Abstract
An effective medium designated phosphate separately autoclaved Reasoner’s 2A supplemented with cycloheximide and tobramycin (PSR2A-C/T) has been developed for the isolation of Flavobacterium and Chryseobacterium strains from the plant rhizosphere. It consists of Reasoner’s 2A agar (R2A) prepared by autoclaving phosphate and agar separately and supplementing with 50 mg L−1 cycloheximide and 1 mg L−1 tobramycin. A comparison was made among the following nine media: PSR2A-C/T, PSR2A-C/T supplemented with NaCl, R2A agar, R2A agar supplemented with cycloheximide and tobramycin, 1/4-strength tryptic soy agar (TSA), 1/10-strength TSA, soil-extract agar, Schaedler anaerobe agar (SAA), and SAA supplemented with gramicidin, for the recovery of Flavobacterium and Chryseobacterium strains from the Welsh onion rhizosphere. Flavobacterium strains were only isolated on PSR2A-C/T, and the recovery rate of Chryseobacterium strains was higher from PSR2A-C/T than from the eight other media. In order to confirm the effectiveness of PSR2A-C/T, bacteria were isolated from onion rhizosphere soil with this medium. Flavobacterium and Chryseobacterium strains were successfully isolated from this sample at a similar rate to that from the Welsh onion rhizosphere.
Keywords: Flavobacterium, Chryseobacterium, Isolation medium, Rhizosphere soil
The plant rhizosphere harbors a large bacterial community consisting of taxonomically and functionally diverse species that play pivotal roles in many processes, such as nitrogen fixation, mineralization, the decomposition of organic matter, and degradation of hazardous and xenobiotic chemicals. In addition, some rhizobacterial isolates affect the growth and ecological fitness of their host plants (2, 46). The biological control of soil-borne plant diseases, bioremediation, and biofertilizers has been attracting increasing attention, and a more detailed understanding of the biological functions of rhizobacteria and plant–rhizobacteria interactions is needed. Recent advances in culture-independent molecular ecological surveys have gradually been revealing bacterial communities in the plant rhizosphere. The phylum Bacteroidetes is one of the bacterial lineages that is abundant in the rhizospheres of various plants (7, 28, 31, 43, 56). For example, Gardner et al. (7) reported that Bacteroidetes bacteria accounted for more than 65% of the total bacterial population in the broccoli rhizosphere.
Among the members of Bacteroidetes, the family Flavobacteriaceae, particularly the genera Flavobacterium and Chryseobacterium, were found to be the most abundant rhizobacteria in the rhizospheres of a wide variety of plants, such as sweet potato (26), tomato (31), cucumber (43), and wheat (56).
The strains of Flavobacterium and Chryseobacterium isolated from soil are known to have beneficial properties for agricultural and industrial use. Plant growth-promoting (PGP) effects are considered to be one of the main agricultural uses of these strains. Several Flavobacterium strains have been reported to possess PGP capabilities, such as the solubilization of inorganic phosphate and production of indole-3-acetic acid (IAA) and 1-aminocyclopropane-1-carboxylate (ACC) deaminase (23, 38). Some Chryseobacterium strains have also been shown to synthesize IAA and ACC deaminase (25).
Another property of rhizobacteria that is of potential agricultural utility is plant protection against pathogens. Several strains of Flavobacterium produce antimicrobial compounds, such as 2,4-di-tert-butylphenol and hydrogen cyanide (HCN) (36, 38). Some Chryseobacterium strains have the ability to produce HCN and antifungal antibiotics and stimulate plant immune systems (17, 25, 32, 56). Strains of Flavobacterium and Chryseobacterium have been industrially used as bioremediation agents. As bioremediation/biodegradation agents, these strains have the ability to produce polysaccharide-degrading enzymes, such as chitinase (14), mannanase (33), and amylase (27, 49). Furthermore, Chryseobacterium strains have been shown to deamidate proteins, such as carboxybenzoxy (Cbz)-Gln-Gly, casein, and keratin, and produce proteases (8, 48, 55). Strains of Flavobacterium and Chryseobacterium are also considered to play important roles in the degradation of aromatic compounds, such as carbendazim (54), polycyclic aromatic hydrocarbons (21), and 2,4-dinitrotoluene in soil (50).
A significantly large number of species assigned to Flavobacterium and Chryseobacterium have more recently been isolated from the rhizospheres of Panax notoginseng (“F. notoginsengisoli”) (57), switchgrass (“F. nitrogenifigens”) (13), bell pepper (Flavobacterium sp. strain F52) (18), eggplant (C. solani) (6), cempedak (C. artocarpi) (45), maize (C. zeae) (12), and peanut (C. arachidis and C. geocarposphaerae) (12), implying that rhizosphere environments potentially harbor novel Flavobacterium and Chryseobacterium strains that may be useful for agricultural and industrial purposes. Our preliminary microbial community analysis using next-generation sequencing technology revealed that the rhizospheres of Allium plants contained a large number of Flavobacterium and Chryseobacterium strains (2.4–10.9% and 0.4–2.4% of total bacterial populations, respectively) (Nishioka, T., et al. 2014. Abstracts for Phytopathological Society of Japan Annual Meeting. p. 103, Hokkaido, in Japanese). Although many Flavobacterium and Chryseobacterium strains have been isolated by commonly used media, such as Reasoner’s 2A (R2A) agar and tryptic soy agar (TSA) (11, 16, 51, 52, 53), the development of selective culture media is necessary for the efficient retrieval of Flavobacterium and Chryseobacterium strains as microbial resources. Therefore, the objective of the present study was to optimize medium compositions for the isolation of Flavobacterium and Chryseobacterium strains from rhizosphere soil.
Materials and methods
Rhizosphere soil samples
Rhizosphere soils of the Welsh onion (Allium fistulosum L.) cv. Kujo-hoso and onion (A. cepa L.) cv. Sharm were used as the source materials for the isolation of bacteria. The Welsh onion and onion were sown in plug trays containing commercial potting medium (Negi-baido: Takii, Kyoto, Japan) and grown for 90 and 40 d, respectively. Seedlings of both plants were transplanted into plastic pots (15 cm in diameter, 8-cm deep) filled with soil collected from a field on the experimental farm of the university and were grown for 70 d in a greenhouse at 20–25°C under natural light with regular watering and fertilizing. Three plants were uprooted and loosely adhering soil was removed from the roots by gentle shaking for the collection of rhizosphere soil. Three grams of the pooled roots of each plant (1 g of the root of each plant body) was then suspended in sterile distilled water (SDW) and shaken for 15 min on a rotary shaker at 150 rpm. Serial soil dilutions of the samples were prepared in SDW and used for the surface plating of agar plates as described below.
Isolation trial 1: Comparisons of basal media
In this trial, six types of agar media were used: R2A agar (34), 1/4-strength TSA (Difco Laboratories, Detroit, MI, USA), 1/10-strength TSA, soil-extract agar (SEA) (22), Schaedler anaerobe agar (SAA) (Oxoid, Basingstoke, UK), and SAA supplemented with gramicidin (17.5 μg mL−1) (SAA-Gr) (37). R2A agar, 1/4-strength TSA, 1/10-strength TSA, and SEA are commonly used media for the isolation of a wide range of bacterial species including Bacteroidetes and SAA-Gr agar has been used to isolate Bacteroidetes bacteria from the gastrointestinal tract of mice.
A 100-μL aliquot of each serial dilution of Welsh onion rhizosphere soil was spread onto the surface of each medium in triplicate plates (90 mm in diameter). R2A agar and SEA plates were incubated at 25°C in the dark for 14 d. Plates of 1/4-strength and1/10-strength TSA were incubated at two different temperatures. One set of each medium plate was incubated at 25°C in the dark for 14 d and the other set was incubated at 30°C in the dark for 3 d. SAA and SAA-Gr plates were incubated at 35°C in the dark for 3 d. Well-isolated, discrete colonies showing a yellow/orange color on plates that yielded 10–100 colonies were selected and streaked using quadrant streaking on R2A agar plates (60 mm in diameter) because Flavobacterium and Chryseobacterium are known to produce bright yellow non-diffusible and non-fluorescent flexirubin pigments (1, 9, 39). After streaking, the plates were incubated at 25°C for 2 d. Single colonies were transferred to newly prepared R2A plates and incubated at 25°C for 3 d. Bacterial cells on the plates were mixed thoroughly with 1 mL of skim milk medium (100 g L−1 skim milk and 16.5 g L−1 L-glutamic acid monosodium salt), transferred into 1.5-mL Eppendorf tubes, and stored at −80°C until used.
Isolation trial 2: The effects of antibiotics
As described below, it was only possible to isolate Bacteroidetes bacteria with a yellow/orange color from R2A medium in isolation trial 1. Accordingly, R2A medium was selected as the basal isolation medium in subsequent isolation trials. Although colonies of Bacteroidetes bacteria appeared on R2A, this medium allowed the growth of contaminants such as fungi and orange pigment-producing sphingomonads belonging to the class Alphaproteobacteria. Accordingly, in this trial, R2A agar was supplemented with the fungicide cycloheximide (50 μg mL−1) and bacteriocide tobramycin (1 μg mL−1) (R2A-C/T). Diluted suspensions of Welsh onion rhizosphere soil were spread onto the surface of each medium in triplicate plates (90 mm in diameter) and incubated at 25°C for 14 d. After being incubated, bacteria with a yellow-orange color were isolated and stored as described above.
Isolation trial 3: The effects of separately autoclaved phosphate
Flavobacterium are sensitive to inhibitory compounds in media in which phosphate compounds are autoclaved together with agar (41). Accordingly, in this experiment, the ingredients of R2A medium were placed into two groups. Solution A was prepared using yeast extract (1.0 g L−1), tryptone (0.5 g L−1), peptone (1.5 g L−1), starch (1.0 g L−1), glucose (1.0 g L−1), and agar (30 g L−1). Solution B consisted of K2HPO4 (0.6 g L−1), MgSO4 (48 mg L−1), and sodium pyruvate (0.6 g L−1). Solutions A and B were autoclaved separately and then mixed at the same quantity in order to prepare the basal medium, designated phosphate separately autoclaved R2A (PSR2A) medium. Two types of R2A agar were prepared: R2A-C/T and PSR2A supplemented with cycloheximide (50 μg mL−1) and tobramycin (1 μg mL−1) (PSR2A-C/T). Diluted suspensions of Welsh onion rhizosphere soil were spread onto the surface of each medium in triplicate plates (90 mm in diameter) and incubated at 25°C for 14 d. After being incubated, bacteria with a yellow-orange color were isolated and stored as described above.
Isolation trial 4: The effects of sodium chloride
Two types of PSR2A were prepared: PSR2A-C/T and PSR2A supplemented with cycloheximide (50 μg mL−1), tobramycin (1 μg mL−1), and 1% NaCl (PSR2A-C/T/N). Diluted suspensions of Welsh onion and onion rhizosphere soil were spread onto the surface of each medium in triplicate plates and incubated at 25°C for 14 d. After being incubated, bacteria with a yellow/orange color were isolated and stored as described above.
16S rRNA gene sequence analysis
The genomic DNA of each isolate was prepared using PrepMan Ultra sample preparation reagent (Applied Biosystems, CA, USA) according to the manufacturer’s instructions. The 16S rRNA gene of each isolate was amplified with 27f and 1492r primers (20) and TaKaRa Ex Taq (Takara Bio, Otsu, Shiga, Japan). The amplification conditions were 94°C for 1 min (initial denaturation), followed by 25 cycles at 94°C for 1 min, at 55°C for 1 min, and at 72°C for 2 min, with a final extension step at 72°C for 8 min. The amplification products were purified using the GenElute PCR Clean-Up Kit (Sigma, MO, USA). Cycle sequencing was performed with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA), 27f or 517r primers (30), and an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The cycle sequencing conditions used were: at 96°C for 1 min (an initial denaturation), followed by 25 cycles at 96°C for 10 s, at 50°C for 5 s, and at 60°C for 4 min. Partial 16S rRNA gene sequences were compared using a BLAST search (http://ddbj.nig.ac.jp/blast/blastn) with the reference sequences in the DNA Data Bank of Japan (DDBJ) database. Information on the closest relatives of the 16S rRNA gene sequences of each strain was provided as supplemental Table 1. The partial 16S rRNA gene sequences (corresponding to bases 1 to 629 of Escherichia coli NBRC102203T of the 16S rRNA gene) obtained from 14 strains related to Flavobacterium and 19 strains related to Chryseobacterium, isolated from Welsh onion and onion rhizosphere soil on PSR2A-C/T in isolation trials 3 and 4, were aligned with reference sequences using CLUSTALW (42). Neighbor-joining trees were constructed with MEGA version 6.06 (40), and 1,000 bootstrap replicates were used to generate a consensus tree.
Nucleotide sequence accession numbers
The 16S rRNA gene sequences of 14 strains related to Flavobacterium and 19 strains related to Chryseobacterium, isolated from Welsh onion rhizosphere soil in isolation trials 3 and 4 and onion rhizosphere soil in isolation trial 4, on PSR2A-C/T were deposited in DDBJ with the following accession numbers: LC096252–LC096256, LC034259–LC034263, and LC034264–LC034267 (for Flavobacterium strains from Welsh onion rhizosphere soil in isolation trials 3 and 4, and onion rhizosphere soil in isolation trial 4, respectively), LC096244–LC096251, LC034273–LC034278, and LC034268–LC034272 (for Chryseobacterium strains from Welsh onion rhizosphere soil in isolation trials 3 and 4, and onion rhizosphere soil in isolation trial 4, respectively).
Results and discussion
Isolation trial 1: Comparisons of basal media
Six types of agar media were used to isolate bacteria from Welsh onion rhizosphere soil. The total bacterial colony counts in the triplicate plates of each medium are summarized in Table 1. Only yellow or orange-colored colonies were selected for further characterization by the 16S rRNA gene sequence analysis. The proportions of bacterial colonies with a yellow/orange color were markedly higher on R2A agar (39.1%) and 1/4-strength TSA (67.3%) than on the other media (0–1.8%). Accordingly, we identified 71 yellow/orange-colored isolates collected from R2A agar and 1/4-strength TSA plates based on partial 16S rRNA gene sequences (Table 2). None of the isolates obtained from 1/4-strength TSA medium were assigned to the phylum Bacteroidetes. In contrast, three genera of Bacteroidetes, including Chryseobacterium, were found on R2A agar medium. Based on the results from isolation trial 1, R2A agar was the most suitable basal medium for the isolation of Bacteroidetes strains.
Table 1.
Number of bacterial colonies on each medium in isolation trial 1.
| Medium | Culture conditions | Number of colonies | ||
|---|---|---|---|---|
|
|
|
|||
| Temperature | Periods | Yellow/orange | Total | |
| R2Aa | 25°C | 2 weeks | 38 | 97 |
| 1/4-strength TSAb | 25°C | 2 weeks | 1 | 59 |
| 1/4-strength TSA | 30°C | 3 d | 33 | 49 |
| 1/10-strength TSA | 25°C | 2 weeks | 2 | 109 |
| 1/10-strength TSA | 30°C | 3 d | 2 | 182 |
| SAAc | 35°C | 3 d | 0 | 66 |
| SAA-Grd | 35°C | 3 d | 0 | 43 |
| SEAe | 25°C | 2 weeks | 0 | 69 |
Reasoner’s 2A agar
Tryptic soy agar
Schaedler anaerobe agar
Schaedler anaerobe agar supplemented with gramicidin
Soil-extract agar
Table 2.
Bacterial isolates obtained from rhizosphere soil of Welsh onion and onion in isolation trial 1–4
| Phylum | Genus | Number of species on medium | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| From Welsh onion rhizosphere | From onion rhizosphere | ||||||||||
|
|
|
||||||||||
| Trial 1 | Trial 2 | Trial 3 | Trial 4 | ||||||||
|
|
|
|
|
||||||||
| R2Aa | 1/4 TSAb | R2A | R2A-C/Tc | R2A-C/T | PSR2A-C/Td | PSR2A-C/T | PSR2A-C/T/Ne | PSR2A-C/T | PSR2A-C/T/N | ||
| Bacteroidetes | Flavobacterium | 5 | 5 | 4 | |||||||
| Chryseobacterium | 1 | 1 | 4 | 8 | 6 | 1 | 5 | 3 | |||
| Niabella | 2 | 2 | 14 | 9 | 2 | 4 | 6 | 6 | |||
| Chitinophaga | 1 | 2 | 1 | 2 | |||||||
| Niastella | 1 | 2 | |||||||||
| Dyadobacter | 1 | ||||||||||
| Taibaiella | 1 | 1 | 1 | 2 | |||||||
| Flavitalea | 1 | ||||||||||
| Proteobacteria | Sphingomonas | 22 | 1 | 6 | 1 | ||||||
| Pseudoxanthomonas | 10 | 1 | |||||||||
| Ralstonia | 3 | 2 | |||||||||
| Sphingopyxis | 1 | 1 | 1 | ||||||||
| Frateuria | 1 | ||||||||||
| Luteibacter | 1 | ||||||||||
| Lysobacter | 1 | ||||||||||
| Rhodanobacter | 1 | ||||||||||
| Pseudomonas | 1 | ||||||||||
| Sphingobium | 1 | ||||||||||
| Stenotrophomonas | 1 | 2 | |||||||||
| Variovorax | 1 | ||||||||||
| Actinobacteria | Microbacterium | 3 | 20 | 1 | 5 | 2 | |||||
| Leifsonia | 1 | 3 | 2 | 5 | |||||||
| Mycobacterium | 1 | 1 | |||||||||
| Aeromicrobium | 1 | ||||||||||
| Cellulosimicrobium | 1 | ||||||||||
| Rhodococcus | 1 | ||||||||||
| Terrabacter | 1 | ||||||||||
| Agrococcus | 1 | ||||||||||
| Total yellow/orange bacterial colonies | 38 | 33 | 14 | 14 | 17 | 19 | 20 | 15 | 23 | 19 | |
| Total bacterial colonies | 97 | 49 | 121 | 106 | 169 | 183 | 248 | 201 | 132 | 155 | |
Reasoner’s 2A agar
1/4-strength tryptic soy agar
Reasoner’s 2A supplemented with cycloheximide and tobramycin
Phosphate separately autoclaved R2A supplemented with cycloheximide and tobramycin
Phosphate separately autoclaved R2A supplemented with cycloheximide, tobramycin, and NaCl
Isolation trial 2: The effects of antibiotics
Although R2A agar medium yielded the maximum number of Bacteroidetes-related strains (11% of yellow/orange-colored bacteria) among the six types of media tested, a significantly large number of strains associated with Proteobacteria (71%) and Actinobacteria (18%) were also retrieved. Furthermore, R2A agar medium allowed fungal contaminants (data not shown). These results led us to use the antibiotics tobramycin and cycloheximide in order to prevent the growth of Gram-negative bacteria and fungi, respectively. Tobramycin is a strong inhibitor of Gram-negative bacteria, including most Sphingomonas strains, but not most strains of Flavobacterium and Chryseobacterium (3–5, 15, 24, 44). Cycloheximide is known to be a strong inhibitor of a wide range of fungi and yeasts (35).
Fourteen yellow/orange-colored colonies appeared on the triplicate plates of R2A agar and R2A-C/T media (Table 2). In accordance with the results of isolation trial 1, six (43%) out of the 14 yellow/orange-colored isolates obtained from R2A agar were assigned to the genus Sphingomonas. In contrast, no Sphingomonas strains were found in R2A-C/T medium, and all of the yellow/orange-colored isolates were assigned to the genus Niabella of Bacteroidetes. Niabella strains are also known to produce flexirubin-type pigments (1, 47); therefore, difficulties are associated with distinguishing Niabella from Flavobacterium and Chryseobacterium based on colony color. Nevertheless, fungal contamination was prevented by using R2A-C/T plates (data not shown), suggesting that the addition of antibiotics is a convenient way by which to prevent the proliferation of rhizosphere-associated Proteobacteria and fungi.
One possible way to improve the recovery rate of target microorganisms on selective medium is to reduce environmental stress factors in the medium. Tanaka et al. (41) previously reported that certain inhibitory substances, such as hydrogen peroxide, were generated in media when phosphate compounds were autoclaved together with agar, and also that Flavobacterium may be highly sensitive to these inhibitory substances. Accordingly, R2A agar medium, prepared by autoclaving phosphate compounds and agar separately (designated PSR2A), was used as the basal medium in isolation trial 3.
Isolation trial 3: The effects of separately autoclaved phosphate
The colony counts of yellow/orange-colored bacteria were 17 and 19 on triplicate plates of R2A-C/T agar and PSR2A-C/T media, respectively (Table 2). Accordingly, no Sphingomonas strains were found on R2A-C/T medium, and all yellow/orange-colored isolates were assigned to Bacteroidetes. Although no isolates assigned to the genus Flavobacterium were recovered from R2A-C/T plates, 5 (26%) out of the 19 isolates obtained from PSR2A-C/T plates were affiliated with the genus Flavobacterium. In addition, the proportion of isolates assigned to the genus Chryseobacterium was higher in PSR2A-C/T plates (42%) than in R2A-C/T plates (24%).
Strains of Flavobacterium and Chryseobacterium are often isolated from seawater (10, 24, 29) whereas, to the best of our knowledge, there have been no reports of marine Niabella strains. Accordingly, we hypothesized that Flavobacterium and Chryseobacterium have a higher salt tolerance than Niabella strains, and, thus, NaCl may be used as an inhibitor of Niabella strains.
Isolation trial 4: The effects of sodium chloride
In this trial, two types of PSR2A media (PSR2A-C/T and PSR2A-C/T/N) were used for bacterial isolation. Serial dilutions of the rhizosphere soil of the Welsh onion were spread onto the surfaces of these media. The colony counts of the yellow/orange-colored bacteria were 20 and 15 on triplicate plates of PSR2A-C/T and PSR2A-C/T/N media, respectively (Table 2). Among the 20 isolates obtained from PSR2A-C/T plates, 16 (80%) were affiliated with the phylum Bacteroidetes. In contrast, nine (60%) out of the 15 isolates obtained from PSR2A-C/T/N plates were affiliated with the phylum Actinobacteria and only the remaining four isolates (27%) were assigned to the phylum Bacteroidetes. The proportion of the isolates assigned to the genus Chryseobacterium was higher in PSR2A-C/T plates (30%) than in PSR2A-C/T/N plates (7%). Isolates assigned to the genus Flavobacterium may only be recovered from PSR2A-C/T plates.
Bacteria were isolated from onion rhizosphere soil, which is also a rich source of Flavobacterium and Chryseobacterium (Nishioka, T., et al. 2014. Abstracts for Phytopathological Society of Japan Annual Meeting. p. 103, Hokkaido, in Japanese), on the above two media. The results obtained showed that 23 and 19 bacterial colonies with a yellow/orange color appeared on triplicate plates of PSR2A-C/T and PSR2A-C/T/N media, respectively (Table 2). Eighteen (78%) and 11 (58%) out of the yellow/orange-colored isolates obtained from the PSR2A-C/T and PSR2A-C/T/N plates, respectively, were affiliated with the Bacteroidetes phylum. The proportion of Chryseobacterium strains isolated from onion rhizosphere soil was higher in PSR2A-C/T plates (22%) than in PSR2A-C/T/N plates (16%), and Flavobacterium strains were only isolated from PSR2A-C/T plates. The reason why Flavobacterium strains did not appear on PSR2A-C/T/N medium was that rhizosphere-associated Flavobacterium may be sensitive to high concentrations of NaCl. Further studies on the physiology of Flavobacterium strains are needed in order to clarify this, and information on optimum NaCl concentrations for the growth of these strains is valuable for the selective isolation of Flavobacterium and Chryseobacterium strains from rhizosphere environments. In short, PSR2A-C/T is suitable as a medium for the isolation of Flavobacterium and Chryseobacterium from the rhizosphere. However, the isolation frequencies of the strains assigned to genus Niabella were relatively high from Welsh onion and onion rhizosphere soil. Further studies to reduce or prevent the growth of Niabella strains will be needed in order to improve the recovery rates of Flavobacterium and Chryseobacterium strains.
Identification of strains recovered using optimized medium
All Flavobacterium strains were clustered with the species originating in soil (F. johnsoniae NBRC 14942T, F. glycines NBRC 105008T, and F. daejeonense NBRC 106390T) (Fig. 1). Kolton et al. (18, 19) previously reported that the genome sizes of soil/rhizosphere-associated Flavobacterium strains (4.0–6.1 Mbp) were nearly twice the size of aquatic Flavobacterium strains (2.86–3.96 Mbp), implying that terrestrial Flavobacterium strains possess higher metabolic capacities (e.g., production of IAA, ACC deaminase, and antimicrobial compounds) than those of aquatic Flavobacterium strains. A comparative genomic analysis of Flavobacterium including the strains obtained in the present study is an important subject awaiting further investigation. Most Chryseobacterium strains were closely related to C. daecheongense NBRC 102008T and C. wanjuense NBRC 106394T (Fig. 1). Depolymerizing enzyme-producing Chryseobacterium strains have recently been discovered; for example, keratinase (8), mannanase (33), protease (48), and α-amylase (49). Biochemical analyses of enzymes produced by Chryseobacterium strains are required in order to ensure the usefulness of PSR2A-C/T medium for exploring beneficial microbial resources.
Fig. 1.
Phylogenetic relationships of partial 16S rDNA gene sequences obtained from strains related to Flavobacterium or Chryseobacterium isolated from Welsh onion and onion rhizosphere soil on phosphate separately autoclaved R2A supplemented with cycloheximide and tobramycin in isolation trials 3 and 4. The dendrogram was generated by the neighbor-joining method. The numbers on the branches represent the confidence intervals generated by bootstrapping with 1,000 replications; only values >60% are shown. The scale bar represents 0.1 substitutions per nucleotide position. Accession numbers are given in parentheses. Escherichia coli NBRC 102203T was used as an outgroup.
Conclusion
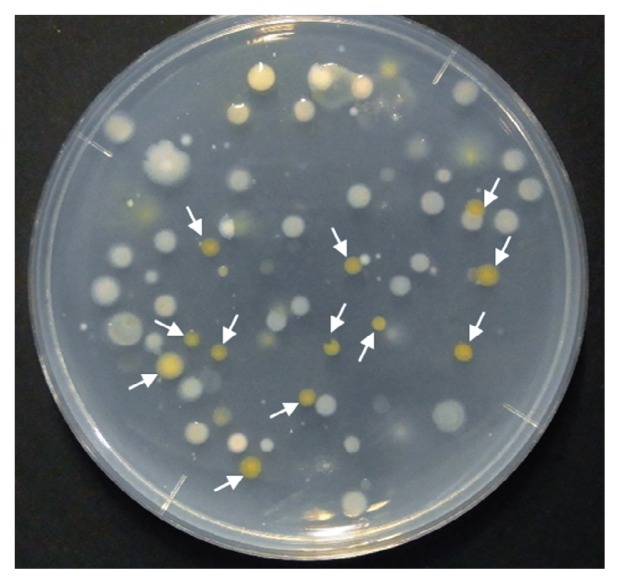
We herein succeeded in developing optimized medium for the isolation of Flavobacterium and Chryseobacterium strains from rhizosphere soil. The following conditions are necessary: 1) the use of R2A agar as the basal medium, 2) supplementation with tobramycin and cycloheximide in order to prevent the growth of Proteobacteria strains and fungi, 3) autoclave phosphate separately to reduce inhibitory substances, 4) an incubation at 25°C for 14 d, and 5) the selection of yellow/orange-pigmented colonies (Fig. 2). This study may open new opportunities for further discovery of the beneficial properties of Flavobacterium and Chryseobacterium strains.
Fig. 2.
Bacterial colonies on phosphate separately autoclaved R2A supplemented with cycloheximide and tobramycin. White arrows indicate the example of bacterial colonies actually selected in this study.
Supplementary Material
Acknowledgements
This work was supported by a Grant-in-Aid for Young Scientists (B) (24780317) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M. S.
References
- 1.Bernardet J.-F., Nakagawa Y., Holmes B. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol. 2002;52:1049–1070. doi: 10.1099/00207713-52-3-1049. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya P.N., Jha D.K. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol. 2012;28:1327–1350. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- 3.Bloom A.H., Perry H.D., Donnenfeld E.D., Davis R.G. Chryseobacterium meningosepticum keratitis. Am J Ophthalmol. 2003;136:356–357. doi: 10.1016/s0002-9394(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 4.Christakis G.B., Perlorentzou S.P., Chalkiopoulou I., Athanasiou A., Legakis N.J. Chryseobacterium indologenes non-catheter-related bacteremia in a patient with a solid tumor. J Clin Microbiol. 2005;43:2021–2023. doi: 10.1128/JCM.43.4.2021-2023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decostere A., Haesebrouck F., Devriese L.A. Shieh medium supplemented with tobramycin for selective isolation of Flavobacterium columnare (Flexibacter columnaris) from diseased fish. J Clin Microbiol. 1997;35:322–324. doi: 10.1128/jcm.35.1.322-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J., Hien T.T.N., Won K., Kim K.-Y., Jin F.-X., Yi T.-H. Chryseobacterium solani sp. nov. isolated from field-grown eggplant rhizosphere soil. Int J Syst Evol Microbiol. 2015 doi: 10.1099/ijs.0.000266. [DOI] [PubMed] [Google Scholar]
- 7.Gardner T., Acosta-Martínez V., Senwo Z., Dowd S.E. Soil rhizosphere microbial communities and enzyme activities under organic farming in Alabama. Diversity. 2011;3:308–328. [Google Scholar]
- 8.Gurav R.G., Jadhav J.P. Biodegradation of keratinous waste by Chryseobacterium sp. RBT isolated from soil contaminated with poultry waste. J Basic Microbiol. 2013;53:128–135. doi: 10.1002/jobm.201100371. [DOI] [PubMed] [Google Scholar]
- 9.Hugo C.J., Segers P., Hoste B., Vancanneyt M., Kersters K. Chryseobacterium joostei sp. nov., isolated from the dairy environment. Int J Syst Evol Microbiol. 2003;53:771–777. doi: 10.1099/ijs.0.02232-0. [DOI] [PubMed] [Google Scholar]
- 10.Imai I., Sunahara T., Nishikawa T., Hori Y., Kondo R., Hiroishi S. Fluctuations of the red tide flagellates Chattonella spp. (Raphidophyceae) and the algicidal bacterium Cytophaga sp. in the Seto Inland Sea, Japan. Mar Biol. 2001;138:1043–1049. [Google Scholar]
- 11.Johansen J.E., Nielsen P., Binnerup S.J. Identification and potential enzyme capacity of flavobacteria isolated from the rhizosphere of barley (Hordeum vulgare L.) Can J Microbiol. 2009;55:234–241. doi: 10.1139/w08-116. [DOI] [PubMed] [Google Scholar]
- 12.Kämpfer P., McInroy J.A., Glaeser S.P. Chryseobacterium zeae sp. nov., Chryseobacterium arachidis sp. nov., and Chryseobacterium geocarposphaerae sp. nov. isolated from the rhizosphere environment. Antonie van Leeuwenhoek. 2014;105:491–500. doi: 10.1007/s10482-013-0101-4. [DOI] [PubMed] [Google Scholar]
- 13.Kämpfer P., Busse H.-J., McInroy J.A., Xu J., Glaeser S.P. Flavobacterium nitrogenifigens sp. nov. isolated from switchgrass (Panicum virgatum) Int J Syst Evol Microbiol. 2015 doi: 10.1099/ijs.0.000330. [DOI] [PubMed] [Google Scholar]
- 14.Kharade S.S., McBride M.J. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol. 2014;196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khianngam S., Akaracharanya A., Lee J.-S., Lee K.C., Kim K.-W., Tanasupawat S. Flavobacterium arsenitoxidans sp. nov., an arsenite-oxidizing bacterium from Thai soil. Antonie Van Leeuwenhoek. 2014;106:1239–1246. doi: 10.1007/s10482-014-0294-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim B.-Y., Weon H.-Y., Cousin S., Yoo S.-H., Kwon S.-W., Go S.-J., Stackebrandt E. Flavobacterium daejeonense sp. nov. and Flavobacterium suncheonense sp. nov., isolated from greenhouse soils in Korea. Int J Syst Evol Microbiol. 2006;56:1645–1649. doi: 10.1099/ijs.0.64243-0. [DOI] [PubMed] [Google Scholar]
- 17.Kim H.-S., Sang M.K., Jung H.W., Jeun Y.-C., Myung I.-S., Kim K.D. Identification and characterization of Chryseobacterium wanjuense strain KJ9C8 as a biocontrol agent of Phytophthora blight of pepper. Crop Prot. 2012;32:129–137. [Google Scholar]
- 18.Kolton M., Green S.J., Harel Y.M., Sela N., Elad Y., Cytryn E. Draft genome sequence of Flavobacterium sp. strain F52, isolated from the rhizosphere of bell pepper (Capsicum annuum L. cv. Maccabi) J Bacteriol. 2012;194:5462–5463. doi: 10.1128/JB.01249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolton M., Sela N., Elad Y., Cytryn E. Comparative genomic analysis indicates that niche adaptation of terrestrial Flavobacteria is strongly linked to plant glycan metabolism. PLoS One. 2014;9 doi: 10.1371/annotation/3eafbdc9-374a-447a-852b-7a64503b8ccb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. Wiley & Sons; Chichester: 1991. pp. 115–175. [Google Scholar]
- 21.Lladó S., Gràcia E., Solanas A.M., Viñas M. Fungal and bacterial microbial community assessment during bioremediation assays in an aged creosote-polluted soil. Soil Biol Biochem. 2013;67:114–123. [Google Scholar]
- 22.Lutton E.M., Schellevis R., Shanmuganathan A. Increased culturability of soil bacteria from Marcellus shale temperate forest in Pennsylvania. J Stud Res. 2013;2:36–42. [Google Scholar]
- 23.Maimaiti J., Zhang Y., Yang J., Cen Y.-P., Layzell D.B., Peoples M., Dong Z. Isolation and characterization of hydrogen-oxidizing bacteria induced following exposure of soil to hydrogen gas and their impact on plant growth. Environ Microbiol. 2007;9:435–444. doi: 10.1111/j.1462-2920.2006.01155.x. [DOI] [PubMed] [Google Scholar]
- 24.Maravić A., Skočibušić M., Šamanić I., Puizina J. Profile and multidrug resistance determinants of Chryseobacterium indologenes from seawater and marine fauna. World J Microbiol Biotechnol. 2013;29:515–522. doi: 10.1007/s11274-012-1205-0. [DOI] [PubMed] [Google Scholar]
- 25.Marques A.P.G.C., Pires C., Moreira H., Rangel A.O.S.S., Castro P.M.L. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol Biochem. 2010;42:1229–1235. [Google Scholar]
- 26.Marques J.M., Silva T.F., Vollu R.E., Blank A.F., Ding G.-C., Seldin L., Smalla K. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol Ecol. 2014;88:424–435. doi: 10.1111/1574-6941.12313. [DOI] [PubMed] [Google Scholar]
- 27.McBride M.J., Xie G., Martens E.C., et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendes R., Kruijt M., de Bruijn I., et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 29.Michel C., Matte-Tailliez O., Kerouault B., Bernardet J.-F. Resistance pattern and assessment of phenicol agents’ minimum inhibitory concentration in multiple drug resistant Chryseobacterium isolates from fish and aquatic habitats. J Appl Microbiol. 2005;99:323–332. doi: 10.1111/j.1365-2672.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 30.Muyzer G., De Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ottesen A.R., Peña A.G., White J.R., et al. Baseline survey of the anatomical microbial ecology of an important food plant, Solanum lycopersicum (tomato) BMC Microbiol. 2013;13:114. doi: 10.1186/1471-2180-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos Solano B., Barriuso Maicas J., Pereyra De La Iglesia M.T., Domenech J., Gutiérrez Mañero F.J. Systemic disease protection elicited by plant growth promoting rhizobacteria strains: relationship between metabolic responses, systemic disease protection, and biotic elicitors. Phytopathology. 2008;98:451–457. doi: 10.1094/PHYTO-98-4-0451. [DOI] [PubMed] [Google Scholar]
- 33.Rattanasuk S., Ketudat-Cairns M. Chryseobacterium indologenes, novel mannanase-producing bacteria. Warasan Songkhla Nakkharin. 2009;31:395. [Google Scholar]
- 34.Reasoner D.J., Geldreich E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;9:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salkin I.F., Hurd N. Quantitative evaluation of the antifungal properties of cycloheximide. Antimicrob Agents Chemother. 1972;1:177–184. doi: 10.1128/aac.1.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sang M.K., Kim K.D. The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J Appl Microbiol. 2012;113:383–398. doi: 10.1111/j.1365-2672.2012.05330.x. [DOI] [PubMed] [Google Scholar]
- 37.Schaedler R.W., Dubos R., Costello R. The development of the bacterial flora in the gastrointestinal tract of mice. J Exp Med. 1965;122:59–66. doi: 10.1084/jem.122.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soltani A.-A., Khavazi K., Asadi-Rahmani H., Omidvari M., Dahaji P.A., Mirhoseyni H. Plant growth promoting characteristics in some Flavobacterium spp. isolated from soils of Iran. J Agric Sci. 2010;2:106–115. [Google Scholar]
- 39.Son H.-M., Kook M., Park S.-Y., Mavlonov G.T., Yi T.-H. Flavobacterium kyungheensis sp. nov., isolated from soil of a ginseng field. Antonie Van Leeuwenhoek. 2013;104:1029–1037. doi: 10.1007/s10482-013-0023-1. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka T., Kawasaki K., Daimon S., Kitagawa W., Yamamoto K., Tamaki H., Tanaka M., Nakatsu C.H., Kamagata Y. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microbiol. 2014;80:7659–7666. doi: 10.1128/AEM.02741-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian Y., Gao L. Bacterial diversity in the rhizosphere of cucumbers grown in soils covering a wide range of cucumber cropping histories and environmental conditions. Microb Ecol. 2014;68:794–806. doi: 10.1007/s00248-014-0461-y. [DOI] [PubMed] [Google Scholar]
- 44.Vaz-Moreira I., Nunes O.C., Manaia C.M. Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl Environ Microbiol. 2011;77:5697–5706. doi: 10.1128/AEM.00579-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venil C.K., Nordin N., Zakaria Z.A., Ahmad W.A. Chryseobacterium artocarpi sp. nov., isolated from the rhizosphere soil of Artocarpus integer. Int J Syst Evol Microbiol. 2014;64:3153–3159. doi: 10.1099/ijs.0.063594-0. [DOI] [PubMed] [Google Scholar]
- 46.Vessey J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255:571–586. [Google Scholar]
- 47.Wang H., Zhang Y.Z., Man C.X., Chen W.F., Sui X.H., Li Y., Zhang X.X., Chen W.X. Niabella yanshanensis sp. nov., isolated from the soybean rhizosphere. Int J Syst Evol Microbiol. 2009;59:2854–2856. doi: 10.1099/ijs.0.010447-0. [DOI] [PubMed] [Google Scholar]
- 48.Wang S.-L., Yang C.-H., Liang T.-W., Yen Y.-H. Optimization of conditions for protease production by Chryseobacterium taeanense TKU001. Bioresour Technol. 2008;99:3700–3707. doi: 10.1016/j.biortech.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 49.Wang S.-L., Liang Y.-C., Liang T.-W. Purification and characterization of a novel alkali-stable α-amylase from Chryseobacterium taeanense TKU001, and application in antioxidant and prebiotic. Process Biochem. 2011;46:745–750. [Google Scholar]
- 50.Wang Z.-Y., Ye Z.-F., Zhang M.-H. Bioremediation of 2,4-dinitrotoluene (2,4-DNT) in immobilized micro-organism biological filter. J Appl Microbiol. 2011;110:1476–1484. doi: 10.1111/j.1365-2672.2011.04997.x. [DOI] [PubMed] [Google Scholar]
- 51.Weon H.-Y., Kim B.-Y., Yoo S.-H., Kwon S.-W., Cho Y.-H., Go S.-J., Stackebrandt E. Chryseobacterium wanjuense sp. nov., isolated from greenhouse soil in Korea. Int J Syst Evol Microbiol. 2006;56:1501–1504. doi: 10.1099/ijs.0.64179-0. [DOI] [PubMed] [Google Scholar]
- 52.Weon H.-Y., Song M.-H., Son J.-A., Kim B.-Y., Kwon S.-W., Go S.-J., Stackebrandt E. Flavobacterium terrae sp. nov. and Flavobacterium cucumis sp. nov., isolated from greenhouse soil. Int J Syst Evol Microbiol. 2007;57:1594–1598. doi: 10.1099/ijs.0.64935-0. [DOI] [PubMed] [Google Scholar]
- 53.Weon H.-Y., Kim B.-Y., Yoo S.-H., Kwon S.-W., Stackebrandt E., Go S.-J. Chryseobacterium soli sp. nov. and Chryseobacterium jejuense sp. nov., isolated from soil samples from Jeju, Korea. Int J Syst Evol Microbiol. 2008;58:470–473. doi: 10.1099/ijs.0.65295-0. [DOI] [PubMed] [Google Scholar]
- 54.Xiao W., Wang H., Li T., Zhu Z., Zhang J., He Z., Yang X. Bioremediation of Cd and carbendazim co-contaminated soil by Cd-hyperaccumulator Sedum alfredii associated with carbendazim-degrading bacterial strains. Environ Sci Pollut Res. 2013;20:380–389. doi: 10.1007/s11356-012-0902-4. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi S., Yokoe M. A novel protein-deamidating enzyme from Chryseobacterium proteolyticum sp. nov., a newly isolated bacterium from soil. Appl Environ Microbiol. 2000;66:3337–3343. doi: 10.1128/aem.66.8.3337-3343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin C., Hulbert S.H., Schroeder K.L., Mavrodi O., Mavrodi D., Dhingra A., Schillinger W.F., Paulitz T.C. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.) Appl Environ Microbiol. 2013;79:7428–7438. doi: 10.1128/AEM.01610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang M.-Y., Xu H., Zhang T.-Y., Xie J., Cheng J., Nimaichand S., Li S.-H., Li W.-J., Zhang Y.-X. Flavobacterium notoginsengisoli sp. nov., isolated from the rhizosphere of Panax notoginseng. Antonie van Leeuwenhoek. 2015;108:545–552. doi: 10.1007/s10482-015-0509-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.