Abstract
Oxidants and canonical Wnt/β-catenin signaling have been shown to decrease cardiac Na+ channel activity by suppressing NaV1.5 expression. Our aims are to determine if hydrogen peroxide (H2O2), one oxidant of reactive oxygen species (ROS), activates Wnt/β-catenin signaling and promotes β-catenin nuclear activity, leading to suppression of NaV1.5 expression and if this suppression requires the interaction of β-catenin with its nuclear partner, TCF4 (also called TCF7L2) to decrease SCN5a promoter activity. The results demonstrated that H2O2 increased β-catenin, but not TCF4 nuclear localization determined by immunofluorescence without affecting total β-catenin protein level. Furthermore, H2O2 exerted a dose- and time-dependent suppressive effect on NaV1.5 expression. RT-PCR and/or Western blot analyses revealed that overexpressing active form of β-catenin or stabilizing β-catenin by GSK-3β inhibitors, LiCl and Bio, suppressed NaV1.5 expression in HL-1 cells. In contrast, destabilization of β-catenin by a constitutively active GSK-3β mutant (S9A) upregulated NaV1.5 expression. Whole-cell recording showed that LiCl significantly inhibited Na+ channel activity in these cells. Using immunoprecipitation (IP), we showed that β-catenin interacted with TCF4 indicating that β-catenin as a co-transfactor, regulates NaV1.5 expression through TCF4. Analyses of the SCN5a promoter sequences among different species by using VISTA tools indicated that SCN5a promoter harbors TCF4 binding sites. Chromatin IP assays demonstrated that both β-catenin and TCF4 were recruited in the SCN5a promoter, and regulated its activity. Luciferase promoter assays exhibited that β-catenin inhibited the SCN5a promoter activity at a dose-dependent manner and this inhibition required TCF4. Small interfering (Si) RNA targeting β-catenin significantly increased SCN5a promoter activity, leading to enhanced NaV1.5 expression. As expected, β-catenin SiRNA prevents H2O2 suppressive effects on both SCN5a promoter activity and NaV1.5 expression. Our findings indicate that H2O2 inhibits NaV1.5 expression by activating the Wnt/β-catenin signaling and β-catenin interacts with TCF4 to transcriptionally suppress cardiac NaV1.5 expression.
Keywords: Oxidant, hydrogen peroxide, β-catenin, TCF4, SCN5a promoter, NaV1.5
1. Introduction
NaV1.5 encoded by the SCN5a gene, the main voltage-gated Na+ channel in the heart of humans and animals, determines cardiac electrical excitability and conduction [1]. Studies have uncovered that altered expression and/or dysfunction of Na+ channel account for arrhythmias in patients with ischemic heart disease (IHD), heart failure, and other heart diseases [1,2]. Both NF-κB and FoxO1 suppress SCN5a promoter activity, leading to the down-regulation of NaV1.5 expression [3,4], indicating that these two signaling pathways play important roles in the regulation of Na+ channel activity in heart diseases.
The canonical Wnt (Wnt/β-catenin) signaling pathway is involved in various physiological and pathological processes, including embryonic development, apoptosis, differentiation, cell cycle arrest, and oxidant stress [5,6]. In the absence of Wnt stimulus, cytoplasmic β-catenin forms the “destruction complex” with APC/Axin/CK-1a/GSK-3β and is then phosphorylated and ubiquitinated [7]. When the Wnt signaling is activated, the β-catenin destruction complex is disassembled, which leads to the stabilization of β-catenin. The stabilized β-catenin accumulates in the nucleus where it interacts with the T-cell factor/lymphoid enhancer factor (TCF/LEF) to transcriptionally regulate gene expression [8]. It has been reported that Wnt/β-catenin activation is associated with a wide range of diseases [9,10]. The Wnt/β-catenin signaling is usually inactivated in the health hearts, but it is activated under abnormal conditions such as cardiac hypertrophy and heart failure [9–11] and myocardial infarction [12]. Wnt ligand stimulation has been shown to decrease Na+ channel activity by suppressing NaV1.5 expression in the neonatal cardiomyocytes of the rat [13,14], but its underlying mechanisms have not been determined.
Hydrogen peroxide (H2O2), one oxidant of reactive oxygen species (ROS) is elevated in the IHD [15] and inhibits cardiac Na+ channel activity by suppressing NaV1.5 expression [4]. H2O2 also activates the Wnt/β-catenin signaling [16], which suggests that H2O2 has a potential role in the regulation of NaV1.5 expression through Wnt/β-catenin signaling.
Presently, we determined the effects of activation of Wnt/β-catenin signaling by H2O2 on NaV1.5 expression. Our studies showed that activation of Wnt/β-catenin signaling by H2O2 inhibits NaV1.5 expression, and TCF4 (also called TCF7L2) is required for β-catenin inhibition of NaV1.5 expression at the transcriptional level. These findings provide new insights into the mechanisms underlying transcriptional suppression of NaV1.5 expression by oxidants.
2. Methods
2.1 Cell Culture
HL-1 cells from the AT-1 mouse atrial cardiomyocyte tumor lineage were kindly provided by Dr. William C. Claycomb at the Louisiana State University Health Sciences Center, New Orleans, USA. These cells were cultured as described previously [17]. HeLa cells (American Type Culture Collection, Manassas, VA, USA) were maintained in 10 cm plates containing Dulbecco’s modified Eagle’s medium (high glucose), 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin.
2.2 Western blotting
Western blots were performed on the proteins extracted from cultured cells and mouse ventricular tissue taken from 10- to 12-week-old adult mouse hearts. The use of mouse heart tissues was authorized by the Chancellor’s Animal Research Committee at the University of California, Los Angeles (ARC # 2013-090-01A) in the studies Thirty micrograms of protein were equally loaded on 8–10% SDS polyacrylamide gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk for 1 hour at room temperature, and then incubated with the indicated primary antibody overnight at 4°C. After one-hour incubation with the appropriate secondary antibody, the specific signals were revealed by the chemiluminescence detection reagent Western lightning plus-ECL. Primary antibodies used in this study include rabbit anti-β-catenin monoclonal (1:1000) (BD Transduction Laboratories™, San Jose, CA, USA),rabbit anti-NaV1.5 polyclonal (1:500) (Alomone Labs, Jerusalem, Israel) and rabbit anti-GSK-3β monoclonal (1:500) anti-TCF4 monoclonal (1:500) (Cell Signaling Technology, Inc. Danvers, MA, USA).
2.3 RT-PCR and Real-Time PCR
Using RNeasy Fibrosis Tissue kit (QIAGEN, Valencia, CA, USA) and following the manufacturer’s instruction, total RNA was extracted from HL-1 cardiomyocytes. The RNA sample was then reversely transcribed to the first-strand cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). The level of β-catenin and NaV1.5 mRNA was measured by using TakaRa EmeraldAMP GT PCR Master Mix (Takara BIO INC. Otsu, Shiga, Japan) or (SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA, USA) with GAPDH as an internal control. Real-time PCR was performed on a 7500 FAST Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The primer sequences used to amplify the cDNA of these genes are as shown in Supplementary Table 1.
2.4 ChIP Assay
Adult mouse ventricular tissues were subjected to the ChIP assay using anti-β-catenin, TCF4 antibodies, or rabbit IgG as a control and the ChIP assay kit materials (Upstate Biotechnology, Lake Placid, NY, USA). The immunoprecipitates were analyzed by immunoblot analysis using anti-β-catenin (BD Transduction Laboratories™, San Jose, CA, USA) and TCF4 (Cell Signaling Technology, Danvers, MA, USA) antibodies and by PCR assay to detect for coimmunoprecipitated DNA using a pair of SCN5a promoter-specific primers (forward 5′-TGGTACATACCGTTTCAGGAC-3′ and reverse 5′-GCACACACACTCACACATAC-3′) that flank the consensus TCF4 binding sites (−1548 to −1473 nt) and a pair of primers (forward 5′-GCCTCATTCGTTGCAGTGCC-3′ and reverse 5′-GCCAGTGCCTTGTGTGGACTCT-3′) that flanked the DNA sequence (−4453 to −4686 nt) lacking TCF4 binding sites in the mouse SCN5a promoter.
2.5 Luciferase promoter Assays
Lipofectamine™ and plus™ reagents (Invitrogen, Carlsbad, CA, USA) were used to transfect SNC5a reporters (SCN5a promoter-Luc reporter plasmid [18] generously provided by Dr. Hideko Kasahara at the University of Florida College of Medicine, Florida, USA), TCF4 siRNA (Santa Cruz Bio., Inc., Dallas, Texas, USA) in the HeLa cells. A Renilla reporter gene (Promega, Madison, WI, USA) was included as an internal control. Cells were seeded in 24-well plates and grew to 90% confluence. Transfections were performed according to the manufacturer’s instructions. Cell lysates were prepared for the luciferase assay 48 hours after transfection as described by the manufacturer (Promega, Madison, WI, USA). Data were analyzed and expressed as the normalized-fold changes to controls.
2.6 Whole cell voltage-clamp recording
Whole-cell voltage clamp recording techniques were used to record Na+ currents in HL-1 cardiomyocytes. The methods used were described previously [4,19]. All whole-cell recordings were conducted at room temperature (22–24°C) using a patch clamp amplifier (Axopatch 200B; Axon Instruments Inc., Foster City, CA, USA). The bath solution for recording membrane currents contained (in mM) 145 NaCl, 4.5 CsCl, 1.5 MgCl2, 1 CaCl2, 5 HEPES, 5 glucose, 0.1 CdCl2 (pH 7.35 with CsOH); the pipette solution contained (in mM) 10 NaF, 110 CsF, 20 CsCl, 10 EGTA, and 10 HEPES (pH 7.35 with CsOH). Electrode resistance ranged from 2.0 to 3.0 MΩ. The experiments were controlled by using pClampex 10.3 software (Axon Instruments Inc., Foster City, CA, USA). Currents were acquired at 20–50 kHz (Digidata 1200 A/D converter; Axon Instruments Inc., Foster City, CA, USA), low-pass-filtered at 5 kHz, and stored on a computer. In all recordings, 80% of the series resistance was compensated. Data analysis was accomplished by using pClampfit 10.3 software (Axon Instruments Inc., Foster City, CA, USA).
2.7 Immunoprecipitation
Total protein was extracted from adult mouse ventricular tissue by using modified RIPA buffer (mmol/L) 50 Tris-HCl, pH 7.4; 150 NaCl; 1 EDTA; 1% Nonidet P-40; 0.1% sodium dodecyl sulfate (SDS); 0.25% Sodium Deoxycholate; 1:100-diluted protease inhibitor cocktail (Sigma, St. Louis, MO, USA); 1 PMSF; and 10 NEM. Immunoprecipitation with mouse monoclonal anti-β-catenin was performed per the manufacturer’s instructions of EZview™ Red Protein G Affinity Gel (Sigma-Aldrich, St. Louis, MO, USA). Proteins were loaded on 10% SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane. The membranes were blocked with 5% non-fat milk for 1 hour at room temperature, and then incubated with the indicated primary antibody overnight at 4°C. After one-hour incubation with the appropriate secondary antibody, the specific signals were revealed by the chemiluminescence detection reagent Western lightning plus-ECL (PerkinElmer, Inc., Waltham, MA, USA) according to the manufacturer’s instruction. Primary antibodies used in this study include rabbit anti-β-catenin monoclonal (1:1000) (BD Transduction Laboratories™, San Jose, CA, USA) and rabbit anti-TCF4 antibody (1:500) (Cell Signaling Technology, Danvers, MA, USA).
2.8 Immunofluorescence
HL-1 cells were dispersed on the glass in the center of the 32 mm dishes and grew to 80% confluence. PBS was used to wash these cells for three times. These cells were immersed in 10% normal goat serum for 60 min to block non-specific binding, and then incubated overnight at 4°C with primary antibody against TCF4 (1:500) (Millipore, Billerica, MA, USA). After washing 6 times with PBS, secondary antibody labeled with DyLight-555 was added. Nuclear counterstain was performed with 4′, 6-Diamidino-2-Phenylindole (DAPI, Sigma-Aldrich, St. Louis, MO, USA).
HL-1 cells expressing β-catenin-GFP were fixed with 4% paraformaldehyde. Nuclear profiles were revealed with a brief incubation in DAPI prior to microscopic observation. Fluorescence was visualized with an inverted ZEISS fluorescence confocal microscope. All imaging was processed in an identical manner to faithfully capture the real time images of each sample.
2.9 Statistics
Group data are presented as mean ± SEM. The statistical significance of differences was assessed by using ANOVA one way analysis or a two-tailed Student t-test and p<0.05 was taken to indicate a statistically significant difference.
3. Results
3.1 H2O2 induced Wnt/β-catenin activation and suppressed NaV1.5 expression in a dose- and time-dependent manner in HL-1 cardiomyocytes
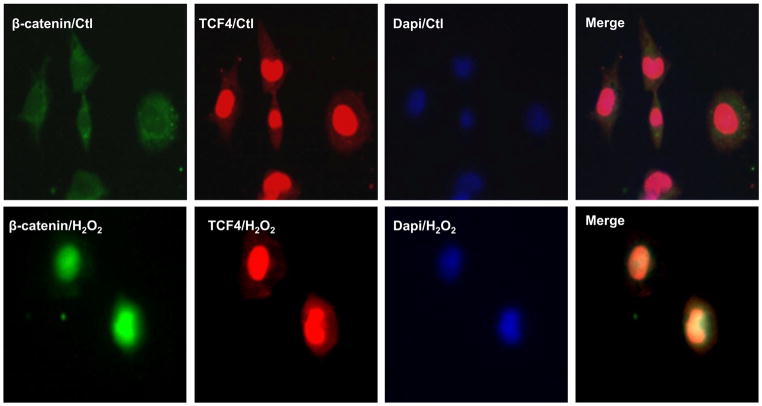
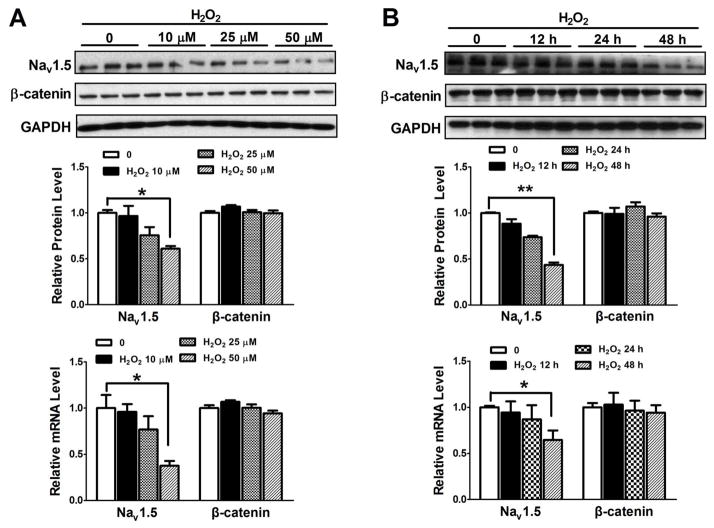
H2O2, one of oxidants, is elevated in the heart after myocardial infarction, and is responsible for structural and electrical cardiac disorders [20]. Therefore, we tested if 50 μM H2O2, which mimics oxidant stress in vivo, was able to induce activation of Wnt/β-catenin signaling and NaV1.5 reduction in HL-1 cardiomyocytes. IF staining showed that treatment with 50 μM H2O2 for one hour led to the accumulation of β-catenin in the nuclei of HL-1 cardiomyocytes (Figure 1) while TCF4 was unaltered (Figure 1). Western blot analysis displayed that 0, 10 μM, 25 μM and 50 μM H2O2 treatment resulted in the significant reduction of NaV1.5 protein in a dose-dependent manner, while the expression of β-catenin was not significantly affected in HL-1 cells (Figure 2A). NaV1.5 mRNA was also significantly reduced by these different concentrations of H2O2 in HL-1 cells (Figure 2A). Western blot analysis was unable to detect the difference of cleaved caspase-3 between 50 μM H2O2 and the control group (Supplementary Figure 1), indicating that the suppression of NaV1.5 expression by this concentration is not due to the cell apoptosis. Moreover, both protein and mRNA levels of NaV1.5 expression were significantly decreased in the HL-1 cells exposed to 50 μM H2O2 for 0, 12 hours, 24 hours in a time-dependent manner while β-catenin expression was not changed (Figure 2B). The relative values of NaV1.5 mRNA to GAPDH were obtained from the real-time PCR amplification curves. As an example shown in Supplemental Figure 2, the amplification curves represented GAPDH and NaV1.5 mRNA levels at the different H2O2 treatment time durations. We also tested if another oxidant, 3-morpholonosydnonimine (SIN-1), which can generate peroxynitrite [21] has the similar effect of H2O2 on NaV1.5 expression in HL-1 cells. As shown in supplemental Figure 2, SIN-1 at different concentrations (0, 2 μM, 10 μM and 20 μM) and durations of culture (0, 24 hours and 48 hours) had no effects on NaV1.5 expression at both protein and mRNA levels (Supplemental Figure 3). These findings indicated that not all the oxidants have effects on NaV1.5 expression.
Figure 1. H2O2 induced β-catenin nuclear localization but TCF4 was not affected.
HL-1 cardiomyocytes were infected with β-catenin-GFP adenovirus for 24 hours, treated with 50 μM H2O2 for 1 hour, and observed by confocal microscopy. β-catenin-GFP adenovirus successfully infected the HL-1 cells and 50 μM H2O2 promoted β-catenin-GFP (green) accumulated in the nuclei. TCF4 were stained with Texas red (red). TCF4 accumulated in the nuclei at the baseline and did not change after 50 μM H2O2 treatment in HL-1 cells. n=3 batches of cells from 3 independent experiments carried on separate occasions for each group.
Figure 2. Dose- and time-dependent effects of H2O2 on NaV1.5 expression in HL-1 cardiomyocytes.
(A) Western blot and real-time PCR were performed to evaluate the expression of β-catenin and NaV1.5 in HL-1 cells after 0, 10 μM, 25 μM or 50 μM H2O2 treatment for 48 hours. NaV1.5 protein and mRNA levels were significantly decreased (p<0.05) with H2O2 in does-dependent manner but β-catenin expression was not significantly affected. (B) Western blot and real time-PCR were performed to evaluate the expression of β-catenin and NaV1.5 expression in HL-1 cells after 50 μM H2O2 treatment for 0, 12 hours, 24 hours or 48 hours. Western blot and real-time PCR showed that NaV1.5 protein and mRNA were significantly decreased with time-dependent manner (p<0.01 and p<0.05) after treatment but β-catenin protein was not significantly altered. *p<0.05 and ** p<0.01; n=3 batches of cells from 3 independent experiments carried on separate occasions for each group.
3.2 Activation of Wnt/β-catenin inhibited NaV1.5 expression in HL-1 cardiomyocytes
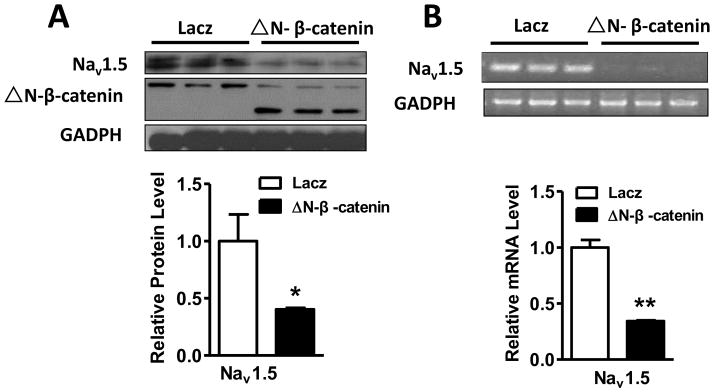
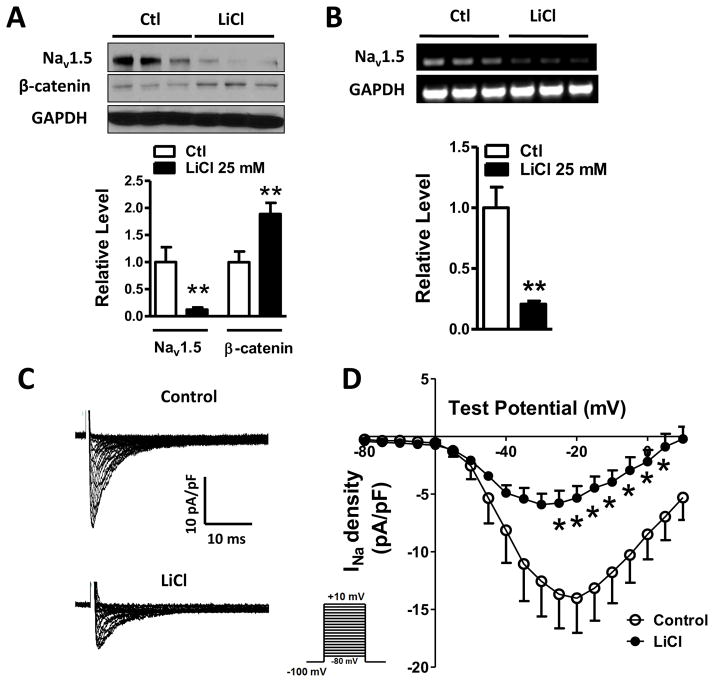
An active form of β-catenin (ΔN-β-catenin, N-terminal deletion of 134 amino acids) lacking GSK-3β phosphorylation sites has frequently been used to mimic the activation of Wnt/β-catenin [22]. Therefore, we infected HL-1 cells with ΔN-β-catenin adenovirus to determine if activation of Wnt/β-catenin affects NaV1.5 expression. Indeed, we found that ΔN-β-catenin significantly suppressed NaV1.5 expression at both protein (p<0.05) and mRNA levels (p<0.01) (Figure 3A and B). Alternatively, we activated the Wnt/β-catenin signaling pathway by two GSK-3β inhibitors, LiCl [23] and Bio [24], in HL-1 cardiomyocytes, and then, determined the NaV1.5 expression. As shown in Figure 4A and B, HL-1 cardiomyocytes treated with 25 mM LiCl for 48 hours had significantly increased β-catenin protein level (p<0.01), and decreased expression of NaV1.5 at both protein and mRNA levels in HL-1 cardiomyocytes as compared to the controls (p<0.01). Bio at a concentration of 5 μM significantly decreased NaV1.5 protein levels in HL-1 cardiomyocytes (p<0.05) (Supplementary Figure 4).
Figure 3. NaV1.5 expression was downregulated by ΔN-β-catenin in HL-1 cells.
HL-1 cells were infected with adenovirus-ΔN-β-catenin and adenovirus-lacz for 48 hours. (A) Western blot was performed on the protein extract from these cells. The result showed that expression of ΔN-β-catenin, a stabilized form of β-catenin significantly suppressed the expression of NaV1.5 protein (p<0.05), compared to the lacz control group. (B) RT-PCR analysis was performed on the total RNA extracted from these cells and the results showed that the mRNA level of NaV1.5 was significantly decreased by ΔN-β-catenin (p<0.01) compared to the lacz control group. *p<0.05 and ** p<0.01; n=3 batches of cells from 3 independent experiments carried on separate occasions for each group.
Figure 4. LiCl inhibited cardiac Na+ channel activity by suppressing NaV1.5 expression through activating Wnt/β-catenin signaling.
(A) Exposure of HL-1 cardiomyocytes to 25 mM LiCl led to a significant increase in β-catenin expression (p<0.01), compared to the control group (p<0.01), accompanied by a significant decrease of NaV1.5 protein (p<0.01). (B) RT-PCR was performed to detect the effect of 25 mM LiCl on NaV1.5 mRNA. NaV1.5 mRNA level was significantly decreased in HL-1 cells (p<0.01) after 25 mM LiCl treatment, compared to the control group. In A and B, Ctl: control; n=3 batches of cells from 3 independent experiments carried on separate occasions for each group. (C) INa currents were recorded in HL-1 cardiomyocytes with or without LiCl treatment. 25 mM LiCl decreased Na+ current densities HL-1 cells. (D) The current-voltage relationship curve of Na+ currents showed that the current densities obtained from peak currents divided by individual cell capacitances were significantly decreased from −25 mV to +5 mV in HL-1 cells (p<0.05), compared to the control group. In C and D, n=7 in the control group and n=7 in the LiCl group. In A, B and D, *p<0.05 and **p<0.01.
3.3 Activation of Wnt/β-catenin signaling pathway inhibited Na+ channel activity in HL-1 cardiomyocytes
To further define the role of Wnt/β-catenin signaling pathway in the regulation of Na+ channel activity, we used LiCl, to treat the HL-1 cells and determine Na+ channel activity. After HL-1 cells were treated with LiCl 25 mM for 48 hours, the whole-cell patch clamp recording was performed to record Na+ currents, and it showed that Na+ currents were largely reduced (Figure 4C). The peak Na+ current densities were significantly decreased at the voltages from −25 mV to 5 mV compared with the control group (Figure 4D).
3.4 Inhibition of Wnt/β-catenin increased NaV1.5 expression in HL-1 cardiomyocytes
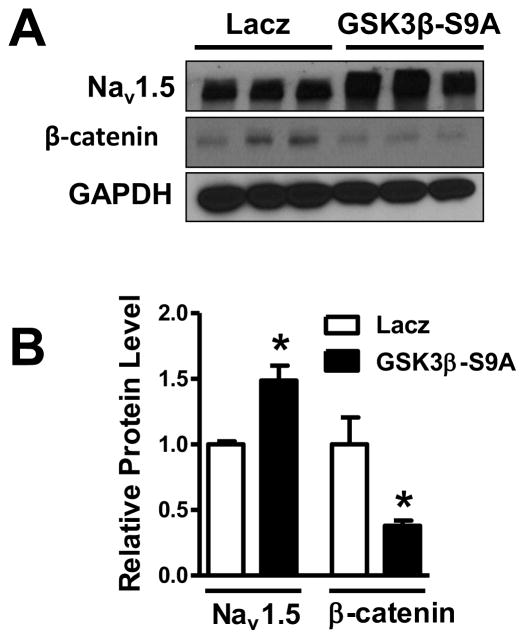
Then, we observed the effects of inhibiting Wnt/β-catenin signal on the expression of NaV1.5 in HL-1 cardiomyocytes. HL-1 cells were infected with adenovirus-lacz and adenovirus-GSK-3β-S9A, respectively, for 48 hours. As expected, overexpression of GSK-3β-S9A, a constitutively active form of GSK-3β with a point mutation at serine 9 [25] in HL-1 cardiomyocytes significantly decreased β-catenin protein (p<0.05) and significantly increased the expression of NaV1.5 protein (p<0.05), compared to the lacz control group (Figure 5A and B). GSK-3β was significantly increased in the cells infected with adenovirus-GSK-3β-S9A (p<0.01), compared to the lacz control group, indicating that GSK-3β-S9A was expressed in the HL-1 cells after infection of GSK-3β-S9A (Supplemental Figure 5).
Figure 5. NaV1.5 expression was upregulated by GSK-3β-S9A in HL-1 cells.
HL-1 cells were infected with adenovirus-lacz and adenovirus-GSK-3β-S9A, respectively, for 48 hours. (A, B) Expression of GSK-3β-S9A (mutated active form of GSK-3β) led to a significant increase of NaV1.5 protein (p<0.05) and was accompanied by significantly decreased β-catenin (p<0.05), compared to the lacz control group. In B, *p<0.05; Ctl: control; n=3 batches of cells from 3 independent experiments carried on separate occasions for each group.
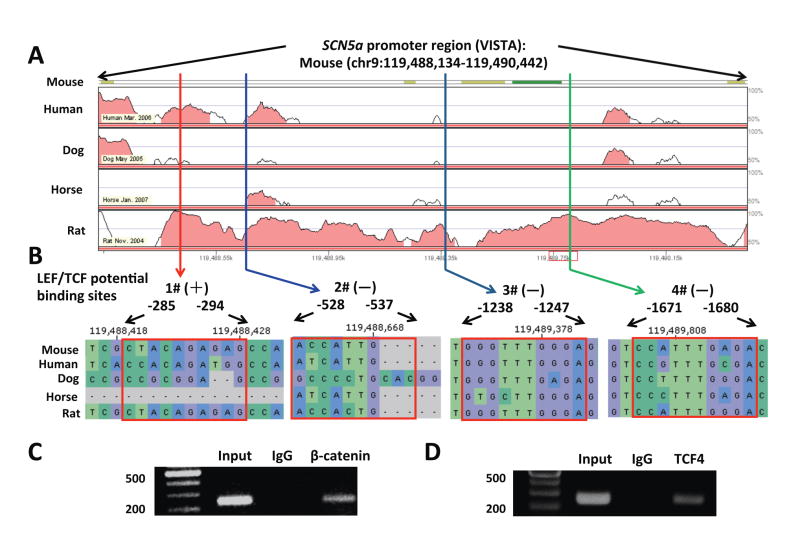
3.5 Conservative TCF4 DNA binding sites in the SCN5a promoter
Analysis by using VISTA tools revealed that the proximal region (2308 bp upstream) of the SCN5a promoter harbors four TCF4 binding sequences (Figure 6A). These sequences are conservative among human, rat, and mouse (Figure 6B), indicating that β-catenin is likely recruited in the proximal region of the SCN5a promoter after binding to TCF4.
Figure 6. Conservative TCF4 DNA binding sites in the SCN5a promoter.
(A) The proximal region (2308 bp upstream) of SCN5a promoter harbors four TCF4 binding sequences. (B) VISTA plot showing the sequences of these four potential binding sites (#1, #2, #3 and #4) in multiple species, including mouse, rat, dog, horse and human. #3 and #4 are more conservative than #1 and #2 potential binding sites.
3.6 β-catenin and TCF4 formed a complex and were recruited in the SCN5a promoter
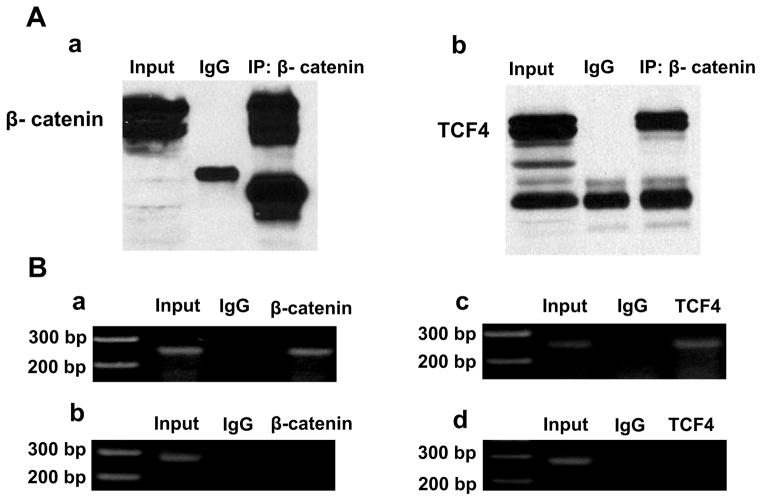
Total protein was extracted from adult mouse ventricular tissues. Antibody against β-catenin was used to do IP; Western blot analysis was performed following IP. As shown in the Figure 7A, antibodies against β-catenin or TCF4 were able to pull down both β-catenin and TCF4, indicating that β-catenin interacts TCF4 in the mouse heart tissues.
Figure 7. β-catenin and TCF4 formed a complex and were recruited in the SCN5a promoter.
(A) Immunoprecipitation (IP) was performed on the total protein extracted from the adult mouse ventricular tissues by using an antibody against β-catenin and Western blot was performed to show this antibody was able to pull down β-catenin itself (a) and TCF4 (b). The results indicated that β-catenin interacts with TCF4. (B) ChIP assay was performed by using antibodies against β-catenin and TCF4, and IgG as control on the adult mouse ventricular tissues. A pair of flanking DNA primer sequences containing (a and c) or lacking TCF4 binding sites (b and d) was used. The results demonstrated that both β-catenin and TCF4 were recruited within the SCN5a promoter region.
Chromatin immunoprecipitation (ChIP) assays were used to determine if both β-catenin and TCF4 are recruited in the SCN5a promoter. As shown in Figure 7B, both β-catenin and TCF4 were recruited in the region of SCN5a promoter in mouse ventricular tissues.
3.7 TCF4 was required for β-catenin inhibition of SCN5a promoter activity
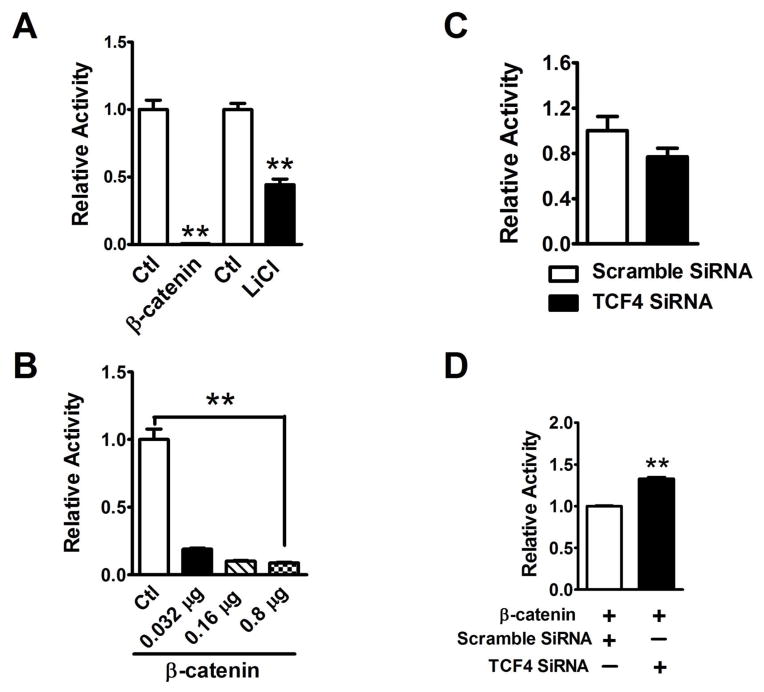
To clarify the transcriptional regulation of NaV1.5 by β-catenin, luciferase promoter activity assay was performed. The results revealed that both β-catenin overexpression and LiCl significantly inhibited the SCN5a promoter activity as compared to the control (p<0.01) (Figure 8A). β-catenin inhibition of SCN5a promoter activity was in a concentration-dependent manner (Figure 8B). β-catenin does not have a DNA binding domain. It functions as a co-transcriptional factor to regulate the target gene expression through TCF4 transcriptional factor [26]. TCF4 SiRNA was used to confirm that β-catenin inhibition of SCN5a promoter activity requires TCF4. In HeLa cells, knockdown of TCF4 by specific targeting SiRNA (Supplementary Figure 6) did not affect the SCN5a promoter activity (Figure 8C), compared with the scrambled SiRNA. However, the SCN5a promoter activity was significantly increased (p<0.01) by TCF4 siRNA in the presence of β-catenin, indicating that β-catenin inhibition of SCN5a promoter activity is dependent on the presence of TCF4 (Figure 8D).
Figure 8. Knockdown of TCF4 attenuated β-catenin inhibition of SCN5a promoter activity.
(A) Both β-catenin overexpression and LiCl significantly inhibited the SCN5a promoter activity (p<0.01), compared to the control groups, respectively. (B) Luciferase promoter assays revealed that β-catenin significantly inhibited SCN5a promoter activity in a concentration-dependent manner (p<0.01). (C) Knockdown of TCF4 by 100 nM SiRNA did not significantly affect the activity of SCN5a promoter, compared to the 100 nM scramble SiRNA group. (D) In the presence of β-catenin, knockdown of TCF4 by SiRNA significantly increased the SCN5a promoter activity (p<0.01), compared to the β-catenin and scramble SiRNA co-transfection group. **p<0.01; Ctl: control; n=3 batches of HeLa cells from 3 independent experiments carried on separate occasions for each group.
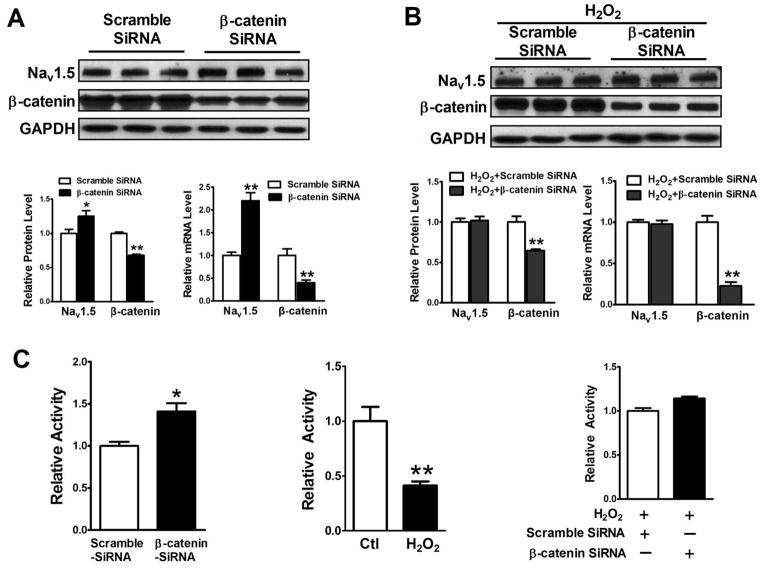
3.8 H2O2 suppression of NaV1.5 expression requires β-catenin
We have found that H2O2 activated Wnt/β-catenin signaling, and decreased NaV1.5 expression. In order to pin down H2O2 suppressive effect on NaV1.5 expression requiring β-catenin, we set up experiments to determine if β-catenin SiRNA prevented H2O2 to inhibit both NaV1.5 expression and SCN5a promoter activity. Total protein and RNA were extracted from HL-1 cells treated with β-catenin and scramble SiRNA for 48 hours or from HL-1 cells treated with β-catenin SiRNA and 50 μM H2O2 in the culture medium for 48 hours. Western blot and real-time PCR were performed. As shown in Figure 9A, SiRNA specifically targeting β-catenin significantly decreased β-catenin and increased NaV1.5 expression at both protein and mRNA levels, compared to the scramble SiRNA group; as illustrated in Figure 9B, H2O2 suppression of NaV1.5 protein and mRNA expression was prevented by β-catenin SiRNA. Luciferase promoter assays were performed on the protein extracts from the Hela cells transfected with SCN5a promoter-Luc reporter plasmid and SiRNA-β-catenin or scramble SiRNA, or from Hela cells transfected with SCN5a promoter-Luc reporter plasmid in the presence or absence of 50 μM H2O2 in the culture medium for 48 hours. As shown in Figure 9C, SiRNA-β-catenin significantly increased SCN5a promoter activity, compared to scramble SiRNA and H2O2 significantly decreased SCN5a promoter activity, compared to the control group; H2O2 inhibitory effect on the SCN5a promoter activity was completely abolished by β-catenin SiRNA.
Figure 9. Knockdown of β-catenin by SiRNA prevented a decrease of both NaV1.5 expression and SCN5a promoter activity induced by H2O2.
HL-1 cells were transfected with 100 nM β-catenin and scramble SiRNA, respectively, for 48 hours without or with 48 hours 50 μM H2O2 treatment. Total Protein and RNA were extracted from these cells. Western blot and real time-PCR were performed. (A) 100 nM β-catenin SiRNA significantly knocked down β-catenin expression at both protein and mRNA levels (p<0.01) and significantly increased NaV1.5 protein and mRNA (p<0.01), compared to the 100 nM scramble SiRNA group. (B) Knockdown of β-catenin completely abolished suppressive effects of 50 μM H2O2 for 48 hours on NaV1.5 expression at both protein and mRNA levels. (C) Luciferase promoter assays were performed on the protein extracts from transfected Hela cells treated with β-catenin or scramble SiRNA or treated with or without 50 μM H2O2 for 48 hours. The results showed that β-catenin SiRNA significantly increased SCN5a promoter activity (p<0.05), compared to the scramble SiRNA control group and that H2O2 significantly inhibited SCN5a promoter activity (p<0.01), compared to the control group. In order to determine if H2O2 effects on the SCN5a promoter activity is related to β-catenin, luciferase promoter assays were performed on the protein extracts from Hela cells transfected with β-catenin or scramble SiRNA and these cells were cultured in the medium containing 50 μM H2O2. The results showed that b-catenin SiRNA completely abolished the H2O2 suppressive effects, compared to the scramble group. *p<0.05 and **p<0.01; n=3 batches of HL-1 or HeLa cells from 3 independent experiments carried on separate occasions for each group.
4. Discussion
The results presented here reveal that the suppression of cardiac NaV1.5 expression caused by activation of Wnt/β-catenin signaling required the presence of TCF4. H2O2, an oxidant, was able to lead to β-catenin nuclear translocation and subsequent suppression of NaV1.5 expression by inhibiting SCN5a promoter activity. The findings provide new insights into the molecular mechanisms of Wnt/β-catenin signaling transcriptionally regulating NaV1.5 expression and H2O2 suppressing NaV1.5 expression. Another oxidant, SIN-1 which can generate peroxynitrite [21] did not affect NaV1.5 expression, indicating that not all oxidants have effects on NaV1.5 expression.
Activation of Wnt/β-catenin signaling by directly manipulating β-catenin cellular levels inhibiting cardiac Na+ channel activity by suppressing NaV1.5 expression was first reported from our laboratory in American Heart Association scientific session 2013 [13]. Recently, a study by Liang W, et al [14] showed that activation of this signaling pathway by Wnt ligand stimulation significantly reduced mRNA and proteins levels of NaV1.5 as well as Na+ channel activity in neonatal rat ventricular myocytes. Consistent with the reduction of Na+ channel activity, activation of Wnt/β-catenin signaling led to the alteration of action potentials with a decrease of the upstroke amplitude and maximum upstroke velocity in these cells [14]. Our study showed that the activation of Wnt/β-catenin signaling induced the suppression of NaV1.5 expression which required TCF4 directly binding to the SCN5a promoter. TCF4 functions as a suppressor in the regulation of NaV1.5 expression. H2O2 did not affect total β-catenin expression, but it was able to promote β-catenin nuclear localization to supress NaV1.5 expression at the transcriptional level. The mechanism of H2O2 augmenting Wnt/β-catenin signaling was proposed by Funato Y, et al [16]. The cytoplasmic proctein Dishevelled (Dvl) usually inteacts with Axin and Frat1/GBP to inhibit GSK3β mediated phosphorylation of β-catenin [27]. This function of Dvl depends on its binding to thrioredoxin-like protein nucleoredoxin [16]. The studies by Funato Y, et al showed that H2O2 inhibted the interaction between thrioredoxin-like protein nucleoredoxin and Dvl, leading to activation of Wnt/β-catenin signaling by increasing nuclear non-phosphorylated β-catenin level [16]. The increase of nuclear non-phosphorylated β-catenin often does not lead to measurable change at total protein levels.
It has been reported that H2O2 suppressed NaV1.5 expression by inhibiting SCN5a promoter activity through NF-κB and FoxO1 signaling pathways [3,4]. We sought to determine if cellular localization of β-catenin protein is altered by ROS and found that 50 μM H2O2 did not trigger the apoptosis of HL-1 cardiomyocytes, but did induce β-catenin accumulation in the nuclei of these cells to suppress SCN5a promoter activity, subsequently leading to the suppression of NaV1.5 expression. β-catenin SiRNA was able to completely abolish the effects of H2O2 on SCN5a promoter activity and NaV1.5 expression. These findings indicate that NF-κB and FoxO1 signaling pathways affecting NaV1.5 expression may be dependent on the activation of Wnt/β-catenin signaling. Indeed, it has been reported that oxidative stress promotes interaction of β-catenin and FoxO that enhances the expression of the target genes [18].
Canonic Wnt/β-catenin signaling is usually silenced in normal adult hearts, but is activated under pathological conditions such as cardiac hypertrophy and heart failure [9–11] and myocardial infarction [12]. Epidemiologic data indicate that ischemic heart disease (IHD), a leading cause of death worldwide, causes 80% of fatal arrhythmias, such as ventricular tachycardia and ventricular fibrillation [28–31]. Abnormal Na+ channel activity affects cardiac excitability and electrical conduction and it has been linked to cardiac arrhythmias in IHD [1]. The expression of NaV1.5, α subunit of the cardiac Na+ channel is significantly decreased in humans with end-stage heart failure due to chronic ischemic and dilated cardiomyopathies [1,2], in the epicardial border zone of the infarcted canine hearts [32], and in the peri-infarction border zone of mouse hearts [19]. However, the mechanisms regulating the Na+ channel expression in IHD, in particular at the transcriptional level, are largely unknown. ROS are significantly increased in IHD [15] and play an important role in cardiac dysfunction [33]. To our best knowledge, our studies first linked H2O2 transcriptionally suppressing NaV1.5 expression to Wnt/β-catenin signaling. The modulation of Na+ channel activity by Wnt/β-catenin signaling could emerge as a novel therapeutic target to tackle the highly relevant problem of fatal cardiac arrhythmias in IHD. Our findings were from the in vitro studies and had some limitations. Studies focusing on the role of H2O2 in the regulation of cardiac NaV1.5 expression and Na+ channel activity in animal models will be pursued.
Supplementary Material
Highlights.
Hydrogen peroxide suppresses NaV1.5 expression by activating Wnt/β-catenin signaling pathway
Activation of Wnt/β-catenin signaling suppresses cardiac Na+ channel activity
Activation of Wnt/β-catenin signaling suppresses NaV1.5 expression through TCF4
β-catenin suppresses NaV1.5 expression by TCF4 inhibiting SCN5a promoter activity
Acknowledgments
Funding
HX is supported by a grant from the AHA (12GRNT9690003) and the NIH (R01 HL122793-01A1); FL is supported by a grant from the AHA and the Lawrence J. and Florence A. DeGeorge Charitable Trust (10GRNT4460014) and the NIH (R01 HL111480-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Drs. William C. Claycomb and Hideko Kasahara for kindly providing HL-1 cells and SCN5a promoter-Luc plasmid, respectively.
Abbreviations
- IHD
Ischemic heart disease
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- ROS
reactive oxygen species
Footnotes
Conflict of Interest
None declared
Parts of this study were presented at the American Heart Association (AHA) Scientific Sessions 2013, Dallas, Texas on November 17, 2013, and AHA Scientific Sessions 2014, Chicago, Illinois on November 18, 2014.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Remme CA, Bezzina CR. Sodium channel (dys)function and cardiac arrhythmias. Cardiovasc Ther. 2010;28:287–294. doi: 10.1111/j.1755-5922.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 2.Hesse M, Kondo CS, Clark RB, Su L, Allen FL, Geary-Joo CT, Kunnathu S, Severson DL, Nygren A, Giles WR, Cross JC. Dilated cardiomyopathy is associated with reduced expression of the cardiac sodium channel Scn5a. Cardiovasc Res. 2007;75:498–509. doi: 10.1016/j.cardiores.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Shang LL, Sanyal S, Pfahnl AE, Jiao Z, Allen J, Liu H, Dudley SC., Jr NF-kappaB-dependent transcriptional regulation of the cardiac scn5a sodium channel by angiotensin II. Am J Physiol Cell Physiol. 2008;294:C372–379. doi: 10.1152/ajpcell.00186.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao W, You T, Ye B, Li X, Dong HH, Hill JA, Li F, Xu H. Reactive oxygen species suppress cardiac NaV1.5 expression through Foxo1. PLoS One. 2012;7:e32738. doi: 10.1371/journal.pone.0032738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer S, Ambrogini E, Bartell SM, Han L, Roberson PK, de Cabo R, Jilka RL, Weinstein RS, O’Brien CA, Manolagas SC, Almeida M. FOXOs attenuate bone formation by suppressing Wnt signaling. J Clin Invest. 2013;123:3409–3419. doi: 10.1172/JCI68049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H, Gruenwald A, Suh JH, Miner JH, Barisoni-Thomas L, Taketo MM, Faul C, Millar SE, Holzman LB, Susztak K. Wnt/beta-catenin pathway in podocytes integrates cell adhesion,differentiation, and survival. J Biol Chem. 2011;286:26003–26015. doi: 10.1074/jbc.M111.223164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao TP, Kuhl M. An updated overview on Wnt signaling pathways: a prelude for more. Circ Res. 2010;106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 8.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, Rose D, Li X, Rosenfeld MG. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125:593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 9.Bergmann MW. WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res. 2010;107:1198–1208. doi: 10.1161/CIRCRESAHA.110.223768. [DOI] [PubMed] [Google Scholar]
- 10.Malekar P, Hagenmueller M, Anyanwu A, Buss S, Streit MR, Weiss CS, Wolf D, Riffel J, Bauer A, Katus HA, Hardt SE. Wnt signaling is critical for maladaptive cardiac hypertrophy and accelerates myocardial remodeling. Hypertension. 2010;55:939–945. doi: 10.1161/HYPERTENSIONAHA.109.141127. [DOI] [PubMed] [Google Scholar]
- 11.Cingolani OH. Cardiac hypertrophy and the Wnt/Frizzled pathway. Hypertension. 2007;49:427–428. doi: 10.1161/01.HYP.0000255947.79237.61. [DOI] [PubMed] [Google Scholar]
- 12.Oerlemans MI, Goumans MJ, van Middelaar B, Clevers H, Doevendans PA, Sluijter JP. Active Wnt signaling in response to cardiac injury. Basic Res Cardiol. 2010;105:631–641. doi: 10.1007/s00395-010-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, Cai B, Li X, Lu Y, Ye B, Li F, Xu HD. β-Catenin inhibition of NaV1.5 requires both TCF4 and FoxO1 in cardiomyocytes (abstract) Circulation. 2013;128:A13278. [Google Scholar]
- 14.Liang W, Cho HC, Marban E. Wnt signalling suppresses voltage-dependent Na(+) channel expression in postnatal rat cardiomyocytes. J Physiol. 2015;593:1147–1157. doi: 10.1113/jphysiol.2014.285551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg K, Jynge P, Bjerve K, Skarra S, Basu S, Wiseth R. Oxidative stress and inflammatory response during and following coronary interventions for acute myocardial infarction. Free Radic Res. 2005;39:629–636. doi: 10.1080/10715760400028027. [DOI] [PubMed] [Google Scholar]
- 16.Funato Y, Michiue T, Asashima M, Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signaling through disheveled. Nat Cell Biol. 2006;8:501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- 17.Xia M, Salata JJ, Figueroa DJ, Lawlor AM, Liang HA, Liu Y, Connolly TM. Functional expression of L- and T-type Ca2+ channels in murine HL-1 cells. J Mol Cell Cardiol. 2004;36:111–119. doi: 10.1016/j.yjmcc.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 19.Cai B, Wang N, Mao W, You T, Lu Y, Li X, Ye B, Li F, Xu H. Deletion of FoxO1 leads to shortening of QRS by increasing Na(+) channel activity through enhanced expression of both cardiac NaV1.5 and beta3 subunit. J Mol Cell Cardiol. 2014;74:297–306. doi: 10.1016/j.yjmcc.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grieve DJ, Byrne JA, Cave AC, Shah AM. Role of oxidative stress in cardiac remodelling after myocardial infarction. Heart Lung Circ. 2004;13:132–138. doi: 10.1016/j.hlc.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Agbani E, Coats P, Wadsworth RM. Threshold of peroxynitrite cytotoxicity in bovine pulmonary artery endothelial and smooth muscle cells. Toxicology in Vitro. 2011;25:1680–1686. doi: 10.1016/j.tiv.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Shevtsov SP, Hsich E, Cui L, Haq S, Aronovitz M, Kerkela R, Molkentin JD, Liao R, Salomon RN, Patten R, Force T. The beta-catenin/T-cell factor/lymphocyte enhancer factor signaling pathway is required for normal and stress-induced cardiac hypertrophy. Mol Cell Biol. 2006;26:4462–4473. doi: 10.1128/MCB.02157-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiyama A, Sakai D, Arai F, Nakajima D, Yokoyama K, Mochida J. Effects of a glycogen synthase kinase-3beta inhibitor (LiCl) on c-myc protein in intervertebral disc cells. J Cell Biochem. 2011;112:2974–2986. doi: 10.1002/jcb.23217. [DOI] [PubMed] [Google Scholar]
- 24.Tseng AS, Engel FB, Keating MT. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem Biol. 2006;13:957–963. doi: 10.1016/j.chembiol.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 26.Kavak E, Najafov A, Ozturk N, Seker T, Cavusoglu K, Aslan T, Duru AD, Saygili T, Hoxhaj G, Hiz MC, Unal DO, Birgul-Iyison N, Ozturk M, Koman A. Analysis of the Wnt/B-catenin/TCF4 pathway using SAGE, genome-wide microarray and promoter analysis: Identification of BRI3 and HSF2 as novel targets. Cell Signal. 2010;22:1523–1535. doi: 10.1016/j.cellsig.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Yuan H, Weaver CD, Mao J, Farr GH, 3rd, Sussman DJ, Jonkers J, Kimelman D, Wu D. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myerburg RJ, Interian A, Jr, Mitrani RM, Kessler KM, Castellanos A. Frequency of sudden cardiac death and profiles of risk. Am J Cardiol. 1997;80:10F–19F. doi: 10.1016/s0002-9149(97)00477-3. [DOI] [PubMed] [Google Scholar]
- 29.Myerburg RJ, Kessler KM, Castellanos A. Sudden cardiac death: Epidemiology, transient risk, and intervention assessment. Ann Intern Med. 1993;119:1187–1197. doi: 10.7326/0003-4819-119-12-199312150-00006. [DOI] [PubMed] [Google Scholar]
- 30.Zipes DP, Wellens HJ. Sudden cardiac death. Circulation. 1998;98:2334–2351. doi: 10.1161/01.cir.98.21.2334. [DOI] [PubMed] [Google Scholar]
- 31.Myerburg RJ, Kessler KM, Castellanos A. Sudden cardiac death. Structure, function, and timedependence of risk. Circulation. 1992;85:I2–10. [PubMed] [Google Scholar]
- 32.Baba S, Dun W, Cabo C, Boyden PA. Remodeling in cells from different regions of the reentrant circuit during ventricular tachycardia. Circulation. 2005;112:2386–2396. doi: 10.1161/CIRCULATIONAHA.105.534784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misra MK, Sarwat M, Bhakuni P, Tuteja R, Tuteja N. Oxidative stress and ischemic myocardial syndromes. Med Sci Monit. 2009;15:RA209–219. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.