Abstract
Mosquitoes are efficient, militarily relevant vectors of infectious disease pathogens, including many RNA viruses. The vast majority of all viruses are thought to be undiscovered. Accordingly, recent studies have shown that viruses discovered in insects are very divergent from known pathogens and that many of them lack appropriate reference sequences in the public databases. Given that the majority of viruses are likely still undiscovered, environ mental sampling stands to provide much needed reference samples as well as genetic sequences for comparison. In this study, we sought to determine whether samples of mosquitoes collected from different sites (the Caribbean and locations on the US East Coast) could be differentiated using metagenomic analysis of the RNA viral fraction. We report here distinct virome profiles, even from samples collected short distances apart. In addition to profiling the previously known viruses from these samples, we detected a number of viruses that have been previously undiscovered.
Keywords: mosquito virus, metagenome sequencing, novel virus, Bunyaviridae, metagenomics, high-throughput sequencing
Introduction
The concept of biological defense research is evolving to include not just the classical biological warfare agents such as Bacillus anthracis and Yersinia pestis but also to overlap somewhat with infectious disease research, especially in the case of emerging infectious diseases. Mosquito viruses are a particularly interesting topic within infectious disease research and biosurveillance for a number of reasons. First, mosquitoes are efficient and militarily relevant vectors of infectious disease. Not only are vector-borne diseases an important issue for international travelers, but historically, they have reduced deployed troops’ fighting capacity or even resulted in suspended operations.1 On the other hand, military campaigns themselves can influence the spread of vector-borne disease as well.1 In fact, nearly one quarter of emerging infectious diseases are vector borne.2 Other than the protozoan diseases malaria and leishmaniasis, many serious vector-borne threats, including Rift Valley fever virus, dengue virus, and chikungunya virus, are viral in nature and mosquito borne. These arthropod-borne viruses are termed arboviruses, and of all the arboviruses of consequence only one of them (African swine fever virus) has a genome made up of DNA; the rest are RNA viruses.
Anthropogenic or natural changes in the environment and/or climate may in future bring people closer to micro-organisms they have not been in contact with previously, potentially bringing novel viruses to the forefront of medical research. The vast majority of viruses are thought to be undiscovered, and recent studies have shown that viruses discovered in insects are very divergent from known pathogens.3–7 In particular, a recent study has shown that when viral particles were purified from wild-caught mosquitoes on the west coast of the United States and only viruses with DNA genomes were sequenced, greater than 48% of the resulting sequences were unidentifiable based on similarity to sequences in public genome databases. This suggests that we have limited knowledge of the microbiota of an important disease vector in the United States, and likely even less knowledge of the microbiota of disease vectors where our armed forces are deployed. The same study also found that a distance of 30–50 km between sampling sites was sufficient to result in a completely distinct mosquito virome, thus illustrating the extent of diversity that exists within these small vectors.
Mosquito viruses are also an interesting subject of research from the aspect of potential use as natural agents for prevention of disease transmission. Virus–virus interactions within a host or vector can manifest in various ways and can have positive or negative effects on either virus.8 Therefore, it stands to reason that infection with some viruses may potentially inhibit a mosquito’s ability to vector other viruses, and such interactions might one day be exploitable to prevent disease transmission. However, to do so successfully could be expected to require having very detailed knowledge of what the normal viral flora in a given mosquito population consists of.
Given that the majority of viruses are likely still undiscovered, environmental sampling stands to provide much needed reference samples for future; to define normal baseline viromes in reservoir, vector, and human populations; and to support the acquisition of basic virological information that will hopefully one day be used to design detection assays as well as therapeutic and prophylactic approaches. In particular, metagenomic sequencing is useful for high-throughput viral sampling studies as it is currently the most unbiased means of detection (requiring no prior knowledge of the organism to be detected, no specific primers or antibodies, etc.) and can therefore detect novel organisms as well as nonculturable organisms and coinfections. In this study, we sought to determine whether the samples of mosquitoes collected from different sites (the Caribbean and locations on the east coast of the United States) could be differentiated using metagenomic analysis of the RNA virus fraction. For this purpose, mosquito samples were collected from several locations, sequenced, and analyzed for taxonomic and functional classification. Herein, we report distinct virome profiles, even from samples collected only short distances apart. In addition to profiling sequences from these samples that correspond to sequences from previously known viruses, we detected sequences that may be derived from a number of viruses that have been previously unreported.
Methods
Mosquito collection
Mosquitoes in Maryland (MD) and West Virginia (WV) were collected using a CDC Mini Light Trap (BioQuip) from multiple outdoor locations in June and July of 2014, between dusk and dawn. Mosquitoes in Puerto Rico (PR) were collected at or after sunset in June of 2014 on walls, furniture, or drying clothes, by using large centrifuge tubes outfitted with a funnel made of Whatman filter paper and set on the outside below the rim of the tube’s mouth. All mosquitoes were killed by freezing and stored at −80 °C until homogenization.9
Mosquito total RNA preparation
Pooled mosquitoes were homogenized using a TissueLyser (Qiagen Sciences Inc.) in 560 µL lysis buffer from the QIAamp Viral RNA kit with a 5 mm stainless steel bead at 25 Hz for two pulses of 30 seconds each. Samples were then centrifuged at 10,000 × g for two minutes and the supernatant removed. Centrifugation and supernatant removal were repeated until a visible pellet no longer formed. A total of 5.6 µL carrier RNA was added and samples were incubated at room temperature for 10 minutes, followed by incubation with 1 µL DNAse at 37 °C for 5 minutes. The QIAamp Viral RNA kit was then used as per manufacturer’s instructions, beginning with the addition of ethanol. For pools of 50 mosquitoes, RNA was eluted from each column with 40 µL of buffer AVE followed by a second elution with 20 µL AVE. For pools of 25 mosquitoes, RNA was eluted from each column with 30 µL AVE, and then 20 µL of the eluate was put back on the column and used for a second elution. Total RNA was quality checked for concentration, for purity using optical density 260/280 and 260/230 ratios, and for integrity (RNA Integrity Number ≥0.8).
Sequencing
Sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Preparation kit with Ribo-Zero Gold (Illumina Inc.) and 1 µg input RNA per pooled sample. Library quality was assessed using an Agilent 2100 Bioanalyzer. Libraries from each location were pooled and sequenced on the MiSeq platform using MiSeq Reagent Kit v2 (500 cycles). All sequence runs were verified as passing internal quality checks for yield and % >Q20. Raw sequencing reads were submitted to NCBI Sequence Read Archive (SRA) under accession SRP070112.
Sequence analyses
Sequences were processed using CLC Genomics Workbench 6.0.5 for Maryland and West Virginia samples and version 6.5 for Puerto Rico samples. Reads were aligned to Aedes aegypti transcripts downloaded from VectorBase (version 3.1; downloaded Aug. 6, 2014) and/or Culex quinquefasciatus transcripts downloaded from NCBI (downloaded Aug. 11, 2014). The resulting set of unaligned reads were de novo assembled into contigs in CLC using default parameters. Contigs 200 bp or larger10 were compared to NCBI’s BLAST databases.11 An e-value cutoff of 1 × 10−5 was used for all searches.3 BLAST results were compared across samples using MEGAN5.12 As a complementary approach to BLAST analysis, LMAT13 was also used to analyze contigs and singleton reads, and the output was visualized using MEGAN. Multiple sequence alignments were performed using Clustal Omega on the EMBL-EBI website (http://www.ebi.ac.uk) and default parameters and/or using CLC with default parameters. Protein modeling was performed using PHYRE2.14 Maximum-likelihood phylogenetic trees were computed in CLC (v.8.5.1) using a neighbor-joining model as the initial tree, Jukes–Cantor parameters for distance measurements and 500 bootstrap replicates.
Results
Total RNA was isolated from mosquitoes collected in various locations from Maryland, West Virginia, and Puerto Rico; pooled by location; and sequenced using the Illumina MiSeq platform. There were roughly 7–29M sequencing reads produced per insect pool (Table 1). The resulting reads were first screened against A. aegypti and C. quinquefasciatus transcripts to reduce the contribution of host-derived reads and then subjected to de novo assembly. Not surprisingly, de novo assembly resulted in a range of assembly sizes from 1936 contigs (Martinsburg, WV, USA) to 65,655 contigs (San German, Puerto Rico). For complex metagenomic samples such as these, the number of contigs can vary based on the sequencing depth, the complexity of sample, and the intrinsic qualities of the sequences themselves (repeat regions, etc.).
Table 1.
Sequencing statistics by location.
| SAMPLE NAME | SAMPLING LOCATIONS (# INSECTS) | TOTAL SEQUENCING READS | MEAN READ LENGTH (NT) | TOTAL CONTIGS | SRA EXPERIMENT |
|---|---|---|---|---|---|
| Frederick | Frederick, MD (50) | 19.5M | 131 | 32,791 | SRR3168926 |
| Germantown | Germantown, MD (60) | 9.1M | 129.6 | 35,345 | SRR3168927 |
| Jefferson | Jefferson, MD (50) | 29.1M | 140.3 | 6,646 | SRR3168918 |
| Martinsburg | Martinsburg, WV (50) | 24.2M | 137.6 | 1,936 | SRR3168919 |
| Cabo Rojo | Cabo Rojo, PR (14) | 10.1M | 131 | 60,178 | SRR3168917 |
| Lajas | Lajas, PR (25) | 10.2M | 131.1 | 57,421 | SRR3168920 |
| Moca Aguadilla | Aguadilla, PR (5); Moca, PR (11); Aguada, PR (9) | 9.9M | 131.1 | 5,999 | SRR3168921 |
| San German | San German, PR (18) | 9.7M | 154.8 | 65,655 | SRR3168922 |
| Arecibo | Arecibo, PR (25) | 6.8M | 145.8 | 26,810 | SRR3168923 |
| Hatillo | Hatillo, PR (25) | 10.0M | 136.5 | 22,487 | SRR3168924 |
| Mayaguez | Mayaguez, PR (25) | 13.0M | 133.4 | 33,800 | SRR3168916 |
| Ponce Adjuntas | Ponce, PR (10); Adjuntas, PR (12) | 8.7M | 146.8 | 4,967 | SRR3168925 |
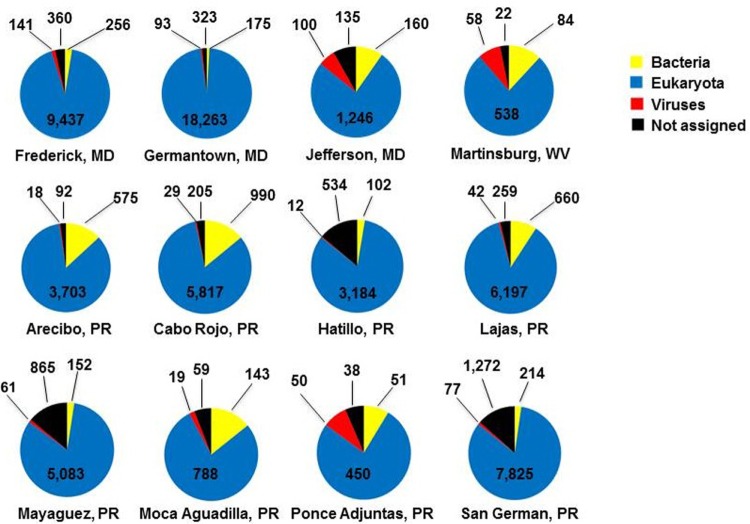
In order to perform taxonomic profiling of each sample, contigs were analyzed by BLASTn and BLASTx against NCBI’s nr/nt. Not unexpectedly, the majority of contigs for each sample was found to be eukaryotic, or mosquito derived (Fig. 1), despite eukaryotic (human, mouse, and rat) ribosomal RNA subtraction probes that are included as part of library preparation and a bioinformatic step aimed at reducing mosquito transcripts. The observation that the majority of identifiable sequences in these samples were eukaryotic in nature is not surprising. For instance, we have previously shown that when RNA from the entire insect is taken as the starting material and unbiased shotgun sequencing is performed post rRNA subtraction, the majority of identifiable sequences are eukaryotic, even for A. aegypti with high-titer dengue virus infection.15 The portions of each sample that were detected as being derived from bacteria and viruses constituted the minority of the sample, and a fair number of sequences could not be assigned to any of these categories by these methods.
Figure 1.
Taxonomic profiling of contigs by location. The numbers indicate raw count of contigs in each category. The de novo assembled contigs were assigned to categories using BLAST.
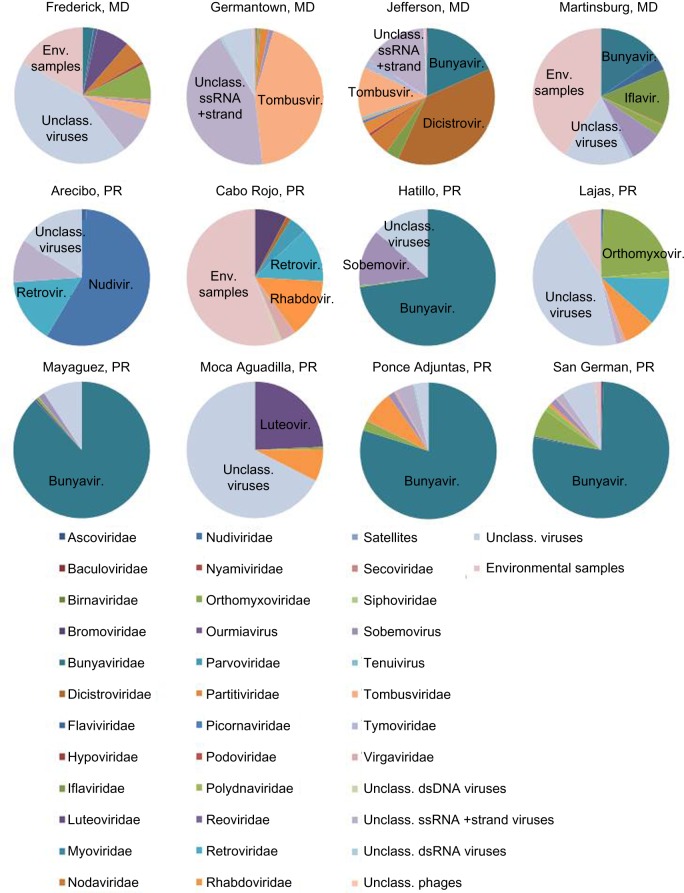
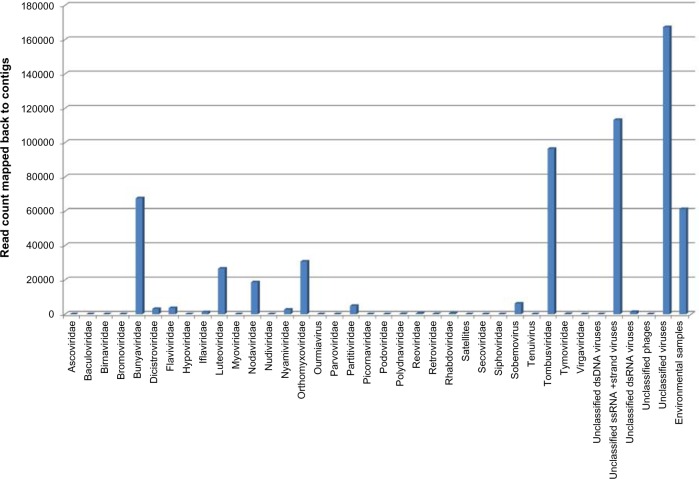
In order to compare the viral content of each sample, the contigs identified as viral by BLASTn and/or BLASTx were categorized by viral family, if the closest sequenced relative was classified (Fig. 2 and Supplementary File 1), or into general categories such as unclassified dsDNA virus. Overall, the viral makeup of samples was found to vary widely across the locations studied, although there were some similarities, particularly among the Puerto Rico samples. The two most common virus groupings identified in the contigs included unclassified viruses (28% of reads mapped back to contigs) and unclassified ssRNA positive-strand viruses (19%), followed by Tombusviridae (16%), Bunyaviridae (11%), and environmental samples (10%; Fig. 3). More than half of the reads making up the assemblies (>57%) were found to be contributed by various types of unclassified viruses and sequences from NCBI classified as environmental samples.
Figure 2.
Virome content by location. The proportion of the identifiable virome contributed by each viral family or group is estimated using a read mapping approach. Briefly, raw reads were mapped back to de novo assembled contigs using CLC Genomics Workbench, and for each of the contigs that was identified by BLAST as being of viral origin, the number of raw reads mapping to that contig was extracted and is presented here.
Figure 3.
Contribution of virus families to total assembled sequences. The total number of raw reads mapped back to viral contigs using CLC Genomics Workbench per virus family or grouping is presented here, for all samples combined.
Overall, sequences were identified corresponding to 33 distinct virus families and 9 other viral groupings, including three genera (Tenuivirus, Sobemovirus, and Ourmiavirus) for which families have not been assigned according to the International Committee on Taxonomy of Viruses (ICTV) Master Species List 2014 v4.16 Among the viral sequences identified was a 10.6 kb contig from the Frederick, MD, pool (Frederick contig 191, sequence provided in Supplementary File 2) with 99% nucleotide identity to Culex flavivirus HOU24518 (FJ502995.1), which was previously isolated in Houston, TX.17 This single contig was covered at an average depth of 34.06× and contained >97.8% of the viral genome, which suggests that this virus may have been present at a relatively high titer as compared with the other viruses identified in that pool. Only 183 and 54 nucleotides were missing from the 5′ and 3′ ends of the genome, respectively. To our knowledge, although Culex flavivirus (CxFV) has been reported from Japan and Indonesia,18 China,19 Guatemala,20 Mexico,21 Argentina, Uganda,22 Trinidad, and the United States (Texas, Colorado, Iowa, Illinois, Mississippi, and Georgia),17,23–26 ours would be the first report of CxFV in the mid-Atlantic region (Maryland) of the United States.
Identified also from the Frederick, MD, pool were multiple sequences (Frederick contigs 89, 350, and 1234; Supplementary File 2) with very high depth of coverage (>800× average depth each) and >99% nucleotide identity to 2 RNA-dependent RNA polymerase (RdRP) genes from the family Bunyaviridae that were detected by metagenomic sequencing of mosquitoes from California (NCBI accession numbers KP642114.1 and KP642115.14) and deposited as sequences from environmental samples at NCBI. Multiple sequence alignment showed that all five sequences are highly similar but for a small number of single-nucleotide polymorphisms.
Among the mosquito pools from Puerto Rico, sequences corresponding to the family Bunyaviridae were found to be very prevalent and the vast majority of the bunyavirus sequences from these pools were found to be derived from Phasi Charoen-like virus. All three genome segments (L, M, and S) were identified in the contigs, and the sequences were found to have ≥95% nucleotide identity to isolate Rio. This virus, which was originally reported in A. aegypti from Thailand,27 was not detected in any of the contigs from the mid-Atlantic pools.
Despite the outbreak of chikungunya virus in the Caribbean, there were no CHIKV sequences detected in either the contigs or the singleton reads, by BLASTn, BLASTx, LMAT, or read mapping. In fact, there were no contigs identified with similarity to this or any other viruses within the Togaviridae family.
Next, contigs with less similarity to sequences within the NCBI databases were more closely examined to determine their possible taxonomic classification and function. In several cases, based on the predicted function and percent identity to their nearest relative, we presume that multiple contigs are likely to be derived from the same virus. In these instances, the sequence and believed function of each contig was used to infer the position in the genome. Novel viral sequences identified included members of the families Bunyaviridae, Orthomyxoviridae, and Dicistroviridae, all of which contain members known to infect arthropods. The novel virus sequences identified in this study are summarized in Tables 2 and 3, and the corresponding assembled sequences are provided in Supplementary File 2.
Table 2.
Novel virus sequences discovered in Maryland and West Virginia mosquitoes.
| POOL | CONTIG DESIGNATION | CONTIG LENGTH (KB) | VIRAL GROUP | CLOSEST SEQUENCED RELATIVE | GENE PRODUCT(S) ENCODED | DEPTH |
|---|---|---|---|---|---|---|
| Frederick, MD | 5105 | 0.26 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | Nucleocapsid | 2.81 |
| 246 | 1.03 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | Glycoprotein | 12.66 | |
| 838 | 0.32 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | Glycoprotein | 4.56 | |
| 4315 | 0.40 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | Glycoprotein | 4.53 | |
| 2895 | 0.37 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | Glycoprotein | 5.76 | |
| 2560 | 0.25 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | RdRp | 5.01 | |
| 562 | 0.34 | Bunyaviridae | Yichang Insect Virus (AJG39334.1) | RdRp | 4.43 | |
| 4733 | 9.8 | Unclassified | Rosy apple aphid virus (DQ286292) | Structural protein, polyprotein | 338.97 | |
| 13,993 | 12.4 | Mononegavirales | Xincheng Mosquito virus (KM81766.1) | Nucleoprotein, Glycoprotein, RdRp | 30.28 | |
| Martinsburg, WV | 825 | 0.59 | Bunyaviridae | Manawa Virus (AFN73042.1) | RdRp | 5.68 |
| 690 | 0.34 | Bunyaviridae | Manawa Virus (AFN73042.1) | RdRp | 4.17 | |
| 464 | 1.24 | Bunyaviridae | Manawa Virus (AFN73042.1) | RdRp | 8.22 | |
| 374 | 0.42 | Bunyaviridae | Manawa Virus (AFN73042.1) | RdRp | 7.35 | |
| 567 | 0.34 | Bunyaviridae | Manawa Virus (AFN73042.1) | RdRp | 6.25 | |
| 295 | 1.74 | Bunyaviridae | Manawa Virus (AFN73042.1) | RdRp | 8.40 | |
| Jefferson, MD | 5816 | 9.68 | Dicistroviridae | Cricket Paralysis virus (NP_647481.1) and Aphid Lethal Paralysis virus (NP_733845.1) | 2 Polyproteins, NS and S | 45.10 |
Table 3.
Novel virus sequences discovered in mosquitoes from Puerto Rico.
| POOL | CONTIG DESIGNATION | CONTIG LENGTH (KB) | VIRAL GROUP | CLOSEST SEQUENCED RELATIVE | GENE PRODUCT(S) ENCODED | DEPTH |
|---|---|---|---|---|---|---|
| Arecibo, Puerto Rico | 14,385 | 0.21 | Unclassified viruses | Dansoman virus (KP714086.1) and/or Chronic Bee Paralysis virus | Replicase | 8.10 |
| 16,219 | 0.36 | Unclassified viruses | Dansoman virus (KP714086.1) and/or Chronic Bee Paralysis virus | Replicase | 2.84 | |
| 24,685 | 0.23 | Flaviviridae | Culex theileri flavivirus RP-2011 | Polyprotein | 1.94 | |
| Cabo Rojo, Puerto Rico | 11,265 | 0.70 | Rhabdoviridae | Wuhan Mosquito Virus 9 (AJG39218.1) | RdRp | 3.41 |
| 36,482 | 0.31 | Rhabdoviridae | Wuhan Mosquito Virus 9 | RdRp | 4.36 | |
| 43,737 | 0.25 | Rhabdoviridae | Sanxia Water Strider virus 5 strain SX5SP11 (KM817634.1) and Wuhan Mosquito Virus 9 (AJG39218.1) | RdRp | 5.95 | |
| 5468 | 0.54 | Rhabdoviridae | Wuhan Mosquito Virus 9 strain JX1-13 (KM817659.1) | RdRp | 3.39 | |
| 40,415 | 0.28 | Parvoviridae | Culex pipiens densovirus (YP_002887624.1) | Nonstructural protein | 3.07 | |
| Hatillo, Puerto Rico | 171 | 0.77 | Unclassified viruses | Ixodes scapularis associated virus 2 (AII01812.1) | RdRp | 50.18 |
| 174 | 0.77 | Unclassified viruses | Ixodes scapularis associated virus 2 (AII01812.1) | RdRp | 46.38 | |
| 8438 | 0.23 | Unclassified viruses | Ixodes scapularis associated virus 1 (AII01797.1) | RdRp | 2.97 | |
| 1161e1 | 1.06 | Sobemovirus | Bat sobemovirus (AGN73380.1) | Capsid | 48.66 | |
| 1162 | 1.35 | Sobemovirus | Bat sobemovirus (AGN73380.1) | Capsid | 19.19 | |
| Mayaguez, Puerto Rico | 213 | 1.78 | Unclassified viruses | Ixodes scapularis associated virus 2 (AII01812.1) | RdRp | 174.41 |
| 371 | 0.52 | Unclassified viruses | Ixodes scapularis associated virus 2 (AII01811.1) | Protease | 181.93 | |
| 372 | 2.74 | Unclassified viruses | Ixodes scapularis associated virus 2 (AII01811.1 and AII01812.1) | RdRp and protease | 68.58 | |
| 10,131 | 0.22 | Orthomyxoviridae | Wuhan Mosquito Virus 5 strain XC1-7 (KM817624.1) | PB1 protein | 2.19 | |
| 12,947 | 0.45 | Orthomyxoviridae | Wuhan Mosquito Virus 5 strain XC1-7 (KM817624.1) | PB1 protein | 3.70 | |
| 15,309 | 0.28 | Orthomyxoviridae | Wuhan Mosquito Virus 5 strain XC1-7 (KM817624.1) | PB1 protein | 2.71 | |
| 17,647 | 0.33 | Orthomyxoviridae | Wuhan Mosquito Virus 5 strain XC1-7 (KM817624.1) | PB1 protein | 4.34 | |
| 1772 | 2.35 | Orthomyxoviridae | Wellfleet Bay virus | Nucleoprotein | 11.73 | |
| San German, Puerto Rico | 3879 | 1.32 | Orthomyxoviridae | Wellfleet Bay virus (YP_00911683.1) | Nucleoprotein | 25.54 |
Notes:
Denotes edited sequence. The portion of chimeric contig that corresponded to eukaryotic mitochondrial RNA was removed.
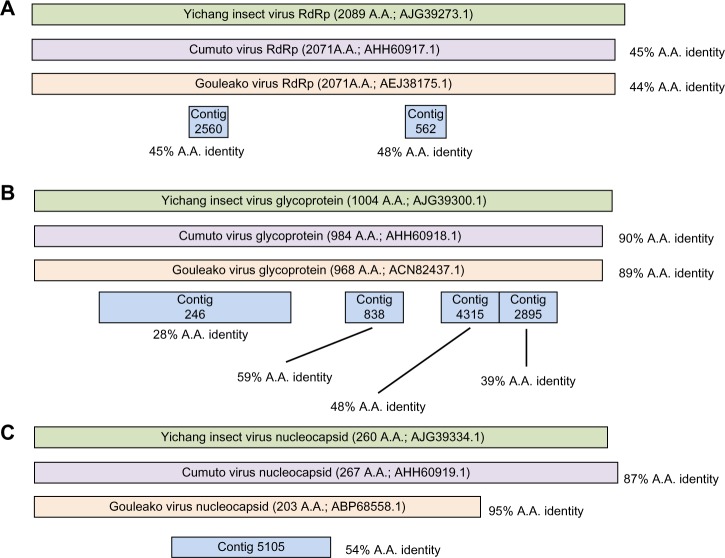
For instance, from the Frederick pool of mosquitoes, sequences that correspond to all three genome segments of a member of the Bunyaviridae were identified. Collectively, Frederick contigs 246, 838, 4315, and 2895 (Supplementary File 2) constitute more than half of the M segment, which in Bunyaviridae encodes the glycoprotein gene (Fig. 4B). One of the two expected conserved domains was detected by BLASTx analysis (Phlebovirus_G1; pfam 07243). Although each of these four contigs was found to have no similarity at the nucleotide level to anything in nt, BLASTx analysis identified 28%, 59%, 48%, and 39% amino acid identity to Yichang insect virus glycoprotein (AJG39300.1; a virus identified in insects from China3). For each of these contigs, the next best hits were to Cumuto virus and Gouleako virus, each of which is more similar to Yichang insect virus than these four Frederick contigs are to any published sequence. Contigs corresponding to the L segment (Frederick contigs 2560 and 562; Supplementary File 2 and Fig. 4A) and S segment (Frederick contig 5105; Supplementary File 2 and Fig. 4C) were also identified, and similar to the M segment contigs, they showed no significant similarity to any sequence in NCBI nt at the nucleotide level, but had protein similarity to Yichang insect virus, followed by Gouleako and Cumuto viruses. The S segment contig that was recovered constitutes roughly one-third of the expected nucleocapsid sequence based on these other three insect viruses and contains a conserved nucleocapsid domain (Tenuivirus/Phlebovirus nucleocapsid protein; pfam 05733). As the M segment contig 246 and the S segment contig 5105 were the largest of the contigs identified for this novel virus candidate, they were each used in multiple sequence alignments and maximum-likelihood phylogenetic trees. Not surprisingly, in both cases, the sequence for this novel virus candidate was found to be more closely related to other insect viruses than to the prototypical human pathogens in this family (Supplementary File 3). Given the apparent divergence of this virus as compared to the available reference sequences, the S segment contig 5105 was used for protein modeling to confirm our supposition that it encodes a novel Bunyaviridae nucleocapsid. Indeed, we found that the predicted three-dimensional structure for this protein is very consistent with that of a Bunyaviridae nucleocapsid protein (Supplementary File 4), confirming our assignment of the sequence.
Figure 4.
Novel bunyavirus example from Frederick, MD. In this schematic, contigs corresponding to various genes of a proposed novel bunyavirus from mosquitoes collected in Frederick, MD, are shown aligned to the three closest sequenced relatives. (A) The contigs corresponding to RdRP sequences, (B) contigs corresponding to glycoprotein sequences, and (C) contigs corresponding to nucleocapsid sequences. Percent amino acid identity for these contigs and for Cumuto and Gouleako sequences is shown with respect to Yichang insect virus protein sequences.
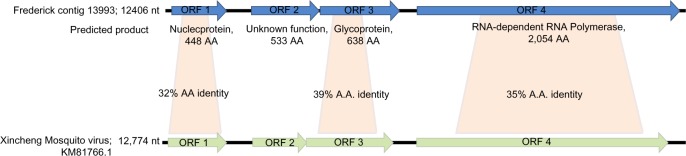
Also from the Frederick pool, we identified a contig of 12.4 kb in length, covered at over 30× average depth. Interestingly, despite its length, this sequence (Frederick contig 13,993; Supplementary File 2 and Fig. 5) was found to have no significant similarity at the nucleotide level to any sequence in NCBI nt. Based on the combination of BLASTx results and RAST annotation, four open reading frames (ORFs) were identified in this contig (BLASTx initially identified two ORFs, one corresponding to the RdRp and one corresponding to a glycoprotein, whereas RAST28 identified two additional ORFs), three of which were found to have significant similarity with ORFs predicted for a virus that was identified by massive parallel sequencing of various insects from China, Xincheng Mosquito virus (KM81766.13). Of these three, ORF 1 encodes the putative nucleoprotein, ORF 3 encodes the putative glycoprotein, and ORF 4 encodes the RdRp. The predicted products of these three ORFs bear 32%, 39%, and 35% amino acid identity to their counterparts in Xincheng Mosquito virus, respectively. A second in silico gene finder, FGENESV (linux1.softberry.com), confirmed the four ORFs.
Figure 5.
Schematic of virus genome encoded by Frederick contig 13,993. The four ORFs encoded by Frederick contig 13,993 are depicted here along with their putative protein products and the amino acid identity of those products to the sequences of the closest sequenced relative, Xincheng Mosquito virus.
As the closest sequenced relative to the novel virus represented by Frederick contig 13,993 is itself a novel virus identified solely from high-throughput genetic characterization of metagenome sequences, we sought to gain more biological information about the virus by using various tools to characterize the features of this new genome further. Additional characterization of the predicted RdRP using DELTA BLAST indicated that the protein contains several conserved domains common to Mononegavirales RdRps, including a domain associated with RNA-capping. Significantly, the nucleotide sequence contains conserved Kozak sequences for all four predicted genes. Homology searches for ORF1 using BLASTp provided little useful information. However, protein modeling using PHYRE2 indicated that ORF1 has 30% sequence similarity to the bornavirus p40 protein. Alignment of the available NCBI sequence for bornavirus p40 (gi_516503) confirmed the modeling results with one long stretch of significant alignment (residues 169–313) and an overall sequence identity of 41% (results not shown). Notably, ORF 2 bears no significant amino acid similarity to ORF 2 of Xincheng Mosquito virus or to anything in NCBI nt, but is predicted by protein modeling in PHYRE2 to be a zinc finger protein possibly involved in transcription and/or RNA binding and to have similar structure to the solved crystal structure of residues 185–199 of herpes simplex virus nuclear egress complex.29
Further sequence analysis of contig 13,993 with ATGPR, a tool for prediction of translation initiation, indicated that the CDS in ORF3 is 228 amino acids larger (638 AA versus 410 AA) than the analogous prediction by RAST. Homology searches using BLASTp indicated that the predicted 638 amino acid protein shared 39% identity with the glycoprotein from Xincheng Mosquito virus. Additional in silico analysis of the predicted protein suggested the presence of a transmembrane domain residing at the C-terminus (TMHMM Server v 2.0 www.cbs.dtu.dk/services/TMHMM). Moreover, the putative glycoprotein includes three potential sites for N-linked glycosylation as well as seven potential sites for O-linked glycosylation. These predictions are overall consistent with the expected properties of a viral glycoprotein (NetNGly 1.0 Server; NetOGly 4.0 Server; http://www.cbs.dtu.dk/).30
In addition, a 9.8 kb contig (Frederick contig 4733; Supplementary File 2) was identified from the Frederick sample that was covered at an average depth of 338.97×. Intriguingly given the relative length of this contig sequence and its deep coverage, BLASTn and megaBLAST searches returned only a short stretch (248 bases) of nucleotide similarity to a positive-strand RNA virus called rosy apple aphid virus (GenBank accession DQ286292),31 and BLASTx revealed limited amino acid identity to that same virus as well. Specifically, the contig encodes a structural protein in the −1 frame that bears 54% amino acid identity to rosy apple aphid virus putative structural protein and 53% amino acid identity to Acyrthosiphon pisum virus (an unclassified virus thought to be distantly related to Picornaviruses,32 NCBI Reference Sequence NC_003780.1) protein P2, and a polyprotein in the −2 frame was found to have 61% amino acid identity to the polyprotein of rosy apple aphid virus and 62% amino acid identity to Acyrthosiphon pisum virus protein P1 by BLASTx. Lending support to the functional assignments, putative conserved domains were detected, specifically 2 for an RdRp and 1 for a helicase. It is likely that this contig consists of the entire or nearly entire genome of this novel virus, as the complete genomes of rosy apple aphid virus and Acyrthosiphon pisum virus are only 9992 and 10,016 nucleotides, respectively.
Given the apparent novelty of some of the viral sequences discovered in this study, we further sought to gage the validity of our findings by looking for evidence of these or related viruses in published insect metagenomes from two recent studies. Specifically, we analyzed reads consisting of 7 mosquito datasets from 3 different species collected in California4 and 15 arthropod datasets plus 1 spider dataset, all 16 of which were collected in China.3 All 23 metagenomic datasets were mapped against Frederick contigs 13,993, 246, 2560, 4733, 5105, and 562, as well as Jefferson contig 5816. Interestingly, in all but one case, there were no mapped reads, indicating the apparent absence of our novel viral sequences within these samples. The one exception is Frederick contig 13,993. In this case, there was one sample from arthropods in China (SRR17457673), from which a positive result was obtained. Specifically, 969 reads from this sample mapped to Frederick contig 13,993, covering 90.28% of the entire sequence length (Supplementary File 5). Even with the 79 single-nucleotide polymorphisms, 1 insertion/deletion, and 18 gaps in the consensus sequence obtained from the Chinese sample, this may represent a virus present in the Chinese insects that is more closely related to the virus represented by Frederick contig 13,993 than any virus whose genome currently exists in the NCBI databases.
Discussion
In this study, we sequenced and analyzed mosquito viromes from four mid-Atlantic locations and eight locations in Puerto Rico, and we found these pooled insect viromes to vary greatly within short distances, even as little as 18 km apart (Supplementary Files 6 and 7). A major limitation of this study is that the genera of the mid-Atlantic mosquitoes were not determined, although the mosquitoes from Puerto Rico were morphologically identified as belonging to the genera Culex and Aedes. In order to have enough material to create sequencing libraries, mosquitoes were pooled by location. Therefore, the virome comparisons presented here were conducted among geographic regions rather than among mosquito species, similar to previously published pooled metagenome analyses of insects3 and of bats of different species roosting together.33
Overall, one major difference observed between the Puerto Rico pools and the mid-Atlantic pools was with respect to the family Bunyaviridae. In the mid-Atlantic mosquitoes, there were novel Bunyaviridae detected, whereas among the Puerto Rico samples there were mainly previously described Bunyaviridae detected, particularly, Phasi Charoen-like virus. Phasi Charoen-like virus was the single most detected, or best represented, virus in all the samples, and it was found to completely dominate the Mayaguez virome. The presence of Phasi Charoen-like virus in mosquitoes from Puerto Rico and not in mosquitoes from Maryland and West Virginia is possibly due to the prevalence of A. aegypti in the tropics and not in the mid-Atlantic states. However, it is currently unclear why the Bunyaviridae sequence fraction from the Maryland and West Virginia mosquitoes is composed of apparently novel sequences, whereas apparently novel Bunyaviridae sequences were not detected in the mosquitoes from Puerto Rico. The lack of good comparator reference sequences in the public databases for many of the viral sequences detected in this study, in particular the Maryland samples, suggests that further exploration of this sequence space via more in-depth study of the mid-Atlantic mosquitoes, particularly cultivation of these viruses in eukaryotic cell culture, might be useful for virus discovery efforts. Interestingly, all the samples from Puerto Rico tested negative for CHIKV and all other viruses from the Togaviridae family. However, confirmed cases have rapidly increased since July 2014 and are now recorded for all the areas included in this study.34 It is likely that samples were collected directly before the virus became prevalent in these areas, or alternatively, perhaps CHIKV was present at levels below the limit of detection or was present in mosquitoes at a low rate (in a proportion less than the number of mosquitoes sequenced in this study).
Herein, we relied completely on shotgun metagenomic sequencing, in the absence of virus cultivation methods, to identify and genetically characterize viruses within insect vectors, including roughly 57 kb of novel viral genomic data. In some cases, nearly complete novel viral genomes were identified. Using the publically available sequence data at NCBI and various analytical methods, including protein modeling tools, we attempted to genetically characterize these newly discovered viruses.
A caveat of our study is that in the absence of any attempt at cultivation, the possibility exists that some of the viral sequences detected in this study may represent endogenous viral elements (EVEs), viral sequences that have been incorporated into the mosquito genome(s). Cui and Holmes recently reported the detection of putative EVEs within published genomes for various insects, including A. aegypti.35 However, we did not detect any direct evidence that the sequences we describe in our study are EVEs. On the contrary, at the time of this work, there are currently 25 mosquito genomes available, and anything correlating to any part of a sequenced mosquito genome would not be detected as a novel virus in our analyses; it would be identified as a mosquito sequence and not studied further. BLAST analysis of the putative novel viral sequences presented here did not reveal hits to viral sequences incorporated into mosquito genomes or any other parts of mosquito genomes, only limited (mostly amino acid) similarity to viral genomes. Additionally, the complete or nearly complete viral genome sequences detected in our study are even more unlikely to be EVEs, or if they are, they must be very recent events, because within them we do not detect nonsense mutations, frame shifts, etc. (as the putative A. aegypti EVEs identified in the work by Cui and Holmes exhibit).35 Furthermore, long RNA viral sequences such as the CxFV sequences, Frederick contig 13,993, and so forth are unlikely to be EVEs since for RNA viruses with no DNA intermediate the mechanism of integration is currently unknown but likely involves viral mRNA36 and Frederick contig 13,993 encodes multiple putative ORFs as well as intergenic regions. It should also be noted that we do not draw a distinction in our study between actively replicating viruses and various forms of nonreplicating viruses because (1) shotgun metagenomic sequencing as was conducted here will detect both replicating and nonreplicating viruses and (2) we sequenced RNA from the entire insect, and in some cases, viruses can be transmitted mechanically, in the absence of replication within the insect.
It is important to note that the novel viral content presented here is very likely an underestimate of the novel virus content in these samples, for two main reasons. First, in order to be detected by our analyses, contigs must bear some homology with published sequences. Extremely divergent viruses would not be detected by our methods. Second, our analysis of novel viruses was limited to assembled sequences; so, viruses present at very low titer and hence represented by very few reads that therefore do not assemble due to lack of overlap would also not be detected. The novel viruses that we were able to identify by our methods, coupled with our finding that more than half of the identified viral reads making up the assemblies were found to be derived from various types of unclassified virus and sequences submitted to NCBI as environmental samples, suggest that a significant fraction of the virome in these tiny vectors is composed of understudied viruses or viruses about which relatively little is known. Therefore, future studies are necessary to fully catalog viral diversity and perform genetic and phenotypic characterization of these viruses. It is clear that the public sequence databases are human–pathogen centric and do not adequately represent environmental sequences from across the entire tree of life. This is very true for viruses, and the majority of them are thought to be as yet unsampled. However, each successive sequencing study has the potential to add to our knowledge base and expand the sequence information that is available to query against, therefore improving detection and aiding our understanding of how insect viromes and human pathogens within them have evolved.
Conclusion
In this study, we sequenced and analyzed mosquito viromes from four mid-Atlantic locations and eight locations in Puerto Rico, and we found these pooled insect viromes to vary greatly within short distances. We identified 57 kb of novel viral sequence data, including several putative intact or nearly intact viral genomes that we present here. Further study is required to sample more environmental niches and add to the viral genome repertoire in public databases. Gaining a better understanding of the microbiota within these tiny disease vectors throughout the world is arguably an important component to both biosurveillance and military force health protection.
Acknowledgments
Ms. Biser was employed at NMRC through the Naval Research Enterprise Internship Program (NREIP). The authors acknowledge Cassie Redden, Jens Andersen, and Kevin Schully for assistance with mosquito collections as well as Joseph Anderson and Regina Cer for helpful discussions. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, nor the U.S. Government. VPM, GP, and TH are military service members of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,804 words, excluding any confidential comments to the academic editor.
FUNDING: This study was supported by Work Unit Number A1417. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: KGF, KAB-L. Performed the sequencing: TB. Analyzed the data: KGF, TB, KAB-L. Wrote the first draft of the manuscript: KGF, KAB-L. Contributed to the writing of the manuscript: TB, VPM, TH, GP, CJS. Agreed with manuscript results and conclusions: KGF, TB, VPM, TH, GP, CJS, KAB-L. Jointly developed the structure and arguments for the article: KGF, TB, KAB-L. Made critical revisions and approved the final version: KGF, KAB-L. All the authors reviewed and approved the final article.
Supplementary Materials
Supplementary File 1. Percentage of viral contigs in each viral family or grouping by location.
Supplementary File 2. Sequences of selected viral contigs.
Supplementary File 3. Maximum likelihood phylogenetic tree from multiple sequence alignment of M segment contig 246 from Frederick sample and known Bunyavirus M segment sequences.
Supplementary File 4. Protein modelling of S segment contig 5105 from Frederick using PHYRE2. (A) Putative 3-dimensional structure of product of Frederick contig 5105. (B) The closest structure in the database to the putative 3-dimensional structure of Frederick contig 5105, which is the solved crystal structure of Rift Valley fever virus nucleocapsid protein.
Supplementary File 5. Mapping of reads from published dataset SRR1745767 against Frederick contig 13,993.
Supplementary File 6. mosquito viromes from Maryland and West Virginia and approximate distance between sampling sites.
Supplementary File 7. Mosquito viromes from Puerto Rico and approximate distance between sampling sites.
REFERENCES
- 1.Pages F, Faulde M, Orlandi-Pradines E, Parola P. The past and present threat of vector-borne diseases in deployed troops. Clin Microbiol Infect. 2010;16(3):209–24. doi: 10.1111/j.1469-0691.2009.03132.x. [DOI] [PubMed] [Google Scholar]
- 2.Junglen S, Kurth A, Kuehl H, et al. Examining landscape factors influencing relative distribution of mosquito genera and frequency of virus infection. Ecohealth. 2009;6(2):239–49. doi: 10.1007/s10393-009-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li CX, Shi M, Tian JH, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015:4. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandler JA, Liu RM, Bennett SN. RNA shotgun metagenomic sequencing of northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Front Microbiol. 2015;6:185. doi: 10.3389/fmicb.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auguste AJ, Carrington CV, Forrester NL, et al. Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex (Culex) declarator mosquitoes in Trinidad. J Gen Virol. 2014;95:481–5. doi: 10.1099/vir.0.058412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasilakis N, Forrester NL, Palacios G, et al. Negevirus: a proposed new Taxon of insect-specific viruses with wide geographic distribution. J Virol. 2013;87(5):2475–88. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffey LL, Page BL, Greninger AL, et al. Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology. 2014;448:146–58. doi: 10.1016/j.virol.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DaPalma T, Doonan BP, Trager NM, Kasman LM. A systematic approach to virus-virus interactions. Virus Res. 2010;149:1–9. doi: 10.1016/j.virusres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Guinn ML, Turell MJ. Effect of triethylamine on the recovery of selected South American alphaviruses, flaviviruses, and bunyaviruses from mosquito (Diptera: Culicidae) pools. J Med Entomol. 2002;39(5):806–8. doi: 10.1603/0022-2585-39.5.806. [DOI] [PubMed] [Google Scholar]
- 10.Stremlau MH, Andersen KG, Folarin OA, et al. Discovery of novel rhabdoviruses in the blood of healthy individuals from West Africa. PLoS Negl Trop Dis. 2015;9(3):e0003631. doi: 10.1371/journal.pntd.0003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21(9):1552–60. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ames SK, Hysom DA, Gardner SN, Lloyd GS, Gokhale MB, Allen JE. Scalable metagenomic taxonomy classification using a reference genome database. Bioinformatics. 2013;29(18):2253–60. doi: 10.1093/bioinformatics/btt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop-Lilly KA, Turell MJ, Willner KM, et al. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl Trop Dis. 2010;4(11):e878. doi: 10.1371/journal.pntd.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Available at: http://talk.ictvonline.org/files/ictv_documents/m/msl/5208.aspx.
- 17.Kim DY, Guzman H, Bueno R, Jr, et al. Characterization of Culex flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology. 2009;386(1):154–9. doi: 10.1016/j.virol.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino K, Isawa H, Tsuda Y, et al. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359(2):405–14. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 19.Liang W, He X, Liu G, et al. Distribution and phylogenetic analysis of Culex flavivirus in mosquitoes in China. Arch Virol. 2015;160(9):2259–68. doi: 10.1007/s00705-015-2492-1. [DOI] [PubMed] [Google Scholar]
- 20.Morales-Betoulle ME, Monzon Pineda ML, Sosa SM, et al. Culex flavivirus isolates from mosquitoes in Guatemala. J Med Entomol. 2008;45(6):1187–90. doi: 10.1603/0022-2585(2008)45[1187:cfifmi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, et al. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80(1):85–95. [PMC free article] [PubMed] [Google Scholar]
- 22.Cook S, Moureau G, Harbach RE, et al. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J Gen Virol. 2009;90(pt 11):2669–78. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolling BG, Eisen L, Moore CG, Blair CD. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am J Trop Med Hyg. 2011;85(1):169–77. doi: 10.4269/ajtmh.2011.10-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blitvich BJ, Lin M, Dorman KS, et al. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa. J Med Entomol. 2009;46(4):934–41. doi: 10.1603/033.046.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crockett RK, Burkhalter K, Mead D, et al. Culex flavivirus and West Nile virus in Culex quinquefasciatus populations in the southeastern United States. J Med Entomol. 2012;49(1):165–74. doi: 10.1603/me11080. [DOI] [PubMed] [Google Scholar]
- 26.Newman CM, Cerutti F, Anderson TK, et al. Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis. 2011;11(8):1099–105. doi: 10.1089/vbz.2010.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandler JA, Thongsripong P, Green A, et al. Metagenomic shotgun sequencing of a Bunyavirus in wild-caught Aedes aegypti from Thailand informs the evolutionary and genomic history of the Phleboviruses. Virology. 2014;46(4–465):312–9. doi: 10.1016/j.virol.2014.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leigh KE, Sharma M, Mansueto MS, et al. Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc Natl Acad Sci U S A. 2015;112(29):9010–5. doi: 10.1073/pnas.1511140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steentoft C, Vakhrushev SY, Joshi HJ, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–88. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryabov EV, Keane G, Naish N, Evered C, Winstanley D. Densovirus induces winged morphs in asexual clones of the rosy apple aphid Dysaphis plantaginea. Proc Natl Acad Sci U S A. 2009;106(21):8465–70. doi: 10.1073/pnas.0901389106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Wilk F, Dullemans AM, Verbeek M, Van den Heuvel JF. Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology. 1997;238(2):353–62. doi: 10.1006/viro.1997.8835. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Victoria JG, Wang C, et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84(14):6955–65. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp TM, Roth NM, Torres J, et al. Chikungunya cases identified through passive surveillance and household investigations – Puerto Rico, May 5–August12, 2014. MMWR. 2014;63(48):1121–8. [PMC free article] [PubMed] [Google Scholar]
- 35.Cui J, Holmes EC. Endogenous RNA viruses of plants in insect genomes. Virology. 2012;427(2):77–9. doi: 10.1016/j.virol.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6(11):e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1. Percentage of viral contigs in each viral family or grouping by location.
Supplementary File 2. Sequences of selected viral contigs.
Supplementary File 3. Maximum likelihood phylogenetic tree from multiple sequence alignment of M segment contig 246 from Frederick sample and known Bunyavirus M segment sequences.
Supplementary File 4. Protein modelling of S segment contig 5105 from Frederick using PHYRE2. (A) Putative 3-dimensional structure of product of Frederick contig 5105. (B) The closest structure in the database to the putative 3-dimensional structure of Frederick contig 5105, which is the solved crystal structure of Rift Valley fever virus nucleocapsid protein.
Supplementary File 5. Mapping of reads from published dataset SRR1745767 against Frederick contig 13,993.
Supplementary File 6. mosquito viromes from Maryland and West Virginia and approximate distance between sampling sites.
Supplementary File 7. Mosquito viromes from Puerto Rico and approximate distance between sampling sites.