Of a subset of 41 patients with advanced non-small cell lung cancer with ALK rearrangements detected by hybrid capture-based comprehensive genomic profiling, 11 previously had tested negative by ALK break-apart fluorescence in situ hybridization. Of these 11 patients, 9 were treated with crizotinib based on comprehensive genomic profiling results; 7 of these achieved a median response duration of 17 months.
Keywords: ALK, Crizotinib, Fluorescence in situ hybridization, Genomic profiling, Fusion
Abstract
Introduction.
For patients with non-small cell lung cancer (NSCLC) to benefit from ALK inhibitors, sensitive and specific detection of ALK genomic rearrangements is needed. ALK break-apart fluorescence in situ hybridization (FISH) is the U.S. Food and Drug Administration approved and standard-of-care diagnostic assay, but identification of ALK rearrangements by other methods reported in NSCLC cases that tested negative for ALK rearrangements by FISH suggests a significant false-negative rate. We report here a large series of NSCLC cases assayed by hybrid-capture-based comprehensive genomic profiling (CGP) in the course of clinical care.
Materials and Methods.
Hybrid-capture-based CGP using next-generation sequencing was performed in the course of clinical care of 1,070 patients with advanced lung cancer. Each tumor sample was evaluated for all classes of genomic alterations, including base-pair substitutions, insertions/deletions, copy number alterations and rearrangements, as well as fusions/rearrangements.
Results.
A total of 47 patients (4.4%) were found to harbor ALK rearrangements, of whom 41 had an EML4-ALK fusion, and 6 had other fusion partners, including 3 previously unreported rearrangement events: EIF2AK-ALK, PPM1B-ALK, and PRKAR1A-ALK. Of 41 patients harboring ALK rearrangements, 31 had prior FISH testing results available. Of these, 20 were ALK FISH positive, and 11 (35%) were ALK FISH negative. Of the latter 11 patients, 9 received crizotinib based on the CGP results, and 7 achieved a response with median duration of 17 months.
Conclusion.
Comprehensive genomic profiling detected canonical ALK rearrangements and ALK rearrangements with noncanonical fusion partners in a subset of patients with NSCLC with previously negative ALK FISH results. In this series, such patients had durable responses to ALK inhibitors, comparable to historical response rates for ALK FISH-positive cases.
Implications for Practice:
Comprehensive genomic profiling (CGP) that includes hybrid capture and specific baiting of intron 19 of ALK is a highly sensitive, alternative method for identification of drug-sensitive ALK fusions in patients with non-small cell lung cancer (NSCLC) who had previously tested negative using standard ALK fluorescence in situ hybridization (FISH) diagnostic assays. Given the proven benefit of treatment with crizotinib and second-generation ALK inhibitors in patients with ALK fusions, CGP should be considered in patients with NSCLC, including those who have tested negative for other alterations, including negative results using ALK FISH testing.
Abstract
摘要
引言. 敏感且特异性地检测到ALK基因组重排为能够从ALK抑制剂治疗获益的非小细胞肺癌 (NSCLC) 患者所必需。ALK分离荧光原位杂交 (FISH) 是获得美国食品和药物管理局批准的标准诊断方法, 但使用其他方法在ALK重排FISH阴性的NSCLC病例中发现了ALK重排, 提示这种方法的假阴性率相当高。本文报告了在临床治疗过程中采用基于杂交捕获法的综合基因组分析 (CGP) 在一个NSCLC病例大型队列中进行检测的结果。
材料与方法. 临床诊疗过程中, 在1 070例晚期肺癌患者中使用应用下一代测序技术的基于杂交捕获法的CGP。对每个肿瘤标本都进行所有种类基因改变的评价, 包括碱基替换、插入/缺失、拷贝数改变和重排, 以及融合/重排。
结果. 共发现47例患者 (4.4%) 携带ALK重排, 其中41例有EML4-ALK融合, 其余6例为其他融合配偶体, 包括3例过去未报告的重排事件: EIF2AK-ALK、PPM1B-ALK和PRKAR1A-ALK。在41例携带ALK重排的患者中, 31例有既往FISH检验结果。其中20例为ALK FISH阳性, 11例 (35%) 为ALK FISH阴性。在后面这11例患者中, 9例依据CGP结果接受了克唑替尼治疗, 其中7例中位缓解时间达到17个月。
结论. 综合基因组分析在既往ALK FISH检验结果为阴性的NSCLC亚组患者中发现了经典ALK重排和非经典融合配偶体。在这一病例系列中, 此类患者对ALK抑制剂治疗产生了持久应答, 缓解率与ALK FISH阳性病例的历史缓解率相当。The Oncologist 2016;21:762–770
对临床实践的提示: 综合基因组分析 (CGP) 包括杂交捕获法以及 ALK 内含子 19 的特异性诱饵, 是在使用标准 ALK 荧光原位杂交法 (FISH) 诊断为阴性的非小细胞肺癌 (NSCLC) 患者中, 鉴别药物敏感性 ALK 融合的可选方法。鉴于已证实克唑替尼和第二代 ALK 抑制剂治疗在 ALK 融合患者中的获益, 应考虑对 NSCLC 患者使用 CGP 检测, 包括那些其他可选方法检验结果阴性的患者 (也包括使用 ALK FISH 检验结果阴性的患者)。
Introduction
The targeted treatment of advanced non-small cell lung cancer (NSCLC) harboring genomic rearrangement of anaplastic lymphoma kinase (ALK) is a paradigm for personalized oncology [1]. The EML4-ALK fusion gene is found in 3%–5% of NSCLCs, and is detected by the U.S. Food and Drug Administration (FDA)-approved companion diagnostic break-apart fluorescence in situ hybridization (FISH) assay from Vysis (Abbott Laboratories, Abbott Park, IL, https://www.abbottmolecular.com), as well as a more recently approved immunohistochemistry (IHC) assay [2, 3]. EML4-ALK fusions in NSCLC occur in at least 8 variants, with all known variants having a breakpoint in intron 19 of ALK, but with a variety of introns in EML4 experiencing rearrangement, resulting in inclusion of differing fragments of the N-terminal region of EML4 [4]. Hybrid-capture-based genomic sequencing that targets intron 19 of ALK can capture all possible breakpoint events involving this intron and unequivocally identify the partner genes. In vitro modeling of EML4-ALK demonstrates that it is an oncogenic driver that is sensitive to multiple ALK kinase inhibitors such as the approved drugs crizotinib, ceritinib, alectinib, and the investigational agent brigatinib, consistent with in vivo studies [5, 6]. Clinical trials have established ALK inhibitors as standard treatment for patients with advanced ALK-rearranged NSCLC, with crizotinib response rates of approximately 74% and median progression-free survival of 10.9 months, compared with approximately 45% and 7.0 months, respectively, with pemetrexed plus either cisplatin or carboplatin [7].
To better estimate the frequency of false-negative results by FISH for ALK rearrangements in the routine practice setting, a review was undertaken of a multi-institutional series of lung carcinoma cases submitted for comprehensive genomic profiling (CGP) in the course of clinical care. Patients harboring ALK rearrangements were identified, and the history of pre-existent ALK FISH testing and response to ALK inhibitors were assessed when available. Local site permissions were used for this study.
Materials and Methods
Comprehensive Genomic Profiling
DNA was extracted from 40 µm of formalin-fixed, paraffin-embedded sections, and CGP was performed on hybridization-captured, adaptor ligation-based libraries to a mean coverage depth of greater than ×650 for at least 3,769 exons of 236 cancer-related genes plus 47 introns from 19 genes frequently rearranged in cancer. CGP is defined as a well-validated molecular assay and interpretive procedure that simultaneously sequences the entire coding sequence of all genes known to be somatically altered and biologically relevant in human cancer, including all clinically relevant classes of genomic alterations (GAs) (i.e., base-pair substitutions, insertions/deletions, copy number alterations and rearrangements), and then matches all clinically relevant alterations to potential targeted treatment options [8]. Clinically relevant genomic alterations (CRGAs) were defined as GAs linked to drugs on the market or under evaluation in mechanism-driven clinical trials. Results from prior ALK FISH testing and response to treatment were obtained by review of available medical records or discussion with the treating physicians in the context of appropriate consent.
Vector Construction and Cell-Line Assays
The gateway entry vector containing fusion sequence PRKAR1A-ALK was synthesized (Thermo Fisher Scientific, Waltham, MA, https://www.thermofisher.com), and the V5 tagged expression clone (pcDNATM 3.2-V5-DEST) was created by performing an LR recombination reaction using gateway LR clonase II enzyme mix (Invitrogen Life Technologies Cat. No.11791-020; Thermo Fisher Scientific) according to the manufacturer’s protocol. Protein expression was verified by transient transfection of the plasmid in 293T cells and Western blot analysis probing with an antibody against V5.
Creation of Stable Cell Lines and Colony-Formation Assay
Mouse embryonic fibroblast cells (NIH3T3) were transfected with the V5-PRKAR1-ALK fusion vector and cells were selected on geneticin (G418) containing DMEM media. Expression of fusion protein was verified by Western blot. The colony-forming ability of cells expressing empty vector or V5-PRKAR1-ALK was evaluated by plating 300 cells per well in a 6-well plate and staining the colonies with crystal violet after 15 days of incubation. To evaluate the effect of crizotinib, PRKAR1-ALK expressing cells were treated with different concentrations of crizotinib (0, 0.01 µM, 0.5 µM, and 1 µM) and allowed to grow for 15 days, supplemented with fresh media every 3 days. Colonies were counted after staining with crystal violet, and the percent survival was plotted compared with untreated cells from data collected from three independent experiments.
Western Blot Analysis
NIH3T3 control cells or cells expressing V5-PRKAR1-ALK were treated overnight with 0.5 µM crizotinib. Proteins extracts were prepared using NTEN buffer (Tris, pH 8.0; NaCl, 150 mM; EDTA, 1 mM; NP40 0.5%; and protease and phosphatase inhibitors) and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Blots were probed with antibodies against V5 (Bethyl Laboratories, Montgomery, TX, https://www.bethyl.com), pERK, and total ERK (Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com).
Results
To validate the efficacy of a hybrid-capture-based CGP assay in detecting clinically relevant oncogenic driver fusion genes, an analytic validation was performed using as standards cell lines known to harbor EML4-ALK, NPM1-ALK, SLC34A2-ROS1, CCDC6-RET, or TMPRSS2-ERG fusions. Pools representing fractions ranging from 20% to 100% of the 5 characterized fusions were analyzed, and in each of 32 total cases, the fusion was appropriately detected using CGP, demonstrating fusion detection sensitivity of 20% cellular fraction and greater (Fig. 1A). In addition, 45 NSCLC cases from a major academic medical center were assayed by CGP on a retrospective research basis by a Clinical Laboratory Improvement Act-certified College of American Pathologists-accredited laboratory. Of 22 cases harboring ALK rearrangements detected by FISH, all were detected by CGP and identified as being EML4-ALK fusions, despite significant variability in breakpoint locations. All but 1 of 23 FISH-negative cases were also negative by CGP; 1 case harbored a previously undetected SOCS5-ALK rearrangement (Fig. 1C, D).
Figure 1.
Analytical validation of fusion gene detection by hybrid-capture-based comprehensive genomic profiling (CGP). (A): Five cell lines harboring characterized fusions were mixed into 22 variably sized pools such that each fusion was represented at 20%, 25%, 33%, 50%, and 100% cellular fraction at least once, for a total of 32 gene-fusion test cases. (B): Repeated testing of clinical formalin-fixed, paraffin-embedded (FFPE) specimens identified to harbor fusions demonstrated detection reproducibility within and across assay batches. (C): Summary of concordance data between fluorescence in situ hybridization and nonhybrid-capture-based CGP for 45 clinical FFPE specimens validated for the presence or absence of ALK rearrangements. (D): Breakpoints in ALK rearrangements detected by CGP.
Samples from 1,070 patients with advanced lung carcinoma were assayed by CGP in the course of clinical care. The median age of the patients was 55 years (range: 17–72 years), and the proportion of women to men was similar (supplemental online Table 1). The frequencies of the most common GAs in this series are shown in supplemental online Figure 1.
A total of 47 cases (4.4%) harboring ALK rearrangements were identified, with each case harboring a single ALK fusion. The ALK fusion partner was EML4 in 41 cases, with the other 6 cases having alternative partners, including KIFB-ALK, CLTC-ALK, TFG-ALK, and the previously undescribed EIF2AK-ALK, PPM1B-ALK, and PRKAR1A-ALK fusions (Fig. 2). Of the 47 patients with ALK rearrangements detected using CGP, 31 had FISH testing data available for review. Of these, 11 cases (35%) were FISH negative (Fig. 3).
Figure 2.
EML4-ALK variant fusions and non-EML4-ALK fusions detected in this series. (A): Of 41 cases harboring EML4-ALK fusions, there was a breakpoint in 17 in intron 13 (variant 1), 16 cases in intron 6 (variant 3), 4 cases in intron 20 (variant 2), 2 cases in intron 2 (rare variant 5a/b), and 2 cases in intron 18 (a heretofore unnamed variant). (B): The 6 cases harboring non-EML4-ALK fusions all had breakpoints in intron 19 of ALK, but breakpoints in the partners were as follows: KIF5-ALK (intron 24), CLTC-ALK (intron 31), TFG-ALK (intron 6), PRKAR1A-ALK (intron 5), PPM1B-ALK (intron 1), and EIF2AK3-ALK (intron 2).
Figure 3.
CONSORT diagram for this study.
Abbreviations: CGP, comprehensive genomic profiling; FISH, fluorescence in situ hybridization.
Of 11 patients with false-negative FISH results, 9 received crizotinib based on the CGP detection of an ALK fusion and 7 responded (6 partial responses and 1 complete response), as identified by investigator assessment of either radiographic and/or metabolic imaging. The response durations of these patients ranged from 5 to 28 (ongoing) months, with the median exceeding 17 months (Table 1). Of the two patients who did not receive crizotinib, one has not yet progressed and the other was lost to follow-up.
Table 1.
Available crizotinib treatment and response data for ALK-rearranged cases
Three cases with ALK rearrangements identified by CGP are highlighted for detailed presentation. Two ALK FISH-negative cases, one harboring the previously undescribed EIF2AK3-ALK (case 1) and another harboring the well-characterized EML4-ALK (case 2), are described. In both of these cases, the patient was switched to crizotinib after CGP results were received, and experienced a durable partial response. The patient in case 3 harbored the novel fusion PRKAR1A-ALK and responded to crizotinib; this fusion was expressed in vitro, promoted growth, and activated the MAPK pathway.
Case Presentation 1
A 71-year-old, female patient who never smoked and who had a history of breast cancer was initially diagnosed with stage IV adenocarcinoma of the lung in 2007, when she presented with a malignant pleural effusion. A pleural biopsy specimen revealed invasive adenocarcinoma with mucin production and malignant cells were positive for TTF-1 IHC. The tumor was negative for both epidermal growth factor receptor (EGFR) mutations and ALK rearrangements, as assessed by a focused “hotspot” test and FISH, respectively. She was initially treated with carboplatin, paclitaxel, and bevacizumab, followed by maintenance bevacizumab. After 4 years, her disease progressed and her treatment was changed to pemetrexed and bevacizumab. Pemetrexed was discontinued because of fatigue; maintenance bevacizumab was administered for another year before disease progression. CT scan of her chest and upper abdomen at progression revealed circumferential, nodular, pleural metastatic disease on the right side with bilateral pulmonary nodules, chest and abdominal wall subcutaneous nodules, peritoneal implants, and mediastinal adenopathy. The patient underwent an excisional biopsy of a chest wall nodule and this confirmed well-differentiated, TTF1-positive adenocarcinoma of the lung with mucinous features.
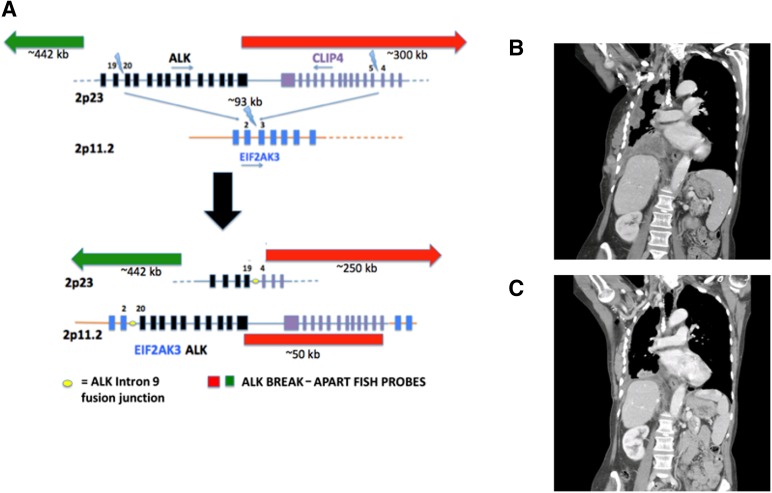
CGP was performed on the most recent biopsy specimen to a depth of ×852, which identified a rearrangement between EIF2AK3 with a breakpoint in intron 2, and with ALK, at a breakpoint in intron 19, both on chromosome 2 (Fig. 4A). The tumor was found to be ALK negative by both IHC (D5F3 and 5A4 antibodies) and FISH.
Figure 4.
A patient harboring the novel FISH-negative EIF2AK4-ALK fusion responded to crizotinib (case 1). (A): EIF2AK4-ALK fusion results from a complex rearrangement leading to insertion of a 92-kb region in 2p23 encoding part of ALK and CLP4 into intron 2 of EIF2AK3 located in 2p11.2. Probe regions are not shown to scale. (B): Coronal image from a CT scan of the chest and upper abdomen before treatment with crizotinib. (C): Coronal image from a CT scan of the chest and upper abdomen 6 weeks after starting treatment with crizotinib, with significant interval decrease in pleural-based disease.
Abbreviation: FISH, fluorescence in situ hybridization.
Based on these results, treatment was initiated with crizotinib. By her 1-month follow-up visit, clinical improvement with decreased dyspnea was observed. A follow-up CT scan done after 6 weeks showed significant reduction in tumor burden in all metastatic sites (Fig. 4B, C). The disease remained controlled for approximately 28 months, after which the patient showed radiographic evidence of disease progression.
Case Presentation 2
A 38-year-old woman was diagnosed with stage IV adenocarcinoma of the lung in 2013, after presenting with cough and flu-like symptoms, and had numerous bilateral pulmonary nodules and lymph nodes positive for metastasis. The tumor was negative for both EGFR mutations by a focused hotspot test and ALK and ROS1 rearrangements by FISH. She was initially treated with four cycles of carboplatin, pemetrexed, and bevacizumab, and had stable disease. She then continued with maintenance pemetrexed and bevacizumab until disease progression after 6 additional cycles. Six months after initial diagnosis, the same biopsy specimen was sent for CGP. Genomic profiling identified an EML4-ALK variant 1 fusion gene; based on these results, treatment was initiated with crizotinib. FISH was repeated after the CGP results on another specimen and the result was positive (72% positivity). A follow-up CT scan done after 8 weeks of treatment showed a partial response (Fig. 5), and the patient retained this response for a total of 12 months before disease progression. The patient was then entered into a trial for alectinib with a good response, and she remains on treatment at this time.
Figure 5.
Response to crizotinib in a patient harboring fluorescence in situ hybridization-negative EML4-ALK (case 2). Axial and coronal images from CT scans (A) 2 weeks before crizotinib was initiated and (B) after 8 weeks of crizotinib therapy.
Case Presentation 3
A 67-year-old man with a 30 pack-year smoking history and a history of early-stage NSCLC diagnosed 6 years before presented with mediastinal adenopathy with probable metastatic disease within multiple lymph nodes in his chest, abdomen, and liver. A right-side paratracheal lymph node biopsy specimen revealed poorly differentiated adenosquamous carcinoma with focal signet ring features morphologically consistent with the initial NSCLC diagnosis. After carboplatin and albumin-bound paclitaxel (Abraxane; Celgene, Summit, NJ, https://www.celgene.com) treatment failed, standard-of-care testing and comprehensive genomic profiling were performed.
The tumor harbored a PRKAR1A-ALK interchromosomal rearrangement and both ALK FISH and ALK IHC were positive. Expression of this novel PRKAR1A-ALK fusion, but not empty vector, led to colony-formation activity in NIH3T3 cells (Fig. 6A), as well as increased basal activation of the MAPK pathway, demonstrated by increased ERK phosphorylation (Fig. 6B). Crizotinib treatment resulted in significant reduction of ERK phosphorylation and colony formation (Fig. 6B-D). These data demonstrate that the PRKAR1A-ALK fusion was both oncogenic and imparted sensitivity to crizotinib.
Figure 6.
Oncogenic PRKAR1A-ALK fusion is sensitive to crizotinib in vitro and in vivo (case 3). (A): NIH3T3 cells stably expressing either empty vector or vector encoding V5-PRKAR1A-ALK fusion were plated for colony formation and stained to show colony growth. (B): NIH3T3 cells stably expressing either empty vector of V5 tagged-PRKAR1A-ALK were treated with crizotinib at doses shown and then processed for Western blotting using antibodies to V5, phosphor-ERK (pERK), or total ERK. (C, D): NIH3T3 cells transformed with V5-PRKAR1A-ALK fusion were treated with increasing concentrations of crizotinib; growth was assayed using colony formation. Representative plates stained for colonies are shown in (C). Quantization of colonies is plotted in (D). (E): Axial image from a CT scan of the chest and upper abdomen before treatment with crizotinib. (F): Axial image from a CT scan of the chest and upper abdomen 3 months after starting crizotinib, with resolution of mediastinal adenopathy.
On the basis of the results, the patient received crizotinib (250 mg p.o. b.i.d.), and experienced significant clinical improvement within 1 month. CT scans obtained after 3 months revealed complete resolution of mediastinal adenopathy (Fig. 6E, F), and after 6 months, his systemic disease remained controlled radiographically. The patient died 7 months after starting crizotinib secondary to multiple cerebral infarcts felt to be related to “cancer coagulopathy” rather than documented radiographic progression of disease or treatment effect.
Discussion
ALK rearrangements have been identified in 5%–6% of lung adenocarcinomas by the FDA-approved FISH companion diagnostic assay, although IHC and reverse transcription-polymerase chain reaction have also been evaluated for this purpose, with the former being FDA approved in June 2015 [3, 9, 10]. Our study demonstrates that in 35% of ALK-rearranged lung carcinomas identified by CGP, ALK FISH was negative. Another recent study showed that for NSCLC that is negative by standard-care-of testing for GAs, namely EGFR mutations and ALK rearrangements, CGP can still identify CRGAs that suggest benefit from targeted therapy, and, within this small series, an ALK rearrangement was identified, and that patient benefitted from crizotinib [11].
The reasons for the failure of FISH to detect ALK rearrangements that were found using CGP involve multiple factors. Failure may reflect complex genomic events that do not provide sufficient separation distance between the break-apart probes to fulfill scoring criteria, other technical limitations of the assay, or constraints on the reporting of such results. For example, where an EIF2AK4-ALK fusion was identified by CGP, analysis of the breakpoint sequencing in intron 19 of ALK revealed that a 94-kb fragment extending from intron 19 of ALK to intron 16 of the neighboring gene CLIP4 was deleted and inserted into a neighboring region of chromosome 2 containing intron 2 of EIFA2AK4 to create the EIF2AK4-ALK fusion. In this case, the flanking genomic break-apart probes (with the 3′ probe covering 300 kb and the 5′ probe covering 500 kb) were largely unperturbed by the 94-kb deletion in this locus (Fig. 4A). ALK IHC was performed on a much smaller portion of the cases reported here, so no meaningful assessment can be made of this strategy, although ALK IHC has been approved as a companion diagnostic.
Importantly, 7 of 9 of the ALK FISH false-negative cases reported here responded to crizotinib, with a median response of 17 months; one response is ongoing, as assessed by the treating physician on the basis of imaging results. As observed here and in many series, the presence of an ALK fusion gene is very strongly mutually exclusive for the presence of other oncogenic drivers of NSCLC. Specifically, in this series of 47 patients who were ALK positive, no concurrent alterations in EGFR, ROS1, RET, BRAF, or KRAS were observed (data not shown). Correspondingly, these patients typically had at least one line of treatment with cytotoxic therapy and progressed, thus highlighting the clinical need for CGP in the course of clinical care.
Of the two ALK FISH-negative cases that did not respond to crizotinib, one case also harbored a TSC2 alteration and initially responded symptomatically to crizotinib but progressed radiographically within 1 month. Activation of the mTOR pathway via mutation and loss of the inhibitory function of TSC2 is well known to be associated with acquired resistance to targeted therapy and could be linked to the de novo resistance observed in this patient [12]. The other nonresponding patient who was ALK FISH negative died within 1 week of receiving crizotinib, and was ultimately deemed “too sick to respond” by the treating physician.
This series of cases also harbored ALK fusions with previously unknown partners. Several non-EML4 ALK fusion partners are known [13–16], and the transforming potential of both KIF5B-ALK and KSC1-ALK has been confirmed by preclinical modeling. We further report three novel fusion partners for ALK, including validation that the in vitro crizotinib-sensitive transforming activity of the PRKAR1A-ALK fusion was consistent with an in vivo response to crizotinib in the corresponding clinical case. In previous trials of crizotinib for treatment of patients with ALK rearrangements, as stratified by FISH, it is possible that such non-EML4-ALK fusion patients may have been excluded, depending on the particulars of the intrachromosomal event in chromosome 2 or the interchromosomal event. Regardless, the clinical benefit of three patients with novel ALK fusions presented in this series strongly suggests that an empiric trial of crizotinib can benefit patients harboring non-EML4-ALK fusions.
The frequency of negative FISH results seen for ALK fusions in this study was surprising, given the fact that the overall frequency of ALK rearrangements in the larger lung carcinoma series was 4.4% (5.1% in lung adenocarcinoma), consistent with previous retrospective studies. This series likely reflects a selection bias, as cases already known to harbor an ALK rearrangement based on prior FISH testing almost certainly will not be submitted for CGP, because ALK inhibitors would thus have been identified as an option. Therefore, the cases here may also represent a result of “clinical intuition,” meaning cases in which the constellation of clinicopathologic features suggests a targetable oncogenic driver may exist despite negative results of prior testing. As such, estimation of a “false-negative” frequency for ALK FISH testing is quite difficult, given how a selection bias may factor in. Nonetheless, the results presented here indicate that the identified ALK FISH false-negative cases are clinically significant and worthy of further investigation. It may be possible that the true frequency of ALK rearrangements in advanced NSCLC as detected by CGP is higher than 4%–5%, but this hypothesis awaits results of ongoing investigation. Correspondingly, the possibility of false-positive results by CGP does exist. At this time, it is unclear what should be the gold standard of such false positives for comparison with any technique against CGP given the high sensitivity of the latter and, for example, the relatively insensitive nature of Sanger sequencing [8].
Recently, false-negative results in the setting of cancer testing have been suggested to carry significant social costs, based on the loss of years of life to society. A metric was proposed for social cost in an FDA white paper on laboratory-developed testing, with false-negative testing for ERBB2 amplification for advanced breast carcinoma carrying a social cost exceeding $700,000 [17]. This cost represents the multiplication of a projected 3-year benefit of overall survival multiplied by value statistical life year (VSLY). Using the midrange estimate of $258,436 for 1 VSLY and multiplying by the average duration of response of 17 months in the false-negative cases in the series suggest a social cost of $366,103 for false-negative results for ALK by standard-of-care testing [18]. This is an approximate measure, as exact measures of overall survival from crizotinib are not readily available because of clinical trials typically incorporating a crossover design. However, a more extensive analysis might suggest an even greater social cost, given the possibility of benefit from second-line ceretinib and investigational ALK inhibitors in the setting of a clinical trial once failure of first-line crizotinib has occurred.
Conclusion
Accurate detection of ALK fusions is imperative in the management of advanced NSCLC, and FISH as standard-of-care testing is insufficient to identify all cases harboring drug-sensitive ALK rearrangements. Current guidelines recommend that EGFR and ALK testing be conducted using next-generation sequencing methods and endorse broader molecular profiling to identify patients harboring unique genomic alterations that may lead to benefit from targeted therapy [19]. Moreover, given the clinical response to crizotinib observed in patients with novel ALK fusions in this study and others [11], patients with non-EML4 ALK translocations should be considered for crizotinib treatment in the first-line setting or after initial chemotherapy, depending on the timing of the molecular testing.
See http://www.TheOncologist.com for supplemental material available online.
This article is available for continuing medical education credit at CME.TheOncologist.com.
Supplementary Material
Acknowledgment
This study was presented in part at ASCO 2014, Chicago, Illinois. Ravi Salgia is currently affiliated with the City of Hope, Duarte, CA.
Footnotes
Editor’s Note: See the related commentary, “Screening for ALK Rearrangements in Lung Cancer: Time for a New Generation of Diagnostics?” by Ibiayi Dagogo-Jack and Alice T. Shaw on page 662 of this issue.
Author Contributions
Conception/Design: Siraj M. Ali, Sai-Hong Ignatius Ou, Vincent A. Miller, Shridar Ganesan
Provision of study material or patients: Siraj M. Ali, Thomas Hensing, James H. Suh, Julia A. Elvin, Kashif Firozvi, Julian R. Molina, Smitha Menon, Julie R. Brahmer, Heber MacMahon, Jan Nowak, Marjorie Zauderer, Neal Fischbach, Vincent A. Miller, Heather Wakelee, Shridar Ganesan, Ravi Salgia
Collection and/or assembly of data: Siraj M. Ali, Thomas Hensing, Eric Sanford, Kyle Gowen, Atul Kulkarni, Jie He, James H. Suh, Julia A. Elvin, Juliann Chmielecki, Nir Peled, Samuel J. Klempner, Garret M. Frampton, Julian R. Molina, Smitha Menon, Julie R. Brahmer, Heber MacMahon, Jan Nowak, Sai-Hong Ignatius Ou, Marjorie Zauderer, Marc Ladanyi, Maureen Zakowski, Neal Fischbach, Jeffrey S. Ross, Vincent A. Miller, Shridar Ganesan, Ravi Salgia
Data analysis and interpretation: Siraj M. Ali, Thomas Hensing, Alexa B. Schrock, Justin Allen, Eric Sanford, Kyle Gowen, Atul Kulkarni, Jie He, James H. Suh, Doron Lipson, Julia A. Elvin, Roman Yelensky, Zachary Chalmers, Juliann Chmielecki, Heather Wakelee, Nir Peled, Samuel J. Klempner, Garret M. Frampton, Smitha Menon, Julie R. Brahmer, Jan Nowak, Sai-Hong Ignatius Ou, Marc Ladanyi, Maureen Zakowski, Jeffrey S. Ross, Phil J. Stephens, Vincent A. Miller, Shridar Ganesan, Ravi Salgia
Manuscript writing: Siraj M. Ali, Thomas Hensing, Alexa B. Schrock, Justin Allen, Eric Sanford, Kyle Gowen, Jie He, Doron Lipson, Roman Yelensky, Sai-Hong Ignatius Ou, Marc Ladanyi, Jeffrey S. Ross, Phil J. Stephens, Vincent A. Miller, Ravi Salgia
Final approval of manuscript: Siraj M. Ali, Alexa B. Schrock, Justin Allen, Doron Lipson, Roman Yelensky, Sai-Hong Ignatius Ou, Marc Ladanyi, Jeffrey S. Ross, Phil J. Stephens, Vincent A. Miller, Ravi Salgia
Disclosures
Siraj M. Ali: Foundation Medicine Inc. (E, OI, IP); Thomas Hensing: Genentech, AstraZeneca (C/A); Alexa B. Schrock: Foundation Medicine Inc. (E, OI); Justin Allen: Foundation Medicine Inc. (E, OI); Eric Sanford: Foundation Medicine Inc. (E, OI); Kyle Gowen: Foundation Medicine Inc. (E, OI); Jie He: Foundation Medicine Inc. (E, OI); James H. Suh: Daicchi Sankyo (C/A), Foundation Medicine Inc. (E, OI), Genentech (H); Doron Lipson: Foundation Medicine Inc. (E, OI); Julia A. Elvin: Foundation Medicine Inc. (E, OI); Roman Yelensky: Foundation Medicine Inc. (C/A); Zachary Chalmers: Foundation Medicine Inc. (E, IP): Juliann Chmielecki: Foundation Medicine Inc. (E, OI); Samuel J. Klempner: Foundation Medicine Inc. (H); Garret M. Frampton: Foundation Medicine Inc. (E, OI); Julian R. Brahmer: Bristol-Myers Squibb, Merck, Celgene (C/A), Bristol-Myers Squibb, Merck, AstraZeneca (RF); Heber MacMahon: Riverain Technology (C/A), Philips Healthcare (RF), University of Chicago (E), medical malpractice cases (ET); Hologic (OI), UCTech (IP); Jan Nowak: OmniSeq (E, OI); Sai-Hong Ignatius Ou: Pfizer, Roche, Boehringer Ingelheim (C/A), Roche, Boehringer Ingelheim (H); Marjorie Zauderer: Verastem, Sellas Life Sciences (RF); Marc Ladanyi: AstraZeneca (C/A); Jeffrey S. Ross: Foundation Medicine Inc. (RF, E, OI); Phil J. Stephens: Foundation Medicine Inc. (E, OI); Vincent A. Miller: Foundation Medicine Inc. (E, OI), Genentech (H); Heather Wakelee: Peregrine, Acea, Helsinn (C/A), Pfizer, Genentech/Roche, Lilly, Xcovery, Exelixis, Clovis, Novartis, Celgene, Gilead, AstraZeneca (RF); Shridar Ganesan: Novartis, Inspirata Inc. (C/A), Inspirata Inc. (OI, IP). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Shaw AT, Hsu PP, Awad MM, et al. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oxnard GR, Binder A, Jänne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;31:1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Food and Drug Administration. List of cleared or approved companion diagnostic devices (in vitro and imaging tools). Available at http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm. Accessed August 22, 2015.
- 4.Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012;18:4682–4690. doi: 10.1158/1078-0432.CCR-11-3260. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim D-W, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 6.Shaw AT, Kim D-W, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon BJ, Mok T, Kim D-W, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 8.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16:e342–e351. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- 10.Thomas A, Liu SV, Subramaniam DS, et al. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–526. doi: 10.1038/nrclinonc.2015.90. [DOI] [PubMed] [Google Scholar]
- 11.Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in “driver-negative” lung adenocarcinomas. Clin Cancer Res. 2015;21:3631–3639. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirazzoli V, Nebhan C, Song X, et al. Acquired resistance of EGFR-mutant lung adenocarcinomas to afatinib plus cetuximab is associated with activation of mTORC1. Cell Reports. 2014;7:999–1008. doi: 10.1016/j.celrep.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou S-HI, Klempner SJ, Greenbowe JR, et al. Identification of a novel HIP1-ALK fusion variant in Non-Small-Cell Lung Cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to Alectinib. J Thorac Oncol. 2014;9:1821–1825. doi: 10.1097/JTO.0000000000000368. [DOI] [PubMed] [Google Scholar]
- 15.Hong M, Kim RN, Song J-Y, et al. HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol. 2014;9:419–422. doi: 10.1097/JTO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 16.Fang DD, Zhang B, Gu Q, et al. HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol. 2014;9:285–294. doi: 10.1097/JTO.0000000000000087. [DOI] [PubMed] [Google Scholar]
- 17.Office of Public Health Strategy and Analysis, US Food and Drug Administration. The public health evidence for FDA oversight of laboratory developed tests. 20 case studies - UCM472777.pdf. Available at http://www.fda.gov/downloads/AboutFDA/ReportsManualsForms/Reports/UCM472777.pdf. Accessed November 29, 2015.
- 18.Murphy K, Topel R. The value of health and longevity. J Polit Econ. 2006;114:871–904. [Google Scholar]
- 19.National Comprehensive Cancer Network. Lung cancer screening, v.2.2016. Available at https://www.nccn.org/professionals/physician_gls/recently_updated.asp
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.