Abstract
We investigated the in situ gene expression profile of sulfur-turf microbial mats dominated by an uncultured large sausage-shaped Aquificae bacterium, a key metabolic player in sulfur-turfs in sulfidic hot springs. A reverse transcription-PCR analysis revealed that the genes responsible for sulfide, sulfite, and thiosulfate oxidation and carbon fixation via the reductive TCA cycle were continuously expressed in sulfur-turf mats taken at different sampling points, seasons, and years. These results suggest that the uncultured large sausage-shaped bacterium has the ability to grow chemolithoautotrophically and plays key roles as a primary producer in the sulfidic hot spring ecosystem in situ.
Keywords: sulfur-turf microbial mats, uncultured Aquificae bacterium, in situ gene expression, sulfide oxidation, primary production
Chemolithoautotrophic Aquificae bacteria are widely distributed and often dominate in various sulfidic geothermal environments such as terrestrial hot springs (5, 11, 27, 29, 35, 36) and deep-sea hydrothermal vents (20, 30, 34). Due to their abundance in sulfidic environments, these Aquificae bacteria are considered to be involved in the sulfur cycle and primary production in these environments.
The white microbial mat, the so-called “sulfur-turf” found in streams of sulfidic, circumneutral, and hypoxic hot spring waters worldwide (13, 17, 33, 40), is an assemblage of white filaments mainly composed of uncultured Aquificae bacteria phylogenetically related to the genus Sulfurihydrogenibium, and elemental sulfur particles or aragonite (13, 17). The dominant organism, often called a large sausage-shaped bacterium (LSSB) because of its conspicuous size (a cell length of 5–40 μm) and shape, has not yet been cultivated. Since the uncultured LSSB thrives in sulfidic (~0.1 mM) and organic-poor (<0.4 mg L−1 of total organic carbon) hot spring water streams and a large amount of elemental sulfur particles are precipitated around the cells (14, 15, 23, 26), sulfide is likely to be the main energy source for growth. However, the physiological features of the uncultured LSSB remain unclear because of its unculturability, which hampers all culturedependent analyses.
In order to clarify the metabolic functions of the uncultured LSSB, we recently conducted a draft genome analysis using the metagenomic library of a sulfur-turf collected from Nakabusa hot spring in Japan (39). The genomic data obtained showed that the uncultured LSSB possesses key genes associated with sulfide, sulfite, and thiosulfate oxidation, microaerobic respiration, and carbon fixation via the reductive TCA (rTCA) cycle, indicating the potential for chemolithoautotrophic growth. However, there has been no evidence to show that these predicted genes of the uncultured LSSB are indeed active in situ. In order to verify this, the present study investigated the in situ expression profiles of the genes associated with inorganic sulfur oxidation and carbon fixation of the uncultured LSSB in the sulfur-turf by using a reverse transcription (RT)-PCR approach based on the metagenomic data we obtained from the environment.
Sulfur-turf samples were collected from a sulfidic hot spring in Nakabusa, Japan (36°23.482N, 137°44.883E) in November 2011 and in July and November 2013. The sulfur-turfs developed on a concrete wall down to 70 cm from the discharge point of hot spring water. The temperature and pH of the stream at sampling points were measured using CT-280WR (CUSTOM, Tokyo, Japan) and a B-211 COMPACT pH METER (HORIBA, Kyoto, Japan), respectively. Samples were collected at three points within the same sulfur-turf using sterilized tweezers (Fig. 1). The collected samples were immediately washed once with 10 mM KH2PO4/K2HPO4 (pH 8.0) buffer and then immersed in RNAlater solution (Ambion, TX, USA). The samples were transported to our laboratory on ice within 8 h and stored in the laboratory at −20°C until used. Each sample was transferred to a Lysing Matrix E tube (MO-BIO, CA, USA), suspended with RLT buffer contained in the RNeasy Mini Kit (Qiagen, Hilden, Germany), and then disrupted by bead-beating using FastPrep FP120 (BIO 101 SAVANT, NY, USA). The suspension was centrifuged at 13,000×g at 4°C for 10 min and the supernatant was mixed with a 0.55 volume of ethanol. The mixture was transferred into an RNeasy mini spin column and subjected to further purification steps using an RNeasy Mini Kit according to the manufacturer’s instructions. A DNase treatment was performed using an RNase-free DNase Set (Qiagen) and TURBO DNase (Ambion) to further remove trace genomic DNA contamination. The reaction product was purified with phenol/chloroform (1:1 [v/v]) and then washed once with chloroform. Total RNA was precipitated with ethanol, washed with 70% (v/v) ethanol, and then resuspended in RNase-free water. Purified total RNA was analyzed by 0.8% (w/v) agarose gel electrophoresis in order to check for RNA degradation. RNA concentrations were quantified using a Qubit fluorometer (Invitrogen, CA, USA) and Quant-iT RNA Assay Kit (Invitrogen) according to the manufacturer’s instructions.
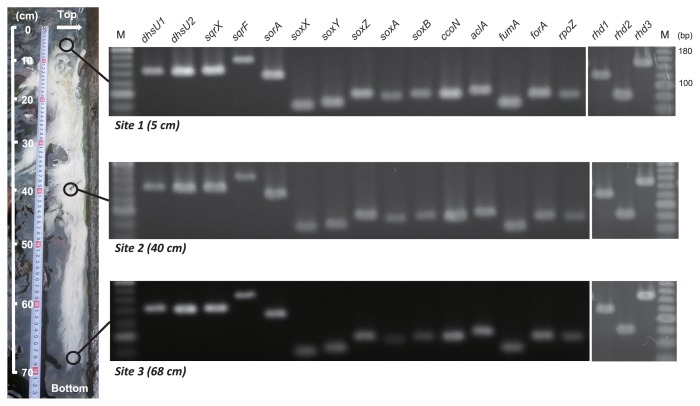
Fig. 1.
Photos of the sulfur-turf (white microbial mat) thriving in the hot spring stream and RT-PCR detection of transcripts of genes related to reduced sulfur compound oxidation, respiration, and carbon fixation. The white arrow shows the discharge point of the spring. Hot spring water flows downward from the top to the bottom in this figure. dhsU1 and dhsU2, sulfide dehydrogenase; sqrX and sqrF, sulfide-quinone reductase; sorA, sulfite dehydrogenase; soxX, soxY, soxZ, soxA and soxB, sox complex; rhd1, rhd2 and rhd3, thiosulfate sulfurtransferase; ccoN, cbb-3 type cytochrome c oxidase; aclA, ATP citrate lyase; fumA, fumarate hydratase; forA, 2-oxoglutarate ferredoxin oxidoreductase; rpoZ, RNA polymerase; M, DNA marker.
In our previous draft genome analysis of the uncultured LSSB, 1,472 ORFs were predicted (39). In order to obtain further insights into its metabolic potential, functional annotations were performed using the KEGG Automatic Annotation Server (KAAS) Ver. 2.0 (21) and Transporter Automatic Annotation Pipeline (TransAAP) (28). The genomic features revealed in this analysis did not positively support the possibility that the uncultured LSSB grows heterotrophically for the following reasons: (i) any gene for a primary enzyme in the glycolysis pathway (i.e. pyruvate kinase) was not found, although all genes associated with glycogenesis and the TCA cycle were identified (Fig. S1). (ii) Any transporters of sugars/oligosaccharides were not annotated by TransAAP (Table S1). (iii) The lack of genes coding pyruvate kinase and the transporters of sugars/oligosaccharides is not likely due to the incompleteness of the draft genome (90% completeness) of the uncultured LSSB because the complete genome of the chemolithoautotrophic sulfur oxidizer Sulfurihydrogenibium azorense, which is phylogenetically closely related to the uncultured LSSB, also lacks the genes for pyruvate kinase and transporters of sugars/oligosaccharides, and this strain is, indeed, unable to grow using any carbohydrates. Considering the in situ environmental conditions (i.e., the small amount of organic carbon in the discharged hot spring water) and rapid growth rate (doubling time of 80–92 min) of sulfur-turf reported previously (15, 23), it is difficult to infer that the uncultured LSSB grows heterotrophically in situ. Therefore, we focused on the in situ autotrophic growth activity of the uncultured LSSB, and attempted to verify whether the genes associated with inorganic sulfur oxidation and carbon fixation of the uncultured LSSB are expressed in the sulfur-turf in situ.
Based on the metagenomic data of the sulfur-turf collected at the same stream in our previous study (39), the targeted genes of the uncultured LSSB for the RT-PCR analysis were selected and specific primers for these genes were designed using Primer3 (16) as follows: sulfide oxidation-related genes (dhsU1 and dhsU2, sulfide dehydrogenase; sqrX and sqrF, sulfide-quinone reductase), a sulfite oxidation-related gene (sorA, sulfite dehydrogenase), thiosulfate oxidation-related genes (soxX, soxY, soxZ, soxA, and soxB, the sulfur oxidation (sox) system without soxC and soxD), a respiration-related gene (ccoN, cbb-3 type cytochrome c oxidase as a terminal oxidase working under microaerobic conditions), the genes responsible for carbon fixation via the reductive TCA cycle (aclA, ATP citrate lyase; fumA, fumarate hydratase; forA, 2-oxoglutarate ferredoxin oxidoreductase), and the gene encoding the RNA polymerase omega subunit (rpoZ) as a positive control (Table S2) (39). In addition to these genes, three genes encoding thiosulfate sulfurtransferase (rhd1, rhd2, and rhd3) were newly annotated in the draft genome in this study (GenBank accession numbers LC021536–LC021538, Table S2) and targeted for the RT-PCR analysis. In order to evaluate the specificity of the designed primers, PCR amplification using DNA extracted from a sulfur-turf as a template with TaKaRa Ex Taq Hot Start Version (TaKaRa, Otsu, Japan) was performed as follows: an initial denaturation step at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and a final extension step at 72°C for 5 min. The PCR product was analyzed by 3% (w/v) agarose gel electrophoresis. The remaining PCR product was purified, cloned, and sequenced, as described previously (39).
Two-step RT-PCR was performed using ReverTra Ace-α-(TOYOBO, Tokyo, Japan) with 0.18–0.25 μg of extracted total RNA and a random primer for RT, according to the manufacturer’s instructions. Following PCR using the gene-specific primers (Table S2), electrophoresis and sequencing of the PCR product were performed according to the above procedure.
The temperature and pH of the stream at three sampling points (5, 40, and 68 cm below the discharge point) were 54.2 to 52.2°C and 7.9 to 8.0, respectively. Using the sulfur-turf samples taken at each of the three points in the hot springs, we investigated in situ transcription profiles using a RT-PCR analysis. We designed gene-specific primers for these genes and confirmed their specificities by PCR amplification and sequencing. As a result, a single PCR band was successfully obtained from sulfur-turf DNA with the corresponding primer sets designed, and the sequences of the respective PCR products were identical to the target sequences in the draft genome of the uncultured LSSB (data not shown).
Using the designed primer sets, a RT-PCR analysis was performed on total RNA extracted from the three points of the sulfur-turf. The expression of all the genes tested was detected at all three positions of the sulfur-turf (Fig. 1). The amplification of negative controls, RT samples without reverse transcriptase, was not detected (data not shown). The sequences (three to eight clones) of each of the RT-PCR products were identical to the corresponding gene sequence in the uncultured LSSB draft genome (except for fumA, 1 mismatch found in 83 nt) (Fig. S2), indicating that the origin of the detected mRNA was the uncultured LSSB. These results suggest that the uncultured LSSB is able to acquire energy using reduced sulfur compounds as electron donors and oxygen as an electron acceptor in the sulfur-turf. Sulfide oxidation by the uncultured LSSB is strongly supported by a previous study demonstrating that sulfide was oxidized to elemental sulfur within a sulfur-turf dominated by the uncultured LSSB (18), while the oxidation of sulfite and thiosulfate by the uncultured LSSB has not yet been demonstrated and needs to be addressed by further physiological and biochemical characterizations. Furthermore, the uncultured LSSB may be capable of fixing CO2 through the rTCA cycle in the sulfur-turf. The expression of the genes related to thioautotrophic growth was observed not only at all three positions within the large sulfur-turf, but also in other sulfur-turf samples harvested in different seasons and years (in July and November 2013, data not shown). Considering this stable expression pattern and the abundance of the uncultured LSSB in the sulfur-turf (>90% of the whole bacterial community) (5, 39), it is likely that the uncultured LSSB plays important roles as a sulfur oxidizer and primary producer in situ.
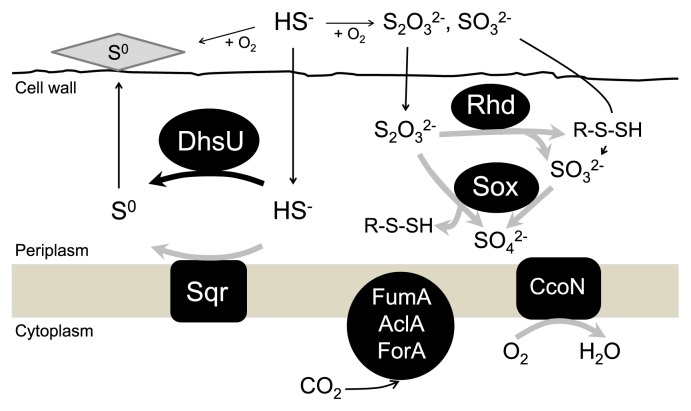
Based on the present in situ gene expression profile and previous genomic and physiological studies on Aquificae and other sulfur-oxidizing bacteria, we hypothesized the active sulfur-oxidation pathways of the uncultured LSSB in the microaerobic and sulfidic environments as follows (Fig. 2): the uncultured LSSB oxidizes sulfide continuously supplied from hot spring water to elemental sulfur by DhsU and Sqr, which have already been characterized biochemically as sulfide-oxidizing enzymes (19, 24, 39). The uncultured LSSB is unlikely to oxidize elemental sulfur in the environment because the abundant precipitation of elemental sulfur was observed around uncultured LSSB cells (39, 40). To date, there has been no evidence for the presence of genes responsible for oxidizing elemental sulfur not only in the draft genome of the uncultured LSSB (39), but also in the complete genomes of Sulfurihydrogenibium isolates (31), which also supports the possibility that the uncultured LSSB is unable to oxidize elemental sulfur in the environment. Since sulfide is constantly supplied from hot spring water, sulfide oxidation to elemental sulfur is considered to be the main sulfur oxidation pathway of the uncultured LSSB. On the other hand, sulfide is abiotically oxidized to thiosulfate, elemental sulfur, and sulfite in the presence of oxygen (3, 4), and the expression of genes involving thiosulfate oxidation was detected. Taken together, in addition to sulfide oxidation, the uncultured LSSB may also utilize thiosulfate as one of the energetic substrates. Two pathways have been proposed for thiosulfate utilization: oxidation to sulfate by an incomplete Sox system (7, 32) and disproportionation into sulfur in the form of sulfane sulfur and sulfite by Rhd (2, 8, 9) (Fig. 2). Thiosulfate oxidation to sulfate mediated by the Sox system likely occurs in situ because all the genes encoding the Sox system were found in the uncultured LSSB genome and expressed in the sulfur-turf. However, sulfite oxidation to sulfate by Sor currently remains unclear because a gene encoding SorA, which is a part of the SorAB two-subunit system, was found in the draft genome, whereas a gene encoding SorB was not annotated. Further biochemical and physiological studies are needed in order to elucidate the sulfite-oxidizing pathway of the uncultured LSSB in more detail. Nevertheless, the potential ability to utilize various reduced sulfur compounds as energetic substrates may provide the uncultured LSSB with an advantage to predominate in the sulfidic environment.
Fig. 2.
Hypothesized model of the active autotrophic sulfur-oxidation pathway in the uncultured LSSB in the in situ sulfur-turf ecosystem. The bold black arrow indicates the reaction that has been biochemically demonstrated in the uncultured LSSB and verified at the transcriptional level in this study. The bold gray arrows show the putative reactions verified at the transcriptional level in this study, but lacking direct biochemical evidence in the uncultured LSSB. The thin black arrows represent the flow of the substrates and products postulated. DhsU, sulfide dehydrogenase; Sqr, sulfide-quinone reductase; Sox, sox complex; Rhd, thiosulfate sulfurtransferase; CcoN, cbb-3 type cytochrome c oxidase; AclA, ATP citrate lyase; FumA, fumarate hydratase; ForA, 2-oxoglutarate ferredoxin oxidoreductase.
Recent metagenomic analyses showed the potential of Aquificae bacteria dominating in microbial mats or streamers to grow autotrophically by inorganic sulfur oxidation (13, 36). In recent years, Hamamura et al. also reported that the expression of some genes involved in thioautotrophic growth under low oxygen conditions was detected in Sulfurihydrogenibium-dominated microbial communities from geothermal springs in Yellowstone National Park in the United States (10). The analogies found in the geographically separated fields in Yellowstone National Park (USA) and Nakabusa hot spring field (Japan) imply that the dominance of the thioautotrophic lifestyle is a common feature of the microbial mat in the geothermal sulfidic ecosystem worldwide.
A number of strains within the phylum Aquificae have been isolated from various sulfidic environments, and reduced sulfur compound-dependent autotrophic growth is observed in some Aquificae isolates (1, 6, 12, 22, 25, 37, 38). The present study clearly demonstrated the in situ expression of the key genes involved in reduced sulfur compound oxidation and carbon fixation in the uncultured LSSB belonging to the phylum Aquificae by a transcriptional analysis. Our results provide insights into the ecophysiological roles of Aquificae bacteria dominating in various sulfidic environments.
Supplementary Material
Acknowledgements
We are grateful to Mr. Takahito Momose (the owner of Nakabusa Hot Spring) for supporting us in the sampling of sulfur-turfs. We greatly thank the guests staying at the Nakabusa hot spring together with us for their profound knowledge of hot springs and heartwarming encouragement.
References
- 1.Aguiar P., Beveridge T.J., Reysenbach A.L. Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int J Syst Evol Microbiol. 2004;54:33–39. doi: 10.1099/ijs.0.02790-0. [DOI] [PubMed] [Google Scholar]
- 2.Aussignargues C., Giuliani M-C., Infossi P., Lojou E., Guiral M., Giudici-Orticoni M-T., Ilbert M. Rhodanese functions as sulfur supplier for key enzymes in sulfur energy metabolism. J Biol Chem. 2012;24:19936–19948. doi: 10.1074/jbc.M111.324863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K.Y., Morris J.C. Kinetics of oxidation of aqueous sulfide by O2. Environ Sci Technol. 1972;6:529–537. [Google Scholar]
- 4.Cline J.D., Richards F.A. Oxygenation of hydrogen sulfide in seawater at constant salinity, temperature, and pH. Environ Sci Technol. 1969;3:838–843. [Google Scholar]
- 5.Everroad R.C., Otaki H., Matsuura K., Haruta S. Diversification of bacterial community composition along a temperature gradient at a thermal spring. Microbes Environ. 2012;27:374–381. doi: 10.1264/jsme2.ME11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores G.E., Liu Y., Ferrera I., Beveridge T.J., Reysenbach A.L. Sulfurihydrogenibium kristjanssonii sp. nov., a hydrogen- and sulfur-oxidizing thermophile isolated from a terrestrial Icelandic hot spring. Int J Syst Evol Microbiol. 2008;58:1153–1158. doi: 10.1099/ijs.0.65570-0. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich C.G., Bardischewsky F., Rother D., Quentmeier A., Fischer J. Prokaryotic sulfur oxidation. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Giuliani M-C., Tron P., Leroy G., Aubert C., Tauc P., Giudici-Orticoni M-T. A new sulfurtransferase from the hyperthermophilic bacterium Aquifex aeolicus. FEBS J. 2007;274:4572–4587. doi: 10.1111/j.1742-4658.2007.05985.x. [DOI] [PubMed] [Google Scholar]
- 9.Giuliani M-C., Jourlin-Castelli C., Leroy G., Hachani A., Giudici-Orticoni M-T. Characterization of a new periplasmic single-domain rhodanese encoded by a sulfur-regulated gene in a hyperthermophilic bacterium Aquifex aeolicus. Biochimie. 2010;92:388–397. doi: 10.1016/j.biochi.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Hamamura N., Meneghin J., Reysenbach A.L. Comparative community gene expression analysis of Aquificales-dominated geothermal springs. Environ Microbiol. 2013;15:1226–1237. doi: 10.1111/1462-2920.12061. [DOI] [PubMed] [Google Scholar]
- 11.Hjorleifsdottir S., Skirnisdottir S., Hreggvidsson G.O., Holst O., Kristjansson J.K. Species composition of cultivated and non-cultivated bacteria from short filaments in an Icelandic hot spring at 88 degrees C. Microb Ecol. 2001;42:117–125. doi: 10.1007/s002480000110. [DOI] [PubMed] [Google Scholar]
- 12.Huber R., Wilharm T., Huber D., et al. Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 13.Inskeep W.P., Rusch D.B., Jay Z.J., et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One. 2010;5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato K., Kobayashi T., Yamamoto H., Nakagawa T., Maki Y., Hoaki T. Microbial mat boundaries between chemolithotrophs and phototrophs in geothermal hot spring effluents. Geomicrobiol J. 2004;21:91–98. [Google Scholar]
- 15.Kimura H., Mori K., Nashimoto H., Hanada S., Kato K. In situ biomass production of a hot spring sulfur-turf microbial mat. Microbes Environ. 2010;25:140–143. doi: 10.1264/jsme2.me09181. [DOI] [PubMed] [Google Scholar]
- 16.Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 17.Maki Y. Study of the “sulfur-turf”: a community of colorless sulfur bacteria growing in hot spring effluent. Bulletin of Japanese Society of Microbial Ecology. 1991;6:33–43. [Google Scholar]
- 18.Maki Y. Rapid biological sulfide oxidation in the effluent of a hot spring. Bulletin of Japanese Society of Microbial Ecology. 1993;8:175–179. [Google Scholar]
- 19.Marcia M., Ermler U., Peng G., Michel H. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mino S., Makita H., Toki T., et al. Biogeography of Persephonella in deep-sea hydrothermal vents of the Western Pacific. Front Microbiol. 2013;4:107. doi: 10.3389/fmicb.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa S., Shtaih Z., Banta A., Beveridge T.J., Sako Y., Reysenbach A.L. Sulfurihydrogenibium yellowstonense sp. nov., an extremely thermophilic, facultatively heterotrophic, sulfur-oxidizing bacterium from Yellowstone National Park, and emended descriptions of the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int J Syst Evol Microbiol. 2005;55:2263–2268. doi: 10.1099/ijs.0.63708-0. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa T., Fukui M. Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. J Gen Appl Microbiol. 2002;48:211–222. doi: 10.2323/jgam.48.211. [DOI] [PubMed] [Google Scholar]
- 24.Nübel T., Klughammer C., Huber R., Hauska G., Schütz M. Sulfide:quinone oxidoreductase in membranes of the hyperthermophilic bacterium Aquifex aeolicus (VF5) Arch Microbiol. 2000;173:233–244. doi: 10.1007/s002030000135. [DOI] [PubMed] [Google Scholar]
- 25.O’Neill A.H., Liu Y., Ferrera I., Beveridge T.J., Reysenbach A.L. Sulfurihydrogenibium rodmanii sp. nov., a sulfur-oxidizing chemolithoautotroph from the Uzon Caldera, Kamchatka Peninsula, Russia, and emended description of the genus Sulfurihydrogenibium. Int J Syst Evol Microbiol. 2008;58:1147–1152. doi: 10.1099/ijs.0.65431-0. [DOI] [PubMed] [Google Scholar]
- 26.Otaki H., Everroad R.C., Matsuura K., Haruta S. Production and consumption of hydrogen in hot spring microbial mats dominated by a filamentous anoxygenic photosynthetic bacterium. Microbes Environ. 2012;27:293–299. doi: 10.1264/jsme2.ME11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell D., Sompong U., Yim L.C., Barraclough T.G., Peerapornpisal Y., Pointing S.B. The effects of temperature, pH and sulphide on the community structure of hyperthermophilic streamers in hot springs of northern Thailand. FEMS Microbiol Ecol. 2007;60:456–466. doi: 10.1111/j.1574-6941.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 28.Ren Q., Chen K., Paulsen I.T. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reysenbach A.L., Wickham G.S., Pace N.R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reysenbach A.L., Longnecker K., Kirshtein J. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl Environ Microbiol. 2000;66:3798–3806. doi: 10.1128/aem.66.9.3798-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reysenbach A.L., Hamamura N., Podar M., et al. Complete and draft genome sequences of six members of the Aquificales. J Bacteriol. 2009;191:1992–1993. doi: 10.1128/JB.01645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano R., Kameya M., Wakai S., Arai H., Igarashi Y., Ishii M., Sambongi Y. Thiosulfate oxidation by a thermo-neutrophilic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Biosci Biotechnol Biochem. 2010;74:892–894. doi: 10.1271/bbb.90948. [DOI] [PubMed] [Google Scholar]
- 33.Skirnisdottir S., Hreggvidsson G.O., Hjorleifsdottir S., Marteinsson V.T., Petursdottir S.K., Holst O., Kristjansson J.K. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl Environ Microbiol. 2000;66:2835–2841. doi: 10.1128/aem.66.7.2835-2841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sylvan J.B., Toner B.M., Edwards K.J. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. MBio. 2012;3:e00279-11. doi: 10.1128/mBio.00279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takacs C.D., Ehringer M., Favre R., Cermola M., Eggertsson G., Palsdottir A., Reysenbach A.L. Phylogenetic characterization of the blue filamentous bacterial community from an Icelandic geothermal spring. FEMS Microbiol Ecol. 2001;35:123–128. doi: 10.1111/j.1574-6941.2001.tb00795.x. [DOI] [PubMed] [Google Scholar]
- 36.Takacs-Vesbach C., Inskeep W.P., Jay Z.J., et al. Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three aquificales lineages. Front Microbiol. 2013;4:84. doi: 10.3389/fmicb.2013.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takai K., Hirayama H., Sakihama Y., Inagaki F., Yamato Y., Horikoshi K. Isolation and metabolic characteristics of previously uncultured members of the order aquificales in a subsurface gold mine. Appl Environ Microbiol. 2002;68:3046–3054. doi: 10.1128/AEM.68.6.3046-3054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takai K., Kobayashi H., Nealson K.H., Horikoshi K. Sulfurihydrogenibium subterraneum gen. nov., sp. nov., from a subsurface hot aquifer. Int J Syst Evol Microbiol. 2003;53:823–827. doi: 10.1099/ijs.0.02506-0. [DOI] [PubMed] [Google Scholar]
- 39.Tamazawa S., Takasaki K., Tamaki H., Kamagata Y., Hanada S. Metagenomic and biochemical characterizations of sulfur oxidation metabolism in uncultured large sausage-shaped bacterium in hot spring microbial mats. PLoS One. 2012;7:e49793. doi: 10.1371/journal.pone.0049793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto H., Hiraishi A., Kato K., Chiura H.X., Maki Y., Shimizu A. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl Environ Microbiol. 1998;64:1680–1687. doi: 10.1128/aem.64.5.1680-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.