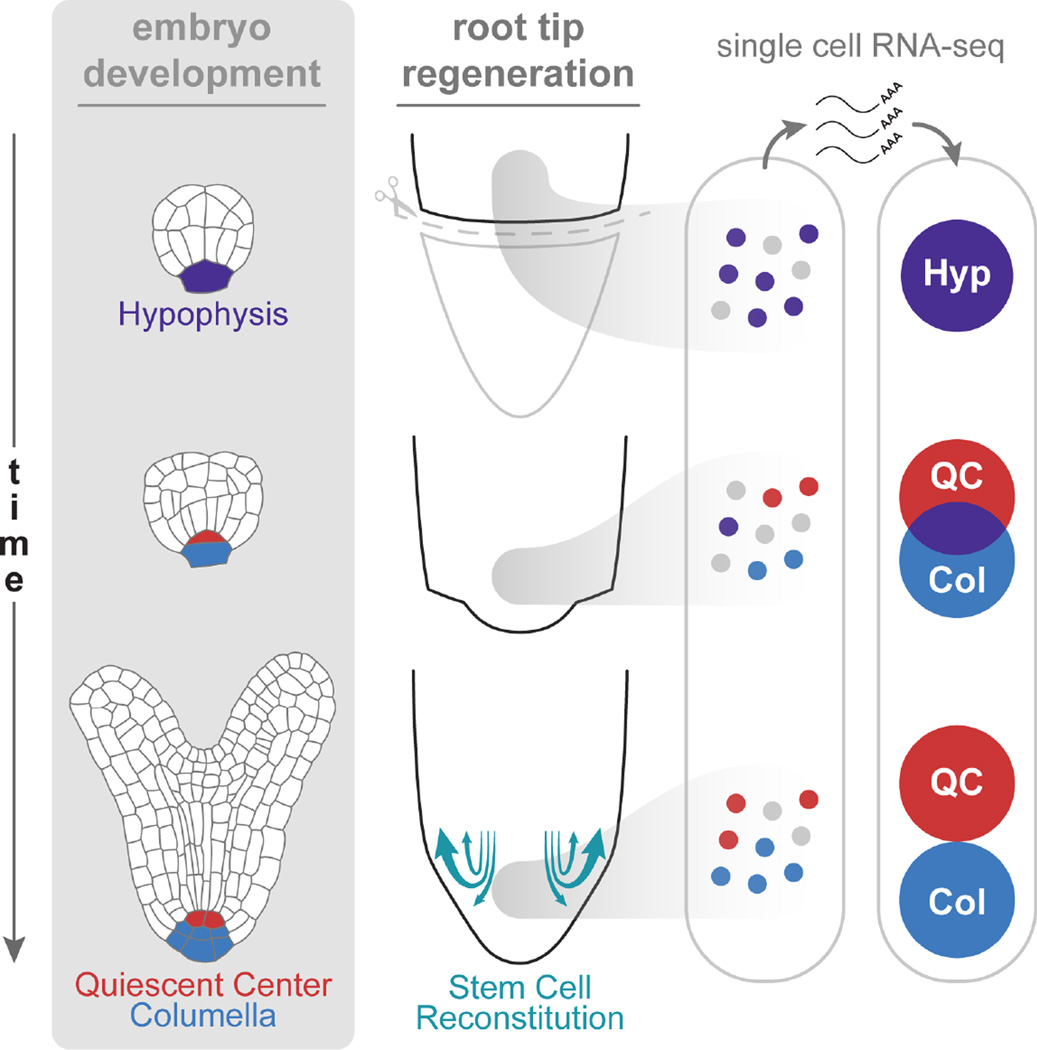
Summary
Plant roots can regenerate after excision of their tip, including the stem cell niche. To determine which developmental program mediates such repair, we applied a combination of lineage tracing, single cell RNA-Seq, and marker analysis to test different models of tissue reassembly. We show that multiple cell types can reconstitute stem cells, demonstrating the latent potential of untreated plant cells. The transcriptome of regenerating cells prior to stem cell activation resembles that of an embryonic root progenitor. Regeneration defects are more severe in embryonic than in adult root mutants. Furthermore, the signaling domains of the hormones auxin and cytokinin mirror their embryonic dynamics, and manipulation of both hormones alters the position of new tissues and stem cell niche markers. Our findings suggest that plant root regeneration follows, on a larger scale, the developmental stages of embryonic patterning and is guided by spatial information provided by complementary hormone domains.
Graphical Abstract
eTOC
Plants have dramatic regenerative capacity, including replacement of their stem cell niche after its complete excision. In a process that recapitulates the steps of embryogenesis, many specialized, transit-amplifying cells can reform stem cells. Complementary hormonal domains provide the spatial cues that play a role in patterning new tissues boundaries and a new stem cell niche.
Introduction
Plants have a wide capacity to regenerate their organs after damage by re-establishing regions of growth and patterning known as meristems (Sugimoto et al., 2011). Remarkably, excision of most of the root meristem, including the entire stem cell niche and its central organizer (the quiescent center; QC), triggers rapid regeneration and resumption of normal growth (Figure 1A; Feldman, 1976; Sena et al., 2009). Here we ask what kind of repair system can restore the root tip’s growth and tissue organization after its complete removal.
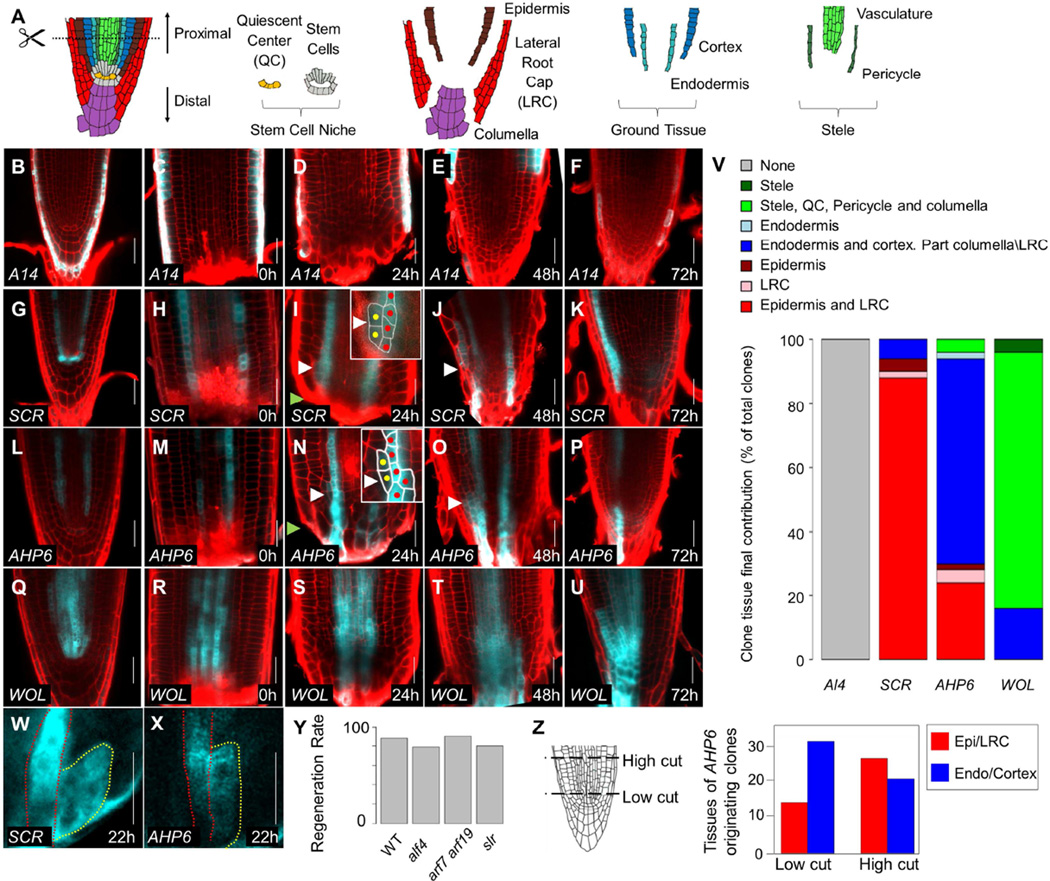
Figure 1. Growth dynamics during root tip regeneration.
A) Schematic representation of root meristem organization. Dotted line marks the cut site used in the study. B–U) Confocal images of tissue specific clones induced using the promoters A14 (B–F), SCR (G–K), AHP6 (L–P), and WOL (Q–U), before (B,G,L,Q),immediately after (C,H,M,R) root cutting, and at 24hpc (D,I,N,S), 48hpc (E,J,O,T), and 72hpc (F,K,P,U). Red channel is propidium iodide staining of cell walls. White arrowheads mark the presumed location of a new stem cell. Green arrowheads mark the cut site. Insets show magnified view of nascent clones. Red and yellow dots mark cells from original clone and new divisions, respectively. V) Proportions of the target tissues in fully regenerated tip for each of the clonal lines. W–X) Part of a time series tracking clones in live roots. Red line marks the original clone and yellow, new growth. See full series in Figure S1. Y) Regeneration rate of mutants in lateral root production. No significant difference was detected. Z) The identity of clones derived from an AHP6 marked tissue at 72hpc from cuts at two different heights. High cuts produced more epidermal clones than low cuts (χ2-test; n=98; p=0.014). Scale bars are 20µm.
Since the stem cell niche is removed with root tip excision, it cannot initiate the regeneration process. However, regeneration may rely on other potent cell types in the remaining stump (Birnbaum and Sánchez Alvarado, 2008; Sugimoto et al., 2011). In particular, the pericycle cell layer has organogenetic capacity and is the source of lateral roots in the adult (Lavenus et al., 2013). Further, under some conditions, it can generate a partially-organized pluripotent tissue known as callus (Atta et al., 2009; Sugimoto et al., 2010), suggesting that the pericycle may serve as a dormant stem cell niche that supports regeneration after damage (Sugimoto et al., 2011). However, plant cells are known to be plastic, and lineage studies show that cells throughout the root meristem can readily change their fate according to their position (Kidner et al., 2000). And while lateral roots are formed from the pericycle, adventitious roots can form from cambium and other vasculature associated cells (Bellini et al., 2014). Thus, an alternative model for regeneration is that missing tissues and stem cells regenerate from any remnant meristematic cell, guided by positional cues.
Tissue repatterning may occur either through the activation of regeneration-specific mechanisms, or by the ‘recapitulation’ of stereotypical organogenesis (Alvarado and Tsonis, 2006). In animals, there is evidence that embryonic gene expression programs and developmental processes are reiterated during regeneration (Chen et al., 2014; Kikuchi et al., 2010; Roensch et al., 2013). Similarly in plants, regeneration is accompanied by activation of key developmental regulators that function in embryogenesis and adult root formation (Kareem et al., 2015; Sena et al., 2009; Xu et al., 2006). However, it is unclear how closely, or if at all, the sequence of early development events is recapitulated during regeneration.
Many plant growth and patterning processes are regulated by the interaction between the phytohormones auxin and cytokinin (Schaller et al., 2015). During embryonic root formation, the two hormones form complementary domains, and perturbation of the signaling pathway of either hormone leads to embryonic root defects (Hamann et al., 2002; Hardtke and Berleth, 1998; Müller and Sheen, 2008). Classic studies demonstrated the importance of the balance between these hormones during in vitro regeneration (Skoog and Miller, 1957), but how this balance mediates tissue formation during regeneration is not well known.
Here, we dissected the early stages of regeneration by combining lineage and marker analysis with gene expression profiling in regenerating cells. We show that new root tissue is formed by the activity of newly specified stem cells recruited from multiple tissues in the remaining stump, ruling out the activity of a cryptic stem population drawn exclusively from pericycle cells. Activation of the new niche is preceded by rapid identity transitions and a sequence of developmental events that closely resembles embryonic root formation. Furthermore, regeneration was impaired in mutants with embryonic root defects but not in mutants that specifically perturbed lateral root development. We further show that altering the auxin and cytokinin domains during a narrow time window causes a coordinated change in the position of multiple root tissues and the stem cell niche. The results suggest that the interaction between these hormones sets up positional information for early tissue patterning and stem cell niche formation and that early events in embryogenesis are replayed within a different cellular organization.
Results
The root regenerates by de novo stem cell niche formation from multiple tissues
To test the different models of regeneration and track the contributions of multiple tissues to root tip regeneration, we generated a lineage tracking system that permanently marks a selected tissue upon induction by dexamethasone (promoter>>CRE:GR 35S:lox-terminator-lox-CFP). Plants carrying lineage constructs for different radial tissues were grown for five days, induced for 24h, checked for robust tissue-specific expression, and then cut and transferred to non-inductive plates. At least 100 plants were examined for each tissue of origin (Figure 1B–V).
Clones generated using the A14 promoter (AT5G43040; Lee et al., 2006), marking the outer layer of the root (Figure 1B–C), did not contribute to the regenerating tip and were pushed upward as the new root tip formed (Figure 1D–F,V). Interestingly, clones generated using the endodermal SCR promoter (Figure 1G–H) mostly produced lineages that occupied the positions of the new epidermis and lateral root cap (LRC; Figure 1I–K,V), overlapping with the WER epidermal/LRC identity marker (Figure S1A). The endodermal clones were continuous with regenerating cells in the epidermal position and converged near the cut site (Figure 1I). Lateral cell divisions characteristic of epidermis/lateral root cap stem cells (Bennett and Scheres, 2010) were observed at this position as early as 24 hours post cut (hpc; Figure 1I), suggesting that an endodermal cell assumed a stem cell identity to generate the new epidermal layer. We verified these endodermis-derived epidermal stem cell-like divisions by live imaging clones over time (Figure 1W, Figure S1B) and by tracking cell division patterns in live roots over 68h (Supplemental Movie 1).
To test whether the pericycle plays a special role in root tip regeneration, we induced and tracked clones using the AHP6 promoter, which marks the xylem pole pericycle and protoxylem (Figure 1A; Figure 1L–M). The AHP6-marked clones mostly produced cells occupying the position of the new cortex/endodermis tissues (Figure 1N–P,V), generated by stem cell like divisions at 24hpc (Figure 1N,X, S1C). These cells produced tissue-specific clones until they were replaced by unmarked cells around 72hpc (Figure 1P). In contrast to lateral root formation (Figure S1D), the contribution of the AHP6-marked clone was limited and did not comprise all new cells of the root tip. It is thus unlikely that root tip regeneration is driven by a lateral root initiation program. As further support, root regeneration frequency was unaffected in mutants severely impaired in the production of pericycle-derived lateral roots or callus (alf4-1 (Sugimoto et al., 2010) 79%, n=49; arf7 arf19 (Okushima et al., 2007) 90%, n=20; slr (Fukaki et al., 2002) 80%, n=50), as compared to wild type (88%, n=100; Figure 1Y).
The coordinated activation of stem cell-like divisions at 24hpc suggested that a new niche may be formed at this time point. Indeed, clones generated using the stele-specific WOL promoter (Figure 1Q–U; Mähönen et al., 2000) gave rise to new distally-growing columella cells, indicating re-establishment of the characteristic bidirectional growth of the niche (Bennett and Scheres, 2010; Figure 1S–U), with cells in the QC position eventually displacing the surrounding stem cells (Figure 1U; Heyman et al., 2013; Kidner et al., 2000). Overall, these transitions indicate that almost all prior cell identities were competent to form new stem cells.
The broad transformations in cell identity rule out a strict model in which remnant cells in the proximal meristem repopulate like identities. However, the different distributions of identity transitions (Figure 1V) may suggest that tissues have restricted competence to take on new fates. To test the role of competence over relative position, we cut the root at two different locations along the tapering tip. The width of the stele and endodermis is 37±2µm just above the QC, but is 56±5µm at 80µm above the QC, due to greater cell numbers in the stele (n=16; Figure 1Z), so that cuts at these locations alter the position of the pericycle in relation to the root center. In agreement with broad competence for fate change, we observed a shift in the identity of clones originating from the AHP6 lineage (Figure 1Z; n= 98; p=0.0135, χ2-test).
Overall, our results reveal that the new root tip is derived from a small population of cells, recruited from multiple tissues, which begin to act in a coordinated stem cell-like manner by 24hpc. These broad fate transitions are guided by the cells’ relative position in the remnant tissue.
Injury triggers a gradual loss of proximal identity near the cut site
To map cell identity transitions, we tracked multiple tissue markers during regeneration. Endodermal marker SCR:YFP (Wysocka-Diller et al., 2000) and stele marker WOL:GFP were already lost near the cut site by 6hpc-12hpc, receding to about 3 cell rows above the cut site by 16hpc (Figure 2A–B). The stele recession was confirmed by loss of xylem marker S4 and phloem marker S32 (Lee et al., 2006; Figure S2A–F). In contrast, expression of the outer layer markers WER and GL2 remained relatively stable (Figure 2C, Figure S2G–I; Lee and Schiefelbein, 1999; Lin and Schiefelbein, 2001). And, the inner endodermal expression of the ground tissue marker J0571 receded more than its outer cortical domain (Figure 2D), capturing a demarcation point in the clearing of cell identities.
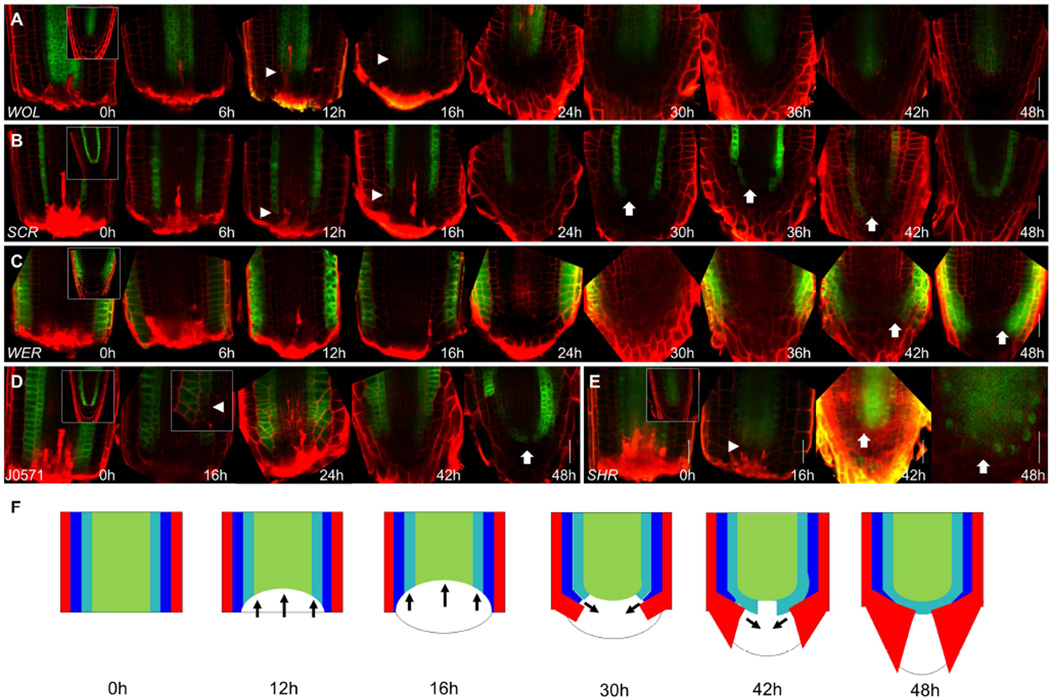
Figure 2. Dynamics of loss and recovery of proximal identities.
A–E) Confocal images of WOL:GFP (A), SCR:YFP (B), WER:GFP (C), J0571 (D) and SHR:SHR:GFP (E) during regeneration. Insets at 0h show uncut roots. Arrowheads mark the receding edge of the proximal identity markers. Arrows mark recovery of identity markers. Inset at (D) 16hpc shows a high magnification of the identity recession region. F) Illustration summarizing identity transition during regeneration. Red - epidermis/lateral root cap; blue – cortex; cyan – endodermis; green – stele. Arrows indicate the directions of identity recession and recovery. Scale bars are 20µm.
Interestingly, while stem cell-like divisions were observed at 24hpc, expression of cell identity markers did not initially correlate with this activity. And although stem cell activity resumed in an inside-out manner, endodermal (SCR) and epidermal (WER) markers began to recover their expression pattern in an outside-in pattern starting at 30hpc, only fully recovering their normal expression in the stem cells by 48hpc (Figure 2B–C). Similarly, the stele marker SHR (Helariutta et al., 2000) only regained its proper distal nuclear localization at 48hpc (Figure 2E). Curiously, in some cases, expression of a small discontinuous SCR domain was visible at the center of the stele.
Together with the identity transitions observed using clonal analysis, these results implicate a dome-shaped region of ~40 cells at the center of the stump as the site of re-patterning. Both the proximodistal and radial axes of the root were reset near the cut site (Figure 2F). Cell identity recovered in the opposite direction to stem cell growth, separating reactivation of stem cells from cellular respecification.
Single cell transcriptomics reveal rapid identity transitions
To characterize the transcriptional dynamics in the region of reorganization, we used single cell RNA-Seq to profile individual stele cells from induced WOL and AHP6 clones in uncut and regenerating roots at three time points – 3hpc, the earliest time point we could collect, 16hpc, prior to stem cell niche activation, and 46hpc, following the recovery of root growth. Cells were collected from dissociated meristems by cell sorting with stringent gates to ensure droplets with only one fluorescent cell, followed by mRNA amplification and sequencing using a modified version of the SMART-Seq2 protocol (Satija et al, 2015)
Cells were classified using the Index of Cell Identity (ICI) algorithm that can identify stable and transitional fates using single cell expression data (Birnbaum and Kussell, 2011; Efroni et al., 2015). We used a reference dataset of 579 identity marker genes (Table S1–S2) to classify cells into 14 root tissue types (Figure S3A; Table S3), which we grouped as stele, QC, columella, epidermis/LRC, and ground tissue (Figure 3A–D).
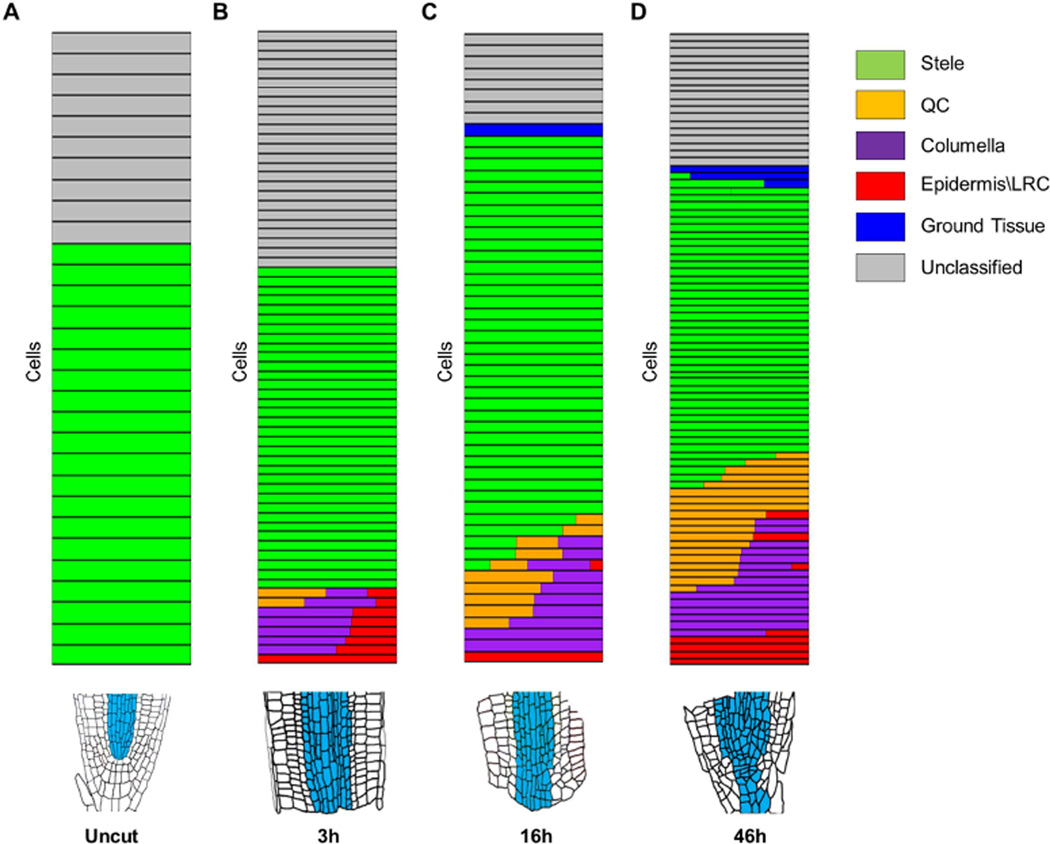
Figure 3. Identity of single cells isolated from regenerating roots.
A–D) Relative cell identity in individual cells isolated from uncut (A) and 3hpc (B), 16hpc (C) and 46hpc (D). Each row represents a single cell. Identity is shown as a color-coded bar consisting of the normalized ICI score for each tissue type. Multiple color bars in a single row indicate mixed-identity within a single cell. Blue sectors in root illustrations (bottom) represent the domains from which single cells were isolated.
We collected 238 cells of which 74% could be classified into one or more reference identity. As expected for WOL and AHP6 marked cells, most (116/177) were classified as stele (Figure 3AD). However, some cells lost their stele identity and gained distal identities as early as 3hpc (Figure 3B). This rapid change in identity is consistent with the observed recession of stele markers near the cut site. The transitioning stele cells scored a surprising mixture of QC, columella, and epidermis/LRC—identities that are either removed by cutting (QC and columella) or are normally absent from internal root tissue (epidermis/LRC). Multidimensional scaling grouped the transdifferentiating cells together, irrespective of their tissue of origin (WOL or AHP6), suggesting cells from different tissue sources converged to a single identity (Figure S3B). The lack of residual stele identity suggested that the mixed identity cells originated from the tip of the stump, where stele markers receded (Figure 2A–D). As regeneration progressed, cells with distinct distal identities, such as QC and columella alone, became more common (Figure 3C–D). Consistent with the clonal analysis, stele-derived cells contributed mainly to the new columella and QC, but also to some of the LRC and ground tissue (Figure 1V).
Single cell analysis thus revealed a rapid change from stele to mixed distal cell identities, which gradually separated into columella and QC during regeneration. This developmental sequence resembles the dynamics of embryonic root formation, during which a single cell – the hypophysis – expresses multiple distal identity markers before dividing to generate distinct QC and columella progenitors (Crawford et al., 2015; ten Hove et al., 2015; Müller and Sheen, 2008; Scheres et al., 1994).
Recovery of distal fates resembles an embryonic developmental sequence
We explored the similarity of regeneration to embryonic root formation by examining the expression of hypophysis-expressed genes (Crawford et al., 2015; Rademacher et al., 2012; Ueda et al., 2011; Wysocka-Diller et al., 2000) in regenerating roots. These genes generally displayed a gradual upregulation in the mixed-identity cells between 3hpc and 16hpc and became differentially expressed as QC and columella identities became distinct (Figure 4A), a pattern we corroborated on a recently identified set of genes expressed in the basal meristem and hypophysis (Wendrich et al., 2015; Figure S4A). In contrast, genes expressed in stages of embryogenesis prior to hypophysis division or that regulate the embryonic formation of other tissues were not induced in the mixed cells (Figure 4B), indicating the activation of a hypophysis-specific rather than a general embryonic program. Hypophysis-expressed genes were consistently upregulated in mixed-identity cells at 3hpc and 16hpc, but their expression was diminished by 46hpc (Figure S4B), further supporting the existence of a transient hypophysislike state during regeneration.
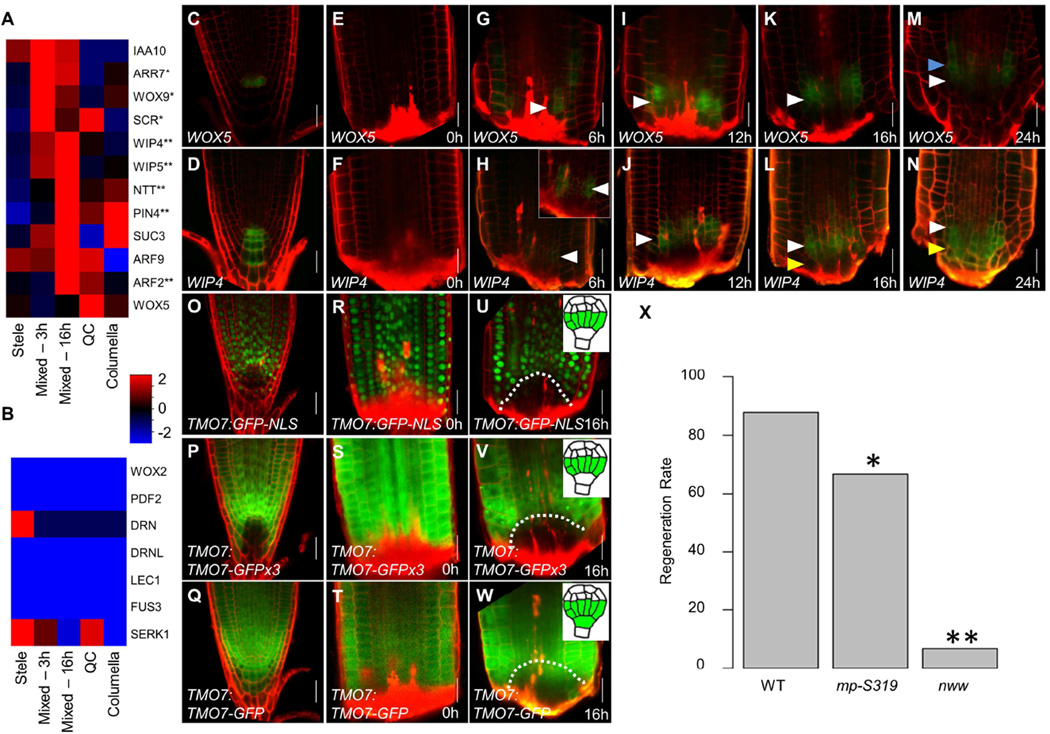
Figure 4. Proximodistal domain separation resembles embryonic hypophysis division.
A–B) Mean expression values of known hypophysis (A) or embryonic but non-hypophysis (B) expressed genes in single cells grouped according to their identity. * and ** marks significant upregulation at 3h or 16h, respectively (p<0.05; Wilcoxon test). C–N) Confocal images of WOX5 (C,E,G,I,K,M) and WIP4 (D,F,H,J,L,N) reporters over the first 24 hours of regeneration. Blue, white, and yellow arrowheads mark the forming proximal, overlapping, and distal domains, respectively. Inset shows magnified and gain-enhanced GFP signal. O–W) Confocal images of transcriptional (O–U) and translational non-mobile (P–V) and mobile (Q–W) reporters of TMO7. Inset at 16h show the embryonic expression of each reporter. Dotted line marks the region of identity loss. X) Regeneration rate of mutants defective in hypophysis division. * p=0.02; ** p<1E-10; χ2-test. Scale bars are 20µm.
To understand the spatial dynamics of distal identity separation, we analyzed reporters that are co-expressed in the hypophysis: WOX5:GFP, which subsequently marks the QC, and WIP4, which later marks both the QC and columella (Figure 4C–D; Crawford et al., 2015; Haecker et al., 2004; Nawy et al., 2005). Starting at 6hpc, both markers were co-expressed within the region of proximal identity loss (Figure 4E–J). Between 16hpc and 48hpc, WOX5 marked a proximal and WIP4 marked a distal domain (Figure 4J–M; Figure S4C–H). Similarly, the hypophysis-expressed marker IAA10 (Rademacher et al., 2012), which is confined to the columella in the adult (Figure S4I), overlapped with the WOX5 domain at 16hpc but then separated into a distal domain (Figure S4J–K). Thus, much like the embryo, these three markers overlap in hypophysislike cells that appear early in regeneration and subsequently separate into distinct domains.
To rule out generally promiscuous expression of QC and columella markers, we examined plants bearing WOX5:mCherry and PET111, which marks differentiated columella (Figure S4L). PET111 never overlapped with WOX5 and was not detected until proximal-distal domain separation at 24hpc (Figure S4M–O).
During hypophysis division, the transcription factor TARGET OF MONOPTEROS7 (TMO7) is expressed in the provascular cells above the hypophysis and moves distally into the hypophysis cell to regulate its division (Schlereth et al., 2010). Analysis of the transcriptional reporter TMO7:GFP-NLS, the translational reporter TMO7:TMO7-GFP, and the non-mobile protein fusion TMO7:TMO-GFPx3, showed that, following tip removal, expression of TMO7 is lost in the stump region by 16 hpc when mixed-distal cell identities were detected in the same domain, while TMO7 protein moves into that region from the surrounding cells (Figure 4O–W). The rapid recovery of this pattern further supports the existence of a hypophysis-like state during early stages of regeneration.
We next used genetic perturbation to test the shared program between embryonic root formation and regeneration. Mutants in JACKDAW (JKD) form a normal embryonic root but fail to maintain expression of QC markers in the adult (Figure S4P–Q; Welch et al., 2007). Consistent with an activation of an embryonic phase of root development, expression of the QC marker WOX5 was re-activated during regeneration of jkd mutants (Figure S4R–S), but diminished when the tip was fully regenerated (Figure S4T–U).
We also tested the regeneration frequency in mutants that exhibit a failure in hypophysis division and subsequently lacked an embryonic root: the auxin response factor MONOPTEROS (MP), and triple mutants of the NO TRANSMITING TRACT gene family (nww; Crawford et al., 2015; Hamann et al., 1999; Hardtke and Berleth, 1998). In the weak mp-S319 allele, ~10% of seedlings fail to form an embryonic root, but those that escape exhibit normal growth (Schlereth et al., 2010; De Smet et al., 2010). The escaped mp-S319 adult roots showed a 25% reduction in the frequency of regeneration compared to wild type. The nww mutants form an embryonic root in only 2.5% of plants but can be rescued with a transient auxin treatment and then grow without supplemental auxin (Crawford et al., 2015). These rescued nww mutant roots had a 92% reduction in regeneration (wild type 88%, n=100; mp-S319 66%, n= 21;nww 7%; n=15;χ2-test, p=0.02 and p<1E-10, respectively; Figure 4X). Thus, both mutant phenotypes are consistent with a reliance on the early phases of embryonic root formation during regeneration.
Hormone and tissue markers follow embryonic dynamics
In the embryo, their signaling domains briefly overlap in the hypophysis before they separate into a proximal cytokinin and a distal auxin domain (Müller and Sheen, 2008). In an apparent recapitulation of the overlap of the two hormones, rapid response genes for cytokinin (ARR family; To et al., 2004) and auxin (AUX/IAA family; Chapman and Estelle, 2009) were coordinately upregulated in single chimeric cells during early stages of regeneration (Figure 5A).
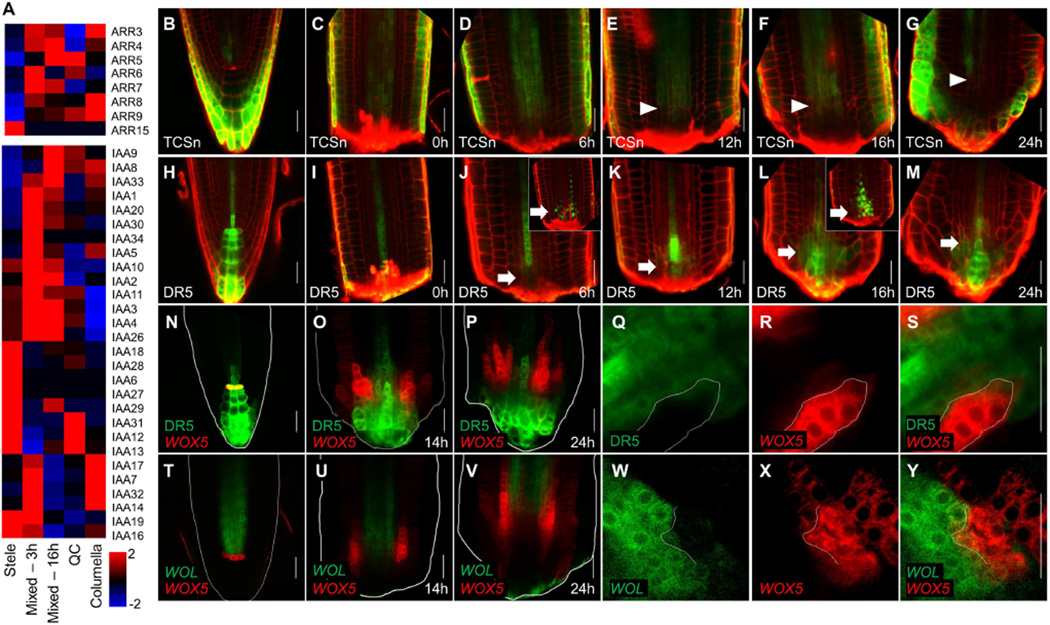
Figure 5. Expression of hormonal response markers during proximodistal domain separation.
A) Mean expression values in single cells, grouped by identity, of A-class ARR (top) or AUX/IAA (bottom) gene families. B–M) Expression of the cytokinin marker TCSn (B–G) and auxin marker DR5 (H–M) during regeneration. Arrowheads mark the recession of the TCSn signal. Arrows mark induction of DR5. Insets in (J) and (L) show expression of the rapid maturing DR5rev:NLS-3xVenus at the corresponding time points. N–P) Dual marker expression of DR5 (green) and WOX5 (red) before cutting (N) and at 14hpc (P) or 24hpc (P). Q–S) An invading sector of WOX5 (red) expression in the DR5 (green) domain in single channel (Q–R) and overlay (S) at 14hpc. T–Y) Confocal image of WOL (green) WOX5 (red) plants, before cut (T) and at 14hpc (U) and 24hpc (V), including high magnification of a WOX5 invading sector (W–Y) at 14hpc. Scale bars are 20µm.
To follow the spatial dynamics of this interaction, we used the cytokinin response reporter TCSn:GFP (TCSn; Zürcher et al., 2013) and the auxin response reporters DR5rev:GFP (Friml et al., 2003) and fast-maturing DR5rev:3xVENUS-N7 (Heisler et al., 2005). In uncut roots, TCSn is expressed in the stele and root cap but is absent from proximal tiers of the columella (Figure 5B; Bishopp et al., 2011; Zürcher et al., 2013). At 6hpc, the remnant TCSn expression in the stele remained unchanged (Figure 5C–D), gradually receding proximally at later time points (Figure 5E–G). The distal auxin maximum (Figure 5H) was completely excised during the removal of the tip, leaving only low expression in immature xylem cells (Figure 5I). However, auxin signaling was rapidly induced in a region of stele near the cut site, as shown by DR5rev:3xVENUS-N7 expression (Figure 5J,L,inset; Figure S5A–E), creating a transient overlap with the cytokinin reporter (Figure 5K–M). Thus, the embryonic dynamics of the two hormones – initial overlap, followed by separation into distinct proximal cytokinin and distal auxin domains – were recapitulated during root tip regeneration.
In the adult root, WOX5 expression is restricted to the QC, whose location overlaps with and depends upon a local maximum of auxin signaling (Sabatini et al., 1999; Xu et al., 2006). In contrast, embryonic WOX5 expression begins in the hypophysis but after hypophysis division shifts proximally away from the auxin maximum (Müller and Sheen, 2008). Regeneration in DR5:GFP WOX5:mCherry roots (Figure 5N) showed that, after a brief overlap (compare WOX5 in Figure 4G and DR5 in 5J at 6hr), WOX5:mCherry expression shifted proximally and the two markers then remained mutually exclusive (Figure 5O–P), exhibiting an embryonic expression pattern.
In addition, the patterns were highly suggestive of regulatory relationships. For example, WOX5 and DR5 expression were almost always in mutually exclusive domains when in close proximity (Figure 5Q–S). Mutually exclusive expression was also often detected between WOX5 and the cytokinin receptor WOL (Figure 5T–Y), suggesting that an interaction between hormone signaling and cell identity markers may guide patterning during regeneration.
Auxin-cytokinin interaction guides the establishment of the proximodistal and radial axes
Both cytokinin and auxin are required for proper formation of the embryonic root, but the role of their interaction is not clear. It was suggested that an auxin domain positions the nascent root embryonic meristem, as mutations in HANABA TARANU (HAN) that cause a proximal and lateral shift in the embryonic auxin domain, also led to a corresponding shift in the expression of distal root markers, including the QC marker WOX5 (Nawy et al., 2010). Interestingly, we saw a similar phenomenon during regeneration in the han-16 mutant, where at 16hpc, WOX5 occupied a more proximal and lateral domain than wild type (Figure S6A, student’s t-test p=0.02), indicating that hormonal domains may influence tissue specification similarly both during embryogenesis and regeneration. We therefore sought to determine whether hormone domains could guide tissue positioning in the regeneration process.
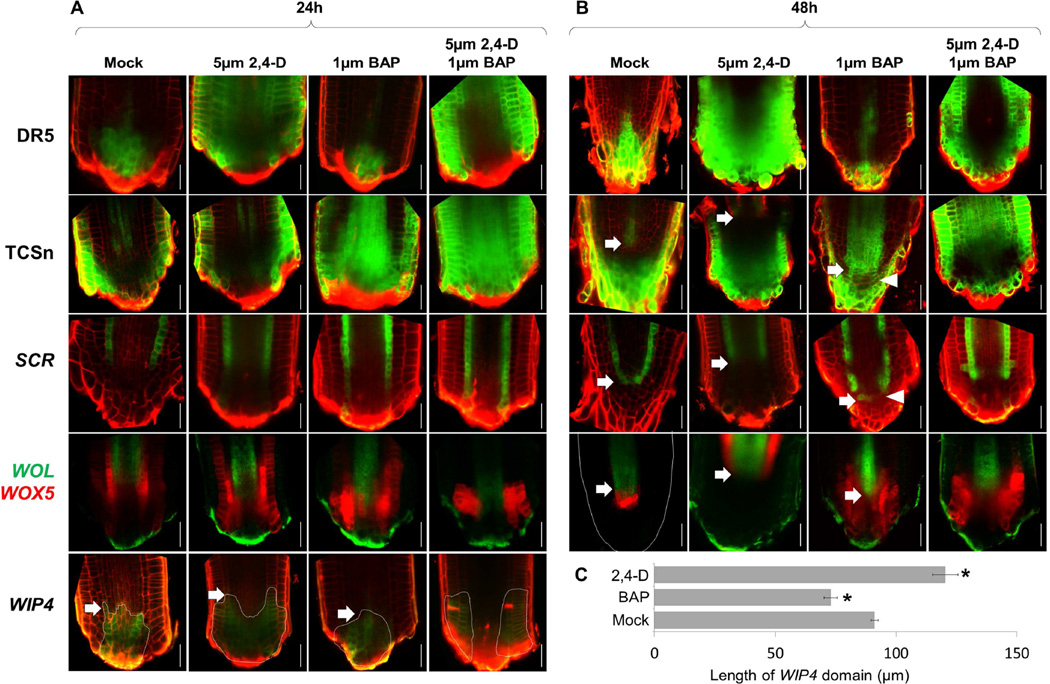
Application of the auxin analog 2,4-D caused expansion of the DR5 signal and a proximal recession of the TCSn signal by 48hpc (Figure 6A–B). In accordance, endodermal and stele markers were expressed in a more proximal position than normal (Figure 6B, Figure S6B), with SCR converging at 72hpc (Figure S6C–D). Strikingly, expression of the distal marker WIP4 was significantly expanded proximally, bounded by an internal stele/cytokinin domain (Figure 6A,C). Our results indicate that auxin treatment causes a coordinated proximal shift in the auxincytokinin border that is accompanied by a coordinated shift in the position of the border between the radial cell files and the cap.
Figure 6. Effects of hormone treatment on meristem patterning.
A–B) Confocal images of mock, 2,4-D, BAP, or 2,4-D and BAP treated regenerating roots at 24hpc (A) and 48hpc (B). Arrows mark proximodistal shifts in position of markers compared to mock treated control. Arrowheads mark sporadic cytokinin signaling expansion and concomitant loss of SCR expression. C) Length of WIP4 domain at 24hpc under different treatments. Error bars are standard error. n=8 for each treatment. * marks significantly different than mock (Student’s ttest, p<1E-4 for BAP, p<1E-3 for 2,4-D). Scale bars are 20µm.
Treatment with cytokinin during regeneration diminished the DR5 domain while causing distal and lateral expansion of the TCSn reporter (Figure 6A–B). Under these conditions, expression of SCR recovered in a more distal position than normal, invading the cap region (Figure 6B). In agreement, the domains of the stele markers WOL (Figure 6B) and SHR (Figure S6E) stabilized at a more distal position than in control plants. Finally, the distal shift in markers coincided with a reduction in the WIP4 domain size (Figure 6A,C), showing that cytokinin application caused a coordinated distal shift in the position of root tissues. To examine the effects of cytokinin treatment on the position of the stem cell niche, we allowed cytokinin treated plants of the SCR lineage line to recover on hormone-free media for 24h. In agreement with the general shift in tissue positioning, we observed that the epidermal stem cell formed at an abnormal distal position (Figure S6F).
We could cause a more dramatic alteration of hormone domains with a dual auxin-cytokinin treatment, which displaced the auxin domain to the flank of the root stump (Figure 6A–B). Accordingly, the WIP4 domain was displaced to a lateral position (Figure 6A). Strikingly, SCR and SHR domains now expanded outward in a radial direction (Figure 6B; S6G), suggesting hormonal interaction can also pattern the radial axis of the root meristem. Indeed, cytokinin treatment by itself caused a patchy loss of SCR (Figure 6B, S6H) with the TCSn domain exhibiting a complementary patchy expansion (Figure 6B).
Interestingly, the effect of hormone treatment on patterning was limited to a narrow early time window, as the same treatments at 24hpc or on intact roots had little effect on patterning (Figure S6I–P). Indeed, a similar narrow window for hormonal response was shown for the embryo (Müller and Sheen, 2008). Thus, the results show that the embryo-like juxtaposition of auxincytokinin domain acts early to set up the position where cell files will converge and the stem cell niche will eventually form. Overall, we conclude that root regeneration involves the de novo formation of the root axis using the same developmental sequence as during embryonic root formation, its position being guided by hormone domains and their interactions.
Discussion
Regeneration through activation of embryonic organogenesis programs
A fundamental question in regeneration biology is that of ‘recapitulation,’ or whether organ regeneration in the adult follows similar programs as those used during embryonic development (Alvarado and Tsonis, 2006). Here, we show that the regeneration of the root tip initiates in an embryonic-like sequence of distal root meristem formation, followed by the activation of a stem cell niche that propagates the root. We have previously reported that the root tip can regenerate even in mutants in which the stem cell niche failed to be maintained in the adult root (Sena et al., 2009). However, these mutants could all properly form an embryonic root (Aida et al., 2004; Sabatini et al., 2003), while rescued nww mutants, which have a functional root meristem, recapitulated the severe embryonic root formation defects when cut. The association between embryonic and regenerative processes is also evident in regeneration of adventitious roots, where mp mutants had reduced capacity to form these roots, and gnom mutants, which are also defective in embryonic root development, could not form them at all (Berleth and Jürgens, 1993; Mayer et al., 1993).
An interesting aspect of root tip regeneration is that, in contrast to the embryo, where the proximal-distal separation involved two cells, the same separation in the root occurs on a larger spatial scale. However, the overall mechanistic similarity between these processes suggests that, even in the embryo, the auxin-cytokinin interaction may act to define distinct spatial domains rather than a specific embryonic cell. Supporting this view is the han mutant, in which a proximal shift in the auxin domain triggers an ectopic formation of a new root at the new auxin boundary, making a specific cellular morphology dispensable for embryonic root formation (Nawy et al., 2010). Indeed, in many regeneration systems, the newly replaced organs exhibit flexibility in scale in order to match the size of the remnant body parts (Oviedo et al., 2003). While genetic modifiers may be required to adapt patterning programs from the small embryo to the larger adult root (Moreno-Risueno et al., 2015), our results suggest that the plant can generate a root on different scales using hormone domains as a patterning mechanism.
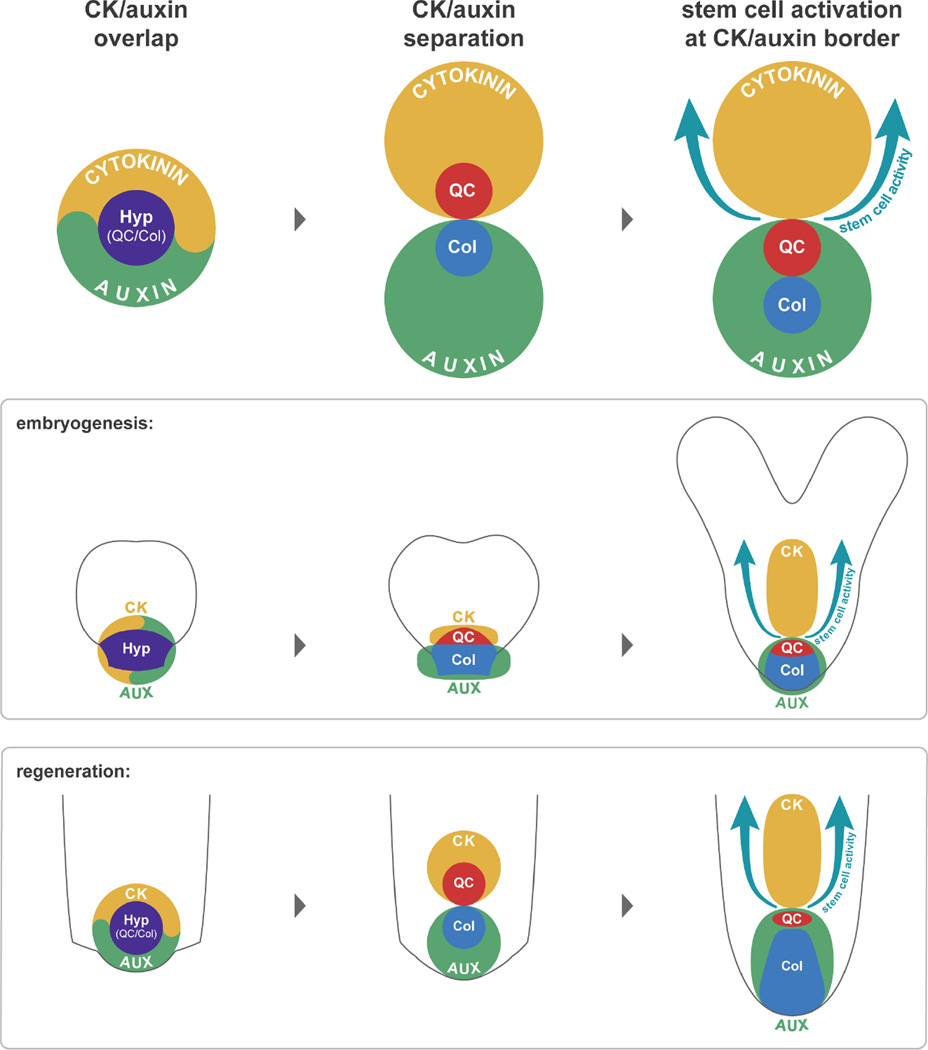
A model for root patterning during early ontogeny
Tissue identities in the reformed meristem appear to be established separately from the activity of the stem cell niche. While stem cells generated new files from the inside out, cell identities all recovered in an outside-in manner. One possibility is that these dynamics reflect a “top-down” flow of cell fate information through cell-to-cell communication (van den Berg et al., 1995). However, cell identity was closely correlated with hormone domain formation, and the position of the root cap boundary and the convergence of radial files could be altered with manipulation of auxin and cytokinin levels. Given the mutual antagonism between these hormones (Bishopp et al., 2011), an attractive hypothesis is that the tendency of auxin and cytokinin to form complementary but juxtaposed domains could be used to position the root cap, internal root tissues, and the stem cell niche in multiple contexts of root formation (Figure 7). Indeed, similar self-organizing auxin-cytokinin interactions provide positional information in other patterning processes in the plant (Bielach et al., 2012; Bishopp et al., 2011; Chang et al., 2015; De Rybel et al., 2014).
Figure 7. A general model for root regeneration.
Similar to embryonic root development, regeneration initiates with a transient overlap of auxin and cytokinin signaling, which then separates to a proximal cytokinin and a distal auxin domain, providing spatial cues for the root cap, stem cell niche, and proximal identities.
It has been shown that mutants defective in pericycle activity and lateral root initiation are also inhibited in some aspects of regeneration (Liu et al., 2014; Sugimoto et al., 2010), but, as shown here, not in root meristem regeneration, demonstrating that regeneration is not absolutely dependent on lateral root programs. Pericycle may be a common source of regenerative tissue but we posit that many cells capable of dividing can form a minimal field of competent cells in which juxtaposed auxin-cytokinin domains serve as positional guides.
The robustness of pattern and developmental plasticity
Our single cell analyses showed that identity transitions are extremely rapid in plants, as early as three hours after injury. Still, despite this plasticity, plant organs are able to maintain robust patterns over long periods of time. How can this contrast between highly responsive organ reorganization and stable organ patterning be reconciled?
Several mechanisms can explain this patterning stability, such as chromatin modifications that limit the cell’s developmental potential (Ikeuchi et al., 2015). Importantly, feedback loops between auxin and cytokinin (Bishopp et al., 2011) and between patterning genes and hormonal pathways have been identified, such as the stele localized transcription factor PHABULOSA, which activates cytokinin synthesis (Dello Ioio et al., 2012), and SHR, which activates cytokinin degradation in the surrounding endodermis (Kurakawa et al., 2007).
Indeed, spatial patterning is strongly buffered during steady-state development as we observed only mild effects when hormonal treatment was applied to intact meristems or to meristems at later stages of regeneration. However, root injury appears to disrupt these stabilizing feedbacks, providing a transient opportunity for hormone signaling to reset tissue boundaries. Accordingly, hormone treatments in the early stages of regeneration could alter major landmarks of the root, such as the extent of the cap and the position of the stem cell niche. The transient destabilization of feedback mechanisms that normally maintain stable patterns in the adult meristem would allow scalable, embryo-like programs to replay during permissive developmental windows, such as instigated by injury.
Experimental Procedures
Plant growth, imaging and regeneration assay
Plants were grown as previously described (Efroni et al., 2015). Mutant alleles of jkd-4, nww and mp-S319 were previously characterized (Crawford et al., 2015; Hassan et al., 2010; Schlereth et al., 2010). Regeneration assays were performed as in Sena et al., (2009). For hormonal treatment, root tips were cut at 80µm above the QC, moved to agar plates (1/2 MS, 0.5% sucrose, 0.8% agar, pH=5.7) containing 2,4-D (2,4-Dichlorophenoxyacetic acid; Sigma), BAP (6-benzylamino purine; Sigma) or both, and placed vertically in a growth chamber for recovery. For confocal imaging, seedlings were stained with propidium iodide (10µg/ml), mounted in water, and visualized using either Leica SPE or Leica SP5 confocal microscopes. For live imaging, cut roots of plants carrying the lineage marker, or the histone marker 35S:HG2B-mCherry, were placed between a coverslip and an agar block and imaged using an inverted SPE confocal at fixed intervals (also see Supplemental Experimental Procedures).
Clonal analysis
The inducible tissue specific lineage lines for SCR, AHP6 and A14 were constructed following the protocols described previously (Efroni et al., 2015). To induce clones, 5 DAG seedlings were placed on agar plates containing 15µm dexamethasone (Sigma) for 24h followed by inspection under a fluorescent microscope. Root tips of uniformly induced plants were cut, and plants were moved to agar plates, placed vertically for recovery, and imaged at specified intervals.
Single cell RNA-Seq and bioinformatic analysis
Cells were isolated from cut roots using a short (1h-2h) cell wall digestion, followed by 3 filtrations through a 40µm screen. CFP positive protoplasts were sorted using FACS (either BD Aria, or Sony SY3200) into 96-well plates containing lysis buffer using gates to ensure a single cell per droplet (Figure S7A,B) Single cells were subject to cDNA synthesis, amplification, library preparation, read alignment and expression calling (Supplemental Experimental Procedures). Data was deposited in GEO (GSE74488). To derive cell identity scores, marker Spec scores were calculated as described previously (Efroni et al., 2015), and 579 tissue markers selected, checking first whether read depth affected cell identity or mixed identity calls (Figure S7C,D). To determine background ICI levels, we used ICI scores for QC, Columella and Epidermis\LRC in uncut, stele-derived cells as threshold values (Figure S7E, Supplemental Experimental Procedures). To produce the multidimensional scaling plot, we used expression of all 579 identity markers, which were z-normalized to reduce outlier effects and scaled using the cmdscale function in R.
Supplementary Material
Highlights.
Removal of root stem cells triggers their reformation from multiple remnant tissues
Stem cell reformation is preceded by embryonic-like development sequence
Antagonistic hormonal signaling domains position regenerated tissues and stem cells
Acknowledgments
We thank Charles W. Melnyk and Bruno Müller for sharing materials, Claude Desplan, Esteban Mazzoni, Phillip Benfey, Kim Gallagher, and Wolfgang Lukowitz for critical reading of this manuscript, and the NYU GenCore for generating RNA-seq data. Funding was provided by NIH R01 GM078279 to KDB, and EMBO LTF185-2010 for IE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, I.E, A.M, T.N., K.D.B; Methodology, I.E., T.N., R.R., R.S., K.D.B; Software, I.E.; Investigation, I.E., A.M., P.L.I., R.R., N.D, A.P.; Resources, R.S.; Writing, I.E., T.N, K.D.B.; Visualization, (Graphical Abstract, Fig. 7, R.R.); Supervision, I.E. K.D.B.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:119–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Alvarado AS, Tsonis Pa. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57:626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I, Bellini Catherine, Pacurar Daniel I, Perrone Irene. Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 2014;65:639–666. doi: 10.1146/annurev-arplant-050213-035645. [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B. Root development-two meristems for the price of one? Curr. Top. Dev. Biol. 2010;91:67–102. doi: 10.1016/S0070-2153(10)91003-X. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryos. Development. 1993;9:299. [Google Scholar]
- Bielach a, Podlesakova K, Marhavy P, Duclercq J, Cuesta C, Muller B, Grunewald W, Tarkowski P, Benkova E. Spatiotemporal Regulation of Lateral Root Organogenesis in Arabidopsis by Cytokinin. Plant Cell. 2012;24:3967–3981. doi: 10.1105/tpc.112.103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum KD, Kussell E. Measuring cell identity in noisy biological systems. Nucleic Acids Res. 2011;39:9093–9107. doi: 10.1093/nar/gkr591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum KD, Sánchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 2011;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. J. Exp. Bot. 2015 doi: 10.1093/jxb/erv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Chen Y, Love NR, Amaya E. Tadpole tail regeneration in Xenopus. Biochem. Soc. Trans. 2014;42:617–623. doi: 10.1042/BST20140061. [DOI] [PubMed] [Google Scholar]
- Crawford BCW, Sewell J, Golembeski G, Roshan C, Long JA, Yanofsky MF. Plant development. Genetic control of distal stem cell fate within root and embryonic meristems. Science. 2015;347:655–659. doi: 10.1126/science.aaa0196. [DOI] [PubMed] [Google Scholar]
- Efroni I, Ip P-L, Nawy T, Mello A, Birnbaum KD. Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 2015;16:9. doi: 10.1186/s13059-015-0580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman LJ. The de novo origin of the quiescent center regenerating root apices of Zea mays. Planta. 1976;128:207–212. doi: 10.1007/BF00393230. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H, Scheres B, Blilou I. JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development. 2010;137:1523–1529. doi: 10.1242/dev.048777. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long Ja, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D, et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science. 2013;342:860–863. doi: 10.1126/science.1240667. [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Lu K-J, Weijers D. Building a plant: cell fate specification in the early Arabidopsis embryo. Development. 2015;142:420–430. doi: 10.1242/dev.111500. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, de Lucas M, De Veylder L, et al. PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants. 2015;1:15089. doi: 10.1038/nplants.2015.89. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK, et al. A PHABULOSA/cytokinin feedback loop controls root growth in arabidopsis. Curr. Biol. 2012;22:1699–1704. doi: 10.1016/j.cub.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al. PLETHORA Genes Control Regeneration by a Two- Step Mechanism PLETHORA Genes Control Regeneration by a Two-Step Mechanism. Curr. Biol. 2015;25:1–14. doi: 10.1016/j.cub.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta. 2000;211:191–199. doi: 10.1007/s004250000284. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich Aa, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae Ca, Stainier DYR, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schiefelbein J. Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development. 2001;128:3697–3705. doi: 10.1242/dev.128.19.3697. [DOI] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell. 2014;7:1–14. doi: 10.1105/tpc.114.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Büttner G, Jürgens G. Apical-basal pattern formation in the Arabidopsis embryo : studies on the role of the gnom gene. Dev. Suppl. 1993;1:149–162. [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardımcı GG, Petricka JJ, Vernoux T, Blilou I, Alonso J, Winter CM, Ohler U, Scheres B, et al. Transcriptional control of tissue formation throughout root development. Science. 2015;350:426–430. doi: 10.1126/science.aad1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Bayer M, Mravec J, Friml J, Birnbaum KD, Lukowitz W. The GATA Factor HANABA TARANU Is Required to Position the Proembryo Boundary in the Early Arabidopsis Embryo. Dev. Cell. 2010;19:103–113. doi: 10.1016/j.devcel.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Newmark Pa, Sánchez Alvarado A. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 2003;226:326–333. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, FreireRios A, Borst JW, Lukowitz W, Jürgens G, et al. Different Auxin Response Machineries Control Distinct Cell Fates in the Early Plant Embryo. Dev. Cell. 2012;22:211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Roensch K, Tazaki A, Chara O, Tanaka EM. Progressive specification rather than intercalation of segments during limb regeneration. Science. 2013;342:1375–1379. doi: 10.1126/science.1241796. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda aS, Wendrich JR, Smit ME, Novak O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, et al. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345 doi: 10.1126/science.1255215. 1255215–1255215. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. An Auxin-Dependent Distal Organizer of Pattern and Polarity in the Arabidopsis Root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. The Yin-Yang of Hormones: Cytokinin and Auxin Interactions in Plant Development. Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;2487:2475–2487. [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu H-Y, Hofhuis H, Birnbaum KD. Organ regeneration does not require a functional stem cell niche in plants. Nature. 2009;457:1150–1153. doi: 10.1038/nature07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 1957;11:118–130. [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell. 2010;18:463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011;21:212–218. doi: 10.1016/j.tcb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T. Transcriptional Activation of Arabidopsis Axis Patterning Genes WOX8/9 Links Zygote Polarity to Embryo Development. Dev. Cell. 2011;20:264–270. doi: 10.1016/j.devcel.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich JR, Möller BK, Uddin B, Radoeva T, Lokerse AS, De Rybel B, Weijers D. A set of domain-specific markers in the Arabidopsis embryo. Plant Reprod. 2015;28:153–160. doi: 10.1007/s00497-015-0266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161:1066–1075. doi: 10.1104/pp.112.211763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.