].
OCR and ECAR measurements
Oxygen consumption rate (OCR) and extra-cellular acidification rate (ECAR) were measured in ES-derived neural progenitor cells with a XF96 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA, USA) as previously described [46]. Cells were seeded in 12 wells of a XF 96-well cell culture microplate (Seahorse Bioscience) at a density of 104 cells/well in 200 μL of DMEM and incubated for 24 h at 37°C in 5% CO2 atmosphere. After replacing the growth medium with 180 μL of bicarbonate-free DMEM prewarmed at 37°C, cells were preincubated for 30 min before starting the assay procedure. Data were expressed as pmol of O2 per minute and normalized to the number of cells.
Statistical analysis
Data are expressed as mean values ± standard deviation (S.D.). At least three independent experiments were done. Statistical analysis was performed by Student’s t-test and one-way ANOVA. A value of P<0.05 was considered to be significant.
Supporting Information
(TIFF)
(TIFF)
(TIF)
(TIFF)
(TIFF)
(TIFF)
Acknowledgments
We are very grateful to Prof. Dr. Erqing Wei for supporting Ca2+ transient measurement.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Natural Science Foundation of China (№ 81573513, № 81173135, № 90813026; http://www.nsfc.gov.cn ), and by Zhejiang Provincial Natural Science Foundation of China (№ LZ12H31001, № Z2110655; http://www.zjnsf.gov.cn ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cheng A, Hou Y, Mattson MP. Mitochondria and neuroplasticity. ASN neuro. 2010;2(5):e00045 10.1042/AN20100019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birket MJ, Orr AL, Gerencser AA, Madden DT, Vitelli C, Swistowski A, et al. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. Journal of cell science. 2011;124(Pt 3):348–58. 10.1242/jcs.072272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nature reviews Neuroscience. 2008;9(7):505–18. 10.1038/nrn2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang P, Galloway CA, Yoon Y. Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLOS ONE. 2011;6(5):e20655 10.1371/journal.pone.0020655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detmer SA, Chan DC. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. The Journal of cell biology. 2007;176(4):405–14. 10.1083/jcb.200611080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130(3):548–62. 10.1016/j.cell.2007.06.026 . [DOI] [PubMed] [Google Scholar]
- 7.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–10. 10.1038/nature07534 . [DOI] [PubMed] [Google Scholar]
- 8.Merkwirth C, Langer T. Mitofusin 2 builds a bridge between ER and mitochondria. Cell. 2008;135(7):1165–7. 10.1016/j.cell.2008.12.005 . [DOI] [PubMed] [Google Scholar]
- 9.Liang X, Mei Y, Huang X, Shen G, Zhu D, Yu Y, et al. Junctophilin 2 knockdown interfere with mitochondrium status in ESC-CMs and cardiogenesis of ES cells. Journal of cellular biochemistry. 2012;113(9):2884–94. 10.1002/jcb.24164 . [DOI] [PubMed] [Google Scholar]
- 10.Zhu DY, Wu JY, Li H, Yan JP, Guo MY, Wo YB, et al. PPAR-beta facilitating maturation of hepatic-like tissue derived from mouse embryonic stem cells accompanied by mitochondriogenesis and membrane potential retention. Journal of cellular biochemistry. 2010;109(3):498–508. 10.1002/jcb.22426 . [DOI] [PubMed] [Google Scholar]
- 11.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. Journal of medicinal chemistry. 2000;43(4):527–50. . [DOI] [PubMed] [Google Scholar]
- 12.Ding L, Liang X, Zhu D, Lou Y. Peroxisome proliferator-activated receptor alpha is involved in cardiomyocyte differentiation of murine embryonic stem cells in vitro. Cell biology international. 2007;31(9):1002–9. 10.1016/j.cellbi.2007.03.013 . [DOI] [PubMed] [Google Scholar]
- 13.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135(2):798–800. 10.1210/endo.135.2.8033830 . [DOI] [PubMed] [Google Scholar]
- 14.Cimini A, Benedetti E, Cristiano L, Sebastiani P, D'Amico MA, D'Angelo B, et al. Expression of peroxisome proliferator-activated receptors (PPARs) and retinoic acid receptors (RXRs) in rat cortical neurons. Neuroscience. 2005;130(2):325–37. 10.1016/j.neuroscience.2004.09.043 . [DOI] [PubMed] [Google Scholar]
- 15.Aleshin S, Strokin M, Sergeeva M, Reiser G. Peroxisome proliferator-activated receptor (PPAR)beta/delta, a possible nexus of PPARalpha- and PPARgamma-dependent molecular pathways in neurodegenerative diseases: Review and novel hypotheses. Neurochemistry international. 2013;63(4):322–30. 10.1016/j.neuint.2013.06.012 . [DOI] [PubMed] [Google Scholar]
- 16.Hall MG, Quignodon L, Desvergne B. Peroxisome Proliferator-Activated Receptor beta/delta in the Brain: Facts and Hypothesis. PPAR research. 2008;2008:780452 10.1155/2008/780452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Loreto S, D'Angelo B, D'Amico MA, Benedetti E, Cristiano L, Cinque B, et al. PPARbeta agonists trigger neuronal differentiation in the human neuroblastoma cell line SH-SY5Y. Journal of cellular physiology. 2007;211(3):837–47. 10.1002/jcp.20996 . [DOI] [PubMed] [Google Scholar]
- 18.D'Angelo B, Benedetti E, Di Loreto S, Cristiano L, Laurenti G, Ceru MP, et al. Signal transduction pathways involved in PPARbeta/delta-induced neuronal differentiation. Journal of cellular physiology. 2011;226(8):2170–80. 10.1002/jcp.22552 . [DOI] [PubMed] [Google Scholar]
- 19.Yu S, Levi L, Siegel R, Noy N. Retinoic acid induces neurogenesis by activating both retinoic acid receptors (RARs) and peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta). The Journal of biological chemistry. 2012;287(50):42195–205. 10.1074/jbc.M112.410381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nature biotechnology. 2004;22(7):833–40. 10.1038/nbt987 . [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453(7193):338–44. 10.1038/nature07042 . [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Wang H, Wu J, Zhu D, Zhang X, Ou L, et al. Enhanced co-expression of beta-tubulin III and choline acetyltransferase in neurons from mouse embryonic stem cells promoted by icaritin in an estrogen receptor-independent manner. Chemico-biological interactions. 2009;179(2–3):375–85. 10.1016/j.cbi.2008.12.007 . [DOI] [PubMed] [Google Scholar]
- 23.Wang DY, Hu YZ, Kong SS, Yu YP, Zhu DY, Lou YJ. Promoting effects of isobavachin on neurogenesis of mouse embryonic stem cells were associated with protein prenylation. Acta pharmacologica Sinica. 2011;32(4):425–32. 10.1038/aps.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou L, Han S, Ding W, Chen Z, Ye Z, Yang H, et al. Design, synthesis and 3D-QSAR study of cytotoxic flavonoid derivatives. Molecular diversity. 2011;15(3):665–75. 10.1007/s11030-010-9289-7 . [DOI] [PubMed] [Google Scholar]
- 25.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116(1):201–11. . [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Zhang X, Wang H, Qi L, Lou Y. Neuroprotective effects of icaritin against beta amyloid-induced neurotoxicity in primary cultured rat neuronal cells via estrogen-dependent pathway. Neuroscience. 2007;145(3):911–22. 10.1016/j.neuroscience.2006.12.059 . [DOI] [PubMed] [Google Scholar]
- 27.Fletcher TL, Cameron P, De Camilli P, Banker G. The distribution of synapsin I and synaptophysin in hippocampal neurons developing in culture. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1991;11(6):1617–26. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams DR, Kim GH, Lee MR, Shin I. Fluorescent high-throughput screening of chemical inducers of neuronal differentiation in skeletal muscle cells. Nature protocols. 2008;3(5):835–9. 10.1038/nprot.2008.47 . [DOI] [PubMed] [Google Scholar]
- 29.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Developmental cell. 2001;1(4):515–25. . [DOI] [PubMed] [Google Scholar]
- 30.Parekh A. Calcium signalling: mitofusins promote interorganellar crosstalk. Current biology: CB. 2009;19(5):R200–3. 10.1016/j.cub.2009.01.012 . [DOI] [PubMed] [Google Scholar]
- 31.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CAt, Ramalho-Santos J, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLOS ONE. 2011;6(6):e20914 10.1371/journal.pone.0020914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teslaa T, Teitell MA. Pluripotent stem cell energy metabolism: an update. The EMBO journal. 2015;34(2):138–53. 10.15252/embj.201490446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Pitta M, Mattson MP. Preventing NAD(+) depletion protects neurons against excitotoxicity: bioenergetic effects of mild mitochondrial uncoupling and caloric restriction. Annals of the New York Academy of Sciences. 2008;1147:275–82. 10.1196/annals.1427.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solanki I, Parihar P, Mansuri ML, Parihar MS. Flavonoid-based therapies in the early management of neurodegenerative diseases. Advances in nutrition. 2015;6(1):64–72. 10.3945/an.114.007500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. The Journal of biological chemistry. 2003;278(19):17190–7. 10.1074/jbc.M212754200 . [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Yin R, Liu J, Wang P, Wu S, Luo J, et al. Peroxisome proliferator-activated receptor delta regulates mitofusin 2 expression in the heart. Journal of molecular and cellular cardiology. 2009;46(6):876–82. 10.1016/j.yjmcc.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu DY, Lou YJ. Inducible effects of icariin, icaritin, and desmethylicaritin on directional differentiation of embryonic stem cells into cardiomyocytes in vitro. Acta pharmacologica Sinica. 2005;26(4):477–85. 10.1111/j.1745-7254.2005.00076.x . [DOI] [PubMed] [Google Scholar]
- 38.Qiu GT, Huang B, Tang SB, Jie HU, Liu ZX. The Investigation of Cell Culture Conditions Maintained Embryonic Stem Cells as Totipotent Cells. Acad J SUMS. 2000;21:100–3. [Google Scholar]
- 39.Wu J, Pan Z, Cheng M, Shen Y, Yu H, Wang Q, et al. Ginsenoside Rg1 facilitates neural differentiation of mouse embryonic stem cells via GR-dependent signaling pathway. Neurochemistry international. 2013;62(1):92–102. 10.1016/j.neuint.2012.09.016 . [DOI] [PubMed] [Google Scholar]
- 40.Yan H, Zhang X, Hu W, Ma J, Hou W, Zhang X, et al. Histamine H3 receptors aggravate cerebral ischaemic injury by histamine-independent mechanisms. Nature communications. 2014;5:3334 10.1038/ncomms4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams DR, Lee MR, Song YA, Ko SK, Kim GH, Shin I. Synthetic small molecules that induce neurogenesis in skeletal muscle. Journal of the American Chemical Society. 2007;129(30):9258–9. 10.1021/ja072817z . [DOI] [PubMed] [Google Scholar]
- 42.Kleiner S, Nguyen-Tran V, Bare O, Huang X, Spiegelman B, Wu Z. PPAR{delta} agonism activates fatty acid oxidation via PGC-1{alpha} but does not increase mitochondrial gene expression and function. The Journal of biological chemistry. 2009;284(28):18624–33. 10.1074/jbc.M109.008797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fonteriz RI, de la Fuente S, Moreno A, Lobaton CD, Montero M, Alvarez J. Monitoring mitochondrial [Ca(2+)] dynamics with rhod-2, ratiometric pericam and aequorin. Cell calcium. 2010;48(1):61–9. 10.1016/j.ceca.2010.07.001 . [DOI] [PubMed] [Google Scholar]
- 44.Coatesworth W, Bolsover S. Spatially organised mitochondrial calcium uptake through a novel pathway in chick neurones. Cell calcium. 2006;39(3):217–25. 10.1016/j.ceca.2005.10.012 . [DOI] [PubMed] [Google Scholar]
- 45.Fornazari M, Nascimento IC, Nery AA, da Silva CC, Kowaltowski AJ, Ulrich H. Neuronal differentiation involves a shift from glucose oxidation to fermentation. Journal of bioenergetics and biomembranes. 2011;43(5):531–9. 10.1007/s10863-011-9374-3 . [DOI] [PubMed] [Google Scholar]
- 46.Invernizzi F, D'Amato I, Jensen PB, Ravaglia S, Zeviani M, Tiranti V. Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells. Mitochondrion. 2012;12(2):328–35. 10.1016/j.mito.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(TIF)
(TIFF)
(TIFF)
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.