Abstract
The effects of pharmaceuticals and personal care products (PPCPs) on aquatic organisms represent a significant current concern. Herein, a targeted metabolomics approach using liquid chromatography-high resolution mass spectrometry (LC-HRMS) is presented to characterise concentration changes in 29 selected metabolites following exposures of aquatic invertebrates, Gammarus pulex, to pharmaceuticals. Method performance revealed excellent linearity (R2 > 0.99), precision (0.1–19%) and lower instrumental limits of detection (0.002–0.20 ng) for all metabolites studied. Three pharmaceuticals were selected representing the low, middle and high range of measured acute measured toxicities (of a total of 26 compounds). Gammarids were exposed to both the no-observed-adverse-effect-level (NOAEL) and the lowest-observed-adverse-effect-level (LOAEL) of triclosan (0.1 and 0.3 mg L− 1), nimesulide (0.5 and 1.4 mg L− 1) and propranolol (100 and 153 mg L− 1) over 24 h. Quantitative metabolite profiling was then performed. Significant changes in metabolite concentrations relative to controls are presented and display distinct clustered trends for each pharmaceutical. Approximately 37% (triclosan), 33% (nimesulide) and 46% (propranolol) of metabolites showed statistically significant time-related effects. Observed changes are also discussed with respect to internal concentrations of the three pharmaceuticals measured using a method based on pulverised liquid extraction, solid phase extraction and LC-MS/MS. Potential metabolic pathways that may be affected by such exposures are also discussed. This represents the first study focussing on quantitative, targeted metabolomics of this lower trophic level benthic invertebrate that may elucidate biomarkers for future risk assessment.
Keywords: Pharmaceuticals, LC-HRMS, Gammarus pulex, Aquatic toxicology, Metabolomics
Graphical abstract
Highlights
-
•
Metabolites of Gammarus pulex were determined following liquid chromatography coupled high resolution mass spectrometry.
-
•
The toxicity study of 26 pharmaceuticals in Gammarus pulex show values between 0.57 mg L− 1 to > 250 mg L− 1.
-
•
Variations in the metabolite concentrations were detected in the pharmaceuticals exposed samples respect to control samples.
-
•
Pathway alterations related to protein synthesis, oxidative stress and signaling cascades were observed in exposed samples.
1. Introduction
In the last decade, pharmaceuticals have been recognised as an emerging class of environmental contaminants and their fate, occurrence and physicochemical behaviour in the aquatic environment have been extensively studied and reviewed (Daughton and Ternes, 1999, Evgenidou et al., 2015, Monteiro and Boxall, 2010). The ecotoxicological consequences of incomplete removal of pharmaceuticals or their metabolites in wastewater or drinking water treatment plants (WWTP/DWTP) are a matter of current environmental concern which still requires further research (Gómez-Canela et al., 2013). Furthermore, pharmaceuticals designed for hospital use are suspected to have more adverse effects than other pharmaceuticals regarding their effect on the aquatic environment (Gómez-Canela et al., 2014, Franquet-Griell et al., 2015). As such, in the last decade, much research has focused on understanding the occurrence and effects (Heberer, 2002) of these contaminants in exposed organisms. Recently, we developed a multi-residue analytical method and determined occurrence of pharmaceuticals in tributaries of the River Thames and in Gammarus pulex (G. pulex) at ng L− 1 and ng g− 1 concentrations respectively across eight sites (Miller et al., 2015). G. pulex is a small, low trophic level species of amphipod crustacean found in freshwaters across Europe and is very common throughout the United Kingdom (UK). G. pulex has many attributes for use in biomonitoring studies including its important role in freshwater food chains where they serve as a food source for other invertebrates, fish and birds (Maltby et al., 2002). This organism has also been extensively used in contaminant monitoring including toxicity assays for various pollutants such as metals, pharmaceuticals, PAHs/PCBs and natural stressors which has shown the importance of this species in environmental risk assessment (Bourgeault et al., 2013, Coulaud et al., 2011, De Lange et al., 2006, De Lange et al., 2009, Lebrun et al., 2012, Maltby et al., 2002, Pellet et al., 2014, Schaller et al., 2011, Vellinger et al., 2012).
Studies of the effects of pharmaceuticals on low trophic level invertebrate organisms such as G. pulex using high resolution confirmatory chemical analysis techniques are lacking. In particular, metabolomics may reduce this knowledge gap by directing effect-based studies that reveal alternative markers or end-points to assess potential toxicity of contaminants. Advances in mass spectrometry (MS) over the last decade have enabled better characterization of the links between the metabolome and phenotype (Dettmer et al., 2007, Villas-Bôas et al., 2005). Targeted liquid chromatography coupled to mass spectrometry (LC-MS) is the technique of choice for the reliable quantitation of known, pre-selected metabolites. It is an approach that will increasingly be used to apply knowledge discovered by non-targeted metabolomics; i.e. the eventual targeted measurement of a metabolic biomarker signature that can predict exposure to a specific environmental stress (Viant, 2008, Viant and Sommer, 2013). Metabolomics studies have made use of high resolution, confirmatory analytical techniques such as nuclear magnetic resonance (NMR) or hyphenated MS technologies to characterise large numbers of compounds for metabolic profiling (targeted) (Zhang et al., 2012). With the development of high resolution mass spectrometry (HRMS), non-targeted screening of several thousand compounds has become possible for studying larger portions of the metabolome in contrast to NMR, which is often limited by low sensitivity (Dunn et al., 2005, Zhang et al., 2012). Early investigations focused on human, plant and microbial metabolomes (Frisvad and Filtenborg, 1983, Horning and Horning, 1971, Taylor et al., 2002, Tweeddale et al., 1998). Other studies have identified changes in metabolomic profiles in mussels resulting from hypoxic conditions; biomarkers associated with withering syndrome in abalone sea snails; and responses to ethinylestradiol (EE2) by rainbow trout (Hines et al., 2007, Samuelsson et al., 2006, Viant et al., 2003). Approaches to characterise the metabolome can involve non-targeted or targeted strategies where for quantitative purposes, targeted analysis offers greater accuracy and precision (Griffiths et al., 2010, Lei et al., 2011).
The aim of the present study was to develop a targeted multi-residue method for the determination of 29 metabolites pertaining to different biochemical classes (amino acids, organic acids, nucleosides, nucleotides, and sugars) using LC-HRMS. As a full scan method also suitable for non-target analysis, the analytical performance of the method was evaluated quantitatively in terms of comprehensive mass spectral characterization, selectivity, sensitivity, intra- and inter-day precision, range and linearity. Acute toxicity of 26 pharmaceuticals in G. pulex was also assessed as an initial screen for compound selection for metabolomics. Three pharmaceuticals showing low, median and high measured LC50 values were then selected for exposures at no-observed-adverse-effect-level (NOAEL) and lowest-observed-adverse-effect-level (LOAEL) concentrations to evaluate alterations in the metabolite profile at 2, 6 and 24 h. This represents the first environmental metabolomics-based investigation of G. pulex by determining changes in endogenous metabolites in response to pharmaceutical residue exposure.
2. Materials and methods
2.1. Reagents, chemicals and consumables
All pharmaceuticals were purchased from Sigma-Aldrich (Steinheim, Germany) and Fluka (Buchs, Switzerland) with a purity of ≥ 97%. HPLC grade acetone, dimethylsulfoxide (DMSO), ethanol (EtOH) and methanol (MeOH) were purchased from Fischer Scientific (Loughborough, UK). Ultra-pure water was sourced from a Millipore Milli-Q water purification system with a specific resistance of 18.2 MΩ cm or greater (Millipore, Bedford, MA, USA). Stock solutions (40 mg mL− 1) were prepared in ultrapure water, acetone, MeOH, EtOH or DMSO, respectively. All stock solutions were stored in silanised amber vials (40 mL) and at − 20 °C in the dark for optimum stability. Organic solvent concentration (acetone, MeOH, EtOH and DMSO) in aqueous solutions used for toxicity testing was negligible. Metabolite standards (organic acids, nucleosides, nucleotides, sugars and amino acids) were supplied by Sigma Aldrich (Steinheim, Germany) and Fluka (Buchs, Switzerland). In addition, the isotopically-labelled algal amino acid mixture (98 atom% as 13C, 98 atom% as 15N) was provided from Sigma Aldrich. The targeted metabolome studied was comprised of 15 amino acids (AAs), 4 nucleosides, 2 nucleotides, 1 sugar, 3 organic acids and 4 compounds related to other families. Full details of the target metabolites and the labelled compounds are shown in Table 1. Finally, the 26 pharmaceutical compounds belonging to 12 different therapeutic classes are listed in Table S1.
Table 1.
Target analytes for metabolic profiling. Kegg number, molecular formula, molecular weight (Mw) and mass spectral characterization (ordered by metabolic group) by LC-HRMS. Instrumental method performance. F: slope; R2: regression coefficient; IDL: instrumental detection limit; RSD: relative standard deviation; MDL: method detection limit.
| Target compounds | KEGG number | Formula | Mw | Exact mass | [M-H]− | Linearity (ng μL− 1) | Calibration type | F | R2 | IDL (ng) | Intra-day precision (5 ng μL− 1) | Inter-day precision (5 ng μL− 1) | % Recovery ± RSD, n = 3 | MDL (ng g− 1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| l-2-Amino-n-butyric acid (AABA) | C00334 | C4H9NO2 | 103.06 | 103.0633 | 102.0560 | 0.05–15 | External | 1e6 | 0.993 | 0.06 | 9 | 2 | 107 ± 14 | 13.9 |
| l-Alanine | C00041 | C3H7NO2 | 89.05 | 89.0477 | 88.0404 | 0.05–15 | External | 1e6 | 0.998 | 0.03 | 8 | 8 | 92 ± 9 | 0.99 |
| l-Aspartic acid | C00049 | C4H7NO4 | 133.04 | 133.0375 | 132.0302 | 0.05–15 | External | 4e5 | 0.995 | 0.20 | 4 | 14 | 87 ± 11 | 22.7 |
| l-Citrulline | C00327 | C6H13N3O3 | 175.09 | 175.0957 | 174.0884 | 0.1–15 | External | 1e6 | 0.999 | 0.15 | 2 | 6 | 87 ± 1 | 23.9 |
| L-Isoleucine | C00407 | C6H13NO2 | 131.09 | 131.0946 | 130.0873 | 0.1–15 | Internal | 4.5 | 0.998 | 0.03 | 12 | 6 | 42 ± 8 | 0.28 |
| l-Leucine | C00123 | C6H13NO2 | 131.09 | 131.0946 | 130.0873 | 0.1–15 | Internal | 1.6 | 0.994 | 0.02 | 13 | 9 | 99 ± 1 | 0.47 |
| l-Methionine | C00073 | C5H11NO2S | 149.05 | 149.0510 | 148.0437 | 0.05–15 | Internal | 21 | 0.997 | 0.02 | 5 | 6 | 67 ± 14 | 2.66 |
| L-Ornithine hydrochloride | C00077 | C5H12N2O2 | 132.10 | 132.0899 | 131.0826 | 0.05–15 | Internal | 0.83 | 0.994 | 0.01 | 4 | 8 | 109 ± 6 | 3.51 |
| l-Phenylalanine | C00079 | C9H11NO2 | 165.08 | 165.0790 | 164.0717 | 0.1–15 | Internal | 1.6 | 0.996 | 0.02 | 14 | 14 | 97 ± 4 | 0.42 |
| l-(−)-Proline | C00148 | C5H9NO2 | 115.06 | 115.0633 | 114.0560 | 0.05–15 | Internal | 1.2 | 0.997 | 0.01 | 5 | 11 | 87 ± 8 | 1.13 |
| l-Serine | C00065 | C3H7NO3 | 105.04 | 105.0426 | 104.0353 | 0.05–15 | Internal | 1.1 | 0.996 | 0.01 | 13 | 13 | 88 ± 14 | 9.50 |
| l-Threonine | C00188 | C4H9NO3 | 119.05 | 119.0582 | 118.0509 | 0.1–15 | Internal | 2.0 | 0.999 | 0.2 | 13 | 7 | 103 ± 11 | 3.59 |
| l-Tryptophan | C00078 | C11H12N2O2 | 204.09 | 204.0899 | 203.0826 | 0.1–15 | External | 7e6 | 0.998 | 0.02 | 8 | 6 | 102 ± 5 | 1.35 |
| l-Tyrosine | C00082 | C9H11NO3 | 181.07 | 181.0739 | 180.0666 | 0.1–15 | External | 2.1 | 0.996 | 0.03 | 7 | 12 | 80 ± 8 | 0.35 |
| l-Valine | C00183 | C5H11NO2 | 117.08 | 117.0790 | 116.0717 | 0.05–15 | Internal | 1.3 | 0.991 | 0.05 | 12 | 13 | 91 ± 6 | 0.80 |
| Cytidine | C00475 | C9H13N3O5 | 243.08 | 243.0855 | 242.0777 | 0.05–15 | External | 3e6 | 0.999 | 0.01 | 8 | 1 | 110 ± 9 | 1.94 |
| Inosine | C00294 | C10H12N4O5 | 268.09 | 268.0808 | 267.0734 | 0.01–15 | Internal | 1.01 | 0.995 | 0.002 | 1 | 14 | 108 ± 8 | 0.04 |
| Thymidine | C00214 | C10H14N2O5 | 242.20 | 242.0903 | 241.0829 | 0.05–15 | Internal | 0.7 | 0.991 | 0.01 | 7 | 5 | 99 ± 7 | 0.50 |
| Uridine | C00299 | C9H12N2O6 | 244.07 | 244.0695 | 243.0622 | 0.05–15 | External | 2e6 | 0.997 | 0.03 | 7 | 5 | 138 ± 2 | 0.33 |
| ADP | C00008 | C10H15N5O10P2 | 427.03 | 427.0294 | 426.0221 | 3–15 | External | 7e5 | 0.993 | 0.15 | 9 | 11 | 25 ± 11 | 162 |
| NADH | C00004 | C21H27N7O14P2 | 663.07 | 663.1091 | 662.1018 | 0.1–15 | External | 1e6 | 0.990 | 0.06 | 2 | 19 | 162 ± 10 | 7.80 |
| Trehalose | C01083 | C12H22O11 | 342.12 | 342.1162 | 341.1089 | 0.05–15 | External | 2e6 | 0.996 | 0.01 | 10 | 0.2 | 81 ± 7 | 1.03 |
| Creatine | C00300 | C4H9N3O2 | 131.07 | 131.0695 | 130.0622 | 0.1–15 | External | 3e5 | 0.991 | 0.11 | 3 | 0.5 | 107 ± 2 | 37.3 |
| Phthalic acid | C01606 | C8H6O4 | 166.14 | 166.0266 | 165.0193 | 0.1–15 | External | 1e6 | 0.997 | 0.01 | 6 | 7 | 102 ± 2 | 13.6 |
| Taurine | C00245 | C2H7NO3S | 125.01 | 125.0147 | 124.0073 | 0.05–15 | Internal | 2.03 | 0.992 | 0.03 | 8 | 14 | 92 ± 20 | 0.23 |
| 1,7-Dimethylxanthine | C13747 | C7H8N4O2 | 180.20 | 180.0647 | 179.0574 | 0.05–15 | Internal | 1.19 | 0.997 | 0.01 | 10 | 13 | 72 ± 4 | 2.33 |
| Hypoxanthine | C00262 | C5H4N4O | 136.04 | 136.0385 | 135.0312 | 0.01–15 | External | 8e6 | 0.991 | 0.004 | 4 | 9 | 102 ± 7 | 0.47 |
| Pyridoxine | C00314 | C8H11NO3 | 169.20 | 169.0739 | 168.0666 | 0.05–15 | External | 3e6 | 0.94 | 0.07 | 6 | 0.1 | 105 ± 10 | 21.6 |
| (−)-Riboflavin | C00255 | C17H20N4O6 | 376.37 | 376.1383 | 375.1310 | 0.1–15 | Internal | 0.9 | 0.996 | 0.04 | 12 | 9 | 89 ± 7 | 6.50 |
| 13C,15N-Isoleucinea | – | C6H13NO2 | 138.12 | 138.1115 | 137.1041 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Leucinea | – | C6H13NO2 | 138.12 | 138.1115 | 137.1041 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Methioninea | – | C5H11NO2S | 155.17 | 155.0646 | 154.0573 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Phenylalaninea | – | C9H11NO2 | 175.12 | 175.1057 | 174.0984 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Prolinea | – | C5H9NO2 | 121.09 | 121.0769 | 120.0695 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Serinea | – | C3H7NO3 | 109.08 | 109.0495 | 108.0422 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Threoninea | – | C4H9NO3 | 124.08 | 124.0685 | 123.0611 | – | – | – | – | – | – | – | – | – |
| 13C,15N-Valinea | – | C5H11NO2 | 123.10 | 123.0925 | 122.0852 | – | – | – | – | – | – | – | – | – |
Internal standards used to quantify (Algal AA mixture).
2.2. Sample collection and preparation
Adult G. pulex were collected several times between September 2014 and April 2015 from the River Cray, UK, a tributary of the River Darent that feeds into the River Thames (51°23′10.5″N 0°06′34.8″E). Adult specimens were collected via the kick sampling netting method. Samples were transported back to the laboratory in Nalgene™ flasks containing a 500 mL grab sample of freshwater obtained from the River Cray. This site has previously been demonstrated to have low pharmaceutical contamination (< LOQ) (Miller et al., 2015). After collection, G. pulex were stored in different artificial freshwater tanks and acclimatized for a minimum of 2–3 days at 15 ± 2 °C under a 12 h:12 h light:dark cycle to allow depuration of any residual contamination. The freshwater crustaceans were fed ad libitum with a minimum of three horse-chestnut leaf discs (Ashauer et al., 2011). Artificial freshwater (AFW) was prepared following United States Environmental Protection Agency (USEPA) regulation (USEPA, 2002). Briefly, 1.20 g MgSO4, 1.92 g NaHCO3 and 0.080 g KCl were added to 19 L of ultra-pure water that was aerated overnight. In parallel, 1.20 g of CaSO4·2H2O was added to 1 L of Milli-Q water and mixed with the previous salt solution to make a total 20 L of AFW (USEPA, United States Environmental Protection Agency USEPA, 2002).
2.3. Pharmaceutical exposures
To select specific pharmaceuticals, the concentrations for exposures and subsequent metabolite profiling, a series of acute toxicity tests to 26 pharmaceuticals were initially performed following the Organization for Economic Co-operation and Development (OECD) 1488/94 guideline (European Communities Commission, 1996). Lethal median concentration effects and its 95% confidence interval (CI), were estimated by fitting immobility concentration responses to the Hill regression model (Eq. 1).
| (1) |
Where, I(Ci) is the proportion of immobile animals at concentration Ci; Ci is the concentration of the respective compound (i); LC50 is the median lethal concentration to the 50% of population and Hill is the shape constant, which depends on the parameters adjusted in the regression model. From these toxicity profiles the NOAEL and the LOAEL were determined. Exposure experiments were performed in Pyrex® beakers, each containing 200 mL of AFW at 15 °C and ten adult animals (> 5 mg wet weight, ww). Live/dead animals were counted after 24 h by gently prodding and observing movement of appendages. The pharmaceutical concentrations tested were in the range of 0.01 to 250 mg L− 1. Control (AFW only) and solvent controls showed no measurable toxicity. Concentrations where 100, 50 and 0% of the animals died were repeated in duplicate or triplicate.
From the initial toxicity tests on 26 pharmaceuticals, 3 were chosen for metabolomics studies representing the high (triclosan), medium (nimesulide) and low (propranolol) range of measured acute toxicity. Each exposure consisted of G. pulex exposed at the NOAEL (C1) and a second higher concentration at the LOAEL (C2) as follows: triclosan, C1 = 0.1 mg L− 1/C2 = 0.3 mg L− 1; nimesulide, C1 = 0.5 mg L− 1/C2 = 1.4 mg L− 1; and propranolol, C1 = 100 mg L− 1/C2 = 153 mg L− 1. Approximately 100 specimens were introduced in a tank containing 1 L of AFW (control), 1 L of AFW spiked with C1 and 1 L of AFW with C2. At three different times (2, 6 and 24 h), 4 replicates of live G. pulex (a pool of 6–7 animals for each replicate) were collected, frozen on dry ice and then stored at − 80 °C before metabolite profiling was performed. A preliminary extraction protocol with 1, 3, 6 and 10 animals showed that a pool of 6–7 G. pulex provided the best method recoveries.
Quantification of pharmaceutical concentrations in G. pulex at the 24 h exposure interval was performed. Separate exposures were set up in triplicate in beakers containing 200 mL of exposure solution (AFW spiked with the respective C1 pharmaceutical dose) and 20 organisms. At 24 h, animals were immediately rinsed with ultra-pure water and then frozen at − 20 °C for 24 h. Full analytical method details are described in Miller et al., 2015. Prior to extraction, frozen G. pulex samples were lyophilised at − 50 °C under vacuum for 48 h and milled in an agate mortar to a fine powder. For each analysis, 20 mg of a lyophilised composite sample were transferred to 2 mL polypropylene tubes (Eppendorf®, Hamburg, Germany), for solid-liquid extraction (SLE). After the addition of 80 μL of stable isotope-labelled internal standards at concentrations between 3.29 and 43.11 ng μL− 1 (final concentration dependent on the AA concentration in the stock reference material mixture), 800 μL of MeOH:HPLC water (90:10) mixture was added and the samples were thoroughly mixed using a Vortex mixer. Then, samples were shaken for 25 min on a vibration plate (IKA® KS 260 basic) and centrifuged at 14,000 rpm for 25 min at 0 °C. Finally, the supernatant was transferred to a chromatographic vial by using a 0.20 μM syringe filter (Whatman, GE Healthcare Life Sciences, Buckinghamshire, UK). All samples were stored at − 80 °C until LC-HRMS analysis.
2.4. Internal pharmaceutical residue determination and metabolite profiling
Internal concentrations of pharmaceuticals in G. pulex were determined using a previously described method (Miller et al., 2015). Briefly, 50 mg of lyophilised G. pulex were extracted in 5 mL of acetonitrile and pre-concentrated on Oasis HLB SPE cartridges (6 mL, 200 mg sorbent). The extract was eluted, dried-down and reconstituted in starting LC mobile phase. The chromatographic separation followed a 75 min gradient (including 12.5 min re-equilibration) using water and acetonitrile with 10 mM ammonium acetate salt. Separation was achieved using a Waters Sunfire C18 reversed-phase column (2.1 × 150 mm, 2.5 μM particle size) and detection was performed by a Waters Quattro triple quadrupole mass analyser using positive and negative electrospray ionization polarity switching (Waters Corporation, Milford, MA, USA). Organic acids, nucleosides, nucleotides, sugars and AAs were measured using LC–HRMS. An Exactive™ mass spectrometer equipped with heated electrospray ionization (H-ESI) source was used (Thermo Fisher Scientific, Bremen, Germany). The system was equipped with a HTC PAL autosampler and a Surveyor MS Plus pump. A TSK Gel-Amide 80 column (2 × 250 mm, 5 μm) for analyte separation was purchased from Sigma Aldrich. Mobile phases were binary mixtures of acetonitrile (A) and 5 mM ammonium acetate (pH 5) in HPLC-grade water (B). Gradient elution started at 75% A and 25% B, which was increased linearly to 30% B in 8 min, increased linearly to 60% B to 12 min and then held for a further 5 min. Initial conditions were returned in 3 min and the system was stabilized after a total equilibration time of 10 min (total run time = 30 min). The flow rate was set at 150 μL min− 1 and the injection volume was 5 μL. Metabolites were analysed under positive/negative ESI mode, but better resolution was obtained in negative ionization mode and so this was used for all experiments. Full scan acquisition over a mass range of 80–800 Da was performed at 70,000 full width at half maximum (FWHM) and spray voltage at 3.00 kV, capillary voltage at 30 V, skimmer voltage at 28 V and tube lens voltage at 130 V were used. A sheath gas flow rate of 45 arbitrary units (au), an auxiliary gas flow rate of 10 au and a capillary temperature at 300 °C were selected.
2.5. Method performance and quantification
Method performance for internal pharmaceutical residue concentrations in G. pulex are described elsewhere (Miller et al., 2015). Here, pharmaceutical residue quantification in exposed G. pulex was performed by a 3-point matrix-matched calibration curve. For nimesulide, spiking concentrations were 2.5, 5 and 10 μg g− 1; for triclosan, these were 1, 2.5 and 5 μg g− 1; and for propranolol these were 1, 2.5 and 5 mg g− 1. The calibration range for endogenous metabolites was from 0.01 to 15 ng μL− 1, using 9 calibration points. The algal amino acid mixture-13C,15N (see Table 1) was used as internal standard (IS) for extraction and analytical control. l-ornithine hydrochloride, inosine, thymidine, taurine, 1.7-dimethylxanthine and (−)-riboflavin were quantified using either 13C,15N-proline or 13C,15N-tyrosine as the internal standard. The remaining 15 compounds were quantified using external calibration and the target compound itself was used as external standard. The instrumental detection limit (IDL) was initially calculated as that concentration giving a signal intensity of 1 × 103, and afterwards measured experimentally by injecting a standard concentration that gave this signal intensity. The method detection limit (MDL) was calculated following the same procedure, using spiked lyophilised G. pulex samples at a concentration of 1 μg g− 1. Intra-assay variation was assessed using five consecutive injections of 5 ng μL− 1 standard solution, and inter-assay variation was determined by measuring the same standard solution on four different days. Solvent blanks did not contain any of the investigated analytes, indicating no carry-over between LC-HRMS runs. Recovery studies were performed in triplicate, using lyophilised G. pulex samples spiked at 1 μg g− 1 with the metabolites mixture and the algal amino acid mixture-13C, 15N. Five replicates of a pool with 6–7 G. pulex were analysed first and the traces of target compounds were subtracted.
2.6. Statistical analysis
Two-way ANOVA considering significant p values ≤ 0.05 was used as a first step to select metabolites with significant changes and to further explore their concentration and time trends. Thus, p values were derived and examined to determine any differences between exposed and control specimens, and to evaluate the effect of the exposure time. All data satisfied the assumptions of normality and homoscedasticity. In addition, all calculations were performed in MATLAB software version R2013b.
3. Results and discussion
3.1. Analytical method performance
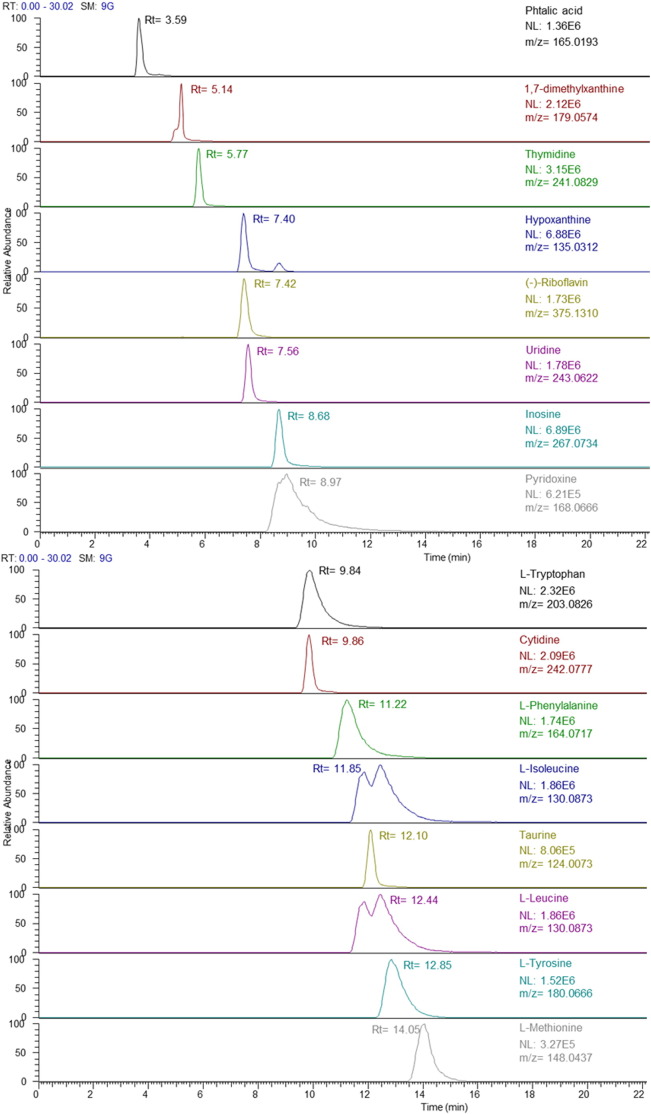
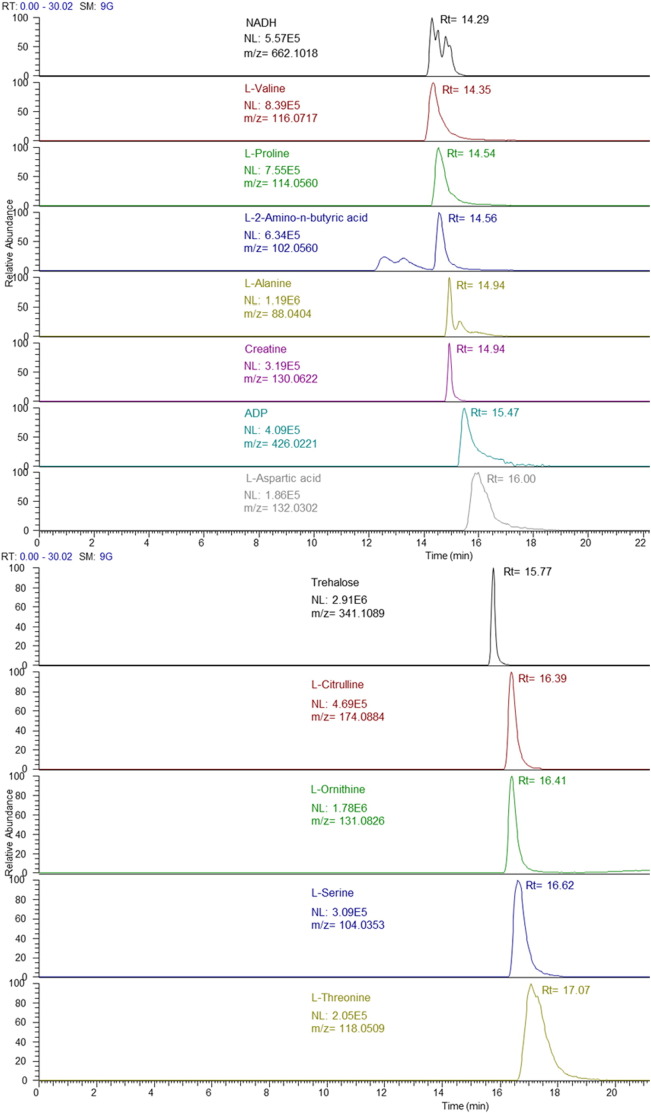
Good correlation coefficients (R2 ≥ 0.99) were obtained for 29 metabolites (Table 1). Responses for fifteen metabolites were linear from 0.05 to 15 ng μL− 1; and for 11 other metabolites, linearity ranged from 0.1 to 15 ng μL− 1. Signals for inosine and hypoxanthine were linear over the range of 0.01 to 15 ng μL− 1; and ADP in the range 3 to 15 ng μL− 1. Therefore, given that this represents a lower concentration range of 10 to 1500 ng g− 1, these were considered fit for purpose for this study. IDL ranged from 0.002 to 0.20 ng, and intra and inter-day precision ranges were from 1 to 14% and from 0.1 to 19%, respectively at the 5 ng μL− 1 concentration level. Twenty seven compounds showed recoveries within the range of 42 ± 8% to 138 ± 2% (Table 1). Overall, recoveries were acceptable and showed excellent precision. However, the two nucleotides ADP and NADH displayed poorer recoveries of 25 ± 11% and 162 ± 10%, respectively. Finally, the MDL ranged from 0.04 (inosine) to 37.3 ng g− 1 (creatine), with the exception of ADP, for which sensitivity was low as would be expected due to the lower recovery observed (MDL: 162 ng g− 1). The extracted ion chromatograms of a target metabolite mixed solution at 5 ng μL− 1 using the TSK Gel-Amide 80 HILIC column is shown in Fig. 1. In summary, the method performance of this analytical method indicated that reliable quantitative measurements could be made for most metabolites and with minimal variance contribution from the matrix. Moreover, as this method incorporated HRMS in full-scan mode, post hoc untargeted data analysis of the metabolome remains possible if required.
Fig. 1.
LC–HRMS extracted ion chromatogram of 29 target compounds from a full scan LC-Orbitrap-MS spectrum using a 5 ppm extraction window.
3.2. Acute toxicity, 24-h pharmaceutical exposures and observed changes in target metabolite concentrations
Acute toxicity tests revealed LC50 at the mg L− 1 level for most of the 26 tested pharmaceuticals. Similar values have been obtained in other freshwater crustaceans (see Table S3). Kim et al. (2009) studied the acute toxicity of pharmaceuticals and personal care products in the freshwater crustacean, Thamnocephalus platyurus (T. platyurus). They reported a similar LC50 value to the present study for triclosan (0.57 mg L− 1), but not for propranolol which lay at 10.31 mg L− 1 (Kim et al., 2009). These values provide a means of ranking the toxic risk, but such values are not environmentally relevant as pharmaceuticals are generally found at concentrations approximately one to two orders of magnitude less (Miller et al., 2015). Therefore, it is unlikely that the majority of these compounds will display any significant acute toxicity. Nonetheless, as a starting point and based on these data, exposures were performed at NOAEL and LOAEL concentrations for triclosan, nimesulide and propranolol to represent compounds at the high, median and low range of measured LC50 (Figs. S1–S3).
3.2.1. Triclosan
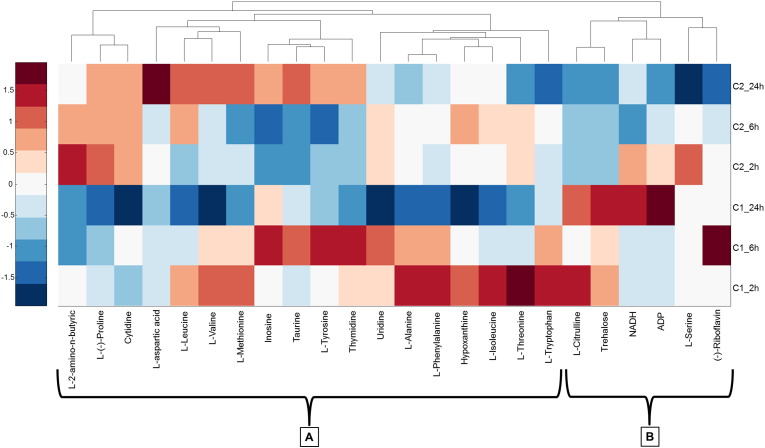
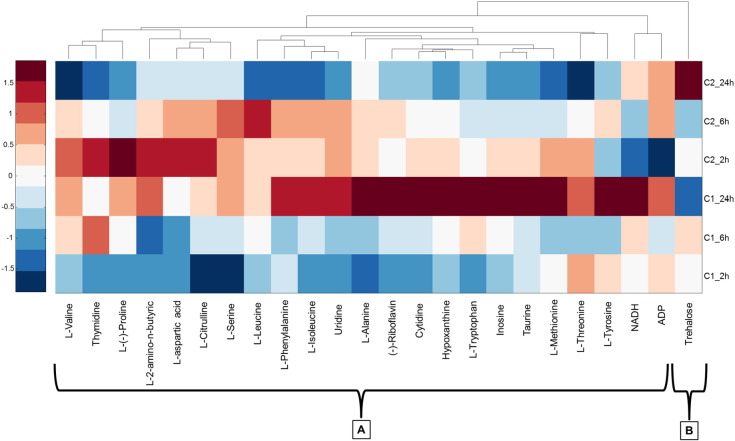
Triclosan, the most toxic of the selected pharmaceuticals (LC50 0.57 mg L− 1) measured here, caused changes in the metabolite concentrations in G. pulex at 0.1 mg L− 1 (C1) and 0.3 mg L− 1 (C2), and was dose responsive. Exposure to triclosan produced significant changes (Two-way ANOVA, p < 0.05) in 19 metabolites: 13 amino acids (l-alanine, cytidine, l-citrulline, l-isoleucine, l-leucine, l-methionine, l-phenylalanine, l-proline, taurine, l-threonine, l-tryptophan, l-tyrosine and l-valine), inosine and uridine (nucleosides) and others like trehalose, hypoxanthine, riboflavin and thymidine (Table 2 and Figure S4). However, only the concentration of 37% of metabolites changed significantly (up or down) across the exposure time. l-isoleucine, l-phenylalanine, l-(−)-proline, taurine, l-threonine, l-tyrosine, l-valine, inosine and thymidine concentrations varied significantly (p < 0.05) with exposure duration (Table 2). A 2-way ANOVA was used to evaluate the interaction between dose and time factors. Cytidine, l-methionine, l-phenylalanine and l-(−)-proline were the only metabolites that had significant interactions (Table 2). As shown in Figure S4, all metabolites except l-citrulline changed in their mean concentrations at C1 relative to the controls. Some metabolites increased more than two to three-fold in comparison to controls such as l-tryptophan, hypoxanthine, l-alanine, l-isoleucine, l-threonine, l-valine and l-thymidine. In other cases, measured concentrations between the exposed and control samples decreased over time (ADP and l-proline). Similar trends occurred at C2 where the 75% of the metabolites increased in their concentration, with the remaining metabolites showing no significant changes (also see Table S1). Table S2 shows the metabolite concentrations determined from all conditions studied. To more conveniently highlight trends, Fig. 2 represents the fold change values at each exposure concentration with respect to controls and plotted in a heat map and hierarchical analysis revealed two distinct clusters (A & B). In general, and for each metabolite in Cluster A of Fig. 2, the fold changes in gammarid metabolites exposed at C1 decreased across the 24-h period. On the contrary, Cluster B generally showed the opposite trend where concentrations of metabolites mostly increased. In the case of samples exposed at C2, eight metabolites of Cluster A (l-aspartic acid, l-leucine, l-valine, l-methionine, inosine, taurine, l-tyrosine and thymidine) had a fold increase and the remainder showed a slight decrease in concentration. The metabolites of Cluster B also showed fold decreases along the 24-h period and the opposite effect to those exposed at C1.
Table 2.
Two-way ANOVA indicating p values for changes in metabolite concentrations based on time and dose factor. The significant changes are in bold where p < 0.05.
|
p-Value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Triclosan |
Nimesulide |
Propanolol |
|||||||
| Metabolite | Dose factora | Time factorb | Interaction dose × time | Dose factora | Time factorb | Interaction Dose × Time | Dose factora | Time factorb | Interaction dose × time |
| AABA | 0.49 | 0.23 | 0.91 | – | 3e − 4 | 0.003 | – | 0.24 | 0.32 |
| l-Alanine | – | 0.19 | 0.15 | 0.53 | – | 6e − 4 | – | 0.001 | 0.03 |
| l-Aspartic acid | 0.17 | 0.37 | 0.25 | 2e − 4 | 0.005 | 0.004 | 0.28 | 0.04 | 0.44 |
| Cytidine | 0.001 | 0.08 | 0.02 | – | 0.02 | – | 5e − 6 | 1e − 5 | 1e − 6 |
| l-Citrulline | 0.01 | 0.81 | 0.95 | 4e − 6 | 2e − 5 | 8e − 6 | 0.53 | 0.29 | 0.07 |
| l-Isoleucine | – | 9e − 4 | 0.12 | – | 0.29 | – | 0.06 | 3e − 4 | 0.003 |
| l-Leucine | 0.01 | 0.47 | 0.73 | – | 0.003 | – | 0.25 | – | – |
| l-Methionine | – | 0.18 | 0.003 | – | 0.15 | 1e − 4 | 0.003 | 0.005 | 0.007 |
| l-Ornithine hydrochloride | Not detected in Gammarus pulex organism | ||||||||
| l-Phenylalanine | – | 0.001 | 0.001 | – | 0.53 | 0.0001 | 0.69 | – | 0.001 |
| l-(−)-Proline | – | 0.001 | 0.01 | 4e − 7 | 6e − 13 | 8e − 9 | – | – | 2e − 4 |
| l-Serine | 0.21 | 0.05 | 0.23 | 0.04 | 0.02 | 0.03 | – | 0.73 | 0.01 |
| Taurine | – | 0.002 | 0.10 | – | 0.58 | 0.002 | 0.03 | 0.05 | 0.75 |
| l-Threonine | – | 0.03 | 0.07 | – | 0.07 | 0.001 | 0.34 | – | 0.001 |
| l-Tryptophan | – | 0.28 | 0.53 | – | 0.08 | – | 0.0012 | – | – |
| l-Tyrosine | – | 0.01 | 0.40 | 0.003 | 0.97 | 0.12 | 0.70 | 0.01 | 0.27 |
| l-Valine | – | 0.01 | 0.05 | – | 0.42 | – | 5e − 4 | 2e − 4 | – |
| Inosine | – | 0.01 | 0.11 | – | 0.17 | 0.03 | 0.12 | 0.002 | 0.27 |
| Uridine | 8e − 4 | 0.45 | 0.26 | – | 0.98 | – | 0.002 | 6e − 4 | 0.009 |
| ADP | 0.21 | 0.25 | 0.74 | 0.04 | 0.14 | 0.01 | 0.02 | 0.87 | 0.64 |
| NADH | 0.05 | 0.41 | 0.31 | 0.49 | 0.72 | 0.12 | 0.52 | 0.58 | 0.83 |
| Trehalose | 0.03 | 0.11 | 0.08 | 0.07 | – | – | 0.21 | – | – |
| Creatine | Not detected in Gammarus pulex organism | ||||||||
| 1,7-dimethylxanthine | Not detected in Gammarus pulex organism | ||||||||
| Hypoxanthine | – | 0.08 | 0.07 | – | 0.14 | – | 1e − 6 | 4e − 5 | 8e − 7 |
| Phtalic acid | Not detected in Gammarus pulex organism | ||||||||
| Pyridoxine (vitamin B6) | Not detected in Gammarus pulex organism | ||||||||
| (−)-Riboflavin | 0.01 | 0.49 | 0.36 | – | 0.46 | 0.002 | 3e − 4 | 0.69 | 0.008 |
| Thymidine | – | 0.01 | 0.12 | – | 7e − 4 | – | 4e − 4 | 0.009 | 2e − 4 |
Dose factor corresponds at the two different concentrations used for each pharmaceutical.
Time factor corresponds at the different times studied (2, 6 and 24 h).
Fig. 2.
Heat map of triclosan exposure representing the fold change of targeted metabolites in each exposure subgroup (C1, C2) relative to controls. All data have been auto scaled by column and dendrograms represent hierarchical analysis for clustering (A & B) of metabolite responses.
3.2.2. Nimesulide
Exposure to nimesulide produced significant changes (Two-way ANOVA, P < 0.05) for 83% of the metabolites analysed, including most amino acids (except l-alanine), such as inosine, uridine and other metabolites including hypoxanthine, riboflavin and thymidine (Table 2 and Fig. S5). l-2-amino-n-butyric (AABA), l-alanine, l-aspartic acid, cytidine, l-citrulline, l-leucine, l-(−)-proline, l-serine, trehalose and thymidine showed a time-related effect (Table 2), affecting 42% of the metabolites studied. On the other hand, considering the interaction between time and dose factor, significant differences (p < 0.05) in all metabolites except for l-tyrosine and NADH were detected (Table 2). Nimesulide exposures induced changes in the metabolome of G. pulex in comparison to controls (Fig. S5). A heat map of nimesulide was prepared as before and hierarchical analysis again revealed two clusters (Fig. 3). In this case, at C1, all the metabolites except trehalose had fold increases along the 24-h period (Cluster A, Fig. 3). Specifically, l-alanine, (−)-riboflavin, cytidine, hypoxanthine, l-tryptophan, inosine, taurine, l-tyrosine and NADH suffered the more important changes (garnet colour). On the other hand, trehalose (Cluster B, Fig. 3) showed the opposite trend and, at 24-h exposure time, the fold change had decreased (blue). In the samples exposed at C2, the metabolites in Cluster A began with positive fold changes (red/orange) at 2-h exposure time and decreased (blue) at 24-h exposure time, with the exception of NADH, ADP and trehalose (Cluster B, Fig. 3).
Fig. 3.
Heat map of nimesulide exposure representing the fold change of targeted metabolites in each exposure subgroup (C1, C2) relative to controls. All data have been auto scaled by column and dendrograms represent hierarchical analysis for clustering (A & B) of metabolite responses.
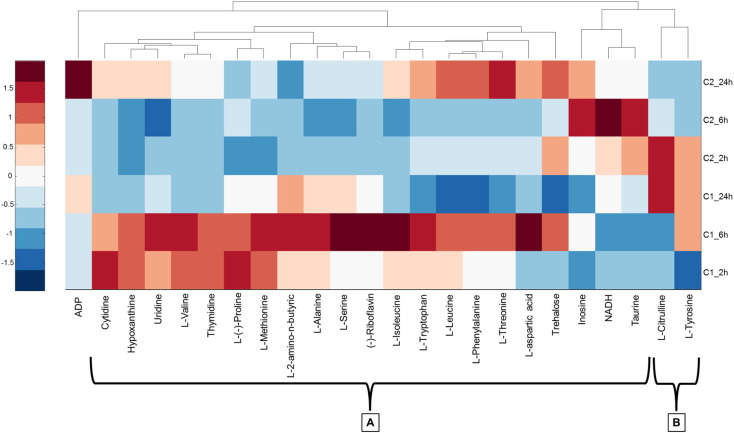
3.2.3. Propranolol
Exposure to propranolol produced significant changes (based on two-way ANOVA, p < 0.05 analysis) in 8 out of 15 AAs (AABA, cytidine, l-methionine, l-proline, l-serine, taurine, l-tryptophan and l-valine), uridine (nucleoside), ADP (nucleotide) and others like hypoxanthine, riboflavin and thymidine (Table 2 and Fig. S6). Moreover, the exposure time affected 71% of the metabolites studied. l-alanine, l-aspartic acid, cytidine, l-isoleucine, l-leucine, l-methionine, l-phenylalanine, l-(−)-proline, l-threonine, l-tryptophan, l-tyrosine, l-valine, inosine, uridine, trehalose, hypoxanthine and thymidine displayed significant changes (p values < 0.05) across the 24 h experiments, see Table 2. Exposures at C1, a concentration below LC10, showed significant changes in the concentrations of the metabolites with respect to controls. For example, AABA increased its concentration by more than two-fold (Fig. S6). In other metabolites, decreasing metabolite concentrations were observed such as taurine, an essential amino acid for cardiovascular function and the central nervous system, or inosine, a nucleoside formed when hypoxanthine is attached to a ribose ring. Table S2 shows the concentrations of the metabolites determined from all exposures. At the higher exposure concentration, 153 mg L− 1 (C2), similar metabolite concentration changes occurred for AABA, taurine and inosine. Additionally, hypoxanthine and thymidine concentrations were different compared to controls (Fig. S6). In the samples exposed at C1, all the metabolites in Cluster A (Fig. 4) had fold decreases along the 24-h exposure time. However, l-citrulline and l-tyrosine (Cluster B, Fig. 4) and ADP had fold increases at 24-h. Finally, in the samples at C2, the general trend in Cluster A plus ADP is that the metabolites had fold increases along 24-h, with the exception of NADH and taurine. Moreover, the metabolites in Cluster B of the Fig. 4 (l-citrulline and l-tyrosine) also decreased their fold changes at 24-h exposure time.
Fig. 4.
Heat map of propranolol exposure representing the fold change of targeted metabolites in each exposure subgroup (C1, C2) relative to controls. All data have been auto scaled by column and dendrograms represent hierarchical analysis for clustering (A & B) of metabolite responses.
3.3. Metabolic pathways potentially affected by selected single pharmaceutical exposures
Little or no reported metabolomics or pathway-based analysis data exists for G. pulex in the literature to our knowledge. However, changes in its metabolic profile following exposure to pharmaceuticals may result in processes that are suggestive of either metabolic (i.e. detoxification) or toxic responses. Firstly, and to support this, the internal concentrations were determined for the two compounds nimesulide and propranolol at the C1 concentration at 24-h time interval. Triclosan was detected, but was unfortunately not quantifiable due to poor standard addition linearity (and similarly poor method performance as a whole). The C1 propranolol dose resulted in measured concentrations up to 4.9 ± 0.3 mg g− 1 (dry weight) and nimesulide reached a mean concentration of 12.2 ± 4.1 μg g− 1 which are both significantly higher concentrations than the estimated dose required for therapeutic effects in humans. When comparing to environmental occurrence concentrations, these compounds did not exceed 36 ng g− 1 in G. pulex (Miller et al., 2015) and other studies in aquatic invertebrates often report concentrations of < 200 ng g− 1 (Dodder et al., 2014, Huerta et al., 2015). However, and although the concentrations determined here are unlikely to be seen in the environment, measurement using such analytical methods enables interpretation of metabolic responses and potentially metabolic pathways indicative of adverse effects in the future for risk assessment purposes. The different responses may be elicited through numerous complex biochemical pathways such as nucleic acid expression, protein synthesis, enzymatic processes and signaling cascades. Thus, identifying and quantifying metabolite change is a useful starting point perhaps to direct metabolomics and extended pathway-based research in the future. The advantage of operating HRMS mass analysers in full-scan mode (as in the present study) is that such further qualitative meta-analysis of the data is still possible using untargeted and/or chemometric approaches at a later time (Farrés et al., 2014). The unbiased nature of non-target metabolomics would also allow the interpretation of metabolic responses in relation to known mode-of-action pathways for pharmaceuticals. Nonetheless, this was beyond the scope of this work and future work will be pursued in this direction. A general observation was that control levels of metabolites showed relatively large scatter in specific cases. During the study the organisms were collected from the same site and were of similar size, but the variances in metabolite profiles of the controls are likely to have resulted from differences between individuals such as their age, moult cycle stage and gender. Furthermore, acclimatisation of mussel populations in laboratory conditions has also been demonstrated to lead to increased metabolic variability (Hines et al., 2007).
3.3.1. Protein synthesis
Exposure to all three pharmaceuticals resulted in an increase in thymidine and inosine concentrations when compared to controls. Amongst other potential reasons, their increase in concentration could be related to increases in protein synthesis as they are associated with tRNA. Cytidine concentrations varied between exposure concentrations and time points for all compounds. Triclosan had elevated cytidine concentrations at both 2 and 6-h time intervals relative to controls in the C1 exposure. The same effect was not observed in C2, often remaining close to the control levels except at the 6-h time interval. Cytidine in the nimesulide exposure was upregulated relative to controls at C1, whereas C2 showed a steady increase in cytidine concentrations over the 24-h exposure period. For propranolol, cytidine concentrations remained below controls at C2 and were initially upregulated at C1 before decreasing below control levels at 24 h. Uridine showed no obvious differences between controls for either exposure concentration in propranolol. Uridine was upregulated in the nimesulide C2 exposure by 24-h whereas in the C1 remained higher than controls at all time points. Triclosan showed increased uridine concentrations in both exposures at the 2 and 6-h intervals when compared with control levels, the C2 uridine concentrations returned to the control level at 24-h. As uridine is absent from DNA and only present in RNA, it is plausible that together these four nucleosides are generally indicative of upregulation of protein synthesis which is potentially induced by all three pharmaceuticals. The upregulation could be considered as a general metabolic response to such xenobiotic exposure, for example, via the production of P450 enzymes (Marionnet et al., 1997, Ortiz-Delgado et al., 2008). Triclosan has also been shown to increase the P450 content of rat liver microsomes (Kanetoshi et al., 1992, Liang et al., 2013).
3.3.2. Xenometabolic pathways
The internal concentrations reached in G. pulex exceed the human therapeutic doses and thus are likely to induce xenometabolism by means of enzymes associated with phase I and II metabolism. It is possible that the increased content of uridine is also related to phase II metabolic processes. Uridine, when converted to a triphosphate nucleotide (UTP), is involved in the biosynthesis of uridine diphosphate glucose (UDPG) which serves as a precursor of uridine glucuronic acid, the primary substrate for phase II glucuronidation reactions catalysed by uridine 5′-diphospho-glucuronosyltransferase (UGT). Major metabolites associated with these three pharmaceuticals are glucuronide conjugates, which support this argument (Macpherson et al., 2013, Walle et al., 1985, Wu et al., 2010). Riboflavin also showed statistically significant 2–3 fold increases in concentration relative to control concentrations. This metabolite is essential for xenometabolic processes as it forms part of flavin adenine nucleotide (FAD) and flavin mononucleotide (FMN), which are essential cofactors for redox reactions involving P450 enzymes and flavin-containing monooxygenases (FMOs). These cofactors are also required for regeneration of reduced glutathione (GSH) from its oxidized form glutathione disulfide (GSSG).
At C1 and C2 nimesulide exposures, AABA showed a significant reduction in concentration and also decreased over the course of the experiment. This metabolite is a precursor to ophthalmic acid, which is associated with oxidative stress and the induction of ophthalmic acid pathways is shown when glutathione levels are reduced (Soga et al., 2006). As nimesulide has been shown to reduce GSH levels as well as induce oxidative stress, it is suggestive that the low levels of AABA are a result of its conversion to ophthalmic acid, but this requires confirmation (Chatterjee et al., 2006, Mingatto et al., 2002, Singh et al., 2010). It has been suggested that the physiological significance in the production of AABA is that it is a cofactor in the transport of glucuronide metabolites in the multi-drug resistance protein 1 (MRP-1) and thus required for elimination of xenobiotics (Soga et al., 2006). Lastly, perturbations in methionine concentrations may also be indicative of xenometabolic pathways. The concentrations of methionine in all exposures were elevated relative to control with the exception of the 2 h C2 sampling point in the propranolol exposure. Methionine acts as a precursor to S-adenosyl methionine (SAM) that is involved in methylation reactions for xenobiotic metabolism. Triclosan, for example, undergoes methylation during metabolism to methyltriclosan and therefore may explain its elevated concentrations in both triclosan exposures (Wu et al., 2010). Finally, this compound also serves as a precursor to GSH biosynthesis and therefore may be elevated even when xenobiotics are not methylated during detoxification.
3.3.3. Signaling cascade
Tryptophan was significantly expressed at increased concentrations in all three pharmaceutical exposures at both C1 and C2. This AA acts as a precursor to serotonin that is often released as a stress response. It is possible that the exposure to these pharmaceuticals induced a stress response. Indeed, C2 is set at the LOAEL with mortality as the adverse effect, which upregulated the synthesis of tryptophan for which serotonin synthesis is dependent (Joseph and Kennett, 1983). Xenobiotics have been previously shown to induce the release of serotonin in rats (Yokogoshi, 1989). However, it is also possible that tryptophan is produced in response to reactive oxygen and nitrogen species (ROS/RNS) produced by the metabolism of the pharmaceuticals (Peyrot and Ducrocq, 2008). Triclosan displayed the highest concentrations of tryptophan reaching up to 142 μg g− 1 when exposed at C2. The higher toxicity of this compound may lead to added stress in comparison to propranolol and nimesulide. The amino acid proline also showed statistically significant changes during the three exposures. In particular, the C1 exposures showed a decrease in proline over time until they approximately reached control levels at the 24-h time interval. The same trend is not observed in the C2 exposures and is more variable when compared to the controls. Proline has been shown in previous studies to be important during stress responses as this amino acid has roles in preventing oxidative stress, maintenance of osmoregulation, energy production, and many other biological functions in plants and amphipods (Maity et al., 2012, Choudhary et al., 2007).
4. Conclusions
A comprehensive optimisation of a targeted multi-residue method for the quantitative determination of 29 metabolites from different biochemical classes using LC-HRMS was performed. The acute toxicity (LC50) was also estimated for 26 pharmaceuticals and revealed that triclosan was the most toxic compound to G. pulex. However, the reliance on acute toxicity data such as LC50 for pharmaceutical risk assessment may ultimately not be realistic as these were at least one–two orders of magnitudes higher than concentrations typically found in the aquatic environment. Exposures performed at the NOAEL and LOAEL with three pharmaceuticals across the range of measured toxicity resulted in quantifiable changes in the metabolome in G. pulex. Measured internal concentrations of nimesulide and propranolol were far higher than the daily recommended doses for therapeutic effects in humans. Alterations in metabolite concentrations were observed, which could be involved in several different pathways relating to protein synthesis, oxidative stress and signaling cascades. However, further efforts are required to fully characterise and understand the effects these contaminants have on any specific pathway. In greater knowledge using such analytical methods for quantitative determinations of endogenous metabolites in aquatic organisms can now be acquired. In addition, the use of full-scan HRMS detection enables non-target meta-analysis to be performed in the future using chemometric tools that could identify biomarkers of exposure and effect for use in the environmental risk assessment of pharmaceuticals.
Role of funding source
Funding bodies played no role in the design of the study or decision to publish.
Conflict of interest
The authors declare no financial conflict of interest.
Acknowledgements
G. McEneff is acknowledged for laboratory assistance and with G. pulex sampling, as well as staff at the Centre for Excellence in Mass Spectrometry at King's College London for LC-HRMS support. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement n. 320737. A JWT Jones Travelling Fellowship was also awarded to C. Gómez-Canela by the Royal Society of Chemistry for his work to be performed at KCL. T. Miller's collaboration was funded by the Biotechnology and Biological Sciences Research Council scholarship scheme (Reference BB/K501177/1).
Editor: D. Barcelo
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.scitotenv.2016.03.181.
Appendix A. Supplementary data
Supplementary material
References
- Ashauer R., Hintermeister A., Potthoff E., Escher B.I. Acute toxicity of organic chemicals to Gammarus pulex correlates with sensitivity of Daphnia magna across most modes of action. Aquat. Toxicol. 2011;103:38–45. doi: 10.1016/j.aquatox.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Bourgeault A., Ciffroy P., Garnier C., Cossu-Leguille C., Masfaraud J.F., Charlatchka R., Garnier J.M. Speciation and bioavailability of dissolved copper in different freshwaters: comparison of modelling, biological and chemical responses in aquatic mosses and gammarids. Sci. Total Environ. 2013;452-453:68–77. doi: 10.1016/j.scitotenv.2013.01.097. [DOI] [PubMed] [Google Scholar]
- Chatterjee M., Sarkar K., Sil P.C. Herbal (Phyllanthus niruri) protein isolate protects liver from nimesulide induced oxidative stress. Pathophysiology. 2006;13:95–102. doi: 10.1016/j.pathophys.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Choudhary M., Jetley U.K., Khan M.A., Zutshi S., Fatma T. Effect of heavy metal stress on proline, malondialdehyde, and superoxide dismutase activity in the cyanobacterium Spirulina platensis-S5. Ecotoxicol. Environ. Saf. 2007;66:204–209. doi: 10.1016/j.ecoenv.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Coulaud R., Geffard O., Xuereb B., Lacaze E., Quéau H., Garric J., Charles S., Chaumot A. In situ feeding assay with Gammarus fossarum (Crustacea): modelling the influence of confounding factors to improve water quality biomonitoring. Water Res. 2011;45:6417–6429. doi: 10.1016/j.watres.2011.09.035. [DOI] [PubMed] [Google Scholar]
- Daughton C.G., Ternes T.A. Pharmaceuticals and personal care products in the environment: agents of subtle change? Environ. Health Perspect. 1999;107:907–938. doi: 10.1289/ehp.99107s6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lange H.J., Sperber V., Peeters E.T.H.M. Avoidance of polycyclic aromatic hydrocarbon-contaminated sediments by the freshwater invertebrates Gammarus pulex and Asellus aquaticus. Environ. Toxicol. Chem. 2006;25:452–457. doi: 10.1897/05-413.1. [DOI] [PubMed] [Google Scholar]
- De Lange H.J., Peeters E.T.H.M., Lürling M. Changes in ventilation and locomotion of Gammarus pulex (Crustacea, Amphipoda) in response to low concentrations of pharmaceuticals. Hum. Ecol. Risk. Assess. 2009;15:111–120. [Google Scholar]
- Dettmer K., Aronov P.A., Hammock B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodder N.G., Maruya K.A., Ferguson P.L., Grace R., Klosterhaus S., La Guardia M.J., Lauenstein G.G., Ramirez J. Occurrence of contaminants of emerging concern in mussels (Mytilus spp.) along the California coast and the influence of land use, storm water discharge, and treated wastewater effluent. Mar. Pollut. Bull. 2014;81(2):340–346. doi: 10.1016/j.marpolbul.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Dunn W.B., Bailey N.J.C., Johnson H.E. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606–625. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- European Communities Commission . 1996. Technical guidance document in support of commission directive 93/67/EEC on risk assessment for new notified substances and commission regulation. [Google Scholar]
- Evgenidou E.N., Konstantinou I.K., Lambropoulou D.A. Occurrence and removal of transformation products of PPCPs and illicit drugs in wastewaters: a review. Sci. Total Environ. 2015;505:905–926. doi: 10.1016/j.scitotenv.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Farrés M., Piña B., Tauler R. Chemometric evaluation of Saccharomyces cerevisiae metabolic profiles using LC–MS. Metabolomics. 2014;11:210–224. doi: 10.1007/s11306-014-0689-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquet-Griell H., Gómez-Canela C., Ventura F., Lacorte S. Predicting concentrations of cytostatic drugs in sewage effluents and surface waters of Catalonia (NE Spain) Environ. Res. 2015;138:161–172. doi: 10.1016/j.envres.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Filtenborg O. Classification of Terverticillate penicillia based on profiles of mycotoxins and other secondary metabolites. Appl. Environ. Microbiol. 1983;46:1301–1310. doi: 10.1128/aem.46.6.1301-1310.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Canela C., Cortés-Francisco N., Ventura F., Caixach J., Lacorte S. Liquid chromatography coupled to tandem mass spectrometry and high resolution mass spectrometry as analytical tools to characterize multi-class cytostatic compounds. J. Chromatogr. A. 2013;1276:78–94. doi: 10.1016/j.chroma.2012.12.031. [DOI] [PubMed] [Google Scholar]
- Gómez-Canela C., Ventura F., Caixach J., Lacorte S. Occurrence of cytostatic compounds in hospital effluents and wastewaters, determined by liquid chromatography coupled to high-resolution mass spectrometry. Anal. Bioanal. Chem. 2014;406:3801–3814. doi: 10.1007/s00216-014-7805-9. [DOI] [PubMed] [Google Scholar]
- Griffiths W.J., Koal T., Wang Y., Kohl M., Enot D.P., Deigner H.-P. Targeted metabolomics for biomarker discovery. Angew. Chem. Int. Ed. 2010;49:5426–5445. doi: 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- Heberer T. Occurrence, fate, and removal of pharmaceutical residues in the aquatic environment: a review of recent research data. Toxicol. Lett. 2002;131:5–17. doi: 10.1016/s0378-4274(02)00041-3. [DOI] [PubMed] [Google Scholar]
- Hines A., Oladiran G.S., Bignell J.P., Stentiford G.D., Viant M.R. Direct sampling of organisms from the field and knowledge of their phenotype: key recommendations for environmental metabolomics. Environ. Sci. Technol. 2007;41:3375–3381. doi: 10.1021/es062745w. [DOI] [PubMed] [Google Scholar]
- Horning E.C., Horning M.G. Human metabolic profiles obtained by GC and GC/MS. J. Chromatogr. Sci. 1971;9:129–140. [Google Scholar]
- Huerta B., Jakimska A., Llorca M., Ruhí A., Margoutidis G., Acuña V., Sabater S., Rodriguez-Mozaz S., Barcelò D. Development of an extraction and purification method for the determination of multi-class pharmaceuticals and endocrine disruptors in freshwater invertebrates. Talanta. 2015;132:373–381. doi: 10.1016/j.talanta.2014.09.017. [DOI] [PubMed] [Google Scholar]
- Joseph M.H., Kennett G.A. Stress-induced release of 5-HT in the hippocampus and its dependence on increased tryptophan availability: an in vivo electrochemical study. Brain Res. 1983;270:251–257. doi: 10.1016/0006-8993(83)90598-x. [DOI] [PubMed] [Google Scholar]
- Kanetoshi A., Katsura E., Ogawa H., Ohyama T., Kaneshima H., Miura T. Acute toxicity, percutaneous absorption and effects on hepatic mixed function oxidase activities of 2,4,4′-trichloro-2′-hydroxydiphenyl ether (Irgasan® DP300) and its chlorinated derivatives. Arch. Environ. Contam. Toxicol. 1992;23:91–98. doi: 10.1007/BF00226000. [DOI] [PubMed] [Google Scholar]
- Kim J.W., Ishibashi H., Yamauchi R., Ichikawa N., Takao Y., Hirano M., Koga M., Arizono K. Acute toxicity of pharmaceutical and personal care products on freshwater crustacean (Thamnocephalus platyurus) and fish (Oryzias latipes) J. Toxicol. Sci. 2009;34:227–232. doi: 10.2131/jts.34.227. [DOI] [PubMed] [Google Scholar]
- Lebrun J.D., Perret M., Geffard A., Gourlay-Francé C. Modelling copper bioaccumulation in Gammarus pulex and alterations of digestive metabolism. Ecotoxicology. 2012;21:2022–2030. doi: 10.1007/s10646-012-0955-7. [DOI] [PubMed] [Google Scholar]
- Lei Z., Huhman D.V., Sumner L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011;286:25435–25442. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Nie X., Ying G., An T., Li K. Assessment of toxic effects of triclosan on the swordtail fish (Xiphophorus helleri) by a multi-biomarker approach. Chemosphere. 2013;90:1281–1288. doi: 10.1016/j.chemosphere.2012.09.087. [DOI] [PubMed] [Google Scholar]
- Macpherson D., Best S.A., Gedik L., Hewson A.T., KD R., Parisi S. The biotransformation and pharmacokinetics of 14C-nimesulide in humans following a single dose oral administration. J. Drug Metab. Toxicol. 2013 [Google Scholar]
- Maity S., Jannasch A., Adamec J., Nalepa T., Höök T.O., Sepúlveda M. Starvation causes disturbance in amino acid and fatty acid metabolism in Diporeia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012;161:348–355. doi: 10.1016/j.cbpb.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Maltby L., Clayton S.A., Wood R.M., McLoughlin N. Evaluation of the Gammarus pulex in situ feeding assay as a biomonitor of water quality: robustness, responsiveness, and relevance. Environ. Toxicol. Chem. 2002;21:361–368. [PubMed] [Google Scholar]
- Marionnet D., Taysse L., Chambras C., Deschaux P. 3-methylcholanthrene-induced EROD activity and cytochrome P450 in immune organs of carp (Cyprinus carpio) Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1997;118:165–170. doi: 10.1016/s0742-8413(97)00101-1. [DOI] [PubMed] [Google Scholar]
- Miller T.H., McEneff G.L., Brown R.J., Owen S.F., Bury N.R., Barron L.P. Pharmaceuticals in the freshwater invertebrate, Gammarus pulex, determined using pulverised liquid extraction, solid phase extraction and liquid chromatography-tandem mass spectrometry. Sci. Total Environ. 2015;511:153–160. doi: 10.1016/j.scitotenv.2014.12.034. [DOI] [PubMed] [Google Scholar]
- Mingatto F.E., Rodrigues T., Pigoso A.A., Uyemura S.A., Curti C., Santos A.C. The critical role of mitochondrial energetic impairment in the toxicity of nimesulide to hepatocytes. J. Pharmacol. Exp. Ther. 2002;303:601–607. doi: 10.1124/jpet.102.038620. [DOI] [PubMed] [Google Scholar]
- Monteiro S.C., Boxall A.B.A. In: Occurrence and Fate of Human Pharmaceuticals in the Environment. D.M. Whitacre., editor. 2010. (Rev. Environ. Contam. Toxicol). [DOI] [PubMed] [Google Scholar]
- Ortiz-Delgado J.B., Behrens A., Segner H., Sarasquete C. Tissue-specific induction of EROD activity and CYP1A protein in Sparus aurata exposed to B(a)P and TCDD. Ecotoxicol. Environ. Saf. 2008;69:80–88. doi: 10.1016/j.ecoenv.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Pellet B., Ayrault S., Tusseau-Vuillemin M.H., Gourlay-Francé C. Quantifying diet-borne metal uptake in Gammarus pulex using stable isotope tracers. Ecotoxicol. Environ. Saf. 2014;110:182–189. doi: 10.1016/j.ecoenv.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Peyrot F., Ducrocq C. Potential role of tryptophan derivatives in stress responses characterized by the generation of reactive oxygen and nitrogen species. J. Pineal Res. 2008;45:235–246. doi: 10.1111/j.1600-079X.2008.00580.x. [DOI] [PubMed] [Google Scholar]
- Samuelsson L.M., Förlin L., Karlsson G., Adolfsson-Erici M., Larsson D.G.J. Using NMR metabolomics to identify responses of an environmental estrogen in blood plasma of fish. Aquat. Toxicol. 2006;78:341–349. doi: 10.1016/j.aquatox.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Schaller J., Brackhage C., Mkandawire M., Dudel E.G. Metal/metalloid accumulation/remobilization during aquatic litter decomposition in freshwater: a review. Sci. Total Environ. 2011;409:4891–4898. doi: 10.1016/j.scitotenv.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Singh B.K., Tripathi M., Pandey P.K., Kakkar P. Nimesulide aggravates redox imbalance and calcium dependent mitochondrial permeability transition leading to dysfunction in vitro. Toxicology. 2010;275:1–9. doi: 10.1016/j.tox.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Soga T., Baran R., Suematsu M., Ueno Y., Ikeda S., Sakurakawa T., Kakazu Y., Ishikawa T., Robert M., Nishioka T., Tomita M. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- Taylor J., King R.D., Altmann T., Fiehn O. Application of metabolomics to plant genotype discrimination using statistics and machine learning. Bioinformatics. 2002;18:S241–S248. doi: 10.1093/bioinformatics/18.suppl_2.s241. [DOI] [PubMed] [Google Scholar]
- Tweeddale H., Notley-McRobb L., Ferenci T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 1998;180:5109–5116. doi: 10.1128/jb.180.19.5109-5116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA, United States Environmental Protection Agency USEPA . 2002. Methods for measuring the acute toxicity of effluents and receiving waters to freshwater and marine organisms. [Google Scholar]
- Vellinger C., Felten V., Sornom P., Rousselle P., Beisel J.N., Usseglio-Polatera P. Behavioural and physiological responses of Gammarus pulex exposed to cadmium and arsenate at three temperatures: individual and combined effects. PLoS One. 2012:7. doi: 10.1371/journal.pone.0039153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant M.R. Recent developments in environmental metabolomics. Mol. Biol. 2008;4:980–986. doi: 10.1039/b805354e. [DOI] [PubMed] [Google Scholar]
- Viant M.R., Sommer U. Mass spectrometry based environmental metabolomics: a primer and review. Metabolomics. 2013;9:144–158. [Google Scholar]
- Viant M.R., Rosenblum E.S., Tjeerdema R.S. NMR-based metabolomics: a powerful approach for characterizing the effects of environmental stressors on organism health. Environ. Sci. Technol. 2003;37:4982–4989. doi: 10.1021/es034281x. [DOI] [PubMed] [Google Scholar]
- Villas-Bôas S.G., Mas S., Åkesson M., Smedsgaard J., Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005;24:613–646. doi: 10.1002/mas.20032. [DOI] [PubMed] [Google Scholar]
- Walle T., Walle U.K., Olanoff L.S. Quantitative account of propranolol metabolism in urine of normal man. Drug Metab. Dispos. 1985;13:204–209. [PubMed] [Google Scholar]
- Wu J.-1., Liu J., Cai Z. Determination of triclosan metabolites by using in-source fragmentation from high-performance liquid chromatography/negative atmospheric pressure chemical ionization ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 2010;24:1828–1834. doi: 10.1002/rcm.4558. [DOI] [PubMed] [Google Scholar]
- Yokogoshi H. Effects of some xenobiotics on the disposition of brain serotonin and catecholamine in rats. Agric. Biol. Chem. 1989;53:1609–1615. [Google Scholar]
- Zhang A., Sun H., Wang P., Han Y., Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material