Abstract
Measuring the severity and progression of facioscapulohumeral muscular dystrophy (FSHD) is particularly challenging because muscle weakness progresses over long periods of time and can be sporadic. Biomarkers are essential for measuring disease burden and testing treatment strategies. We utilized the sensitive, specific, high-throughput SomaLogic proteomics platform of 1129 proteins to identify proteins with levels that correlate with FSHD severity in a cross-sectional study of two independent cohorts. We discovered biomarkers that correlate with clinical severity and disease burden measured by magnetic resonance imaging. Sixty-eight proteins in the Rochester cohort (n = 48) and 51 proteins in the Seattle cohort (n = 30) had significantly different levels in FSHD-affected individuals when compared with controls (p-value ≤ .005). A subset of these varied by at least 1.5 fold and four biomarkers were significantly elevated in both cohorts. Levels of creatine kinase MM and MB isoforms, carbonic anhydrase III, and troponin I type 2 reliably predicted the disease state and correlated with disease severity. Other novel biomarkers were also discovered that may reveal mechanisms of disease pathology. Assessing the levels of these biomarkers during clinical trials may add significance to other measures of quantifying disease progression or regression.
Keywords: FSHD, Facioscapulohumeral, D4Z4, DUX4, Biomarker, Proteomics, Muscular dystrophy, Creatine kinase, Carbonic anhydrase, Troponin
1. Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is an adult onset, dominantly inherited, slowly progressive muscular dystrophy with no treatment. Individuals are often diagnosed in the second decade of life with symptoms of scapular winging, foot drop, and facial muscle weakness [1]. These symptoms are almost always progressive and spread to other muscle groups resulting in debilitating weakness and reduced quality of life [2]. The primary molecular event involves epigenetic de-repression of a macrosatellite array of D4Z4 DNA repeats on chromosome 4 resulting in the expression of the double homeobox 4 (DUX4) gene in a small fraction of myonuclei [3–7]. The DUX4 transcription factor is toxic to human [8] and mouse [9,10] cells when overexpressed from viral vectors, and cultured human myoblasts expressing endogenous DUX4 die within 24 hours of DUX4 activation [11]. MRI studies of FSHD-affected individuals show normal appearing muscle adjacent to muscles with short-TI inversion recovery (STIR) bright signal and muscles with fatty replacement of tissue [12–15]. Although there is a predilection for certain muscle groups, different muscles can be affected in different individuals, or in different extremities of the same individual, adding to the complexity of assessing disease burden. Several therapeutic approaches are being developed [16,17] but tools for assessing efficacy in the absence of animal models are currently lagging the development of therapies [18].
Assessing therapeutic efficacy in a FSHD clinical trial is challenging because the disease is highly variable and slowly progressive, so strength measurements need to be taken in large numbers of subjects over long time intervals to reveal significant effects [2]. Biomarkers assessed in blood plasma or serum that reflect disease pathology could provide a quick, objective, and quantitative assessment of disease severity, allowing meaningful changes to be assessed in a time period where functional changes may not yet be realized. Because all muscles are exposed to the circulation, muscle-derived serum proteins should reflect an average disease burden throughout the body.
We used the SOMAscan assay, based on Slow Off-rate Modified Aptamers, to assess the levels of 1129 target proteins. The technology utilizes nucleic acid secondary structures to recognize 3D epitopes on proteins with very high sensitivity and specificity [19,20]. This platform has been used for the discovery and validation of biomarkers in a number of diseases including DMD [21], Alzheimer’s [22,23], non-small cell lung cancer [24], and pulmonary tuberculosis [25]. We identified 4 proteins with blood levels that independently correlated with the presence and severity of disease in two separate FSHD cohorts and correlate with MRI STIR bright signal in subjects from the Seattle cohort. The serum levels of these biomarkers will be an important parameter for validation in future longitudinal studies and should allow quantification of disease severity in combination with MRI or assessments of strength and movement.
2. Materials and methods
2.1. Standard protocol approvals, registrations, and patient consents
Venipuncture was performed with informed consent using documents and protocols approved by the Human Subjects Independent Review Board (IRB) at the University of Washington, Seattle, WA, and University of Rochester, Rochester, NY. Subjects in the Seattle cohort also consented to MRI studies as approved by the Seattle Children’s Hospital IRB. Written informed consent was obtained from all subjects participating in the study.
2.2. Study design
The Seattle cohort was composed of 18 FSHD-affected individuals and 12 control subjects whom we obtained demographic data, performed a brief physical exam, and calculated clinical severity scores [26]. A blood sample was obtained in an EDTA-containing tube (purple top) and plasma was prepared from approximately 2 ml of uncoagulated whole blood by centrifugation at 1500 × g for 10 min and freezing samples at −80 °C. All samples were thawed once for proteomic analysis. Additional sample was used to confirm a FSHD diagnosis by pulse field gel electrophoresis (PFGE), and Southern blots that were probed with the P13E11, “A”, and “B” probes [27]. When a contracted D4Z4 fragment was not observed, sequencing of SMCHD1 was performed and mutations were confirmed to be causative by quantifying D4Z4 CpG methylation [28]. Subjects’ ages ranged from 20 to 79, with 13 male and 5 female in the FSHD group and ages 21–72 with 6 male and 6 female in the control group.
Eleven of the same subjects with FSHD were recruited for a study evaluating whole body MRI data. All MRI data acquisition occurred at Seattle Children’s Hospital on a 3 T Siemens Trio scanner running software VB17. Flexible array coils were employed to collect data in the thigh, calf, upper body (including shoulders, back, pectoral muscles) and abdominal region. The following sequences were collected: 3-plane localizers, T1 (TE = 8.9 ms, TR = 510 ms, 320 × 224, 5 mm thick, 50 slices upper body/40 slices lower body), and STIR (TE = 37 ms, TR = 4300, flip 150°, same resolution).
Likewise, physical exam, clinical severity scores and demographic data were obtained from 24 FSHD-affected and 23 control subjects that comprise the Rochester cohort. Ages ranged from 18 to 69, with 11 male and 13 female in the FSHD group and 10 male and 13 female in the control group. A diagnosis of FSHD was confirmed by PFGE. Serum was prepared from clotted blood samples collected in vacu-tubes without anti-coagulant (red top) by centrifugation at 1500 × g for 10 min and freezing at −80 °C. All samples were thawed once for proteomic analysis. The levels of 1129 proteins were determined using the SomaLogic technology [20] by sending a small frozen aliquot to SomaLogic Inc. for testing.
Subjects affected by diabetes, chronic obstructive pulmonary disease, chronic heart failure, current malignancy, previous treatment with chemotherapy or radiation, use of corticosteroids for a period exceeding 2 weeks in the last 5 years, use of statins in the past year, wheel chair dependence, or pregnancy were excluded.
2.3. SomaLogic proteomic methods
SomaLogic Inc. (Boulder, CO) performed analysis on serum or plasma samples that had been stored at −80 °C or below and had never been previously thawed. The SOMAmer-based proteomic assay consists of equilibrium binding of fluorophore-tagged SOMAmers and proteins in plasma in solution and automated partitioning steps to capture only the SOMAmers that are in complexes with their cognate proteins [20]. The assay transforms the measurement of proteins into the measurement of the corresponding SOMAmers via hybridization of the SOMAmer DNA oligonucleotides to an array of antisense probes using a hybridization gasket slide (Agilent Technologies, Santa Clara, CA, USA). The liquid handling steps of the assay (protein binding) are performed by a Biomek robot, and the fluorescent signal generated in the hybridization step is captured. Protein concentrations were reported in relative fluorescence units (RFU).
2.4. Data analysis and statistical method
Data were analyzed using the R environment for statistical computing. The Kolmogorov–Smirnov (KS) test [29] was used for unpaired comparisons between FSHD and control subjects. Correction for multiple comparisons was performed according to the procedure of Benjamini and Hochberg [30] and adjusted p-values (q-values) are shown in Tables 1 and 2. The list of statistically significant biomarkers (p < .005) was limited to those that had a 1.5 fold or greater change in concentration in FSHD-affected subjects when compared with controls.
Table 1.
Aptamer targets with significantly different plasma concentrations in the Seattle cohort.
| Targeta | Target protein description | RFUb (Control) |
RFU (FSHD) |
FCc | p-valued | Adjusted p-valuee |
|---|---|---|---|---|---|---|
| CA-III | Carbonic anhydrase 3 | 2,241.9 | 8,530.0 | 3.8 | 2.31E–08 | 2.61E–05 |
| CK-MB | Creatine kinase M:Creatine kinase B | 877.4 | 3,598.8 | 4.1 | 2.47E–06 | 1.40E–03 |
| TNNI2 | Troponin I, fast skeletal muscle | 6,439.7 | 19,193.7 | 3.0 | 6.54E–06 | 2.46E–03 |
| CK-MM | Creatine kinase M-type | 699.6 | 1,860.6 | 2.7 | 4.07E–05 | 7.66E–03 |
| HSP 70 | Heat shock 70 kDa protein 1A/1B | 7,168.3 | 10,794.2 | 1.5 | 4.49E–04 | 2.67E–02 |
| KI2L4 | Killer cell immunoglobulin-like receptor 2DL4 | 1,110.1 | 2,113.4 | 1.9 | 8.89E–04 | 4.02E–02 |
| MMP-9 | Matrix metalloproteinase-9 | 18,897.5 | 35,708.6 | 1.9 | 1.74E–03 | 4.90E–02 |
| FABP | Fatty acid-binding protein | 1,553.5 | 4,202.8 | 2.7 | 1.74E–03 | 4.90E–02 |
| TNFRSF11A | Tumor necrosis factor receptor superfamily member 11A | 1,036.6 | 2,840.1 | 2.7 | 3.13E–03 | 7.35E–02 |
| CAT | Catalase | 12,738.6 | 24,318.3 | 1.9 | 3.13E–03 | 7.35E–02 |
| METAP1 | Methionine aminopeptidase 1 | 4,173.2 | 1,398.7 | −3.0 | 4.07E–05 | 7.66E–03 |
| BTK | Tyrosine-protein kinase BTK | 7,640.7 | 2,621.1 | −2.9 | 1.01E–04 | 1.63E–02 |
| PTPN6 | Tyrosine-protein phosphatase non-receptor 6 | 3,872.8 | 2,579.8 | −1.5 | 2.11E–04 | 1.84E–02 |
| CSK | Tyrosine-protein kinase CSK | 6,546.6 | 2,537.4 | −2.6 | 2.11E–04 | 1.84E–02 |
| PKCB | Protein kinase C beta type (variant beta-II) | 8,577.8 | 3,186.4 | −2.7 | 2.11E–04 | 1.84E–02 |
| PPIF | Peptidyl-prolyl cis-trans isomerase F, mitochond. | 18,723.6 | 5,525.1 | −3.4 | 2.11E–04 | 1.84E–02 |
| PRKCA | Protein kinase C alpha type | 28,408.3 | 7,684.1 | −3.7 | 2.11E–04 | 1.84E–02 |
| PRKACA | cAMP-dependent protein kinase cat. subunit α | 3,604.3 | 952.4 | −3.8 | 2.11E–04 | 1.84E–02 |
| AKT1 | Protein kinase B alpha/beta/gamma | 3,155.0 | 1,787.9 | −1.8 | 3.29E–04 | 2.66E–02 |
| PDIA3 | Protein disulfide-isomerase A3 | 8,103.7 | 5,002.3 | −1.6 | 4.49E–04 | 2.67E–02 |
| ITGA2B | Integrin alpha-IIb: beta-3 | 24,449.8 | 8,392.3 | −2.9 | 4.49E–04 | 2.67E–02 |
| CAXIII | Carbonic anhydrase 13 | 2,880.9 | 859.8 | −3.4 | 4.49E–04 | 2.67E–02 |
| PRKCQ | Protein kinase C theta type | 3,472.1 | 1,976.9 | −1.8 | 8.89E–04 | 4.02E–02 |
| DBNL | Drebrin-like protein | 2,187.7 | 1,507.2 | −1.5 | 1.74E–03 | 4.90E–02 |
| YWHAB | 14-3-3 protein family | 6,280.4 | 3,031.9 | −2.0 | 1.74E–03 | 4.90E–02 |
| PPID | Peptidyl-prolyl cis-trans isomerase D | 4,003.2 | 1,753.3 | −2.3 | 1.74E–03 | 4.90E–02 |
| PDE5A | cGMP-specific 3′,5′-cyclic phosphodiesterase | 19,191.8 | 6,694.4 | −2.9 | 1.74E–03 | 4.90E–02 |
| HSPD1 | 60 kDa heat shock protein, mitochondrial | 23,223.0 | 7,291.5 | −3.2 | 1.74E–03 | 4.90E–02 |
| SRC | Proto-oncogene tyrosine-protein kinase Src | 34,491.7 | 8,094.9 | −4.3 | 1.74E–03 | 4.90E–02 |
| EIF4G2 | Eukaryotic translation initiation factor 4 gamma 2 | 2,692.3 | 468.6 | −5.8 | 1.74E–03 | 4.90E–02 |
| HSD17B10 | 3-hydroxyacyl-CoA dehydrogenase type-2 | 4,863.7 | 1,714.4 | −2.8 | 3.13E–03 | 7.35E–02 |
| PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | 2,110.9 | 525.3 | −4.0 | 3.13E–03 | 7.35E–02 |
| PPP3CA | Calcineurin | 1,925.5 | 1,002.9 | −1.9 | 3.32E–03 | 7.36E–02 |
| LYN | Tyrosine-protein kinase Lyn | 8,987.8 | 3,100.4 | −2.9 | 3.32E–03 | 7.36E–02 |
| FER | Tyrosine-protein kinase Fer | 1,285.4 | 207.9 | −6.2 | 3.32E–03 | 7.36E–02 |
Significant targets were defined as those having a FC ≥ 1.5 and p-value ≤ .005.
RFU = Relative fluorescence units.
FC = Fold change.
p-values are from the D-statistic generated with the Kruskal–Wallis test.
False discovery rate was determined by adjusting p-values for multiple testing using the formula of Benjamini and Hochberg.
Table 2.
Aptamer targets with significantly different serum concentrations in the Rochester cohort.
| Targeta | Target protein description | Mean RFUb (Control) |
Mean RFU (FSHD) |
FCc | p-valued | Adjusted p-valuee |
|---|---|---|---|---|---|---|
| CK-MB | Creatine kinase M:Creatine kinase B heterodimer | 2,153.3 | 4,867.5 | 2.3 | 2.65E−05 | 5.99E−03 |
| CA- III | Carbonic anhydrase 3 | 2,687.2 | 6,040.8 | 2.2 | 3.21E−05 | 6.05E−03 |
| TCTP | Translationally-controlled tumor protein | 3,984.9 | 6,907.3 | 1.7 | 8.12E−05 | 1.02E−02 |
| CK-MM | Creatine kinase M-type | 1,108.8 | 2,006.3 | 1.8 | 2.84E−04 | 1.69E−02 |
| CSK | Tyrosine-protein kinase CSK | 1,018.7 | 1,612.0 | 1.6 | 2.37E−04 | 1.69E−02 |
| PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | 776.5 | 1,478.3 | 1.9 | 2.37E−04 | 1.69E−02 |
| NUDCD3 | NudC domain-containing protein 3 | 550.5 | 2,059.8 | 3.7 | 6.76E−04 | 3.02E−02 |
| FER | Tyrosine-protein kinase Fer | 325.1 | 653.4 | 2.0 | 8.29E−04 | 3.02E−02 |
| NSF1C | NSFL1 cofactor p47 | 1,043.7 | 1,952.4 | 1.9 | 8.29E−04 | 3.02E−02 |
| DUS3 | Dual specificity protein phosphatase 3 | 1,000.4 | 2,009.8 | 2.0 | 7.46E−04 | 3.02E−02 |
| UFM1 | Ubiquitin-fold modifier 1 | 3,708.3 | 5,593.3 | 1.5 | 6.62E−04 | 3.02E−02 |
| SHC1 | SHC-transforming protein 1 | 8,151.0 | 15,477.8 | 1.9 | 8.29E−04 | 3.02E−02 |
| IF4G2 | Eukaryotic translation initiation factor 4 gamma 2 | 1,087.2 | 1,737.5 | 1.6 | 9.88E−04 | 3.38E−02 |
| C3b | Complement C3b | 4,002.6 | 6,241.6 | 1.6 | 1.97E−03 | 5.30E−02 |
| DRG-1 | Vacuolar sorting-associated protein VTA1 homolog | 14,957.0 | 28,488.9 | 1.9 | 2.36E−03 | 5.33E−02 |
| GPVI | Platelet glycoprotein VI | 10,462.9 | 16,710.3 | 1.6 | 2.36E−03 | 5.33E−02 |
| SBDS | Ribosome maturation protein SBDS | 1,796.2 | 2,894.3 | 1.6 | 2.03E−03 | 5.33E−02 |
| PSMA | Glutamate carboxypeptidase 2 | 2,917.7 | 4,791.0 | 1.6 | 2.55E−03 | 5.48E−02 |
| TNNI2 | Troponin I, fast skeletal muscle | 8,282.0 | 15,461.1 | 1.9 | 3.02E−03 | 5.48E−02 |
| CA-XIII | Carbonic anhydrase 13 | 3,102.6 | 7,303.8 | 2.4 | 2.63E−03 | 5.48E−02 |
| BTK | Tyrosine-protein kinase BTK | 1,483.7 | 3,241.0 | 2.2 | 3.02E−03 | 5.48E−02 |
Significant targets were defined as those having a FC ≥ 1.5 and p-value ≤ .005.
RFU = Relative fluorescence units.
FC = Fold change.
p-values are from the D-statistic generated with the Kruskal–Wallis test.
False discovery rate was determined by adjusting p-values for multiple testing using the formula of Benjamini and Hochberg.
Unsupervised hierarchical clustering analysis was performed using the Euclidean distance between the log2 of the median expression ratio for a particular set of biomarkers and calculating a set of dissimilarities between samples using Lance–Williams dissimilarity update formula according to the method of Murtagh and Legendre.
Locally weighted scatterplot smoother (LOESS) curve fitting line approximations were calculated using the R environment and the ggplot2 package with the geom-smooth function set to “loess”.
2.4.1. MRI scoring
One of the authors (MRF) blinded to subject phenotypes assessed muscle change using a modified scale based on a previously described rating scale for fat [31,32]. Briefly, 0: normal; 1: scattered small areas of abnormality; 2: numerous discrete areas of increased signal intensity, less than 30% of the muscle volume; 3: numerous discrete areas with early confluence, 30–60% volume; 4: >60% with patchy loss of fascial structure; or 5: pronounced fatty replacement throughout with complete fascial structure loss. In addition, the STIR+ intensity was rated according to the following categories: 0: none; 1: minimal interfascicular edema; 2: minimal inter- and intrafascicular edema; or 3: moderate inter- and intrafascicular edema. Data were compiled for each subject and displayed as a heat map to demonstrate the overall disease burden per subject.
3. Results
3.1. Selection and exclusion of subjects
Study subjects from the Seattle cohort consisted of members of local support groups or FSHD research fund-raising activity participants. Control individuals were identified randomly or were asymptomatic and test-negative relatives of FSHD subjects. Both control and FSHD-affected individuals who were taking medications or had medical conditions that would be predicted to alter muscle physiology were excluded (see the section “Study design”). The Seattle cohort included sixteen individuals with FSHD1 and two individuals with FSHD2. Subjects grouped under the Rochester cohort were ascertained similarly however serum was prepared from blood samples. All FSHD-affected individuals in the Rochester cohort carried a diagnosis of FSHD1 and the disease causing D4Z4 array contraction.
3.2. Biomarkers with statistically significant differences in serum or plasma concentration were identified in both the Seattle and Rochester cohorts
The concentrations of 1129 different proteins were determined for each subject in both cohorts. Because blood samples were prepared slightly differently between cohorts, the analysis could not be combined but independent analyses allowed verification of significant findings within each cohort. We found 51 proteins with plasma concentrations that differed significantly between FSHD-affected and control groups in the Seattle cohort (p value < .005). Sixty-eight proteins had significantly different distributions of serum concentrations comparing FSHD-affected and controls in the Rochester cohort (p value < .005). Limiting these lists to proteins with at least a 1.5 fold difference between FSHD-subjects and controls revealed 35 proteins in the Seattle cohort (Table 1) and 21 proteins in the Rochester cohort (Table 2), allowing us to focus on biomarkers with maximum sensitivity and specificity.
3.3. Classification of biomarkers
Biomarkers found in both cohorts could be grouped into several broad categories. Members of both lists included skeletal muscle specific proteins found in the serum of subjects with Duchenne muscular dystrophy (DMD) [21] and can be characterized as proteins that leak into the blood as muscles break down. These included creatine kinase isoforms MM and MB, carbonic anhydrase types 3 and 13, Troponin I from fast twitch skeletal muscle, and fatty acid binding protein (FABP3) found both in heart and skeletal muscle.
Proteins involved in protein folding and maintenance of aberrant cellular translation also represent a broad category found in both lists. Members of this group include ubiquitin-fold modifier 1 (UFM1), eukaryotic translation initiation factor 4 gamma 2 (IF4G2), vacuolar protein sorting-associated protein VTA1 homolog (DRG-1), ribosome maturation protein (SBDS), and heat shock 70 kDa protein 1A/1B (HSP70) and 60 kDa heat shock protein, mitochondrial (HSPD1). With the exception of HSP70, these proteins appear to be specific for FSHD pathology as none were found to be significantly altered in a recently published DMD study using the same technology [21].
Consistent with a recent report of novel pathways identified in FSHD cells [11], proteins involved in cell adhesion, fusion, and migration were also found to be significantly elevated in FSHD-affected subjects. Members of this category include 2-phosphoinositide-dependent protein kinase 1 (PDPK1), tyrosine-protein kinase (FER), and NSFL1 cofactor p47 (NSFL1C). Therefore this proteomics approach lends further support to the importance of these pathways in FSHD pathology [11].
Finally, proteins with a possible inflammatory role were also present in both lists. These included complement 3b (C3b), B-cell tyrosine-protein kinase (BTK), killer cell immunoglobulin-like receptor 2DL4 (KIR2DL4), tumor necrosis factor receptor superfamily member 11A (TNFRSF11A), catalase (CAT), and matrix metalloproteinase 9 (MMP9). However, acute phase reactants that accompany both acute and chronic inflammatory states were not among the proteins found to be significantly elevated in FSHD-affected individuals, suggesting that inflammation may not be a primary component of the disease process.
3.4. Biomarker levels predict disease status
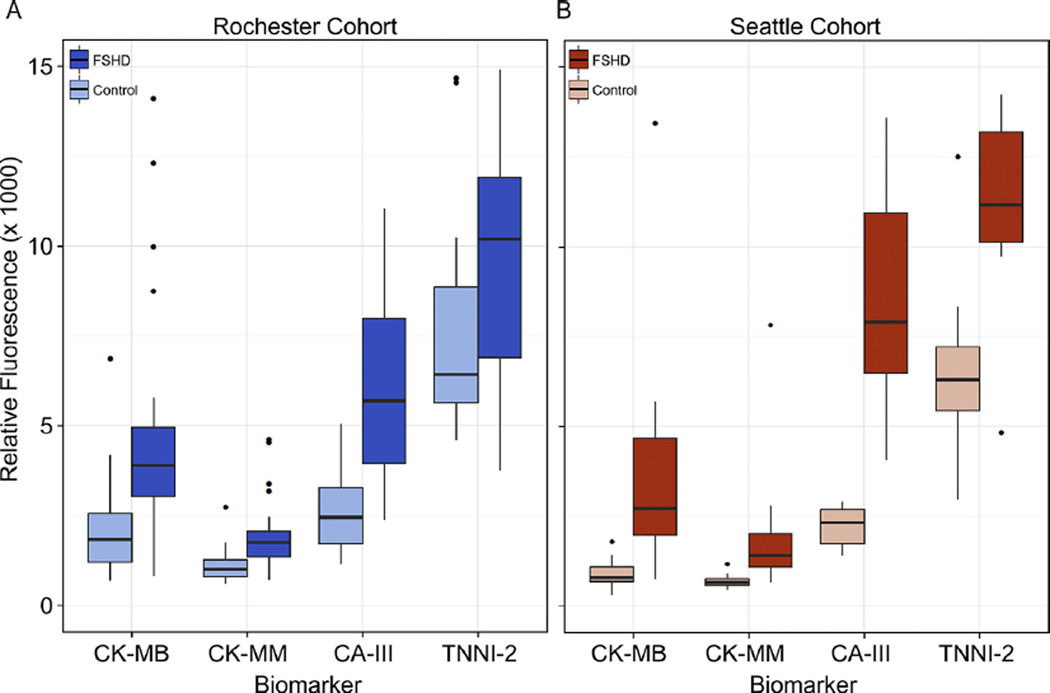
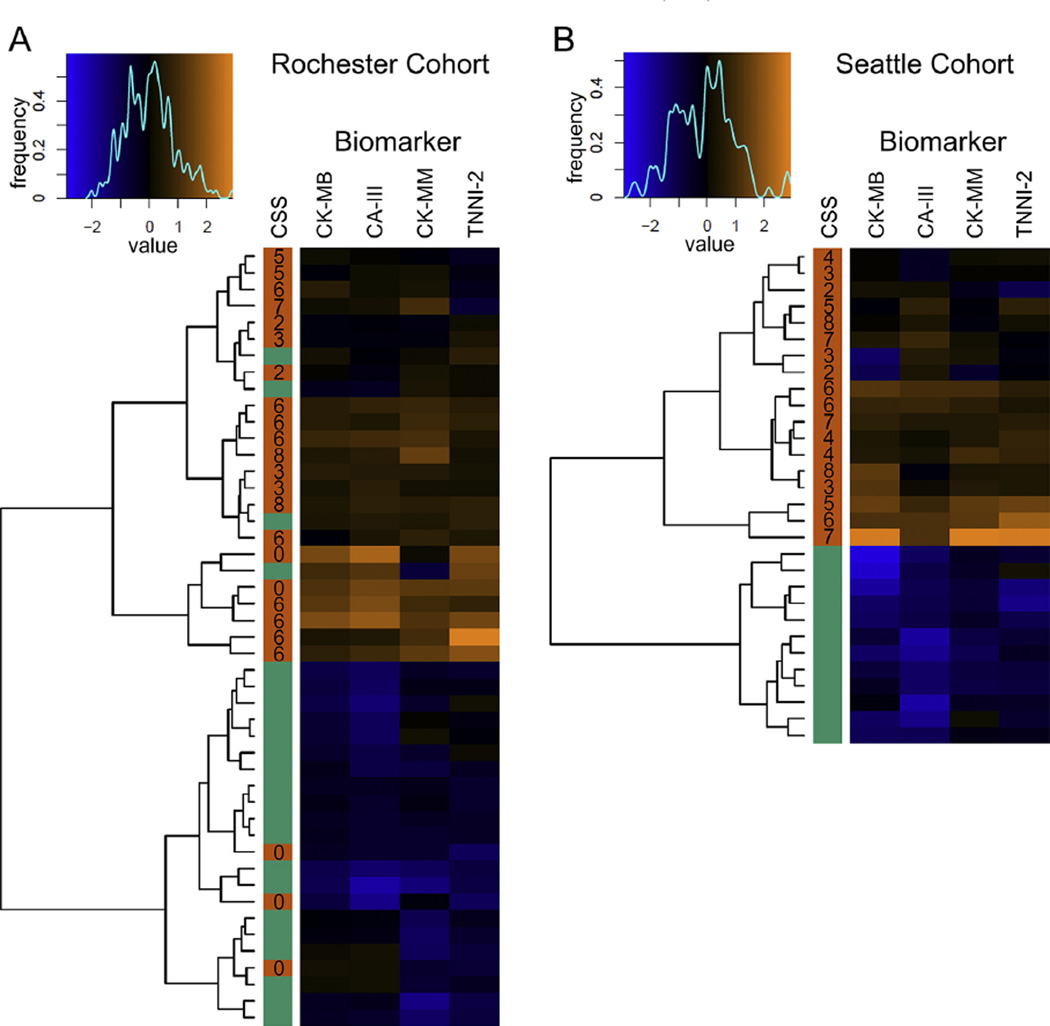
We noted several biomarkers that were present in both the Seattle and Rochester lists (Tables 1 and 2). These included the MM and MB isoforms of creatine kinase, carbonic anhydrase III, and troponin I type 2. As expected, the variability was narrow in control groups and larger in FSHD-affected groups that had varying disease severities/burdens (Fig. 1).We focused on the levels of these proteins to determine if they could predict the disease state in subjects of either cohort. Utilizing the technique of unsupervised hierarchical clustering, the samples were computationally arranged into blocks of similarity and the log of the median expression ratio is indicated by color, with blue being low and orange being high (Fig. 2). The disease status is indicated by a solid bar on the left side of each map and demonstrates that samples from FSHD-affected subjects group together 93% of the time. Mis-groupings were limited to the Rochester cohort where multiple individuals with very mild disease symptoms (clinical severity score [CSS] = 0) were included.
Fig. 1.
Biomarker concentrations in controls and FSHD-affected subjects from the Rochester and Seattle cohorts. Box plots (25th–75th percentile) and whiskers (Tukey’s method) showing comparisons of plasma concentrations for each of four biomarkers in control and FSHD-affected subjects. The black bar indicates median value, outliers (more or less than 3/2 times the upper and lower quartiles) are shown as single dots. (A) Rochester cohort with control group in light blue and FSHD group in dark blue. (B) Seattle cohort with control group in light red and FSHD group in dark red.
Fig. 2.
Heat maps of Rochester and Seattle cohorts showing unsupervised clustering. Each map shows the relationship of the Euclidean distances between the log2 of the median expression ratio for each biomarker. Log2 median expression ratios are assigned a color based on the value and the color bin’s are plotted in the spectral graph above each map (x axis). The line drawn over the spectrum shows the frequency of samples with each particular color or bin (y axis). A dendrogram drawn to the left of each map shows relative similarities between samples. The color coded column labeled CSS shows the disease status (orange = FSHD and green = Control) with the clinical severity score of the FSHD subject adjacent to the respective sample row.
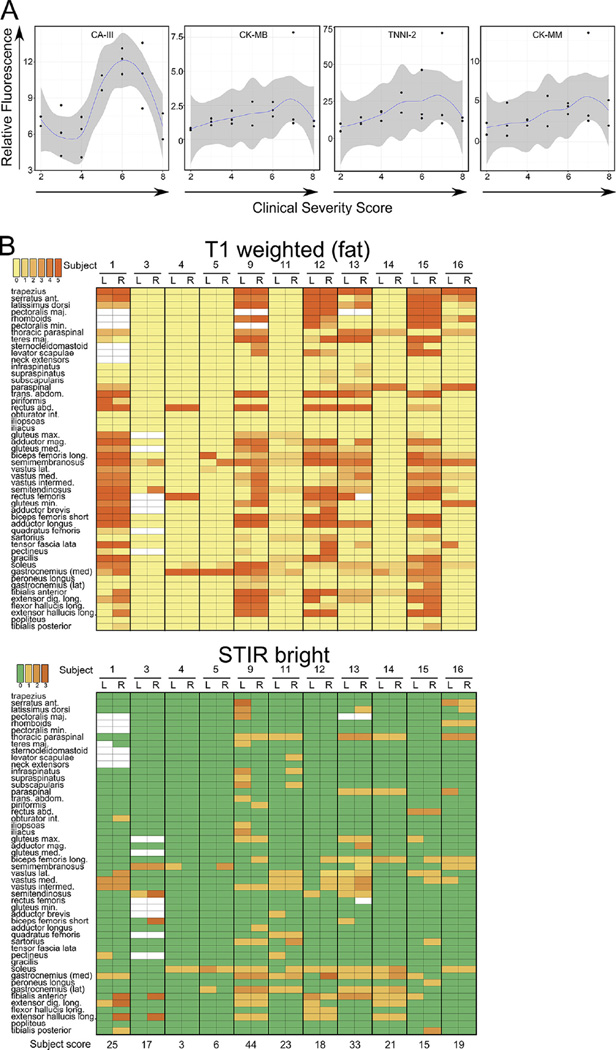
3.5. Plasma levels of four biomarkers correlate with disease severity
Optimally, biomarkers should correlate with disease severity so that their levels can be used to determine the efficacy of a treatment during a clinical trial. We determined disease severity using a previously published scoring criteria [26] and generated a clinical severity score for each FSHD-affected subject (CSS, Fig. 3A). Because the Seattle cohort contained a relatively even distribution of subjects with CSS values ranging from 2 to 8, the plasma concentration of each biomarker relative to the CSS of the subjects within the Seattle cohort was plotted. Plasma levels for all four markers correlated directly with CSS when the CSS was ≤6 (Fig. 3A). Above a CSS of 6 biomarker levels began to decline as might be expected when muscle volume has decreased due to fatty replacement typically seen on the MRI of subjects with advanced disease [14]. Carbonic anhydrase III appears to have the largest dynamic range of the biomarkers identified here and may be the most useful moving forward. These data also provide information for the selection of optimal clinical severities for entering clinical trials where disease improvement or progression will be measured. Additional metrics such as MRI and strength/movement measurements are likely to add sensitivity to measuring disease severity in the context of a clinical trial.
Fig. 3.
Biomarker concentrations correlate with clinical severity of FSHD. (A) Individual plots of the plasma concentration of each biomarker (relative fluorescence on the y axis) relative to clinical severity score (x axis) from the Seattle cohort. A LOESS polynomial regression was performed to draw a best fit line and the region in dark gray indicates the 95% confidence interval of the mean. (B) Heat maps showing relative amounts of fat infiltration (left) and STIR bright signal (right) from the right and left sides of 50 different muscles in 11 subjects from the Seattle cohort. The T1 weighted signal (fat bright) was scored on a scale of 0–5, with 0 being normal and 5 being complete fatty replacement of the indicated muscle. STIR bright signal was scored from 0 (no signal) to 3 (maximum brightness). White boxes indicate that the radiologist was unable to assign a score to that muscle. The active disease burden was quantified by summing the left and right side STIR signal scores for each subject and is shown below the STIR bright heat map.
3.6. Biomarker levels correlate with STIR bright muscle involvement determined by whole body MRI
Whole body MR imaging was performed on 11 of the 18 FSHD-affected subjects from the Seattle cohort. A radiologist scored the intensity of the signal produced by T1 weighted images (Fig. 3B, left panel) and short-T1 inversion recovery (STIR, Fig. 3B, right panel) for each of 50 muscles (right and left sides). The STIR scores were totaled for each subject to produce a score that loosely approximates active disease burden. Subjects were ranked by disease severity using either biomarker levels or the STIR score, and the rank score determined by plasma biomarker concentrations was plotted versus the rank score determined by STIR bright quantification. Despite the lack of radiographic information on muscle volume, a positive correlation between these ranks was observed that approached significance (rho = 0.354, p = 0.14).
4. Discussion
FSHD is caused by epigenetic and genetic elements that are unique to the primate lineage [33], a feature that makes development of animal models challenging [34]. Therefore, it may be necessary to test treatments in humans after toxicology studies have been completed in animals. Because FSHD is slowly progressive, conventional testing of muscle strength and function to measure disease progression in the context of a treatment requires large numbers of subjects with measurements taken over long periods of time [2]. Given the limited number of individuals able to participate in clinical trials, and the length of time that may be necessary to show a significant effect, having sensitive acutely-changing biomarkers may allow quick assessment of a number of therapeutic approaches. All skeletal muscles are exposed to the circulation so biomarker levels may also reflect overall disease burden in ways not possible by limited examination, biopsy, or imaging techniques. We used an emerging technology to detect novel disease biomarkers in the serum of people affected by FSHD and identified several that correlate with the disease state, severity, and MR imaging. These biomarkers may be more responsive to disease progression than conventional clinical outcome measures and therefore may be crucial for the efficient conduct of future clinical trials.
Unlike the muscle destruction seen in DMD, the smoldering prolonged muscle destruction typical of FSHD-affected muscles necessitates precise measurement of biomarker concentrations so that subtle changes can be detected. The SomaLogic technology used here has high specificity and sensitivity over 4 logs providing a large dynamic range for assessment [20]. Importantly the dynamic range exceeds that of traditional methods of measurement and suggests that biomarkers that may have traditionally seemed too variable for disease assessment may be worth re-evaluating. Several studies have now shown the variation of markers such as CK in large numbers of control and dystrophy-affected subjects and the trends appear to be more reliable than previously thought [21,35,36]. In addition, following biomarker levels in a single individual over time establishes a subject-specific baseline and thus allows one to determine directional variation from that line regardless of the absolute level.
We saw a weak correlation of the number of STIR (+) muscles determined by MRI with biomarker levels. The weak correlation was not surprising given that not all muscles could be scored (including facial muscles that are significantly affected in FSHD), quantification of MRI signals is subjective, MRI changes may not be seen for some damaged muscle states, and MRI scoring as performed here did not take into account the volume of the affected muscle (so the quadriceps contributed to the score equally to the tibialis anterior).
In addition to allowing measurement of disease burden, biomarker discovery provides insight into disease pathology. Although not particularly surprising, the pattern of biomarker levels as they are plotted against disease severity suggests a decrease in muscle tissue at the severe end of the clinical severity spectrum. Decreased muscle tissue with fat replacement is seen when severely affected individuals are imaged by MRI and assessment of these biomarkers allows quantification of MRI findings. Also, a unique group of biomarkers (proteins involved in protein folding and translational processing) were discovered that have not been observed in a larger DMD study and lend further support to the ideas that cell migration [11] and translation of transcripts normally degraded by nonsense mediated decay (NMD) [37] or mis-spliced transcripts [11] are important features of FSHD molecular pathology.
Likely multiple modalities of disease assessment will be necessary to evaluate changes in clinical presentation over the course of a clinical trial. Serum biomarkers, MRI, facial movement, muscle strength, and dynamic assessments of gait and activity can be combined to increase the power of assessment of disease progression. It will be important to follow the levels of biomarkers over time to measure disease progression in the same individuals and to correlate plasma concentrations with other sensitive measures of disease burden such as MRI imaging of muscle pathology.
Supplementary Material
Acknowledgments
We would like to thank the subjects who participated in the study and Colleen Donalin-Smith for collection and serum preparation of samples from the Rochester cohort. We also appreciate the advice and helpful comments from the SomaLogic team including Robert Kirk DeLisle, Darryl Perry, and Malti Nikrad.
Disclosures Supported by NIH-NIAMS 5R01AR064197 to DGM and by Friends of FSH Research to RT, SDF and DGM, Seattle Children’s Translational Research Ignitions Project Program to SDF and DGM, and by Kacy Murray and the Anderson Family Foundation to DGM.
Footnotes
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.nmd.2016.04.012.
References
- 1.Padberg GW, Lunt PW, Koch M, Fardeau M. Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 1991;1:231–234. doi: 10.1016/0960-8966(91)90094-9. [DOI] [PubMed] [Google Scholar]
- 2.The FSH-DY Group. A prospective, quantitative study of the natural history of facioscapulohumeral muscular dystrophy (FSHD): implications for therapeutic trials. Neurology. 1997;48:38–46. doi: 10.1212/wnl.48.1.38. [DOI] [PubMed] [Google Scholar]
- 3.van Overveld PG, Lemmers RJ, Sandkuijl LA, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 4.Snider L, Geng LN, Lemmers RJ, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckers M, Gabriels J, van der Maarel S, et al. Active genes in junk DNA? Characterization of DUX genes embedded within 3.3 kb repeated elements. Gene. 2001;264:51–57. doi: 10.1016/s0378-1119(00)00602-8. [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka G, Goto K, Ishihara T, et al. FSHD-like patients without 4q35 deletion. J Neurol Sci. 2004;219:89–93. doi: 10.1016/j.jns.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Gabriels J, Beckers MC, Ding H, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 8.Kowaljow V, Marcowycz A, Ansseau E, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Bosnakovski D, Xu Z, Gang EJ, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace LM, Liu J, Domire JS, et al. RNA interference inhibits DUX4-induced muscle toxicity in vivo: implications for a targeted FSHD therapy. Mol Ther. 2012;20:1417–1423. doi: 10.1038/mt.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickard AM, Petek LM, Miller DG. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum Mol Genet. 2015;24:5901–5914. doi: 10.1093/hmg/ddv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman SD, Poliachik SL, Carter GT, Budech CB, Bird TD, Shaw DW. The magnetic resonance imaging spectrum of facioscapulohumeral muscular dystrophy. Muscle Nerve. 2012;45:500–506. doi: 10.1002/mus.22342. [DOI] [PubMed] [Google Scholar]
- 13.Kan HE, Scheenen TW, Wohlgemuth M, et al. Quantitative MR imaging of individual muscle involvement in facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2009;19:357–362. doi: 10.1016/j.nmd.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Friedman SD, Poliachik SL, Otto RK, et al. Longitudinal features of STIR bright signal in FSHD. Muscle Nerve. 2014;49:257–260. doi: 10.1002/mus.23911. [DOI] [PubMed] [Google Scholar]
- 15.Olsen DB, Gideon P, Jeppesen TD, Vissing J. Leg muscle involvement in facioscapulohumeral muscular dystrophy assessed by MRI. J Neurol. 2006;253:1437–1441. doi: 10.1007/s00415-006-0230-z. [DOI] [PubMed] [Google Scholar]
- 16.Wallace LM, Garwick-Coppens SE, Tupler R, Harper SQ. RNA interference improves myopathic phenotypes in mice over-expressing FSHD region gene 1 (FRG1) Mol Ther. 2011;19:2048–2054. doi: 10.1038/mt.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Himeda CL, Jones TI, Jones PL. CRISPR/dCas9-mediated transcriptional inhibition ameliorates the epigenetic dysregulation at D4Z4 and represses DUX4-fl in FSH muscular dystrophy. Mol Ther. 2016;24:527–535. doi: 10.1038/mt.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Statland JM, McDermott MP, Heatwole C, et al. Reevaluating measures of disease progression in facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2013;23:306–312. doi: 10.1016/j.nmd.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraemer S, Vaught JD, Bock C, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS ONE. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hathout Y, Brody E, Clemens PR, et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2015;112:7153–7158. doi: 10.1073/pnas.1507719112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sattlecker M, Kiddle SJ, Newhouse S, et al. Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement. 2014;10:724–734. doi: 10.1016/j.jalz.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Kiddle SJ, Sattlecker M, Proitsi P, et al. Candidate blood proteome markers of Alzheimer’s disease onset and progression: a systematic review and replication study. J Alzheimers Dis. 2014;38:515–531. doi: 10.3233/JAD-130380. [DOI] [PubMed] [Google Scholar]
- 24.Mehan MR, Williams SA, Siegfried JM, et al. Validation of a blood protein signature for non-small cell lung cancer. Clin Proteomics. 2014;11:32. doi: 10.1186/1559-0275-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Groote MA, Nahid P, Jarlsberg L, et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS ONE. 2013;8:e61002. doi: 10.1371/journal.pone.0061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricci E, Galluzzi G, Deidda G, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol. 1999;45:751–757. doi: 10.1002/1531-8249(199906)45:6<751::aid-ana9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 27.Lemmers RJ, de Kievit P, Sandkuijl L, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 28.Lemmers RJ, Tawil R, Petek LM, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrick RM. A short-cut solution for the Kolmogorov-Smirnov test. NADC-MR-6504. NADC-MR Rep. 1965:1–4. [PubMed] [Google Scholar]
- 30.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 31.Kim HK, Merrow AC, Shiraj S, Wong BL, Horn PS, Laor T. Analysis of fatty infiltration and inflammation of the pelvic and thigh muscles in boys with Duchenne muscular dystrophy (DMD): grading of disease involvement on MR imaging and correlation with clinical assessments. Pediatr Radiol. 2013;43:1327–1335. doi: 10.1007/s00247-013-2696-z. [DOI] [PubMed] [Google Scholar]
- 32.Tasca G, Iannaccone E, Monforte M, et al. Muscle MRI in Becker muscular dystrophy. Neuromuscul Disord. 2012;22(Suppl 2):S100–S106. doi: 10.1016/j.nmd.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Leidenroth A, Hewitt JE. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol Biol. 2010;10:364. doi: 10.1186/1471-2148-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lek A, Rahimov F, Jones PL, Kunkel LM. Emerging preclinical animal models for FSHD. Trends Mol Med. 2015;21:295–306. doi: 10.1016/j.molmed.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CH, Leung M, Liang WC, Hsieh TJ, Chen TH, Jong YJ. Correlation between muscle involvement, phenotype and D4Z4 fragment size in facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2012;22:331–338. doi: 10.1016/j.nmd.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Lunt PW, Harper PS. Genetic counselling in facioscapulohumeral muscular dystrophy. J Med Genet. 1991;28:655–664. doi: 10.1136/jmg.28.10.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Q, Snider L, Jagannathan S, et al. A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. Elife. 2015;4 doi: 10.7554/eLife.04996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.