Abstract
Aim
Potent risk factors at both genetic and non-genetic levels are accountable for susceptibility and instigation of different cardiovascular phenotypes. Recently, homocysteine is being identified as an important predictor for cardiovascular diseases. Homocysteine remethylation plays a key role in the synthesis of methionine and S-adenosine methionine. Methylenetetrahydrofolate reductase (MTHFR) and methionine synthase (MTR) genes are known to regulate the homocysteine remethylation reaction and higher homocysteine level is significantly associated with diverse cardiovascular phenotypes. In this context, we aimed to carry out a study on the association of MTHFR (C677T) and MTR (A2756G) gene polymorphism with CVD in population of Jammu region of J&K state.
Materials and methods
A total of 435 individuals were enrolled (195 CVD patients and 240 controls) for the case–control study. Genotyping of MTHFR C677T and MTR A2756G gene polymorphism was done by PCR-RFLP technique. Biochemical parameters were estimated by biochemical analyser.
Results
Metabolic variables such as serum LDL-C, TC and TG were significantly higher in patients (p < 0.0001), whereas serum HDL-C was higher in controls. Majority of the patients were having history of hypertension (57.44%; p < 0.0001) as a concomitant condition. The evaluation of genetic association showed that, MTHFR C6877T (OR: 8.89, 95% CI: 2.01–39.40) and MTR A2756G (OR: 1.48, 95% CI: 1.09–2.00) polymorphisms associated with higher risk of CVD.
Conclusion
The present study reveals significant differences in nongenetic variables among patients and control as well as association of gene polymorphisms with CVD risk.
Keywords: CVD, MTHFR, MTR, Polymorphism
1. Introduction
Notwithstanding the immense advancement and sophistication in healthcare system, the challenge to check the incidence of cardiovascular diseases (CVDs) still remains, be it a developed nation like the USA or a developing country like India. It is a matter of great concern that CVDs alone would soon be the single largest cause of mortality accounting for more than a third of all deaths worldwide.1 Besides well established risk factors such as hypertension (HTN), diabetes mellitus (DM), obesity, smoking, male gender, dyslipidemia, sedentary lifestyle and family history,2, 3, 4, 5 genetic alterations in the genes controlling homocysteine and folate metabolism are also linked to onset of CVDs.6, 7 Several investigators have implicated elevated plasma homocysteine as an independent marker for atherosclerotic cardiovascular condition.8, 9, 10, 11 Homocysteine and folate metabolism is dependent on couple of genes performing their specific role, but two genes namely MTR and MTHFR genes are considered critical genes for development of diseased cardiovascular phenotypes particularly congenital heart diseases12, 13 and coronary artery diseases.14, 15, 16 MTHFR enzyme is a co-factor for conversion of 5,10-methylenetetrahydrolate to 5-methyltetrahydrofolate. It has been demonstrated that C677T polymorphism in exon 4 of MTHFR gene reduces enzyme activity, which may be a plausible reason for elevated concentration of plasma homocysteine.17, 18, 19, 20, 21 MTR is engaged in dual performance of both demethylation of 5-methyltetrahydrofolate to tetrahydrofolate and also, remethylation of homocysteine to methionine by utilizing methyl group donated by 5-methyltetrahydrofolate. A common polymorphism in the MTR gene (A2756G) is associated with hyperhomocystenemia and DNA hypomethylation.22, 23, 24 The distribution of MTHFR and MTR gene SNPs project a higher degree of heterogeneity not among worldwide populations25, 26, 27, 28 but a transient difference was observed in allelic distribution among different Indian populations due to diverse ethnicity.29, 30 The state of Jammu & Kashmir represents diverse cultural and genetic heritage belonging to Kashmiri (Kashmir), Dogra (Jammu) and Ladakhi populations (Ladakh). The ethnicity, genetic makeup, basic lifestyle and dietic pattern of an individual can greatly influence the onset and pathogenesis of disease. The lack of any molecular level study in context to CVD, targeting the populations of Jammu, called for undertaking the research work documented herein. We have attempted to assess link of MTHFR (C677T) and MTR (A2756G) gene variation and risk of CVD in populace of Jammu region of J&K state.
2. Materials and methods
2.1. Study population
The study population comprised of 195 CVD cases (men = 128; women = 67) and 240 healthy controls (men = 145; women = 95), who were genetically unrelated and belonging to Jammu region of Jammu & Kashmir state. Eligible cases were patients with major CVD phenotypes encompassing coronary artery disease, acquired arrhythmia (associated with atherosclerotic coronary condition) and myocardial infarction (including acute coronary syndrome).
2.2. Ethical approval
The present study design was duly approved by Animal and Human Experimentation Ethical Committee (AHEEC), University of Jammu, Jammu and Kashmir, India. Each study participant was made aware of the nature and scope of the study. Data and blood collection from study individuals was effected after having their informed written consent.
2.3. Data collection and clinical evaluation
Non-genetic data including age, dwelling, age of onset and duration of disease, history of HTN/DM, habit of smoking/alcohol intake, sedentary lifestyle, family history, diet pattern, anthropometric (body mass index and waist hip ratio) and physiometric variables (SBP, DBP, PR and PP) were recorded in pre-designed health datasheet from each study participant.
2.3.1. Demographic profile
The dwelling pattern of the study participants was divided into urban (who had the advantage of instant access to urbane facilities), sub-urban (who dwelled at a distance) and rural (who had remote access to urbane settlements) areas.
2.3.2. Behavioral determinants
Smokers were categorized as current smoker (who smokes one or more cigarette/tobacco per day), ex-smoker (person with former smoking habit for at least 1 year) and non-smoker (person who never smoked). Alcohol intake was assessed in three categories as: current alcoholics (person who drinks per day or week), former alcoholics (subjects with previous history of alcohol intake) and non-alcoholics (person who never drunk). Physical activity was assessed as performers (person involved in any significant bodily activity like walking/jogging/exercise as a minimum of 30 min) and sedentary (person not involved in any bodily exercise).
2.3.3. Anthropometric characteristics
Body mass index (BMI) was calculated as ratio of weight and height (weight in kg/height in m2) and the values were defined according to the recommendations proposed by WHO for Asians.31 Waist hip ratio (WHR) was obtained as waist circumference divided by hip circumference and was defined as ≥0.90 for men and ≥0.80 for women.32
2.3.4. Physiometric characteristics
According to JNC7 guidelines patient on antihypertensive medications or having a systolic blood pressure (SBP) of 140 mmHg or greater and a diastolic blood pressure (DBP) of 90 mmHg or greater were considered as having HTN.33 Pulse rate (PR) was counted by feeling radial artery at the wrist over one minute. Pulse pressure (PP) was calculated by applying formula: PP = Systolic blood pressure (SBP) − Diastolic blood pressure (DBP).
2.4. Biochemical profiling
Five milliliters of peripheral blood was collected in EDTA vacutainers from each fasting study individual. Lipid profiling (LDL-C, HDL-C, TC and TG) was done on automated biochemical analyzer (Roche, Cobas CIII).
2.5. DNA isolation and genotyping
Genomic DNA was isolated from peripheral blood lymphocytes using phenol–chloroform–isoamyl extraction.34 Qualification and quantification of extracted DNA were done by using agarose gel electrophoresis and spectrophotometry. The MTHFR C677T (rs1801133) and MTR A2756G (rs1805087) gene polymorphisms were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) genotyping technique. The MTHFR C677T polymorphism was detected by target DNA amplification using the forward primer 5′-TGA AGG AGA AGG TGT CTG CGG GA-3′ and the reverse primer 5′-AGG ACG GTG CGG TGA GAG TG-3′ described by Matsuo et al. (2001).23 For detection of genotypes of MTR A2756G variants following site specific oligonucleotide primers were used: forward primer 5′-TGT TCC AGA CAG TTA GAT GAA AAT C-3′ and the reverse primer 5′-GAT CCA AAG CCT TTT ACA CTC CTC-3′.23
2.5.1. PCR conditions
Standard PCR reactions for both MTHFR and MTR genes were performed in Applied Biosystems thermal cycler (Make Veriti by Life Technology, Singapore) using 25 μl of final reaction mixture containing 2 μl DNA (50 ng–100 of genomic DNA), 5 μl flexi buffer (5×), 0.5 μl dNTPs (10 mM), 0.3 μl Taq polymerase (5 U/μl), 0.5 μl each primers (100 pmol/μl) and 13.7 μl sterile distilled water to make up the final volume. PCR thermal cycling conditions for MTHFR 677 were: pre-denaturation – 2 min at 94 °C, denaturation – 30 s at 94 °C, annealing – 1 min at 62 °C, extension – 30 s at 72 °C and final extension – 7 min at 72 °C. The PCR was carried out for 40 cycles. For MTR gene, the conditions were as follows: pre-denaturation – 4 min at 95 °C, followed by 35 cycles with denaturation – 1 min at 95 °C, annealing – 1.5 min at 61 °C, extension – 1 min at 72 °C and final extension – 7 min at 72 °C. The amplified PCR products were 198 bp for MTHFR gene and 211 bp for MTR gene.
2.5.2. RFLP conditions
The restriction digestion was performed by using 0.3 μl of restriction enzyme (10 U/μl), 2 μl NEB restriction buffer 4, 10 μl of amplified PCR product and 7.7 μl of sterile distilled water to reach total volume of 20 μl. The restriction endonuclease used for detection of MTHFR C677T polymorphism was Hinf I (New England Biolabs) and for MTR, it was Hae III (New England Biolabs). The restriction digestion mixture was given overnight incubation at 37 °C.
2.5.3. Detection of genotypes
The products of restriction digestion were separated on a 4% agarose gel pre-stained with ethidium bromide and analyzed under UV light. Genotypes of all individuals were noted as:
MTHFR C677T polymorphism: CC (wild) = 198 bp; CT (hetero) = 198 bp + 175 bp + 23 bp; and TT (mutant) = 175 bp + 23 bp (Fig. 1). MTR A2756G polymorphism: AA (wild) = 211 bp; AG (hetero) = 211 bp + 131 bp + 80 bp; and GG (mutant) = 131 bp + 80 bp (Fig. 2).
Fig. 1.
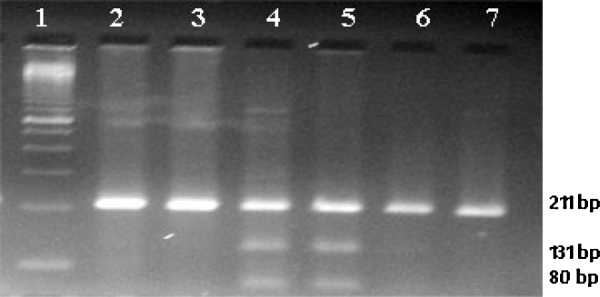
Showing the restriction digestion of MTR PCR product with HaeII enzyme. Lane no. 1 – marker, Lanes 2, 3, 6 and 7 – homozygous wild, Lanes 4 and 5 – heterozygous.
Fig. 2.
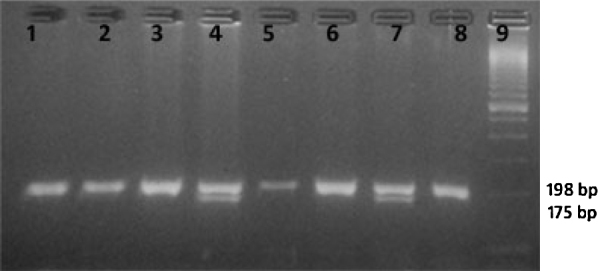
Showing the restriction digestion of MTHFR PCR product with HinfI enzyme. Lane no. 9 – marker, Lanes 1, 2, 3, 5, 6 and 8 – homozygous wild, Lanes 4 and 7 – heterozygous.
2.6. Statistical analysis
For non-genetic variables, mean and standard deviation were calculated and Student's t-test was performed to calculate the difference between the patients and controls. The allele frequencies were calculated by allele counting for each genetic marker. Hardy–Weinberg equation and the differences in genotypic frequencies were examined by using Chi-square test. The odd-ratios (OR) for different MTHFR and MTR genotypes/alleles were calculated with 95% confidence interval (CI) by unconditional logistic regression and concurrently, p values were calculated to conclude risk association with CVD. A value of p < 0.05 was considered as statistically significant. Statistical analysis was performed by using Statistical Package for Social Sciences (SPSS) software version 17.
3. Results
3.1. Non-genetic factors
Different non-genetic variables including physiometric, anthropometric and biochemical variables along with personal history of study participants are enlisted in Table 1, Table 2.
Table 1.
Prevalence of CVD risk factors in study groups.
| Parameters | Patients |
Controls |
OR | p-Value | 95% CI | ||||
|---|---|---|---|---|---|---|---|---|---|
| Men (n = 128) | Women (n = 67) | Total (n = 195) | Men (n = 145) | Women (n = 95) | Total (n = 240) | ||||
| Age (years) | 58.98 ± 15.08 | 53.40 ± 13.94 | 57.06 ± 14.90 | 42.03 ± 12.58 | 41.73 ± 12.51 | 41.91 ± 12.53 | – | – | – |
| Dwelling | |||||||||
| Urban | 61 (47.66%) | 24 (35.82%) | 85 (43.59%) | 28 (19.31%) | 25 (26.31%) | 53 (22.08%) | – | – | – |
| Sub-urban | 49 (38.28%) | 23 (34.33%) | 72 (36.92%) | 65 (44.83%) | 41 (43.16%) | 106 (44.17%) | – | – | |
| Rural | 18 (14.06%) | 20 (29.85%) | 38 (19.49%) | 52 (35.86%) | 29 (30.53%) | 81 (33.75%) | – | ||
| Physical activity | |||||||||
| Performers | 67 (52.34%) | 23 (34.33%) | 90 (46.15%) | 102 (70.34%) | 60 (63.16%) | 162 (67.5%) | Ref | ||
| Sedentary | 61 (47.66%) | 44 (65.67%) | 105 (53.85%) | 43 (29.66%) | 35 (36.84%) | 78 (32.5%) | 2.42 | <0.0001* | 1.64–3.58 |
| Smoking | |||||||||
| Current | 25 (19.53%) | 1 (1.5%) | 26 (13.33%) | 34 (23.45%) | 2 (2.11%) | 36 (15%) | 1.09 | 0.76 | 0.63–1.89 |
| Formers | 38 (29.69%) | 1 (1.5%) | 39 (20%) | 8 (5.52%) | – | 8 (3.33%) | 7.35 | <0.001* | 3.33–16.23 |
| Non-smokers | 65 (50.78%) | 65 (97.0%) | 130 (66.67%) | 103 (71.03%) | 93 (97.89%) | 196 (81.67%) | Ref | ||
| Duration of smoking (years) | 24.51 ± 13.63 | 13.5 ± 9.19 | 24.17 ± 13.60 | 11.90 ± 13.82 | 30.5 ± 34.65 | 12.75 ± 15.01 | – | – | – |
| Alcohol intake | |||||||||
| Current | 43 (33.59%) | – | 43 (22.05%) | 27 (18.62%) | – | 27 (11.25%) | 2.49 | 0.0007* | 1.47–4.22 |
| Former | 16 (12.5%) | – | 16 (8.21%) | – | – | – | – | – | – |
| Never | 69 (53.91%) | 67 (100%) | 136 (69.74%) | 118 (81.38%) | 95 (100%) | 213 (88.75%) | Ref. | ||
| Duration of alcohol intake (years) | 22.12 ± 14.55 | – | 21.14 ± 13.75 | 9.07 ± 5.05 | – | 9.07 ± 5.05 | – | – | – |
| Diet pattern | |||||||||
| Vegetarian | 25 (19.53%) | 33 (49.25%) | 58 (29.74%) | 52 (35.86%) | 55 (57.89%) | 107 (44.58%) | Ref | ||
| Non-vegetarian | 103 (80.47%) | 34 (50.75%) | 137 (70.26%) | 93 (64.14%) | 40 (42.11%) | 133 (55.42%) | 1.90 | 0.002* | 1.27–2.83 |
| History of HTN | |||||||||
| Yes | 81 (63.28%) | 31 (46.27%) | 112 (57.44%) | – | – | – | – | <0.0001* | – |
| No | 47 (36.72%) | 36 (53.73%) | 83 (42.56%) | – | – | – | – | ||
| History of DM | |||||||||
| Yes | 25 (19.53%) | 13 (19.40%) | 38 (19.49%) | – | – | – | – | 0.0001* | – |
| No | 103 (80.47%) | 54 (80.60%) | 157 (80.51%) | – | – | – | |||
HTN, hypertension; DM, diabetes mellitus.
Significant values.
Table 2.
Physiometric, anthropometric and biochemical variables in the study groups.
| Parameters | Patients |
Controls |
p-Value | ||||
|---|---|---|---|---|---|---|---|
| Men (n = 128) | Women (n = 67) | Total (n = 195) | Men (n = 145) | Women (n = 95) | Total (n = 240) | ||
| Blood pressure (mmHg) | |||||||
| Systolic (SBP) | 143.75 ± 19.53 | 138.19 ± 23.66 | 141.84 ± 21.15 | 125.45 ± 7.31 | 122.73 ± 7.94 | 124.37 ± 7.67 | <0.0001* |
| Diastolic (DBP) | 88.63 ± 9.46 | 87.04 ± 10.58 | 88.09 ± 9.86 | 86.04 ± 10.16 | 83.82 ± 8.74 | 85.16 ± 9.67 | 0.002* |
| Pulse pressure (PP) | 55.19 ± 15.72 | 51.15 ± 18.03 | 53.80 ± 16.62 | 39.41 ± 8.93 | 38.90 ± 6.73 | 39.21 ± 8.12 | <0.0001* |
| Pulse rate (PR) | 80.53 ± 11.38 | 85.12 ± 17.34 | 82.11 ± 13.85 | 73.43 ± 4.16 | 72.94 ± 2.76 | 73.24 ± 3.67 | <0.0001* |
| BMI | 24.11 ± 4.30 | 24.67 ± 5.65 | 24.30 ± 4.80 | 23.01 ± 3.80 | 22.46 ± 4.50 | 22.79 ± 4.07 | 0.0004* |
| WHR | 1.00 ± 0.07 | 0.99 ± 0.10 | 1.00 ± 0.08 | 0.95 ± 0.05 | 0.95 ± 0.05 | 0.95 ± 0.05 | <0.0001* |
| Total cholesterol (TC) (mg/dl) | 184.26 ± 58.85 | 185.30 ± 65.23 | 184.62 ± 60.95 | 131.44 ± 31.17 | 124.82 ± 31.67 | 128.82 ± 31.47 | <0.0001* |
| Triglycerides (TG) (mg/dl) | 207.95 ± 86.16 | 198.13 ± 71.69 | 204.58 ± 81.42 | 124.13 ± 26.75 | 122.04 ± 26.45 | 123.30 ± 26.6 | <0.0001* |
| HDL-C (mg/dl) | 35.62 ± 9.95 | 37.24 ± 9.54 | 36.18 ± 9.82 | 50.81 ± 7.03 | 52.28 ± 6.25 | 51.39 ± 6.75 | <0.0001* |
| LDL-C (mg/dl) | 126.66 ± 69.75 | 135.76 ± 74.53 | 129.78 ± 71.37 | 78.26 ± 18.56 | 75.58 ± 20.37 | 77.20 ± 19.30 | <0.0001* |
Values are mean ± SD.
SBP, systolic blood pressure; DBP, diastolic blood pressure, BMI, body mass index; WHR, waist hip ratio, HDL, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol.
Significant values.
3.1.1. Demographic characteristics
During the study course, we enrolled a good deal of male patients (n = 128) as compared to females (n = 67). Majority of the patients were natives of urban (43.59) and sub-urban (36.92%) areas of Jammu region. The dwelling pattern of controls showed that most of them belong to sub-urban (44.17%) areas of Jammu region followed by rural (33.75%) and urban (22.08%) vicinity.
3.1.2. Behavioral determinants and diet pattern
The CVD patients were elder than the controls, with a mean age of 57.06 ± 14.90 years compared to 41.91 ± 12.53 years in the control group. Sedentary behavior was significantly exhibited by patient group (53.85%; p < 0.0001) as compared to controls (32.5%). The prevalence of physical inactivity was higher in female patients in contrast to male patients in the present study (67.12% vs 48.18%). Smoking appeared to be potent risk factor for CVD in our population with 13.33% patients engaged in current smoking habit [OR = 1.12, 95% CI (0.6531–1.9238); p = 0.6787], 21.90% were former smokers [OR = 7.35, 95% CI (3.33–16.23); p < 0.001] and 66.67% were non-smokers. On the contrary, frequency of non-smokers was higher among controls. The pattern of alcohol intake was higher in cases (22.05% current alcoholics and 8.21% former alcoholics) than controls (11.25% current alcoholics). Women participants of our study were not reported to be engrossed in drinking habit owing to cultural perspective. Moreover, female subjects of the present study were not found to be actively engaged in smoking as like male subjects. 70.26% of patients were reportedly non-vegetarian, whereas 55.42% controls were also found to be keen for non-vegetarian diet. Vegetarian diet was more prevalent in female participants.
3.1.3. Anthropometric, physiometric, metabolic characteristics and co-morbid disease
BMI and WHR were calculated for both study groups as increment in these anthropometric variables adds to risk of CVD in an individual. It was seen that patients had significantly higher BMI and WHR values (24.30 ± 4.80 and 1.00 ± 0.08) than controls (22.79 ± 4.07 and 0.95 ± 0.05) respectively. The SBP, DBP, PP, and PR were significantly higher in CVD patients as compared with controls. Significant distinction was observed in serum LDL-C, HDL-C, TC and TG levels among cases and controls. The mean total cholesterol of cases and controls was 184.62 ± 60.95 and 128.82 ± 31.47, those of triglycerides were 204.58 ± 81.42 and 123.30 ± 26.6 and those of LDL-C were 129.78 ± 71.37 and 77.20 ± 19.30 respectively. However, HDL-C level appeared to be in lower range in patients (36.18 ± 9.82) and higher in controls (51.39 ± 6.75, p < 0.0001). In comparison to DM (18.1%), history of HTN (53.33%) was found to be higher in patients as a co-morbid condition. The percentage of male patients with HTN as a co-morbid disease was more than DM, whereas sex-specific comparison showed that frequency of CVD and DM was almost similar in both male (19.53%) and female (19.40%) patients.
Among different non-genetic risk factors, lifestyle parameters namely sedentary behavior, history of previous smoking and alcohol intake; SBP; WHR; high levels of serum LDL-C, TG and low levels of HDL-C were highly significant (p < 0.0001) to be associated with CVD.
3.2. Genetic factors
Genotypic and allelic allocation of both MTHFR (C677T) and MTR (A2756G) gene polymorphisms among cases and controls are summarized in Table 3. The MTHFR genotype distribution was in the range of Hardy–Weinberg equilibrium for both study participants (cases: χ2 = 0.27, p = 0.05; controls: χ2 = 0, p = 0.05). The frequencies of CC vs CT and CC vs CT + TT [OR = 9.20, 95% CI (2.06–41.01); p = 0.00047] genotypes differed significantly between the two groups. OR for C vs T allele showed that ‘T’ allele of MTHFR gene was adding 8.89 folds (p = 0.00053) risk for the development of CVD in our population. There was a statistical significant difference between genotypic distribution for MTR (A2756G) polymorphism between patients (χ2 = 22.1, p = 0.001) and controls (χ2 = 20.72, p = 0.001). Logistic regression for AA vs AG [OR = 1.63, 95% CI (1.11–2.39); p = 0.01] and AA vs AG + GG [OR = 1.69, 95% CI (1.15–2.47); p = 0.007] genotypes appeared to have a significant association. The mutant “G” allele of MTR (A2756G) polymorphism is having a significant role for the development of CVD phenotype in the study population [OR = 1.48, 95% CI (1.09–2.00); p = 0.01] (Table 4).
Table 3.
Genotypic and allelic allocation of MTHFR (C677T) and MTR (A2756G) polymorphism in study groups.
| Genotypes/alleles/genetic model | CVD cases (n = 195) (%) | Controls (n = 240) (%) | OR (95% CI)* | p-Value |
|---|---|---|---|---|
| MTHFR C677T | ||||
| 677CC | 181 (92.82%) | 238 (99.17%) | 1 (Reference) | |
| 677 CT (CC vs CT) | 14 (7.18%) | 2 (0.83%) | 9.20 [2.06–41.01] | 0.00047* |
| 677 TT (CC vs TT) | 0 (0%) | 0 (0%) | Not possible† | – |
| 677 CT + TT (CC vs CT + TT) | 14 (7.18%) | 2 (0.83%) | 9.20 [2.06–41.01] | 0.00047* |
| 677C | 0.96 | 0.996 | 1 (Reference) | |
| 677T (C vs T) | 0.04 | 0.004 | 8.89 [2.01–39.40] | 0.00053* |
| MTRA2756G | ||||
| 2756AA | 81 (41.54%) | 131 (54.58%) | 1 (Reference) | |
| 2756AG (AA vs AG) | 110 (56.41%) | 109 (45.42%) | 1.63 [1.11–2.39] | 0.01* |
| 2756GG (AA vs GG) | 4 (2.05%) | 0 (0%) | Not possible† | – |
| 2756AG + GG (AA vs AG + GG) | 114 (58.46%) | 109 (45.42%) | 1.69 [1.15–2.47] | 0.007* |
| 2756A | 0.7 | 0.8 | 1 (Reference) | |
| 2756G (A vs G) | 0.3 | 0.2 | 1.48 [1.09–2.00] | 0.01* |
OR and 95% CI were calculated with MTHFR 677CC and MTR 2756AA genotype as reference group.
Some genotype combinations were not observed, so it was not possible to calculate odds ratio.
Significant values.
Table 4.
Combined genotype frequencies of MTHFR and MTR gene polymorphisms in study groups.
| Variant MTHFR C677T/MTR A2756G | CVD cases (n = 195) (%) | Controls (n = 240) (%) | OR (95% CI)* | p-Value |
|---|---|---|---|---|
| 677CC/2756AA | 73 (37.44%) | 130 (54.16%) | 1 (Reference) | |
| 677CT/2756AA | 8 (4.10%) | 1 (0.42%) | 14.25 [1.75–116.17] | 0.01* |
| 677TT/2756AA | 0 | 0 | Not possible† | |
| 677CC/2756AG | 104 (53.33%) | 108 (45%) | 1.71 [1.16–2.54] | 0.007* |
| 677CT/2756AG | 6 (3.08%) | 1 (0.42%) | 10.68 [1.26–90.48] | 0.03* |
| 677TT/2756AG | 0 | 0 | Not possible† | |
| 677CC/2756GG | 4 (2.05%) | 0 | Not possible† | |
| 677CT/2756GG | 0 | 0 | Not possible† | |
| 677TT/2756GG | 0 | 0 | Not possible† |
Some genotype combinations were not observed, so it was not possible to calculate odds ratio.
Significant values.
4. Discussion and conclusion
Study of both non-genetic and genetic determinants can give an apparent delineation of disease pathophysiology and the later generates possible options for disease management. The classical non-genetic risk factors, which were undertaken in the present study included dwelling, physical inactivity, smoking, alcohol intake, diet pattern, any history of concomitant disease including HTN, DM, etc. Main portion of the patient participants of our study belonged to urban counterparts of Jammu region. Sedentary behavior was adopted by higher percentage of patient population as their basic lifestyle pattern. Smoking emerged to be potent risk factor for CVD in our population. Men, when compared to women, were reportedly more prone to smoking. Strong association of smoking with CAD was also reported by Achari and Thakur (2004).35 A significant pattern of fluctuations in biochemical variables (LDL, HDL, TC and TG) from the normal range was observed in cases. In comparison to DM, the present study revealed that history of HTN in patients was a predominant co-morbid state. Iqbal and researchers (2012) suggested HTN as one of major contributors for CVD phenotype.36 On the contrary, a previous report37 revealed a higher incidence of HTN and DM as concomitant conditions than HTN and CAD, in Jammu region of J&K state. Aggarwal et al. (2012) reported high prevalence of smoking, dyslipidemia and HTN in Indian patients with CAD.38 Study undertaken by Iyer et al. (2011)4 and De et al. (2004)39 are in concordance with the present study.
Every individual has a unique identity in terms of both phenotype and genotype. Of course, it is a universal truth that 99.9% of human genome architecture is identical with a difference of only 0.1%.40 In the last couple of decades, a tremendous effort has been made by researchers worldwide to elucidate a functional link between such human genetic variations particularly SNPs and complex traits including CVDs. Since, MTHFR and MTR genes are the key genes of homocysteine pathway, any variation in these genes could bring a deviation in the said pathway from routine functioning and is correlated to the appearance of complex cardiovascular phenotypes. The present study has been designed to elucidate the association of MTHFR (C677T) and MTR (A2756G) gene variants in susceptibility of CVDs in populace of Jammu region.
In our study, we found higher frequency of T allele in cases as compared to controls, and a significant association of the said polymorphism was observed in CVD patients. Similarly, Dhar et al. (2010) found T allele to be significantly related with CAD in eastern Indian population.41 Matam et al. (2014) depicted that T allele greatly influences the risk of CAD in south Indian population (T vs C: OR = 3.1; p = 0.00001).42 Research on other Indian populations has shown MTHFR (C677T) gene polymorphism to be a strong candidate for CVDs.16, 43, 44 A non-significant association with CAD was formulated by Pandey et al. (2011) despite having higher frequency of T allele in cases than the controls.45 Vasisht and colleagues (2002) also found higher frequency trend for ‘T’ allele in north Indian CAD patients than in controls but these associates failed to proclaim any significant association of the said polymorphism with the atherosclerotic coronary phenotype.46 Study conducted by other researchers showed negative association of MTHFR gene polymorphism with CAD.29, 47 Regarding different worldwide population groups strong positive relationship between TT genotype of MTHFR (C677T) SNP and risk of CVD has been described by some eminent researchers,48, 49, 50 whereas a couple of case–control studies presented contradictory results.51, 52, 53, 54 Recently, it has been proposed that evaluation of CVD risk in T2DM patients cannot be carried out on the presence of either MTHFR C677T polymorphism or mutant genotypes.55 Investigations on MTHFR gene polymorphisms by various distinguished researchers worldwide reported MTHFR (C677T) gene variant as a feeble marker for CAD,29, 52, 56, 57, 58 arrhythmia,53 MI50 and stroke.59 Conversely, Kluijtmans and associates (1996) reported positive association of homozygous TT genotype with a three-fold increase in risk for premature CVD.19 Several research studies have assessed an apparent association of MTHFR (677-TT) genotype with hyper-homocystinuria and CVD.60, 61, 62 A comparison on genotypic and allelic frequency of MTHFR gene polymorphism in relation to CVD phenotypes between present study and other studies is depicted in Table 5.
Table 5.
MTHFR (C677 T) gene polymorphism in different ethnic groups.
| Ethnicity | Frequency of genotypes (%) |
Frequency of alleles (%) |
Type of CVD phenotype | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases |
Controls |
Cases |
Controls |
|||||||||
| CC | CT | TT | CC | CT | TT | C | T | C | T | |||
| North Indian | 92.82 | 7.18 | 0 | 99.17 | 0.83 | 0 | 96.41 | 3.59 | 99.5 | 0.5 | CAD, MI, Arrhythmia | Present study |
| North Indian | 70.9 | 24.6 | 4.4 | 73.59 | 25 | 1.4 | 83.3 | 16.7 | 86.1 | 13.9 | CAD | Pandey et al. (2011)47 |
| East Indian | 51.6 | 21.6 | 26.72 | 72.94 | 14.11 | 12.9 | 61.96 | 38.03 | 63.97 | 36.12 | CAD | Dhar et al. (2010)43 |
| South Indian | 73.3 | 26.7 | 0 | 80 | 20 | 0 | 86 | 14 | 90 | 10 | MI and DM | Angeline et al. (2009)66 |
| Arabs | 64.2 | 32.1 | 3.7 | 72.2 | 25.8 | 2 | 80 | 20 | 85 | 15 | CAD | Abu-Amero et al. (2003)54 |
| Turkish | 48.7 | 40 | 11.3 | 55.9 | 40.7 | 3.4 | 69 | 31 | 76 | 24 | CAD | Caner et al. (2008)60 |
| Italian | 29.2 | 48.7 | 22.1 | 24.7 | 52.5 | 22.8 | 53.5 | 46.5 | 50.93 | 49.07 | Arrhythmia | Giusti et al. (2007)55 |
CAD, coronary artery disease; EH, essential hypertension; MI, myocardial infarction; DM, diabetes mellitus.
Regarding MTR gene, the present study has figured out higher frequency of G allele in cases than in controls. Similarly, Jemaa et al. (2008) revealed significant and dependent association between the G allele of the MTR gene and MI in Tunisian population.63 Huang and colleagues (2008), observed a striking difference between MTHFR (C677T), MTR (A2756G) gene polymorphisms and hyperlipidemia in Northern Chinese subjects.64 These investigators reported that the subjects with G allele of MTR gene and increased levels of homocysteine had higher risk of combined hyperlipidemia. On the contrary, insignificant association was observed by Lakshmi et al. (2011)43 and Kanth et al. (2011).29 Singh and Lele (2012), in their meta-analysis declared MTR (A2756G) gene (OR = 1.61; p = 0.06) as a weak marker for CAD.7 Salomon and co-authors (2001) suggested that homozygosity for MTR 2756G genotype did not confer risk for idiopathic venous thromboembolism.65 As in the results concerning MTHFR gene, controversial results were also shown by MTR (A2756G) gene polymorphism and risk of CVD among different populations (Table 6).
Table 6.
MTR (A2756G) gene polymorphism in different ethnic groups.
| Ethnicity | Frequency of genotypes (%) |
Frequency of alleles (%) |
Type of CVD phenotype | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases |
Controls |
Cases |
Controls |
|||||||||
| AA | AG | GG | AA | AG | GG | A | G | A | G | |||
| North Indian | 41.54 | 56.41 | 2.05 | 54.58 | 45.52 | 0 | 69.74 | 30.26 | 77 | 23 | CAD, MI, Arrhythmia | Present study |
| South Indian | 94 | 6 | 0 | 210 | 0 | 0 | 97 | 3 | 100 | – | CAD | Kanth et al. (2011)29 |
| South Indian | 170 | 149 | 31 | 126 | 141 | 12 | 69.8 | 31.2 | 70.4 | 29.6 | CAD | Lakshmi (2011)45 |
| Chinese | 88.9 | 10.7 | 0.39 | 84.6 | 15 | 0.4 | 94.2 | 5.8 | 92.1 | 7.9 | Cerebrovascular diseases | Aifan et al. (2009)67 |
| Tunisian | 72 | 26.2 | 1.9 | 80.5 | 18.7 | 0.9 | 85 | 15 | 90 | 10 | MI | Jemaa et al. (2008)65 |
| Italian | 70 | 28 | 2 | 68.9 | 29 | 2.1 | 83.99 | 16.01 | 83.39 | 16.61 | Arrhythmia | Giusti et al. (2007)55 |
CAD, coronary artery disease; EH, essential hypertension; MI, myocardial infarction.
In the work under report, we observed a significant association of both MTHFR (C677T) and MTR (A2756G) gene polymorphisms with CVD. The possible limitations of the present study may include small sample size, sample enrolment from single region of J&K state and lack of homocysteine measurements. Plasma homocysteine levels appear to increase with age and sex as are CVDs. In the present study the mean age of the patients also differs significantly from the controls, which speculates the attribution of risk of CVD to difference in plasma homocysteine levels in the study participants. Since, we were not able to measure homocysteine levels in the study participants hence; it is difficult to reach a definite conclusion. The study needs to be further replicated by taking into consideration the above-mentioned limitations, which may help in ascertaining the results. We intend to plan a wider research study on diversity of gene variations of homocysteine metabolism in CVD among populations of three regions of the J&K state in future.
Conflicts of interest
The authors have none to declare.
Acknowledgements
The authors are thankful to study participants for providing their blood samples and the medical staff of ASCOMS hospital for their medical assistance and guidance. The authors would like to acknowledge the J&K State Council for Science and Technology for giving financial support.
References
- 1.World Health Organization; 2008. Global Burden of Disease. 2004 Update. [Google Scholar]
- 2.Cutter J., Tan B.Y., Chew S.K. Levels of cardiovascular disease risk factors in Singapore following a national intervention programme. Bull World Health Organ. 2001;79:908–915. [PMC free article] [PubMed] [Google Scholar]
- 3.Tanuseputro P., Manuel D.G., Leung M., Nguyen K., Johansen H. Risk factors for cardiovascular disease in Canada. Can J Cardiol. 2003;19:1249–1260. [PubMed] [Google Scholar]
- 4.Iyer U.M., Bhoite R.M., Shah T. Risk factor analysis in coronary heart diseases and identifying at risk patients using a simple risk score test. Asian J Exp Biol Sci. 2011;2:40–46. [Google Scholar]
- 5.Panwar R.B., Gupta R., Gupta B.K. Atherothrombotic risk factors and premature coronary heart disease in India: a case–control study. Indian J Med Res. 2011;134:26–32. [PMC free article] [PubMed] [Google Scholar]
- 6.Blom H.J., Smulders Y. Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J Inherit Metab Dis. 2011;34:75–81. doi: 10.1007/s10545-010-9177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh P.R., Lele S.S. Folate gene polymorphisms MTR A2756G, MTRR A66G, and BHMT G742A and risk for coronary artery disease: a meta-analysis. Genet Test Mol Biomark. 2012;16:471–475. doi: 10.1089/gtmb.2011.0237. [DOI] [PubMed] [Google Scholar]
- 8.Graham I.M., Daly L.E., Refsum H.M. Plasma homocysteine as a risk factor for vascular disease: the European Concerted Action Project. JAMA. 1997;277:1775–1781. doi: 10.1001/jama.1997.03540460039030. [DOI] [PubMed] [Google Scholar]
- 9.Nygard O., Nordrehaug J.E., Refsum H. Plasma homocysteine levels and mortality in patients with coronary artery disease. N Engl J Med. 1997;337:230–236. doi: 10.1056/NEJM199707243370403. [DOI] [PubMed] [Google Scholar]
- 10.Cattaneo M. Hyperhomocysteinemia, atherosclerosis and thrombosis. Thromb Haemost. 1999;81:165–176. [PubMed] [Google Scholar]
- 11.Dwivedi M.K., Tripathi A.K., Shukla S., Khan S., Chauhan U.K. Homocysteine and cardiovascular disease. Biotechnol Mol Biol Rev. 2011;5:101–107. [Google Scholar]
- 12.García-Fragoso L., García-García I., Leavitt G. MTHFR polymorphisms in Puerto Rican children with isolated congenital heart disease and their mothers. Int J Genet Mol Biol. 2010;2:43–47. [PMC free article] [PubMed] [Google Scholar]
- 13.Mohamad N.A., Vasudevan R., Ismail P. Analysis of homocysteine metabolism enzyme gene polymorphisms in non-syndromic congenital heart disease patients among Malaysians. Life Sci J. 2014;11:318–326. [Google Scholar]
- 14.Andreassi M.G., Botto N., Cocci F. Methylenetetrahydrofolate reductase gene C677T polymorphism, homocysteine, vitamin B12, and DNA damage in coronary artery disease. Hum Genet. 2003;112:171–177. doi: 10.1007/s00439-002-0859-3. [DOI] [PubMed] [Google Scholar]
- 15.Djuric D., Jakovljevic V., Rasic-Markovic A., Djuric A., Stanojlovic O. Homocysteine, folic acid and coronary artery disease: possible impact on prognosis and therapy. Indian J Chest Dis Allied Sci. 2008;50:39–48. [PubMed] [Google Scholar]
- 16.Tripathi R., Tewari S., Singh P.K., Agarwal S. Association of homocysteine and methylene tetrahydrofolate reductase (MTHFR C677T) gene polymorphism with coronary artery disease (CAD) in the population of North India. Genet Mol Biol. 2010;33:224–228. doi: 10.1590/S1415-47572010005000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frosst P., Blom H.J., Milos R. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 18.Ma J., Stampfer M.J., Hennekens C.H. Methylenetetrahydrofolate reductase polymorphism, plasma folate, homocysteine, and risk of myocardial infarction in US physicians. Circulation. 1996;94:2410–2416. doi: 10.1161/01.cir.94.10.2410. [DOI] [PubMed] [Google Scholar]
- 19.Kluijtmans L.A., van den Heuvel L.P., Boers G.H. Molecular genetic analysis in mild hyperhomocysteinemia: a common mutation in the methylenetetrahydrofolate reductase gene is a genetic risk factor for cardiovascular disease. Am J Hum Genet. 1996;58:35–41. [PMC free article] [PubMed] [Google Scholar]
- 20.Brattstrom L., Wilcken D.E., Ohrvik J., Brudin L. Common methylenetetrahydrofolate reductase gene mutation leads to hyperhomocysteinemia but not to vascular disease: the result of a meta-analysis. Circulation. 1998;98:2520–2526. doi: 10.1161/01.cir.98.23.2520. [DOI] [PubMed] [Google Scholar]
- 21.Hustad S., Midttun Ø., Schneede J. The methylenetetrahydrofolate reductase 677C–> T polymorphism as a modulator of a B vitamin network with major effects on homocysteine metabolism. Am J Hum Genet. 2007;80:846–855. doi: 10.1086/513520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leclerc D., Campeau E., Goyette P. Human methionine synthase: cDNA cloning and identification of mutations in patients of the cblG complementation group of folate/cobalamin disorders. Hum Mol Genet. 1996;5:1867–1874. doi: 10.1093/hmg/5.12.1867. [DOI] [PubMed] [Google Scholar]
- 23.Matsuo K., Suzuki R., Hamajima N. Association between polymorphisms of folate- and methionine-metabolizing enzymes and susceptibility to malignant lymphoma. Blood. 2001;97:3205–3209. doi: 10.1182/blood.v97.10.3205. [DOI] [PubMed] [Google Scholar]
- 24.Sampaio-Neto L.F., Allen R.H., Guerra-Shinohara E.M. Association between decreased vitamin levels and MTHFR, MTR and MTRR gene polymorphisms as determinants for elevated total homocysteine concentrations in pregnant women. Eur J Clin Nutr. 2008;62:1010–1021. doi: 10.1038/sj.ejcn.1602810. [DOI] [PubMed] [Google Scholar]
- 25.Cronin S., Furie K.L., Kelly P.J. Dose-related association of MTHFR 677T allele with cumulative meta-analysis. Stroke. 2005;36:1581–1587. doi: 10.1161/01.STR.0000169946.31639.af. [DOI] [PubMed] [Google Scholar]
- 26.Klerk M., Verhoef P., Clarke R. MTHFR 677C-->T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA. 2002;288:2023–2031. doi: 10.1001/jama.288.16.2023. [DOI] [PubMed] [Google Scholar]
- 27.Laraqui A., Allami A., Carrié A. Influence of methionine synthase (A2756G) and methionine synthase reductase (A66G) polymorphisms on plasma homocysteine levels and relation to risk of coronary artery disease. Acta Cardiol. 2006;61:51–61. doi: 10.2143/AC.61.1.2005140. [DOI] [PubMed] [Google Scholar]
- 28.Laraqui A., Allami A., Carrie A. Relation between plasma homocysteine, gene polymorphisms of homocysteine metabolism-related enzymes, and angiographically proven coronary artery disease. Eur J Intern Med. 2007;18:474–483. doi: 10.1016/j.ejim.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Kanth V.V.R., Golla J.P., Sastry B. Genetic interactions between MTHFR (C677T), methionine synthase (A2756G, C2758G) variants with vitamin B12 and folic acid determine susceptibility to premature coronary artery disease in Indian population. J Cardiovasc Dis Res. 2011;2:156–163. doi: 10.4103/0975-3583.85262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellampalli R., Phani N.M., Bhat K.G. Significance of 5,10-methylenetetrahydrofolate reductase gene variants in acute lymphoblastic leukemia in Indian population: an experimental, computational and meta-analysis. Leuk Lymphoma. 2014:1–10. doi: 10.3109/10428194.2014.953154. [Early Online] [DOI] [PubMed] [Google Scholar]
- 31.WHO Expert Consultation Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363:157–163. doi: 10.1016/S0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization . WHO; Geneva: 2011. Waist Circumference and Waist–Hip Ratio Report of a WHO Consultation. [Google Scholar]
- 33.Chobanian A.V., Bakris G.L., Black H.R. The seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J., Russell D.W. 2nd ed. Cold Spring Harbour Laboratory Press; Cold Spring Harbor: 2001. Molecular Cloning, A Laboratory Manual. [Google Scholar]
- 35.Achari V., Thakur A.K. Association of major modifiable risk factors among patients with coronary artery disease – a retrospective analysis. JAPI. 2004;52:103–108. [PubMed] [Google Scholar]
- 36.Iqbal R., Ahmad Z., Malik F. A statistical analysis of hypertension as cardiovascular risk factor. Middle-East J Sci Res. 2012;12:19–22. [Google Scholar]
- 37.Rani R., Mengi V., Verma A., Sharma H.K. Prevalence study of hypertension among adults in an urban area of Jammu. JSIR. 2014;3:143–147. [Google Scholar]
- 38.Aggarwal A., Aggarwal S., Goel A., Sharma V., Dwivedi S. A retrospective case–control study of modifiable risk factors and cutaneous markers in Indian patients with young coronary artery disease. JRSM Cardiovasc Dis. 2012 doi: 10.1258/cvd.2012.012010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De A., Podder G., Adhikari A. Comparative study of risk factors of cardiac diseases among urban and rural population. Int J Hum Genet. 2013;13:15–19. [Google Scholar]
- 40.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;7011:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 41.Dhar S., Chatterjee S., Ray S. Polymorphisms of methylenetetrahydrofolate reductase gene as the genetic predispositions of coronary artery diseases in eastern India. JCDR. 2010;1:152–157. doi: 10.4103/0975-3583.70922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matam K., Khan I.A., Hasan Q., Rao P. Coronary artery disease and the frequencies of MTHFR and PON1 gene polymorphism studies in a varied population of Hyderabad, Telangana region in south India. J King Saud Univ – Sci. 2014 [Google Scholar]
- 43.Lakshmi S.V.V., Naushad S.M., Rupasree Y., Rao D.S., Kutala V.K. Interactions of 5′ UTR thymidylate synthase polymorphism with 677C/T methylene tetrahydrofolate reductase and 66 A/G methyltetrahydrofolate homocysteine methyl-transferase reductase polymorphisms determine susceptibility to coronary artery disease. J Atheroscler Thromb. 2011;18:56–64. doi: 10.5551/jat.5702. [DOI] [PubMed] [Google Scholar]
- 44.Gupta S.K., Kotwal J., Kotwal A., Dhall A., Garg S. Role of homocysteine & MTHFR C677T gene polymorphism as risk factors for coronary artery disease in young Indians. Indian J Med Res. 2012;135:506–512. [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey U., Kumari R., Nath B. Association of angiotensin-converting enzyme, methylene tetrahydrofolate reductase and paraoxonase gene polymorphism and coronary artery disease in an Indian population. Cardiol J. 2011;18:385–394. [PubMed] [Google Scholar]
- 46.Vasisht S., Gulati R., Narang R.O. Polymorphism (C677T) in the 5,10-methylenetetrahydrofolate reductase (MTHFR) gene: a preliminary study on North Indian men. Indian J Clin Biochem. 2002;17:99–107. doi: 10.1007/BF02867949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalal A.B., Tewari D., Tewari S. Association of coronary artery disease with polymorphisms of angiotensin converting enzyme and methylenetetrahydrofolate reductase gene. Indian Heart J. 2006;58:330–335. [PubMed] [Google Scholar]
- 48.Aleyasin A., Ghaedi M., Davoodi S., Abbasi S.H., Madani M. Methylenetetrahydrofolate reductase (MTHFR) gene C677T polymorphism is associated with coronary atherosclerosis disease in a sample of Iranian patients. J Tehran Heart Cent. 2006;1:77–81. [Google Scholar]
- 49.Ilhan N., Kucuksu M., Kaman D., Ilhan N., Ozbay Y. The 677C/T MTHFR polymorphism is associated with essential hypertension, coronary artery disease, and higher homocysteine levels. Arch Med Res. 2008;39:125–130. doi: 10.1016/j.arcmed.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Nasiri M., Roostaei A., Ehsanian Z. Association of methylenetetrahydrofolate reductase (MTHFR) gene C677T and A1298C polymorphisms with myocardial infarction from North of Fars Province. Res Mol Med. 2014;2:37–41. [Google Scholar]
- 51.Wilcken D.E.L., Wang X.L., Sim A.S., McCredie R.M. Distribution in healthy and coronary populations of the methylenetetrahydrofolate reductase (MTHFR) C677T mutation. Arterioscler Thromb Vasc Biol. 1996;16:878–882. doi: 10.1161/01.atv.16.7.878. [DOI] [PubMed] [Google Scholar]
- 52.Abu-Amero K.K., Wyngaard C.A., Dzimiri N. Prevalence and role of methylenetetrahydrofolate reductase 677 C/T and 1298 A/C polymorphisms in coronary artery disease in Arabs. Arch Pathol Lab Med. 2003;127:1349–1352. doi: 10.5858/2003-127-1349-PAROMR. [DOI] [PubMed] [Google Scholar]
- 53.Giusti B., Gori A.M., Marcucci R. Role of C677T and A1298C MTHFR, A2756G MTR and −786 C/T eNOS gene polymorphisms in atrial fibrillation susceptibility. PLoS ONE. 2007;2:e495. doi: 10.1371/journal.pone.0000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ibrahiem M.M., Gaber O., Mohammed S.H., Salem E.M. Methylenetetrahydrofolate reductase C677T polymorphism and relationship with coronary artery disease. Egypt J Chem. 2009;27:177–194. [Google Scholar]
- 55.Bahadır A., Eroz R., Türker Y. Does the MTHFR C677T gene polymorphism indicate cardiovascular disease risk in type 2 diabetes mellitus patients? Anatol J Cardiol. 2015;15:524–530. doi: 10.5152/akd.2014.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu L.A., Ko Y.L., Wang S.M. The C677T mutation of the methylene tetrahydrofolate reductase gene is not associated with the risk of coronary artery disease or venous thrombosis among Chinese in Taiwan. Hum Hered. 2001;51:41–45. doi: 10.1159/000022958. [DOI] [PubMed] [Google Scholar]
- 57.Yilmaz H., Isbir S., Agachan B. C677T mutation of methylenetetrahydrofolate reductase gene and serum homocysteine levels in Turkish patients with coronary artery disease. Cell Biochem Funct. 2006;24:87–90. doi: 10.1002/cbf.1206. [DOI] [PubMed] [Google Scholar]
- 58.Caner M., Bircan R., Sevinç D. MTHFR, prothrombin and factor V gene variants in Turkish patients with coronary artery stenosis. Genet Mol Biol. 2008;31:836–838. [Google Scholar]
- 59.Kalita J., Srivastava R., Bansal V., Agarwal S., Misra U.K. Methylenetetrahydrofolate reductase gene polymorphism in Indian stroke patients. Neurol India. 2006;54:260–263. doi: 10.4103/0028-3886.27148. [DOI] [PubMed] [Google Scholar]
- 60.Clarke R., Daly L., Robinson K. Hyperhomocysteinemia: an independent risk factor for vascular disease. N Engl J Med. 1991;324:1149–1155. doi: 10.1056/NEJM199104253241701. [DOI] [PubMed] [Google Scholar]
- 61.Nair K.G., Nair S.R., Ashavaid T.F., Dalal J.J., Eghlim F.F. Methylenetetrahydrofolate reductase gene mutation and hyperhomocysteinemia as a risk factor for coronary heart disease in the Indian population. J Assoc Phys India. 2002;50:9–15. [PubMed] [Google Scholar]
- 62.Yakub M., Moti N., Parveen S. Polymorphisms in MTHFR, MS and CBS genes and homocysteine levels in a Pakistani population. PLoS ONE. 2012;7:e33222. doi: 10.1371/journal.pone.0033222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jemaa R., Achouri A., Kallel A., Ali S.B., Mourali S. Association between the 2756A> G variant in the gene encoding methionine synthase and myocardial infarction in Tunisian patients. Clin Chem Lab Med. 2008;46:1364–1368. doi: 10.1515/CCLM.2008.306. [DOI] [PubMed] [Google Scholar]
- 64.Huang L., Song X.M., Zhu W.L., Li Y. Plasma homocysteine and gene polymorphisms associated with the risk of hyperlipidemia in Northern Chinese subjects. Biomed Environ Sci. 2008;21:514–520. doi: 10.1016/S0895-3988(09)60011-8. [DOI] [PubMed] [Google Scholar]
- 65.Salomon O., Rosenberg N., Zivelin A. Methionine synthase A2756G and methylenetetrahydrofolate reductase A1298C polymorphisms are not risk factors for idiopathic venous thromboembolism. Hematol J. 2001;2:38–41. doi: 10.1038/sj.thj.6200078. [DOI] [PubMed] [Google Scholar]
- 66.Angeline T., Thiruvarutselvi G., Isabel W. MTHFR (Ala222Val) polymorphism and AMI in patients with type II diabetes mellitus. Indian J Clin Biochem. 2009;24:137–141. doi: 10.1007/s12291-009-0025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aifan L., Hong Z., Yuming X. Research on the relationship between the polymorphisms of Methionine synthase (MS A2756G) gene and ischemic cerebrovascular disease. Life Sci J. 2009;6:27–31. [Google Scholar]


