Abstract
Resistance to anoikis is a pre-requisite for tumor metastasis. The epithelial-to-mesenchymal transition (EMT) allows tumor cells to evade anoikis. The wound healing regulatory transcription factor Grainyhead-like 2 (GRHL2) suppresses/reverses EMT, accompanied by suppression of the cancer stem cell (CSC) phenotype and by re-sensitization to anoikis. Here, the effects of GRHL2 upon intracellular metabolism in the context of reversion of the EMT/CSC phenotype, with a view toward understanding how these effects promote anoikis sensitivity were investigated. EMT enhanced mitochondrial oxidative metabolism. While this was accompanied by higher accumulation of superoxide, the overall level of Reactive Oxygen Species (ROS) declined, due to decreased hydrogen peroxide. Glutamate Dehydrogenase 1 (GLUD1) expression increased in EMT, and this increase, via the product α-ketoglutarate (α-KG), was important for suppressing hydrogen peroxide and protecting against anoikis. GRHL2 suppressed GLUD1 gene expression, decreased α-KG, increased ROS and sensitized cells to anoikis.
Implications
These results demonstrate a mechanistic role for GRHL2 in promoting anoikis through metabolic alterations.
Introduction
When deprived of connections to their extracellular matrix, normal epithelial cells trigger a process of programmed cell death referred to as anoikis (1). The ability to metastasize depends critically upon a cancer cell’s ability to evade anoikis (2–4). The transcriptional reprogramming event known as the Epithelial to Mesenchymal transition (EMT) confers anoikis resistance and ultimately, increased metastatic potential (5). The increased phenotypic plasticity underlying transitions between epithelial and mesenchymal states is thought to be epigenetically driven (6–10). One additional manifestation of this increased plasticity, aside from EMT, is that cancer stem cell subpopulations emerge, that, like cells resulting from EMT, are resistant to anoikis; these contribute crucially to both metastasis and disease recurrence.
The wound healing regulatory transcription factor, Grainyhead-like 2 (GRHL2), plays an indispensable role in the maintenance of the epithelial phenotype and branching morphogenesis (11–14). Previously, we reported that GRHL2 suppresses the oncogenic EMT, i.e., promotes MET (15–17). The loss of GRHL2 expression is associated with aggressive, metastatic breast tumor types that contain an unusually large fraction of EMT-like subpopulations, and in the cancer stem cell-like subpopulation of tumors resulting from EMT and/or drug resistance. Interestingly, the constitutive expression of GRHL2 invariably led also to anoikis-sensitivity (15, 16). The mechanism underlying this effect was unclear, however.
Reactive oxygen species (ROS) are ubiquitously important in apoptosis through mechanisms such as direct mitochondrial peroxidation of cardiolipin, a lipid which sequesters cytochrome c, as well as direct inactivation of the anti-apoptotic Bcl-2 (18). Accordingly, ROS contributes to anoikis as well as the non-apoptotic cell death of detached cells accompanying ATP loss (19, 20). It is unclear at present, however, whether changes in ROS levels occur in EMT or cancer stem cell transitions, and, if so, what drives these changes.
The HMLE cell line is an immortalized mammary epithelial cell line that contains a CD44hi/CD24low subpopulation referred to as the Mesenchymal Sub-Population (MSP) that co-expresses EMT and cancer stem cell phenotypes (21, 22). Previously, we reported that MSP cells have low GRHL2 expression, and are resistant to anoikis (15, 16). Forced expression of GRHL2 in MSP suppressed their cancer stem cell-like as well as EMT phenotypes and sensitized them to anoikis. The possibility that GRHL2 achieves the latter effect by altering intracellular metabolism has not been explored.
Glutaminolysis is a critical metabolic pathway on which tumors rely as a major alternate carbon source to glucose, with significant ramifications for cell proliferation, metabolic adaptability and cell survival (23–25). Following deamination of glutamine to produce glutamate, glutamate dehydrogenase-1 (GLUD1 or GDH1) generates the Krebs cycle intermediate α-ketoglutarate (α-KG), which is converted by the Krebs cycle to fumarate, an important cofactor for glutathione peroxidase enzymes. Accordingly, α-KG is an important protective factor against oxidative stress, and GLUD1 is over-expressed in breast and lung carcinomas (26, 27).
In this paper, we report that, in reversing EMT, GRHL2 suppresses GLUD1 expression, elevating H2O2 ROS levels and promoting anoikis-sensitivity. GRHL2 also reversed the cancer stem cell-like shift to oxidative phosphorylation-based ATP production and cell survival. These results inform a novel connection between EMT, metabolic pathways, ROS and anoikis-sensitivity, regulated by GRHL2.
Materials and Methods
Cell lines
HMLE, and HMLE+Twist-ER cells and generously provided by R. Weinberg (The Whitehead Institute, Cambridge, MA); MCF10A neoT cells were provided by F. Miller (Karmanos Cancer Center). HMLE and HMLE+Twist-ER cells were maintained in Advanced Dulbecco’s Modified Eagle’s Medium (DMEM): Ham’s F-12 (Gibco) + 5% horse serum + 1X penicillin-streptomycin-glutamine (PSG) + 10 μg/mL insulin, 10 ng/mL EGF, 0.5 μg/mL hydrocortisone. MCF10A neoT cells were maintained in the same media as HMLE cells with the addition of 0.1 μg/mL cholera toxin. If indicated, HMLE+Twist-ER cells were growth in the presence of 4-hydroxytamoxifen (10 ng/mL) in order to activate the ER inducible Twist construct. MSP cells were obtained from HMLE cells by sorting for CD44-APC high population/CD24-Cy5.5 LOW. HMLE cells were sorted according to CD24-Cy5.5 high and CD44-APC low.
Generation of stable cell lines by retroviral transduction
The template for Human GRHL2 was purchased from Open Biosystems (MHS4426-99625903). GRHL2 was subcloned by standard molecular biology protocols into the pMXS-IRES-puro retroviral vector in frame with the C-terminal 3X-FLAG tag (vector contribution of Russ Carstens, University of Pennsylvania). Retroviral packaging and amplification was done in GP2+293T cells by transfection of 4.5 μg of retroviral plasmid and 2.5 μg of pCMV-VSV-G on collagen coated 60 mm2 dishes that were pre-coated for >1 hr with 50 μg/mL collagen using Lipofectamine 2000 (Invitrogen). Plates were refed 4–6 hours following transfection and viral supernatants were harvested approximately 36 hours following refeed. Viral stocks were filtered through 0.45 μm filters (Whatman) and 1 mL of supernatant was used to infect 1 well of target cells. This was followed by 1,400 RPM centrifugation for 1 hour at room temperature followed by 6 hour or overnight incubation. Infected cells were passaged to 100 mm dishes, and incubated for 48 hours following infection. Cells were then selected for puromycin (2 μg/mL for HMLE or MCF10A neoT cells).
Generation of stable cell knockdown cell lines by lentiviral transduction
Lentiviral GLUD1 short hairpin RNA (shRNA) was purchased from Open Biosystems in the pLKO vector. ShRNA #1 was Open Biosystems catalogue number RHS3979-201758959 and shRNA#2 was RHS3979-201758962. pLKO scramble control vector was generously contributed by the laboratory of Dr. Scott Weed (WVU). Lentiviral constructs were packaged and amplified in 293T cells by transfection of 3.5 μg shRNA vector, 2.3 μg sPAX2, and 1.2 μg CMV-VSV-G on 60 mm2 dishes that were pre-coated for >1 hr with 50 μg/mL collagen using Lipofectamine 2000. GRHL2 shRNA was described previously and was shown to duplicate the biologic effects of transfected siRNAs (15).
Western Blotting
SDS-PAGE was conducted using 4–20% gradient Tris-Glycine gels (Invitrogen). Proteins were electrophoretically transferred to polyvinylidine difluoride filters (Immobilon) in 5% methanol Tris-Glycine transfer buffer containing 5% methanol. PBS + 0.1% Tween-20 + 5% nonfat milk were used for blocking filters, primary antibodies were incubated in PBS+0.1% Tween-20 + 5% nonfat milk. Primary antibodies were typically incubated between 2 hours at room temperature or overnight at 4 degrees C. Primaries used were: E-cadherin, ms (BD Biosciences), CD44 (HCAM) ms [Santa Cruz Biotech (SCBT)]; GRHL2, rb (Sigma), Akt or pAkt, rb (Cell Signaling); GLUD1, rb (Abcam 168352), α-tubulin, Rb (Cell Signaling), Fibronectin, ms (BD Biosciences), GAPDH, ms (Origene), β-actin, ms (Thermo-pierce), SOD2, Rb (Cell Signaling), HIF1α, ms (BD Biosciences), p110, ms (BD Biosciences), TOMM20, ms (BD Biosciences), VDAC1, Rb (Cell Signaling). Secondary antibodies for chemiluminescence were either anti-mouse or anti-rabbit, conjugated to horseradish peroxidase (HRP) enzyme (Bio-Rad). Secondary antibodies (Biorad) were used at 1:3000 dilution and filters were incubated for approximately 1 hour at room temperature. Western blots were developed via ECL Super Signal West Pico (Thermo-Pierce).
Anoikis Assays
For anoikis assays, caspase activation was measured using the Caspase-Glo 3/7 assay kit (Promega). Cells were dissociated using TrypLE Express (Invitrogen) and a fixed number of cells (1.5 × 105 cells) were placed per 6 well poly-(2-hydroxyethyl methacrylate) (poly-HEMA) coated low attachment plates in 2.0 ml of normal growth medium + 0.5% methylcellulose for the indicated time. Aliquots of cells were mixed 1:1 with caspase glo reagent at indicated time points and assayed for luminescence utilizing a Wallac Envison Perkin Elmer plate reader according to manufacturer’s instructions. Where indicated, time-zero cell death values were subtracted from the data presented to normalize for small loading variation.
ROS Assays
For ROS assay, cells were seeded as above and attached for 48 hours. Cells were detached by trypsinization, and equal numbers of cells were placed in a 15 mL centrifuge tube. Cells were assayed immediately by flow cytometry after staining for 15 min at 37 degrees with 1 uM CM-H2DCFDA (Invitrogen) protected from light. If indicated, 24 hr ROS was examined after plating cells in low attachment polyHEMA (2 mg/mL) coated 100 mm dishes in 5 mL of 0.5% methylcellulose to prevent excess cell clumping. Cells were harvested by diluting well with 10 mL DME/F12 + 10% Horse Serum containing media. Cells were centrifuged at 2000 RPM for 2 minutes. Pellets were washed 2X with HBSS and stained in 1 uM CM-H2DCFDA as indicated previously. In some cases, Amplex Red (Invitrogen) assays were used to measure ROS in order to compare attached and suspended conditions and were used according to manufacturer’s instructions. For measurement of general cellular superoxide, cells were processed as above and stained with 5 uM dihydroethidium (DHE) (Invitrogen) for 15 minutes at 37 degrees C protected from light followed by flow cytometric analysis at 605 nm fluorescence. Mitochondrial specific superoxide was measured using mitoSOX (Invitrogen) dye by staining cells with 5 uM mitoSOX solution for 10 minutes at 37 degrees C protected from light. Samples were analyzed by flow cytometry at 580 nm fluorescence using a BD LSRFortessa Cell analyzer.
ATP Assays
For the measurement of ATP in attached cells, Cell Titer Glo (Promega) reagent was added to 96 well plates. Luminescence was read after 10 minute incubation at room temperature. Values were normalized based on BCA protein assay (Pierce). To measure ATP in detached conditions, cells were detached and placed in 6 well poly-HEMA dishes. Aliquots were mixed 1:1 with Cell Titer Glo reagent. Values were normalized based on BCA protein assay (Pierce).
JC1 Assay
Cells were seeded on poly-d-lysine coated MatTek dishes. Cells were incubated in 2 ug/mL JC1 (5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide) (Invitrogen) at 37 degrees for 30 minutes protected from light. After incubation, dye was washed off of cells and replaced with normal growth medium. Cells were analyzed using an Axiovert 100M LSM510 Zeiss Confocal microscope. Cells were excited with an argon-ion laser source at 488 nm. JC1 monomer emission was observed at 529 nm and J-aggregate emission was observed at 590 nm. Images were acquired using 20X/0.75 Plan-Neofluar objective. Software used was Zeiss AIM software, version 3.2.
Oxygen Consumption and Extracellular Acidification Rate
The oxygen consumption rates (OCR) and extracellular acidification rate (ECAR) of adherent cells were measured utilizing the Seahorse XF Extracellular Flux Analyzer (Seahorse Biosciences Inc., North Billerica, MA). This method of analysis allows for real time monitoring of extracellular acidification rates and oxygen consumption based on two fluorimetric probes inserted into the small volume well. Equal numbers of cells (7500 cells per well) were seeded in a defined small volume in Seahorse XF-96 well plates. The following day, wells were refed with fresh HEPES buffered media approximately 1 hour prior to assay. In short, the sensor cartridge was loaded with metabolic inhibitors to target specific components of the electron transport chain at specific times, while monitoring pH change as well as oxygen consumption. The metabolic inhibitors used are oligomycin (ATP synthase inhibitor), followed by the protonophore FCCP (an uncoupler of oxidative phosphorylation), and finally a combination of rotenone (complex I inhibitor) and antimycin A (complex III inhibitor). Basal OCR and ECAR measurement during time course treatment with the inhibitors listed above allow for calculations of basal OCR, maximum mitochondrial respiratory capacity, OCR due to proton leak, ATP production, spare respiratory capacity, and non-mitochondrial derived OCR. Fatty acid oxidation study was done by treating cells overnight prior to assay with substrate limited media (DME/F12 media + 1% horse serum, 0.5 mM carnitine hydrochloride [pH adjusted], 1X PSG, 0.5 mM D-(+)-Glucose, 5 mM HEPES buffer). On day of assay, substrate limited media was replaced with FAO medium 45 minutes prior to the start of the assay (Homemade Krebs Henseleit Buffer modified with CaCl2: To 500 mL sterile water the following was added: 111 mM NaCl, 5.7 mM KCl, 1.25 mM CaCl2, 2.0 mM MgSO4, 1.2 mM Na2HPO4; This buffer was supplemented with 2.5 mM glucose, 0.5 mM carnitine, 5 mM HEPES, pH adjusted to 7.4 with NaOH). 15 minutes prior to start of assay, 40 μM Etomoxir was added to the appropriate wells. At the beginning of the assay, either palmitate:BSA or BSA control was added to appropriate wells. Mitostress test was the conducted according to manufacturer’s instructions. For glycolytic stress test, cells are deprived of glucose for approximately 1 hour prior to the beginning of the assay. ECAR is measured and glucose is re-introduced to the media. The increase in ECAR is attributed to glucose conversion to pyruvate through glycolysis. Oligomycin is then added to shunt all metabolic function to glycolysis, which results in a large increase in ECAR indicating the glycolytic reserve. Following addition of 2-deoxy-glucose (competitive inhibitor of hexokinase), the non-glucose derived OCR can be calculated.
Flow Cytometry Sorting
Trypsinized cells (1 × 106) were stained with APC tagged mouse anti-human CD44 (BD Biosciences 560890), and PerCP-Cy5.5 tagged mouse anti-human CD24 (BD Biosciences 561647) in 100 uL of flow sorting buffer (1X PBS, 2.5 mmol/L EDTA, 10 mmol/L HEPES, and 2% horse serum) on ice protected from light for 30 minutes. Following staining, cells were washed with 1X HBSS and resuspended in flow sorting buffer. Cells were analyzed and sorted using a FACS-Aria.
Cell Culture Antioxidant Treatments
For suppression of reactive oxygen species, the following antioxidants were used. Where indicated, HMLE cells were treated with N-acetyl cysteine (NAC) at the concentration indicated for 1 hr prior to assay. Reduced glutathione (GSH) was added to HMLE cells after pH correction to 7.4 and incubated for approximately 1–2 hours depending on experiment at the indicated concentration. 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) was added to cells at the indicated concentration for overnight pretreatment of HMLE or HMLE+shGLUD1 cells. HMLE or HMLE+shGLUD1 cells were treated with cell permeable di-methyl-α-ketoglutarate (Sigma) which was pH corrected to 7.4. Cells were treated at the indicated concentration between overnight and 2 hour pre-treatment.
RNA Sequencing
RNA was isolated in triplicate from both MSP cells expressing vector alone or stably expressing GRHL2. For RNA isolation, the RNeasy Plus Kit (Qiagen) was used and was quantified using Nanodrop (Fisher Scientific). The RNA quality check was performed on Bioanalyzer (Agilent). RNA quality check and libraries builds were done by the WVU genomics core. RNA samples were then sent to the University of Illinois Core Facility for 100BP single end data RNA sequencing. FastQC was used to visualize quality of sequencing. After it was determined that no filtering was needed, the reads were mapped to hg38 using STAR. The data was then quantified with featureCounts with the following flag set: —ignore dup—primary. This count data was used as input into DEseq2 to produce a list of differentially expressed genes with some visualizations also included. The list of genes with FPKM was also acquired and was produced from the count data with a simple custom script.
Kinetic α-KG assay
HMLE, MSP, or MSP+GRHL2 cells were seeded in 6 well dishes in duplicate wells and allowed to attach for 24 hours. Wells were washed 2X with PBS, followed by lysis in 300 uL RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton-X100, 1% Sodium Deoxycholate, 0.1% sodium dodecyl sulfate, + pierce miniprotease inhibitor table per 10 mL lysis buffer). Lysates were scraped into microfuge tubes and cleared in cold centrifuge at 4oC at full speed for 10 minutes. Supernatants were saved and assayed in 100 uL reaction buffer (100 mM KPO4 pH 7.2, 10 mM NH4Cl, 5 mM MgCl2, and 0.15 mM NADH added immediately prior to assay). 10 uL of sample were added to 100 uL reaction buffer. Immediately prior to the beginning of the assay, 0.7 uL (0.5 units) of L-Glutamate Dehydrogenase enzyme (bovine liver, sigma G2626) was added to each of the wells using a multi-channel pipettor. Positive control samples were done to ensure linear range of α-KG concentration in samples. OD was read kinetically at 37 oC at 340 nm reading the sample every 10–20 seconds depending on the number of samples in the assay. Depending on the level of α-KG in the sample, the reaction would proceed more quickly or more slowly. The slope of the line created by monitoring the decline in NADH absorbance as it was used up in the reaction was calculated and indicated the relative concentration of α-KG. Calculated slopes were normalized to protein in the samples as determined quantitatively by BCA assay (Pierce).
Extraction of Cells for LC-MS Analysis
All solvents used were high quality HPLC gradient grade and sterile milliQ grade water was used. Cells were plated in triplicate per condition in 6 well dishes. Cells were seeded for at least 24 hours prior to extraction. Wells were washed 3X with PBS. Extract solution was made up of 50% methanol, 30% acetonitrile, and 20% milliQ water. Extraction buffer was pre-chilled in a dry ice ethanol bath. Eppendorf tubes were pre-chilled in a dry ice/ethanol bath placed in a metal heating block for maximum cooling. Intracellular extracts were made by adding 500 uL of pre-chilled extraction buffer to wells and placing plate on a shaker in the cold room for 10 minutes. The solution was then transferred into pre-chilled Eppendorf tubes. Samples were centrifuged at 0 oC at full speed for 10 minutes. Supernatants were transferred to HPLC vials and stored at −80 oC. For LC-MS method, Column was the sequant Zic-pHilic (150 mm × 2.1 mm i.d. 5 μm) with the guard column (20 mm × 2.1 mm i.d. 5 μm) from HiChrom, Reading, UK. Mobile phase A: 20 mM ammonium carbonate plus 0.1% ammonia hydroxide in water. Mobile phase B: acetonitrile. The flow rate was kept at 100 μl min 1 and gradient as follow: 0 min 80% of B, 30 min 20% of B, 31 min 80% of B, 45 min 80% of B. The mass spectrometer (Thermo Exactive Orbitrap) was operated in a polarity switching mode.
Immunofluorescence
Cells were plated on chamber slides (LabTek154526) using ~0.5 ml medium per chamber. Wells were pre-coated with sterile 0.5 mg/ml poly-lysine or 20 ug/ml collagen for ~1 hour, and washed off prior to plating. Cells were washed in HBSS or D-PBS, and fixed in EM grade 4% formaldehyde in 1X PBS for 20 minutes at room temperature. Cells were washed 2X with PBS. Samples were then permeabilized in 0.5% TX100 in PBS at 4 degrees for 10minutes. Permeabilization solution was washed 2X with PBS. Cells were incubated in 100mM glycine in PBS for 10 minutes to quench the formaldehyde. Cells were blocked for one hour in PBS+10% goat serum+0.1% Tween-20+0.1%BSA for 1 hour. Primary antibody was diluted in blocking solution, and incubated 1 hr/RT or overnight/4 degrees (TOMM20, ms; BD Biosciences). Primary antibody was typically diluted 1:250. After probing, cells were washed with PBS+0.1% Tween-20, 3X. Secondary antibody (Anti-mouse Alex Fluor 555; Invitrogen) was diluted in blocking solution, and incubated 1–2 hr/RT (typically 1:1000 dilution). Cells were washed again with PBS+0.1% Tween-20, 3X. Samples mounted in ProLong Gold+DAPI.
Statistical Analysis
Error bars in graphs represent SDs. P-values were calculated using a Student’s two-tailed t-test.
Results
EMT shifts metabolism from glycolysis to oxidative phosphorylation, which is reversed by GRHL2
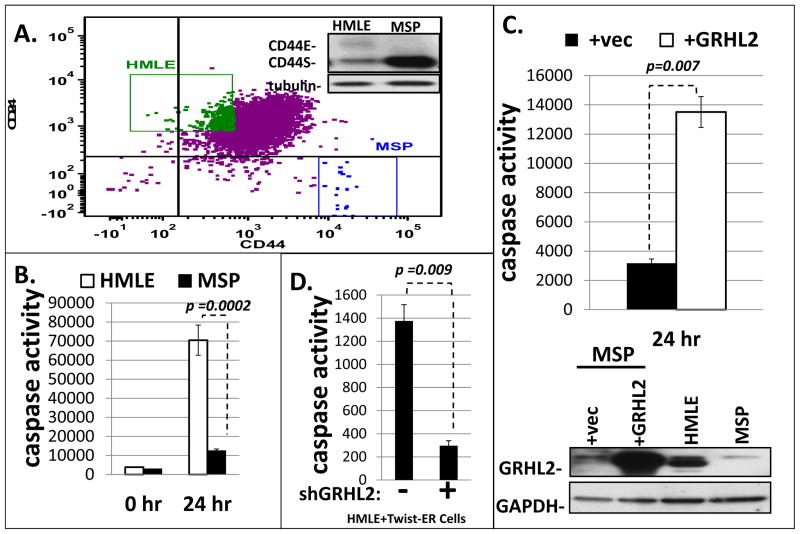
MSP and TGF-β-induced MCF10aneoT cells result from EMT that requires GRHL2 down-regulation (15, 16, 28), accompanied by a CD44highCD24low marker profile (figure 1A). Re-expression of GRHL2 sensitized these cell lines to anoikis significantly, as assessed by a caspase activation assay (figure 1B, 1C, figure S1A). Conversely, the stable knockdown of GRHL2 using shRNA conferred anoikis-resistance in HMLE+Twist-ER cells as well as MCF10A neoT cells (figure 1D, S1B).
Figure 1. MSP cells are an anoikis resistant subpopulation of HMLE cells which have undergone EMT and lost GRHL2 expression.
A. (upper panel): HMLE cells were sorted for CD44 and CD24 by FACS (insert shows up-regulated CD44S in MSP cells); B: Resulting sub-populations were assayed for anoikis sensitivity by caspase 3/7 activation assay; C. MSP cells in which GRHL2 was re-expressed were assayed for anoikis compared to vector control (24 hour time point with zero time subtracted). (lower panel): confirmation of GRHL2 expression by western blotting. D. HMLE+Twist-ER cells depleted of GRHL2 by shRNA transduction were assayed for anoikis; the confirmation of GRHL2 knockdown is shown as part of figure 5C.
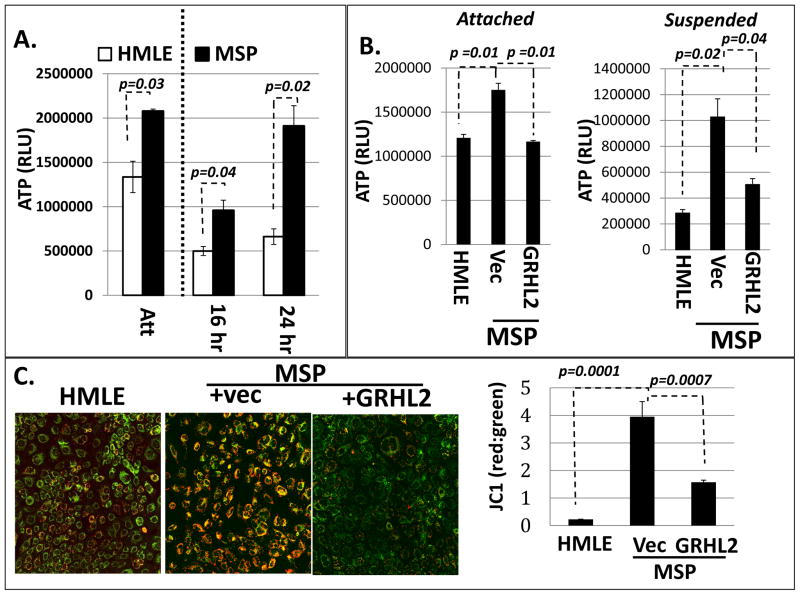
We first endeavored to characterize the effect of EMT and, conversely, the effect of GRHL2-induced MET on metabolism in our system. We found that the ATP level in attached MSP cells was elevated significantly compared to HMLE cells. When cells were suspended, ATP level declined significantly in HMLE cells, but was maintained in MSP cells, even at twenty-four hours of suspension (Figure 2A). GRHL2 reduced the level of ATP in both suspended and attached MSP cells (figure 2B).
Figure 2. ATP levels are elevated in mesenchymal relative to epithelial cells due to increased mitochondrial membrane potential (ΔΨ).
A. HMLE and MSP cells were analyzed for ATP level under attached or suspended (16 h, 24 h) conditions. (The line indicates that the attached level cannot be directly compared to the suspended levels due to proliferation in attached condition). B. GRHL2 suppresses ATP level in MSP cells in both attached and suspended conditions. C. ΔΨ is elevated in MSP cells, which is reduced by GRHL2 overexpression (JC1 fluorescent probe; green=monomers to indicate free JC1, orange=aggregates of JC1 proportional to ΔΨ); (right panel): Image J quantification and ratio calculation of raw intensity pixel density for single phase red and green images.
In light of the higher efficiency of mitochondrial oxidative phosphorylation compared to glycolysis in the production of ATP, these results suggested that EMT perhaps caused a shift to the former. MSP cells had a higher mitochondrial membrane potential (ΔΨ), reflecting increased oxidative phosphorylation. Conversely, GRHL2 overexpression resulted in decreased ΔΨ in MSP cells (Figure 2C). These results, and the marked ATP differences, indicated that EMT shifted cells toward an increased utilization of mitochondrial oxidative phosphorylation to increase basal ATP levels and sustain ATP even in suspended cells.
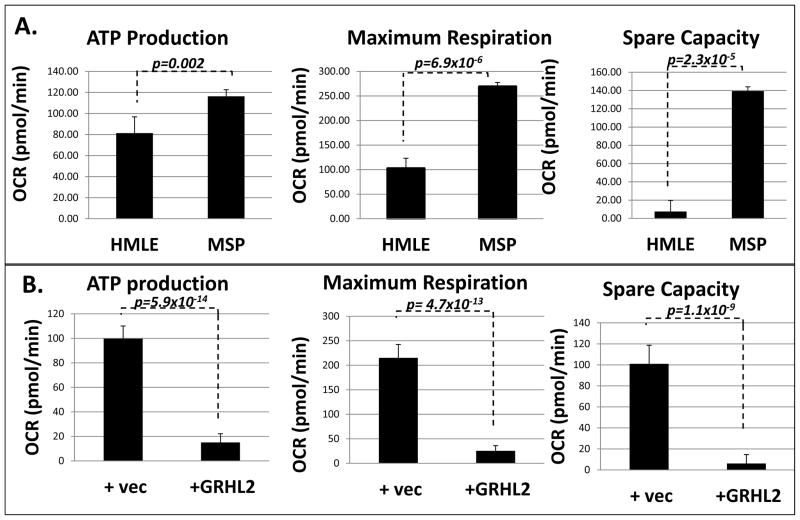
In order to test the effect of EMT or GRHL2-induced MET on regulation of oxidative metabolism specifically, we compared oxygen consumption rate in HMLE, MSP, and MSP+GRHL2 cells using the Seahorse mitochondrial stress test. The Seahorse XF assay system measures extracellular acidification rate (ECAR) as well as oxygen consumption rate (OCR) in order to determine fluxes in metabolic parameters that occur upon addition of various inhibitors and stimulators of the electron transport chain. Our results indicated that MSP cells had greater ATP, maximum respiration, as well as spare oxidative capacity relative to HMLE cells, and re-expression of GRHL2 reversed these effects (figures 3A,B). The increased mitochondrial activity of MSP and reversal by GRHL2 were not limited to cells utilizing glucose and glutamine as carbon sources, as similar effects were seen with the fatty acid carnitine as the sole carbon source (figures S4A,S4B). Moreover, HMLE cells had higher glycolytic activity than MSP cells (figure S2A). Stably depleting GRHL2 in both HMLE+Twist-ER and MCF10A neoT cells partially reversed the mesenchymal metabolic phenotype, increasing ATP and maximum respiration (figure S2B, S2C, S2D). To determine whether the increased OCR observed was due to elevated mitochondrial number, cell lines were stained for a mitochondrial structural protein, TOMM20. Surprisingly, we found that MSP cells had lower mitochondrial staining than HMLE cells (figure S2E, S2F). This may indicate that HMLE cells partially compensate for low mitochondrial activity by up-regulating mitochondrial biogenesis. Taken together, these results confirmed that mesenchymal cells enhanced oxidative phosphorylation, and partially suppressed glycolysis, both of which are counteracted by GRHL2. Confirming this observation, we found that treatment with 2-deoxyglucose (2-DG) resulted in greater decrease in ATP level in HMLE cells (58% of no treatment control) and MSP+GRHL2 cells (59.1% of no treatment control) than in MSP cells (72.1% of no treatment control) (figure S3A). GLUD1 depletion did not directly affect ATP levels, as reported (29). The loss of GLUD1 did, however, result in a decrease in mitochondrial membrane potential (figure S3B).
Figure 3. Shift from glycolysis to oxidative phosphorylation accompanies EMT; reversal by GRHL2.
A. MSP cells have higher ATP production, maximum respiration, and higher spare capacity for oxidative phosphorylation than HMLE; B. Reversal by GRHL2
EMT suppresses the accumulation of ROS; reversal by GRHL2
To identify genes regulated by GRHL2 underlying its metabolic effects, we conducted RNA-seq analysis. Surprisingly, we did not find that major ETC components or Krebs cycle metabolic enzymes were significantly affected by GRHL2 expression, with the possible exception of pyruvate dehydrogenase kinase 4 (PDK4), but its levels were highly sensitive to the initial conditions of each individual experiment, precluding definitive conclusions as to its regulation (data not shown). GRHL2 did, however, regulate several genes that participate in reactive oxygen species (ROS) regulation (Table 1). Interestingly, GRHL2 expression did not significantly affect expression of the major ROS regulatory enzymes superoxide dismutase or catalase (Table 1).
Table 1.
Effect of GRHL2 on expression of ROS-regulatory genes
| Gene ID | Fold Change (MSP+GRHL2/MSP+vec control) | Significance in ROS or Metabolism |
|---|---|---|
|
ALOX15 ALOXE3 ALOX12P2 |
+236.0 +21.0 +18.3 |
Can reportedly generate lipid peroxide radicals from arachidonic acid substrates |
| SLC1A6 | +165.7 | Glutamate/Aspartate transporter which may alter α-KGDH activity resulting in H2O2 radicals and reduce its TCA activity |
| NCF2 (p67-PHOX) | +23.4 | Cytoplasmic subunit of NADPH-Oxidase (NOX) Complex |
| FAT4 | −19.6 | Human homologue of Drosophila Fat. Potentially contains Ftmito fragment which binds complex I and stabilizes it to reduce ROS/increase mitochondrial activity |
| SOD1 | −1.4 | Soluble superoxide dismutase enzyme involved in cytoplasmic O2·− detoxification. Not significantly by GRHL2 overexpression |
| SOD2 | +1.2 | Mitochondrial isoform of superoxide dismutase involved in conversion of mitochondrial O2·− into diffusible H2O2. Not significantly altered by GRHL2 overexpression |
| SOD3 | NA | Extracellular superoxide dismutase |
| Cat | −1.3 | Catalase enzyme involved in detoxification of H2O2 ROS species. Appears to be slightly regulated by EMT, but not significantly altered by GRHL2 overexpression |
| NQO1 | −1.9 | ROS detoxifying enzyme which reduces quinines to hydroquinones – major Nrf2 target gene |
| GLUD1 | −2.5 | Reported be the primary source of cellular regulation of α-KG which feeds to TCA cycle to increase Fumarate and stabilize GPx1 |
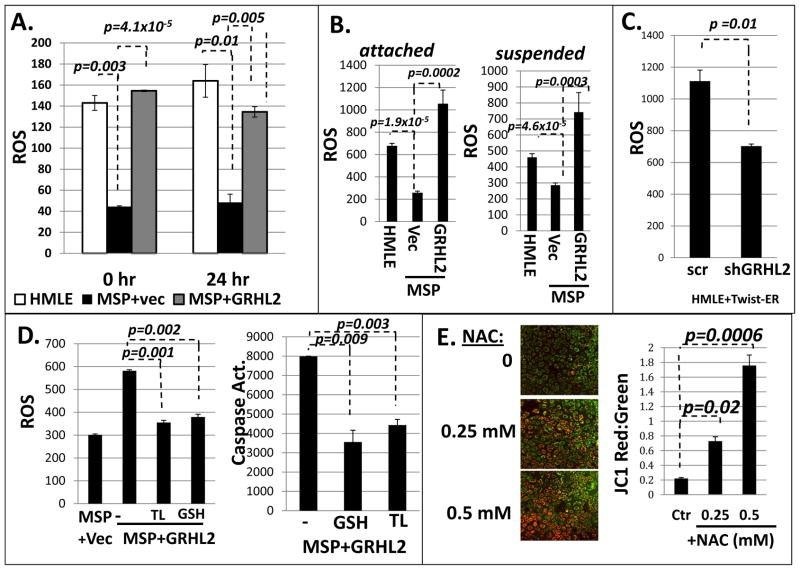
We compared endogenous ROS levels in HMLE, MSP, and MSP+GRHL2 cells. We found that the MSP cells display a significant reduction in ROS by fluorigenic reporter assay (CM-H2DCFDA) in both 0 hour and 24 hour suspended conditions, and that GRHL2 increased ROS to the approximate level detected in HMLE cells (figure 4A). Inducing EMT in the basal mammary epithelial cell line, MCF10A neoT, with TGF-β treatment resulted in decreased ROS; further, overexpression of GRHL2 elevated ROS level (figure S5A). Conversely, shRNA depletion of GRHL2 suppressed ROS levels both in HMLE+Twist-ER cells and MCF10A neoT cells (figures 4C, S5D).
Figure 4. Epithelial cells have higher ROS than mesenchymal cells, and the excess ROS promotes anoikis.
A. HMLE cells have higher ROS compared to MSP cells, which is elevated by GRHL2 expression in MSP cells: CM-H2DCFDA fluorescence at the indicated times of suspension. B. Hydrogen peroxide is the major regulated ROS type (Amplex Red assay); C. shRNA depletion of GRHL2 decreases ROS in HMLE+Twist-ER cells. D. Antioxidant compounds protect against anoikis. MSP+GRHL2 cells were pre-treated overnight with reduced glutathione (GSH, 5 mM) and Trolox (100 uM) followed by ROS and anoikis assay; E. Suppression of ROS resulted in increased JC1 aggregate formation and elevated ΔΨ. HMLE cells were treated with N-acetyl cysteine (NAC) at 0.25 and 0.5 mM for 1 hr before staining with JC-1 fluorimetric dye. (right panel): Image J quantification and ratio calculation of raw intensity pixel density of single phase red and green images.
To identify the species of ROS present, we analyzed cells with a variety of fluorometric dyes with varying species selectivities. CM-H2DCFDA staining is a general ROS indicator that responds well to hydrogen peroxide (H2O2). In order to verify that this was the predominant species, we stained cells with MitoSOX red, dihydroethidium (DHE), and Amplex red-hydrogen peroxide reporter. Amplex Red, which reports hydrogen peroxide specifically, indicated ROS trends similar to those obtained by CM-H2DCFDA staining in both attached and suspended conditions (figure 4B). MitoSOX red and DHE staining report mitochondrial and generalized superoxide, respectively. These probes showed that HMLE cells had lower mitochondrial and general superoxide (O2·−) levels than MSP cells and that overexpression of GRHL2 resulted in a decreased O2·− level (figure S5B, S5C). Mitochondrial electron transport is the major source of cellular superoxide (30). In this light, our superoxide data correlated well with our data on mitochondrial oxidative phosphorylation. These data indicated clearly that mitochondria are not the major source of higher ROS in normal epithelial cells or GRHL2-reverted epithelial cells, as this ROS was hydrogen peroxide, not superoxide, and mitochondria were more active in mesenchymal cells, which had lower overall ROS.
We further sought to examine the role played by ROS in contributing to anoikis sensitivity. HMLE cells were treated with two common antioxidant compounds including reduced glutathione and the vitamin E derivative, Trolox. We found that these compounds reduced ROS level in cells and significantly reduced both ROS and sensitivity to anoikis (Figure 4D). In figure S5A, we show that MCF10a neoT cells experience a large ROS burst between 0 and 24 hours. Speculatively, this burst may occur much earlier in HMLE cells, resulting in little difference between 0 and 24 hours. It is also plausible that in HMLE cells at 24 hours, enough cell death has occurred to obscure the ROS burst. These findings support the crucial role of ROS in triggering anoikis.
We have previously reported that CD44S, which is upregulated in MSP cells, contributes to anoikis resistance (31). CD44 was shown elsewhere to decrease ROS by shifting metabolism from oxidative phosphorylation toward glycolysis and pentose phosphate pathways, thereby increasing reduced glutathione levels (32). Consistent with this, the stable knockdown of CD44 resulted in increased ROS, and increased sensitivity to anoikis (figure S7A, S7B). Our metabolic findings were inconsistent with the mechanism proposed in (32) because we observed a shift toward oxidative phosphorylation in MSP cells (figure 3) and because reduced glutathione levels in HMLE and MSP cells were similar (figure S8).
Interestingly, neutralizing cellular ROS in HMLE cells with the scavenger N-acetyl cysteine (NAC) resulted in a significant increase in ΔΨ (Figure 4E). Neutralizing ROS in MSP+GRHL2 cells replicated this effect (figure S6A), indicating that the low-ROS cellular environment could play a role in sustaining the elevated mitochondrial function that we observed in mesenchymal cells (figure 2).
EMT up-regulates glutamate dehydrogenase-1; reversal by GRHL2
Our RNA-seq data indicated that GRHL2 down-regulated several genes that have well established effects on ROS in other systems (Table 1). Although NQO1 knockdown potentiated anoikis in non-small cell cancer cells (33), manipulation of the levels of NCF2/p67PHOX, ALOX15, FAT4 and NQO1 produced no significant change in ROS in our cells (data not shown). GRHL2 also down-regulated Glutamate Dehydrogenase 1 (GDH1 or GLUD1), an enzyme implicated in the suppression of ROS (27). By western blotting, we found that GLUD1 protein was upregulated in MSP cells relative to HMLE cells, and downregulated in MSP by GRHL2 expression, in agreement with our RNA-seq data (figure 5A). Similarly, EMT that was generated by transient activation of Twist-ER protein also up-regulated GLUD1, which was prevented by GRHL2 (figure 5B); similar effects were found in TGF-β-induced MCF10a neoT cells (Figure S8). Conversely, GLUD1 expression increased following depletion of GRHL2 in both HMLE+Twist-ER and MCF10A neoT cells (figure 5C), indicating that GRHL2 is an important repressor of GLUD1.
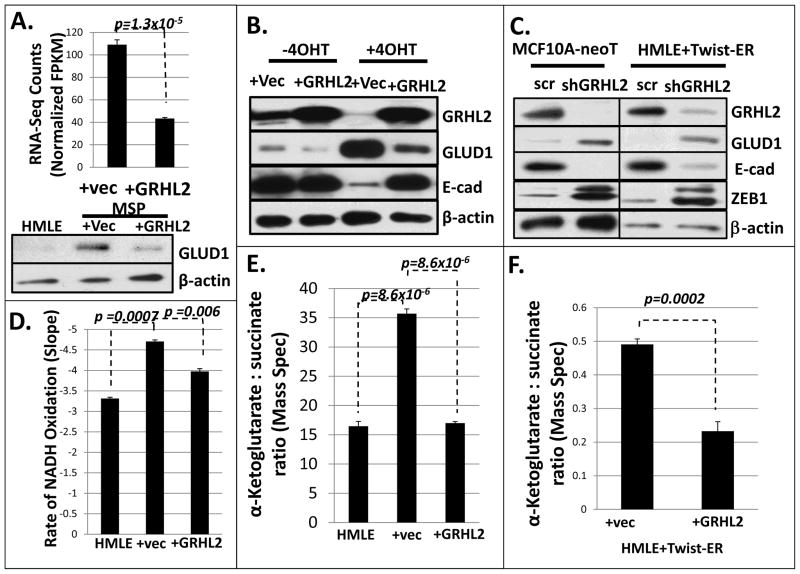
FIGURE 5. GRHL2 regulates Glutamate dehydrogenase-1 and α-ketoglutarate levels.
A, GLUD1 is downregulated by GRHL2 in MSP cells (upper panel: RNA-seq data lower panel: western blot); B. GLUD1 expression is upregulated during EMT and is downregulated by GRHL2 in HMLE-Twist cells; C. shRNA depletion of GRHL2 results in increased GLUD1 protein. D. α-KG is higher in MSP cells relative to HMLE cells, and GRHL2 overexpression decreases it (enzymatic assay measuring loss of NADH absorbance); E. α-KG:succinate ratio is higher in MSP than in HMLE or MSP+GRHL2 cells (mass spectrometric assay). F. α-KG:succinate ratio is decreased by GRHL2 in HMLE cells induced to undergo EMT by Twist-ER activation (mass spectrometric assay).
Depletion of GLUD1 was previously shown to reduce α-ketoglutarate (α-KG) levels, indicating that GLUD1, through glutaminolysis, is a major source of α-KG (27). Interestingly, we found that MSP cells have an elevated α-KG:succinate ratio relative to HMLE cells and that GRHL2 decreased α-KG, using both enzymatic and liquid chromatography/mass spectrometric assays, consistent with the expression levels of GLUD1 (Figure 5D, 5E, 5F).
GLUD1 suppresses ROS and anoikis
Next, we tested the effects of altered GLUD1 and α-KG levels on ROS and anoikis. The stable knockdown of GLUD1 enzyme using shRNA transduction resulted in both an increase in ROS level as well as increased sensitivity to anoikis (figure 6A). A similar effect of GLUD1 knockdown was observed in HMLE+Twist-ER cells (figure S8C).
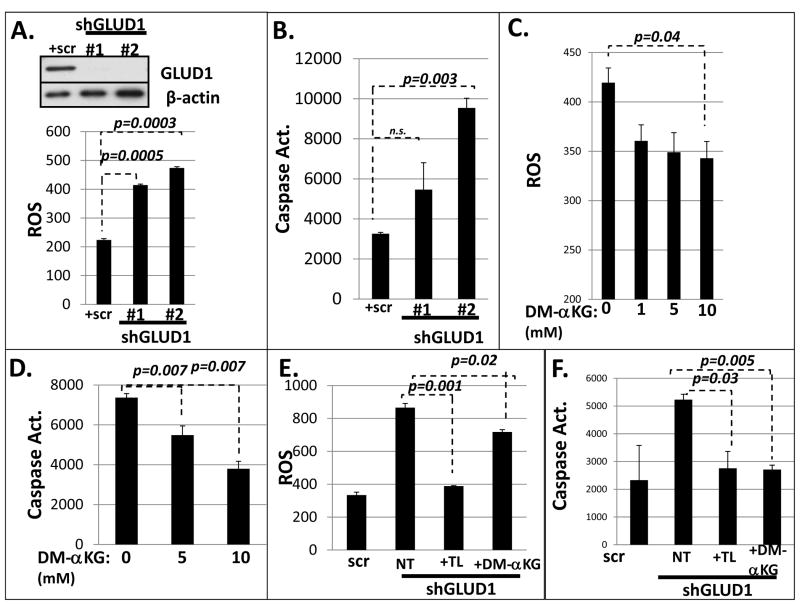
FIGURE 6. GLUD1 and α-KG regulate anoikis through ROS.
A. (upper panel): Western blot analysis of HMLE+shGLUD1 cells. (lower panel): GLUD1 knockdown elevates ROS level; (B) GLUD1 knockdown promotes anoikis as assessed by CM-H2DCFDA assay and caspase 3/7 activation assay (respectively); C. Dimethyl-α-KG lowers ROS; (D) Dimethyl-α-KG protects HMLE cells against anoikis; E. DM-αKG and Trolox lower ROS level; (F) DM-αKG and Trolox protect GLUD1 knockdown cells against anoikis.
To confirm that the GLUD1 effect on ROS was mediated by α-KG, we tested the effect of α-KG supplementation using the cell-permeable di-methyl-α-ketoglutarate derivative in GLUD1-depleted cells. This resulted in a dose dependent protection from anoikis as well as a reduction in ROS in HMLE cells (figure 6B). Moreover, the excess anoikis observed in HMLE cells as well as MSP+GRHL2 cells was suppressed by α-KG replacement, demonstrating that the loss of GLUD1 promoted anoikis through ROS (figure 6C, 6D, S10). Neutralizing ROS with antioxidants or α-KG partially protected HMLE+shGLUD1 cells against anoikis as well (figure 6E, 6F). These data indicate the critical role of GLUD1 up-regulation during EMT, resulting in protection from anoikis by suppressing ROS, and the converse role of GRHL2 in promoting anoikis through GLUD1 down-regulation (summarized in figure 7).
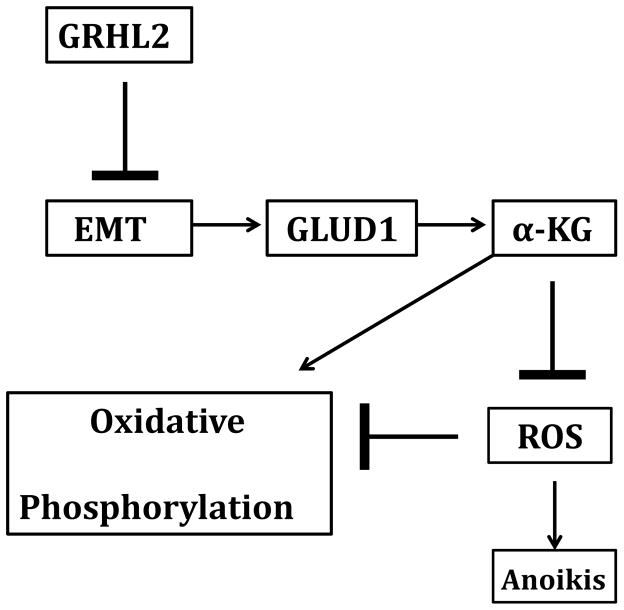
FIGURE 7.
GRHL2 suppresses EMT and results in the loss of GLUD1. This decreases α-KG, suppressing oxidative metabolism, increasing ROS, and sensitizing cells to anoikis.
Discussion
The role and mechanism of the Warburg-like shift to aerobic glycolysis in tumor cells has received extensive investigation, but this effect is now known to be highly context dependent. In fact, recent studies indicate that cancer stem cells (related, in some instances, to EMT) develop an enhanced capacity for and dependence on oxidative phosphorylation ((34), see (35) for an excellent review). Moreover, oxidative phosphorylation can contribute to tumor metastasis (36, 37). This is thought, counter-intuitively, to have advantages for slow-cycling cancer stem cells, such as better maintenance of ATP in nutrient-poor tumor environments due to the higher efficiency of the TCA/ETC pathway (using fatty acids or glutamine as carbon sources); moreover, the tumor microenvironment is rarely, if ever, hypoxic enough to inhibit oxidative phosphorylation (35). Our metabolic results comparing MSP vs. HMLE cells are highly consistent with this new understanding, and they show that GRHL2 reverts MSP cells back to an epithelial type metabolism (higher glycolysis, lower oxidative phosphorylation), accompanying re-sensitization to anoikis.
Metabolic changes due to oncogenic transformation, EMT or transition to cancer stem cells have been reported extensively, with unique effects in each system (36, 38–41). In this paper, we provide a mechanism by which EMT/cancer stem cell transition lowers ROS so at protect cells against anoikis. Under this model, GRHL2 reverses EMT, suppressing mitochondrial oxidative function by repression of the critical enzyme regulating glutaminolysis, GLUD1, resulting in loss of glutamine utilization, increased ROS level, ultimately shifting metabolism resulting in anoikis sensitivity (Figure 6).
There may be multiple mechanisms of ROS neutralization downstream of α-ketoglutarate. First, it can act synergistically with other compounds (e.g., ascorbic acid) as a co-antioxidant (42). Secondly, the reverse GLUD1 reaction yields glutamate, contributing to glutathione synthesis. Thirdly, α-ketoglutarate is a co-factor for both histone demethylase and DNA demethylase enzymes. Changes in the level of this co-factor could affect gene expression epigenetically, as observed in other settings (43), with effects on metabolism and ROS. Finally, a recent report shows that up-regulated GLUD1 in tumor cells led to increases in α-ketoglutarate, increasing fumarate, in turn, through Krebs cycle enzymes. This contributed to the neutralization of cellular ROS, because fumarate activated glutathione peroxidase activity. GLUD1 up-regulation was, accordingly, shown to be pro-tumorigenic, and this effect was mediated by decreased ROS levels (27). In renal cell carcinoma, fumarate hydratase (FH) deficiency was increased fumarate to millimolar levels, contributing to oxidative stress rather than alleviating it (44). However, the major ROS in that study was superoxide rather than hydrogen peroxide, and GRHL2 did not affect FH level in our cells (data not shown). We speculate that the contradictory effects of fumarate in FH-deficient vs. GLUD1-over-expressing tumor cells may be reconciled by these considerations.
Interestingly, autophagy, which may play positive or negative roles in anoikis and non-apoptotic cell death after detachment, is regulated by glutaminolysis as well, through mTORC complexes (25). Speculatively, GLUD1/α-ketoglutarate alterations due to EMT or GRHL2 may affect cell survival through these additional pathways.
The upregulation of GLUD1 would have multiple advantages for EMT/CSC cells, including increased use of glutamine or glutamate as alternative carbon sources in glucose-poor environments, protection against ROS, and the increased ETC/oxidative phosphorylation capability afforded by less ROS-induced mitochondrial damage. Interestingly, the aspartate/glutamate transporter SLC1A6 was down-regulated dramatically by EMT, and, conversely, up-regulated by GRHL2 (table 1). Recently, SLC1A6 was found to act primarily as a glutamate exporter (45). EMT/CSC cells are predicted, therefore, to have increased accumulation of intracellular glutamate, contributing to the pathway that we have emphasized here.
Supplementary Material
Acknowledgments
The authors thank Dr. Stephanie Rellick and the laboratory of Dr. James Simpkins for assistance with Seahorse techniques, Dr. Kathy Brundage for flow cytometry, Drs. John Hollander and John LeMasters for technical advice concerning mitochondrial metabolism, Tyler Calkins for early contributions to the project, Aniello Infante and Ryan Percifield for assistance in RNA library construction and analysis of RNA seq data, Dr. Zachary Schafer for providing constructs and technical advice, Dr. Michael Ochs for significant guidance with bioinformatics analysis, and Dr. Amanda Ammer for expertise in Confocal image acquisition. The work was supported by a grant from the Mary Kay Foundation and a grant from the National Institute Of General Medical Sciences, U54GM104942. The following NIH grants supported the flow cytometry facility: GM103488/RR032138; RR020866;OD016165;GM103434.
Footnotes
The authors have no conflict of interest to disclose.
References
- 1.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–26. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiarugi P, Giannoni E. Anoikis: a necessary death program for anchorage-dependent cells. Biochem Pharmacol. 2008;76:1352–64. doi: 10.1016/j.bcp.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 3.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833:3481–98. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer. 2014 doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 5.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126:21–9. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malouf GG, Taube JH, Lu Y, Roysarkar T, Panjarian S, Estecio MR, et al. Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome biology. 2013;14:R144. doi: 10.1186/gb-2013-14-12-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh B, Shamsnia A, Raythatha MR, Milligan RD, Cady AM, Madan S, et al. Highly adaptable triple-negative breast cancer cells as a functional model for testing anticancer agents. PloS one. 2014;9:e109487. doi: 10.1371/journal.pone.0109487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javaid S, Zhang J, Anderssen E, Black JC, Wittner BS, Tajima K, et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell reports. 2013;5:1679–89. doi: 10.1016/j.celrep.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature structural & molecular biology. 2011;18:867–74. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walentin K, Hinze C, Werth M, Haase N, Varma S, Morell R, et al. A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development. 2015;142:1125–36. doi: 10.1242/dev.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell. 23:2845–55. doi: 10.1091/mbc.E12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 137:3835–45. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH, et al. Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium.[corrected] Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9356–61. doi: 10.1073/pnas.1307589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cieply B, Riley Pt, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, et al. Suppression of the Epithelial-Mesenchymal Transition by Grainyhead-like-2. Cancer research. 2012;72:2440–53. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-Mesenchymal Transition and Tumor Suppression Are Controlled by a Reciprocal Feedback Loop between ZEB1 and Grainyhead-like-2. Cancer research. 2013;73:6299–309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mlacki M, Kikulska A, Krzywinska E, Pawlak M, Wilanowski T. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Experimental biology and medicine. 2015;240:1396–401. doi: 10.1177/1535370215588924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huttemann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, et al. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion. 2011;11:369–81. doi: 10.1016/j.mito.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–13. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamarajugadda S, Stemboroski L, Cai Q, Simpson NE, Nayak S, Tan M, et al. Glucose oxidation modulates anoikis and tumor metastasis. Molecular and cellular biology. 2012;32:1893–907. doi: 10.1128/MCB.06248-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell States in the breast. Cell. 2011;145:926–40. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu W, Pelicano H, Huang P. Cancer metabolism: is glutamine sweeter than glucose? Cancer cell. 2010;18:199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villar VH, Merhi F, Djavaheri-Mergny M, Duran RV. Glutaminolysis and autophagy in cancer. Autophagy. 2015;11:1198–208. doi: 10.1080/15548627.2015.1053680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friday E, Oliver R, 3rd, Welbourne T, Turturro F. Glutaminolysis and glycolysis regulation by troglitazone in breast cancer cells: Relationship to mitochondrial membrane potential. Journal of cellular physiology. 2011;226:511–9. doi: 10.1002/jcp.22360. [DOI] [PubMed] [Google Scholar]
- 27.Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer cell. 2015;27:257–70. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. The Journal of biological chemistry. 2013;288:22993–3008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang SW, Lee S, Lee EK. ROS and energy metabolism in cancer cells: alliance for fast growth. Arch Pharm Res. 2015;38:338–45. doi: 10.1007/s12272-015-0550-6. [DOI] [PubMed] [Google Scholar]
- 30.Mailloux RJ, Harper ME. Mitochondrial proticity and ROS signaling: lessons from the uncoupling proteins. Trends in endocrinology and metabolism: TEM. 2012;23:451–8. doi: 10.1016/j.tem.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Cieply B, Koontz C, Frisch SM. CD44S-hyaluronan interactions protect cells resulting from EMT against anoikis. Matrix biology: journal of the International Society for Matrix Biology. 2015;48:55–65. doi: 10.1016/j.matbio.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer research. 2012;72:1438–48. doi: 10.1158/0008-5472.CAN-11-3024. [DOI] [PubMed] [Google Scholar]
- 33.Madajewski B, Boatman MA, Chakrabarti G, Boothman DA, Bey EA. Depleting Tumor-NQO1 Potentiates Anoikis and Inhibits Growth of NSCLC. Molecular cancer research: MCR. 2016;14:14–25. doi: 10.1158/1541-7786.MCR-15-0207-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon N, Skinner AM, Pommier RF, Schillace RV, O’Neill S, Peckham JL, et al. Gene expression signatures of breast cancer stem and progenitor cells do not exhibit features of Warburg metabolism. Stem cell research & therapy. 2015;6:157. doi: 10.1186/s13287-015-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viale A, Corti D, Draetta GF. Tumors and Mitochondrial Respiration: A Neglected Connection. Cancer research. 2015 doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- 36.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nature cell biology. 2014;16:992–1003. 1–15. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amoedo ND, Rodrigues MF, Rumjanek FD. Mitochondria: are mitochondria accessory to metastasis? Int J Biochem Cell Biol. 2014;51:53–7. doi: 10.1016/j.biocel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Cuyas E, Corominas-Faja B, Menendez JA. The nutritional phenome of EMT-induced cancer stem-like cells. Oncotarget. 2014;5:3970–82. doi: 10.18632/oncotarget.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer cell. 2013;23:316–31. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlashi E, Lagadec C, Vergnes L, Reue K, Frohnen P, Chan M, et al. Metabolic differences in breast cancer stem cells and differentiated progeny. Breast cancer research and treatment. 2014;146:525–34. doi: 10.1007/s10549-014-3051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–12. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang YH, Park SH, Lee YJ, Kang JS, Kang IJ, Shin HK, et al. Antioxidant alpha-keto-carboxylate pyruvate protects low-density lipoprotein and atherogenic macrophages. Free radical research. 2002;36:905–14. doi: 10.1080/1071576021000005348. [DOI] [PubMed] [Google Scholar]
- 43.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–6. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng L, Cardaci S, Jerby L, MacKenzie ED, Sciacovelli M, Johnson TI, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nature communications. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaudin S, Welsh J. 1,25-Dihydroxyvitamin D induces the glutamate transporter SLC1A1 and alters glutamate handling in non-transformed mammary cells. Molecular and cellular endocrinology. 2016 doi: 10.1016/j.mce.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.