Abstract
Mitochondria of mammalian cells contain multiple copies of mitochondrial (mt) DNA. Although mtDNA copy number can fluctuate dramatically depending on physiological and pathophysiologic conditions, the mechanisms regulating mitochondrial genome replication remain obscure. Hypoxia, like many other physiologic stimuli that promote growth, cell proliferation and mitochondrial biogenesis, uses reactive oxygen species as signaling molecules. Emerging evidence suggests that hypoxia-induced transcription of nuclear genes requires controlled DNA damage and repair in specific sequences in the promoter regions. Whether similar mechanisms are operative in mitochondria is unknown. Here we test the hypothesis that controlled oxidative DNA damage and repair in the D-loop region of the mitochondrial genome are required for mitochondrial DNA replication and transcription in hypoxia. We found that hypoxia had little impact on expression of mitochondrial proteins in pulmonary artery endothelial cells, but elevated mtDNA content. The increase in mtDNA copy number was accompanied by oxidative modifications in the D-loop region of the mitochondrial genome. To investigate the role of this sequence-specific oxidation of mitochondrial genome in mtDNA replication, we overexpressed mitochondria-targeted 8-oxoguanine glycosylase Ogg1 in rat pulmonary artery endothelial cells, enhancing the mtDNA repair capacity of transfected cells. Overexpression of Ogg1 resulted in suppression of hypoxia-induced mtDNA oxidation in the D-loop region and attenuation of hypoxia-induced mtDNA replication. Ogg1 overexpression also reduced binding of mitochondrial transcription factor A (TFAM) to both regulatory and coding regions of the mitochondrial genome without altering total abundance of TFAM in either control or hypoxic cells. These observations suggest that oxidative DNA modifications in the D-loop region during hypoxia are important for increased TFAM binding and ensuing replication of the mitochondrial genome.
Keywords: Hypoxia, Reactive oxygen species, Oxidative DNA modifications, D-loop, Mitochondrial DNA replication, TFAM binding, Ogg1 overexpression
INTRODUCTION
Reactive oxygen species (ROS) from mitochondria and other endogenous and exogenous sources constantly generated in living cells have the potential to damage lipids, proteins, RNA and DNA [1]. DNA damage, in particular, may trigger cell death pathways or lead to mutations [2]. Pathogenic oxidant stress is directed not only at the nuclear genome; oxidative damage and resulting mutations in the mitochondrial DNA also are linked to a range of human disorders, including aging, cancer, neurodegenerative and cardiovascular diseases [3, 4].
Traditional concepts hold that maintenance of DNA integrity is indispensable to normal cellular physiology; damage must be repaired, or cells die or harbor potentially deleterious mutations. However, there is emerging evidence that, at least for nuclear genes, controlled DNA damage and repair may be necessary for normal transcriptional regulation. For example, in vitro studies on the p50 subunit of the NF-κB transcription factor binding to the NF-κB promoter showed that oxidation of guanine at sites critical for protein recognition increases p50 binding affinity [5]. In MCF-7 cells, activation of estrogen responsive elements in selected genes leads to transient, oxygen radical-mediated DNA strand breaks that appear to be required for long-range changes in DNA topography and increased mRNA expression [6, 7]. We have shown previously that a similar pathway is operative in pulmonary artery endothelial cells (PAECs) where hypoxia, like other physiologic signals using reactive oxygen species as second messengers [8, 9], causes ROS-dependent modifications at specific bases within hypoxia-response elements of hypoxia- inducible genes [10–12]. The oxidative lesions associated with hypoxic signaling are temporally related to mRNA accumulation [11] and restricted to hypoxia-response elements associated with transcriptionally-active nucleosomes [13]. Mimicking the effect of hypoxia by introducing model oxidative base modifications in the hypoxia-response element of the VEGF promoter leads to enhanced sequence flexibility, altered transcription factor binding and more robust reporter gene expression [10, 14]. Collectively, these findings support the concept that controlled, ROS-mediated nuclear DNA damage and repair are associated with normal physiologic signaling and function to alter the topology and flexibility of key promoter sequences thereby facilitating regulatory protein binding and productive transcription [7, 15]. These observations raise an intriguing question relative to the mitochondrial genome: could a similar model for ROS-dependent transcriptional activation be operative in mitochondria?
In mammalian cells, each of the hundreds-to-thousands of mitochondria per cell harbors 2–10 copies of mitochondrial DNA [16]. Increases in the cellular contents of mtDNA can be stimulated by a variety of metabolic and/or pathophysiologic stresses. For example, increased mtDNA content has been reported in aging tissues [17–19], some cancers [20], in cells treated with lipopolysaccharide [21], and in cells treated with non-lethal concentrations of hydrogen peroxide [22]. In many cell types, including lung cells, both hypoxia and hyperoxia can also stimulate an increase in mtDNA content [23–27]. Importantly, a feature common to all of these diverse conditions is increased oxidant stress. However, the mechanism by which oxidant stress stimulates mtDNA replication remains unknown.
As discussed subsequently, multiple lines of indirect evidence suggest that “DNA damage and repair pathway” believed to govern nuclear gene expression also functions to regulate mtDNA replication. In many cell types, oxidant stress leads to upregulated expression of the key transcription factor driving mitochondrial gene expression and mtDNA replication, mitochondrial transcription factor A (TFAM) [28]. TFAM, being the main structural protein of the nucleoid, plays a significant role not only in mitochondrial genome transcription and replication, but also in packaging and repair of the mtDNA [29]. TFAM initiates mitochondrial transcription by binding to a non-coding regulatory sequence known as the D-loop region, which contains light-strand and heavy-strand promoter sequences. While the fine mechanism of mtDNA replication remains controversial, both existing models of this process hold that mtDNA synthesis requires extension of RNA primers, the generation of which is mediated by transcription machinery [30]. Thus, the TFAM-mediated processes of mtDNA transcription and replication are tightly and inextricably linked [31, 32].
The D-loop region is known to be exquisitely sensitive to oxidant stress. In this regard, while mtDNA is about 30-fold more sensitive to ROS-mediated damage than the nuclear genome [33], the few studies focusing on the D-loop region suggest that it is even more prone to oxidant attack than the coding portion of the mitochondrial genome [34, 35]. It has also been reported that incorporation of oxidative base damage products into model oligonucleotides enhances DNA binding affinity for TFAM [36].
Previously, we have successfully shown that hypoxia induces ROS production in rat pulmonary artery endothelial cell culture [8, 9] and that mitochondria are involved in this process [8, 13, 37]. Against this background, the present study tested the hypothesis that controlled oxidative DNA damage and repair in the D-loop region of the mitochondrial genome are required for mtDNA replication and transcription in hypoxia. To address this issue and to decrease hypoxia-induced mtDNA oxidative damage, we used our previously published strategy – targeting human DNA glycosylase Ogg1 (hOgg1), an enzyme executing the first step in base excision repair, to mitochondria of rat pulmonary artery endothelial cells [38]. Oxidative stress can result in a variety of mtDNA lesions, including single-strand breaks, abasic sites and oxidized DNA bases, among which guanine is the base most susceptible to oxidation [39]. Mitochondria employ multiple DNA repair mechanisms to repair ROS-induced damage, but base excision repair (BER) is the primary pathway used to remove oxidatively-modified DNA bases [39, 40]. Several DNA glycosylases responsible for recognition and removal of base lesions during the first step in the BER pathway have been identified in mitochondria, including Ogg1, NEIL1 and NEIL2, MutY homolog MYH, Endo III homolog NTH1, and uracil DNA glycosylase UNG1 [40]. Ogg1 and NEIL enzymes have overlapping specificity to 8-oxoguanine; nevertheless, Ogg1 remains the primary enzyme for the repair of oxidized purines [40, 41]. In our previous work we showed that cells deficient in Ogg1 demonstrated increased mtDNA damage and oxidant-mediated apoptosis, indicating that Ogg1 may be a rate-limiting enzyme in the mitochondrial BER [42]. On the other hand, overexpression of mitochondria-targeted human DNA glycosylase Ogg1 (hOgg1) in rat PAECs and other cell types significantly enhanced DNA repair capacity and protected mtDNA from oxidant-induced damage [38, 43–45]. In the present study we expected that overexpression of mitochondria-targeted hOgg1 would improve mitochondrial DNA repair and, thus, decrease hypoxia-induced oxidative damage to mitochondrial genome. Then we analyzed replication and transcription of mtDNA in control cells and cells transfected with hOgg1 under hypoxic conditions. Abundance of TFAM, as a key mitochondrial transcription factor, and its binding to the mtDNA were also studied.
MATERIALS AND METHODS
Cell culture and treatment
Rat PAECs were harvested and cultured as described previously [42]. Control, “normoxic” cells were cultured in a water-jacketed incubator purged with air + 5% CO2, while “hypoxic” cells were cultured for the indicated periods in an incubator purged with air, N2, and CO2 to create an environment consisting of 2% O2, 5% CO2, and 93% N2.
Ogg1 overexpression and its assessment
We stably transfected rat PAECs with lentivirus to overexpress hOgg1 in mitochondria. Empty lentiviral vector and vector containing hOgg1 gene were prepared as described previously [38]. Transfected cells were cultured on medium containing 10 μg/ml puromycin for 72 hours to exclude cells without lentiviral construct. Transfection was confirmed by real time and conventional RT-PCR analyses, and Western blot analysis as described earlier [11, 38]. Briefly, for quantitative PCR analysis total RNA was isolated from rat PAECs using PrepEase RNA Spin Kit (Afflymetrix, Santa Clara, CA) and quantitative real-time PCR was then performed using the USB VeriQuest SYBR Green One-Step qRT-PCR Master Kit with Fluorescein (Afflymetrix, Santa Clara, CA) according to the manufacturer’s protocol using sets of primers for rat and human Ogg1 listed in Table 1. Immunoblotting analysis was performed using an antibody to Ogg1 (Abcam, Cambridge, UK).
Table 1.
Primer sequences used for PCR assessment of mRNA abundance, Fpg-sensitive oxidative base damage, and for mtDNA/protein cross-linking and immunoprecipitation analysis
| Gene | Forward primer | Reverse primer |
|---|---|---|
| D-loop | ATTTATCCTCATAGACAAAG | TTTACCAATGCTAAGATTT |
| ATP6 | CGAAACTATCAGCCTATT | AGTAGAAGTAGAATAATAAATGTAA |
| Cox2 | GCTGTCATTCTTATTCTAA | GGATTATGTAGGAGTCAA |
| ND4 | CTCCGCAACAGAACTAAT | GTTGAGTGTTCCTATTGAGT |
| Ogg1, rat | GCTTGATGATGTCACTTATC | CTCTTCTAGGATGGCTTTAG |
| Ogg1, human | GATGTTACCCTGGCTCAA | GATGTTGTTGTTGGAGGAA |
| 28S | GATTCCCACTGTCCCTACC | ACCTCTCATGTCTCTTCACC |
Isolation of subcellular fractions and Western immunoblot analyses for Ogg1 and TFAM
Subcellular fractions were prepared as described previously [46] with minor modifications. In brief, PAECs were rinsed three times with PBS and two times with 0.25 M sucrose and 10 mM triethanolamine-acetic acid, pH 7.8, at room temperature and harvested in ice-cold 0.25 M sucrose, 1 mM EDTA, and 10 mM triethanolamine-acetic acid, pH 7.8. The following steps were carried out at 0–4°C. The cell suspension was transferred to a Dounce grinder and homogenized with 10 strokes. The homogenate was centrifuged on a cushion (5 ml) containing 0.35 M sucrose, 20 mM HEPES-NaOH pH 7.4, and 1 mM EDTA at 700 g for 10 min at 4°C. The fraction around and above the interphase was collected as crude mitochondria and reserved for mitochondrial isolation. The nuclear pellet was suspended in 3 ml of nuclear isolation buffer (0.25 M sucrose, 20 mM HEPES-NaOH pH 7.4, 25 mM KCl, and 5 mM MgCl2) and purified on a 3-ml cushion containing 0.8 M sucrose, 20 mM HEPES-NaOH pH 7.4, 25 mM KCl, and 5 mM MgCl2 at 3,000 g for 15 min at 4°C. The nuclear pellet so obtained was washed with nuclear isolation buffer and centrifuged at 1,000 g for 10 min. The pellet containing purified nuclei was suspended in 300 μl of RIPA buffer (Cell Signaling Technology, Danvers, MA), incubated for 30 min on ice, and centrifuged at 18,000 g for 15 min. The supernatant was designated as the “nuclear fraction.” The crude mitochondrial fraction, collected as described above, was centrifuged at 18,000 g for 20 min to pellet mitochondria, which were suspended in 2 ml of mitochondrial isolation buffer (0.2 M mannitol, 50 mM sucrose, 20 mM HEPES-NaOH pH 7.4, and 1 mM EDTA) and centrifuged under the same conditions. The pellet containing mitochondria was suspended in 300 μl of RIPA buffer (Cell Signaling Technology, Danvers, MA), incubated for 30 min on ice, and centrifuged at 18,000 g for 15 min. This latter supernatant was designated as the mitochondrial fraction. Nuclear and mitochondrial fractions were subjected to Western immunoblot analysis as described previously [38] using antibody against Ogg1 (Abcam, Cambridge, UK) to determine subcellular distribution of the enzyme in transfected cells. To assess total TFAM abundance, cells were harvested after hypoxic exposure, lysed in 2% SDS electrophoresis loading buffer, and subjected to immunoblot analysis using TFAM antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Antibodies against β-actin (Sigma, St. Louis, MO), ATP synthase (Complex V) subunit alpha (Thermo Fisher Scientific, Waltham, MA) and against lamin B (Santa Cruz Biotechnology, Santa Cruz, CA) were used as markers for total protein and mitochondrial and nuclear fractions respectively. The peroxidase-conjugated secondary antibody were detected by chemiluminescence with SuperSignal West Dura substrate (Thermo Fisher Scientific, Waltham, MA) using Gel Logic 1500 Imaging System (Kodak, Rochester, NY).
Assessment of mtDNA replication and transcription
The content of mtDNA in rat PAECs was determined by slot blot analysis as described previously [47]. In brief, isolated DNA was precisely quantified, adjusted to the same concentration with H2O, and treated with 0.4 N NaOH for 10 min at room temperature to denature the DNA. The indicated amounts of total DNA were then blotted onto a nylon membrane (Roche Diagnostics, Mannheim, Germany) using a slot blot apparatus (Hoefer, Holliston, MA), membranes were hybridized with a DIG-labeled mtDNA-specific probe and processed as described below. Mitochondrial DNA content was normalized to the nuclear DNA, using a probe to the VEGF gene. Primers used for PCR-generation of mtDNA probe (coding region) and VEGF probe are listed in Table 2.
Table 2.
Primer sequences used for PCR-generation of probes for the Southern and slot-blot analyses
| Probe | Forward primer | Reverse primer |
|---|---|---|
| D-loop | TATTTTCCCCAAGCATATAAGC | CATTGAAGTTTCAGGTGTAGG |
| Coding region | CCCTACTTACTGGCTTCAATCTAC | CATACCATACCTATATATCCGAAGG |
| VEGF | TCTGTCTGCCAGCTGTCTCT | GAGCTCTTGTCTGATCTTCATAC |
Transcription of mitochondrial genes ATP6, Cox2, and ND4 was assessed by real time RT-PCR analysis as described earlier in Methods, using sets of primers listed in Table 1.
Detection of mtDNA oxidative damage
Two different strategies were employed to detect damage to the mitochondrial genome. First, quantitative Southern blot analysis was used to detect oxidative damage in large sequences of mtDNA. To measure oxidative mtDNA damage separately in two mtDNA regions, we cut cellular mtDNA with restriction enzymes into two fragments of interest: a smaller sequence containing the D-loop region and a larger fragment containing most of the coding sequences. Second, we used a PCR-based assay to detect base modifications in short sequences of selected mitochondrial regions. For both assays, total DNA was isolated immediately after treatment using the DNeasy Blood and Tissue Kit (Qiagen GmbH, Valencia, CA). Before isolation all buffers were purged with nitrogen to prevent DNA oxidation.
Southern blot analysis was performed as published previously with minor modifications [42]. In brief, purified DNA was digested with PpuMI and AhdI restriction enzymes (New England Biolabs, Beverly, MA), 10 U/ μg DNA, overnight at 37 °C. This resulted in cutting mtDNA into two fragments – a small (2.7 kb) sequence containing the D-loop region and a large (13.6 kb) coding sequence. Digested DNA samples were precipitated, dissolved in TE buffer, and precisely quantified on the Bio-Rad Versa Fluor fluorometer (Bio-Rad Laboratories, Hercules, CA) using Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies, Carlsbad, CA). To reveal oxidative base modifications, DNA was treated with formamidopyrimidine glycosylase (Fpg; New England Biolabs, Beverly, MA), a bacterial DNA repair enzyme that cleaves DNA at sites of oxidized purines. Samples containing 500 ng DNA were treated with 8 units of Fpg in 20 μl of reaction volume at 37 °C for 1 h. Subsequently, Fpg-treated and untreated samples were incubated with 0.1 N NaOH for 15 min at 37°C, and resolved in 0.6% agarose alkaline gel. After electrophoresis and DNA transfer to a nylon membrane (Roche Diagnostics, Sigma, St. Louis, MO), portions of the membrane with DNA sequences of interest were hybridized with PCR-generated probes to the corresponding regions of mtDNA. The mtDNA probes, labeled with a DIG-labeling kit (Roche Diagnostics, Sigma, St. Louis, MO), were generated with rat mtDNA sequences used as templates and the primers listed in Table 2. After cross-linking, the membranes were washed and processed according to the manufacturer’s suggestions. Hybridization bands were detected with Amersham Hyperfilm ECL (GE Healthcare, Piscataway, NJ) and a Gel Logic 1500 Imaging System (Kodak, Rochester, NY). Single-strand breaks formed at the sites of oxidized purines by Fpg treatment result in decrease of Southern blot hybridization band intensity that depends on the relative amount and integrity of DNA. Changes in hybridization band intensity between Fpg-treated and untreated DNA indicate the extension of oxidative base damage. These changes in the Fpg-sensitive lesion density were calculated as negative ln of the quotient of hybridization band intensities in Fpg-treated and untreated samples and normalized to 10 kb. This method of calculation is based on the application of a Poisson equation for the undamaged sequences and has been traditionally used for the analyses of DNA damage by Southern blot and quantitative PCR [33, 48].
As a complimentary method to detect mtDNA oxidative damage we employed Fpg-sensitive real time PCR analysis as described earlier [8], using the primers encompassing the sequences in the D-loop region and in the ND4 gene and listed in Table 1. The basis of the assay is treatment of mtDNA with Fpg that removes oxidized purines form DNA, thereby creating single-strand breaks and blocking PCR amplification at these sites. Differences in PCR amplification between Fpg-treated and untreated DNA are thus a specific indicator of the presence of oxidative base damage. The Fpg cleavage reaction was performed by incubating 250 ng of DNA with 8 units of Fpg in 1× NEBuffer 1 (10 mM Bis-Tris propane-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.0) and 100 μg/ml BSA in a volume of 50 μl at 37°C for 1 h. Fpg was then inactivated by heating at 60°C for 5 min. An aliquot containing 10 ng DNA was then used for the PCR assay to detect Fpg-sensitive cleavage sites. Data are presented as the fraction of intact DNA, calculated as the quotient of signal intensities in Fpg-treated and untreated DNA.
Analysis of TFAM binding to mtDNA
TFAM binding to the D-loop and coding regions of mtDNA was studied with mtDNA/protein cross-linking and immunoprecipitation analysis using a standard, commercially-available ChIP assay kit (Active Motif, Carlsbad, CA) and adapted ChIP protocol as described elsewhere [49] with modifications. Briefly, after exposure to hypoxia, ~2×107 PAECs were fixed with 1% formaldehyde (Sigma, St. Louis, MO) for 10 minutes, washed with ice-cold 1x PBS, and fixation reaction terminated by addition of Glycine Stop Fix solution for 5 minutes. PAECs were washed and collected in Cell Scraping Solution supplemented with 0.5 mM PMSF. DNA was sheared to ~500 bp fragments by sonication for ten 20 s pulses at 25% amplitude with a Vibracell VCX 130PB (Sonics & Materials, Newtown, CT). Protein-mtDNA complexes were then immunoprecipitated with TFAM antibody (Santa Cruz Biotechnology, Santa Cruz, CA) according to the manufacturer’s instructions. The yield of target region DNA in each sample after precipitation was analyzed by real-time PCR using USB VeriQuest Fast SYBR Green qPCR Master Kit with Fluorescein (Afflymetrix, Santa Clara, CA). The primers used for the analysis of TFAM binding to the D-loop region and to the ATP6 site in coding region are listed in Table 1. Amplification of input DNA before immunoprecipitation at a dilution of 1:10 was used as a positive control. A companion analysis without any antibody served as a negative control.
Statistical analysis
Reduced data are presented as the mean ± standard error (SE). Depending on the experimental design, differences in mean values were assessed using unpaired t-test or one-way ANOVA combined with Dunnett test. P values <0.05 were taken as evidence of statistical significance.
RESULTS
Hypoxia increases the abundance of mtDNA in rat PAECs
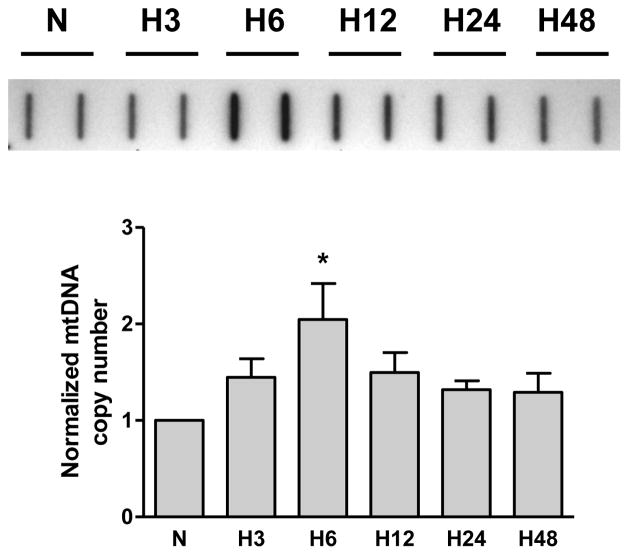
Our initial experiments with rat PAECs using slot-blot analysis showed that hypoxia caused a time-dependent increase in mtDNA abundance. The results of these studies are presented in Figure 1 and show the time course of mtDNA content in the rat PAECs after cell exposure to hypoxia. Mitochondrial DNA copy number in PAECs peaked at 6h of hypoxic exposure and remained elevated till 24h under hypoxia. According to our hypothesis, this elevation in mtDNA content requires increased base oxidation in the D-loop region of mtDNA.
Fig. 1.
Hypoxia increases mtDNA copy number in rat pulmonary artery endothelial cells. TOP: Representative slot-blot of mtDNA from rat pulmonary artery endothelial cells in normoxia (N) and after hypoxic exposure for 3, 6, 12, 24, and 48 hours (H3 – H48). BOTTOM: Reduced data of hybridization band intensities normalized to nuclear DNA. Mean ± SE, N = 4, *P < 0.05, significantly different from normoxic controls.
Hypoxia selectively damages the D-loop region of mtDNA in rat PAECs
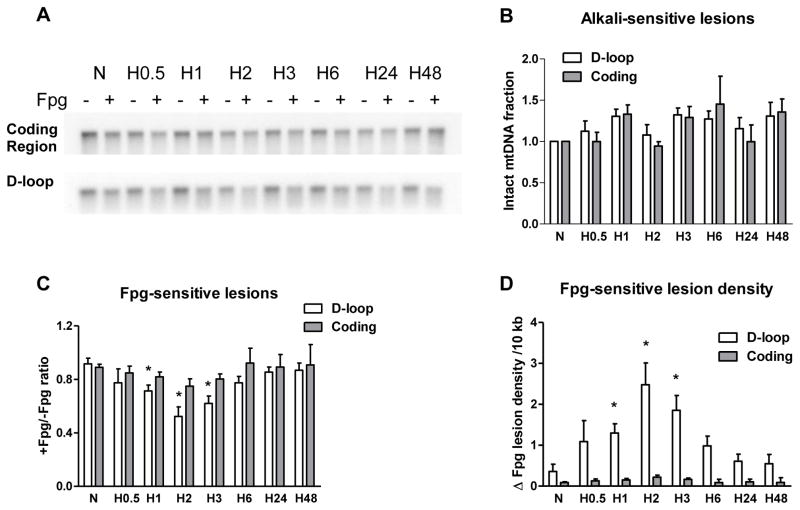
Analysis of mtDNA from pulmonary cells of various origins for oxidative modifications in the coding and D-loop regions during hypoxia revealed significantly different levels of oxidative damage in these two sequences of the mitochondrial genome. To enable detection of oxidative modifications, we treated DNA with Fpg glycosylase, which cleaves DNA at sites with purine base oxidative damage. Application of alkaline gel electrophoresis and Southern hybridization to samples not treated with Fpg was used to reveal alkali-detectable lesions such as strand breaks and abasic sites. Treatment of the samples with Fpg along with alkali was employed to detect oxidative damage to purine bases. In experiments with rat PAECs we found that the hypoxia did not induce alkali-detectable DNA damage in either the coding region or the D-loop region (Figure 2B), but generated oxidative base modifications that were prominent in the mtDNA D-loop region after 1 and 3 hours of hypoxic exposure and were absent in the coding region of the mitochondrial genome (Figure 2C). Quantitative data calculated as increase in purine oxidative lesion density are depicted in Figure 2D and show that hypoxia significantly elevated the density of purine base oxidation products in the D-loop region of mitochondrial genome.
Fig. 2.
Hypoxia-induced oxidative base damage is localized to the D-loop region of the mitochondrial genome in rat pulmonary artery endothelial cells. (A) Representative quantitative Southern blot analysis of oxidative damage in coding and D-loop regions of mtDNA from normoxic (N) rat pulmonary artery endothelial cells and cells after hypoxia exposure for 0.5, 1, 2, 3, 6, 24 and 48 hours (H0.5 – H48). Samples were treated (+) or not treated (−) with Fpg to reveal oxidatively modified purines. (B) Calculated data showing absence of alkali-detectable lesions in hypoxic samples without Fpg treatment normalized to normoxic control. (C) Calculated data expressed as ratio between Fpg treated and not treated samples showing the fraction of mtDNA without oxidative damage in above mentioned experimental groups. (D) Calculated data showing change in Fpg-sensitive lesion density per 10 kb. Mean ± SE, N = 3 – 9, *P < 0.05, significantly different from normoxic controls.
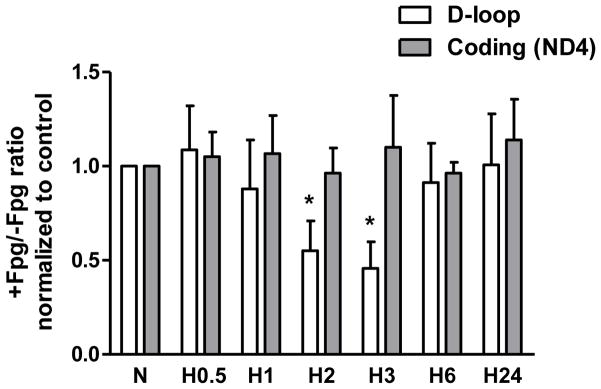
As an additional approach to confirm the detection of oxidative base modifications in sequences of the D-loop and coding regions of the mitochondrial genome, we used Fpg-sensitive conventional and real time PCR analyses. The results of experiments with both PCR techniques confirmed findings obtained in the Southern blot studies; hypoxia-induced damage was restricted to regulatory D-loop region in the mitochondrial genome (Figure 3). Fpg-sensitive lesions were observed only in D-loop region after culturing rat PAECs in hypoxic conditions for 3 hours, whereas the coding region of mtDNA, specifically a sequence nested within the ND4 gene, did not show any oxidative damage after hypoxic exposure. It is important to note that the oxidative lesions to the D-loop region during hypoxia temporally preceded hypoxia-induced increase in mtDNA copy number.
Fig. 3.
Results of Fpg-sensitive quantitative real time PCR analysis of oxidative DNA damage in D-loop and coding regions of mtDNA from normoxic (N) rat pulmonary artery endothelial cells and cells exposed to hypoxia for 0.5, 1, 2, 3, 6 and 24 hours (H0.5 – H24). The ratio of PCR product accumulation in treated and untreated with Fpg samples is indicative of oxidative DNA damage. Mean ± SE, N = 3, *P < 0.05, significantly different from normoxic controls.
Transfection of rat PAECs with mitochondria-targeted hOgg1
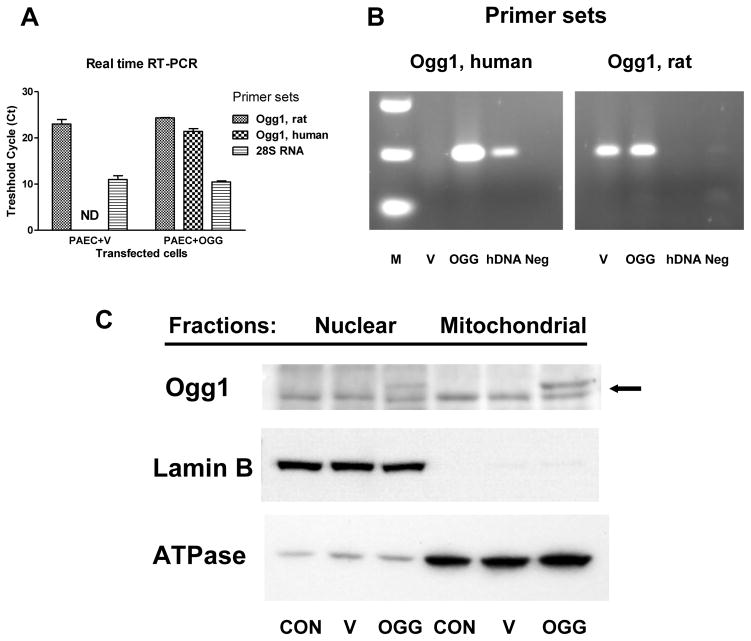
To explore the role of oxidative DNA damage to the D-loop region in mtDNA replication, we overexpressed mitochondria-targeted human DNA repair glycosylase Ogg1 and studied the effect of enhanced DNA repair on mitochondrial DNA replication and transcription during hypoxia. The results of real time and conventional RT-PCR analysis shown in Figure 4A–B confirmed successful overexpression of hOgg1 in rat PAECs. The results of Western blot analysis depicted in Figure 4C show preferential mitochondrial localization of hOgg1. Empty vector- and hOgg1-transfected cells were then exposed to hypoxia and analyzed for oxidative mtDNA damage, mtDNA copy number and transcription of mitochondrial genes.
Fig. 4.
Transfection of rat pulmonary artery endothelial cells with human 8-oxoguanine glycosylase (Ogg1). Abundance of rat and human Ogg1 mRNA in samples from vector- transfected (V) and Ogg1- transfected (OGG) cells revealed by real time (A) and conventional (B) RT-PCR analyses. Sets of primers specific to rat or human Ogg1 genes were used to distinguish between endogenous Ogg1 and the product of lentiviral construct used for transfection. Primers to 28S RNA were used as a loading control. ND – non-detectable value; M – markers; hDNA – positive control human DNA; Neg – negative control. (C) Representative Western blot analysis of nuclear and mitochondrial fractions from non-transfected cells (CON) and cells transfected with empty vector (V) or with Ogg1 construct (OGG). Arrow points to endogenous Ogg1 (lower band, approx. 39 kDa) and to Ogg1 vector product (upper band, approx. 42 kDa).
Overexpression of hOgg1 eliminates hypoxia-induced oxidative damage in D-loop region and attenuates hypoxia-induced mtDNA replication in rat PAECs
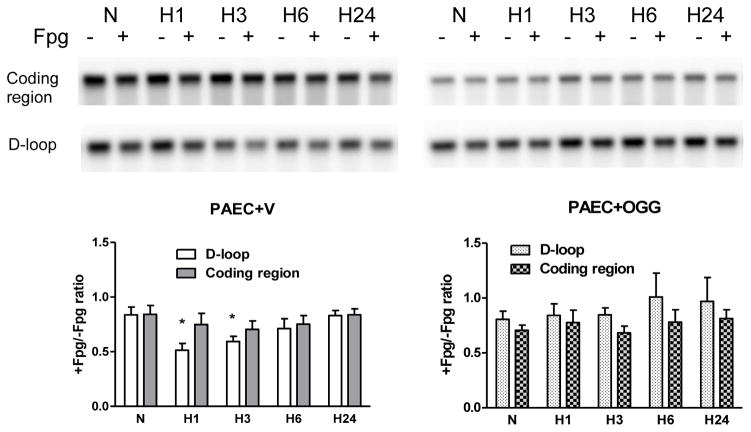
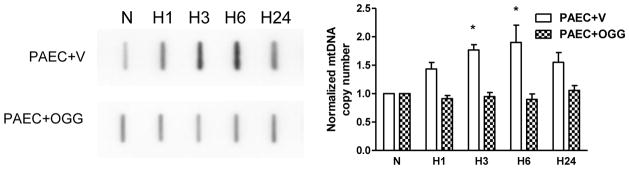
The quantitative Southern blot analysis shown in Figure 5 indicates that overexpression of mitochondria-targeted hOgg1 in PAECs suppressed hypoxia-induced oxidative modifications to the regulatory D-loop region of the mitochondrial genome, but did not change the baseline level of oxidation in the coding region of the mtDNA. If our hypothesis is correct, then elimination of D-loop oxidation should prevent changes in mtDNA replication and transcription. Slot blot analysis was used to assess the mtDNA content in normoxic and hypoxic vector- and hOgg1-transfected cells. Results shown in Figure 6 revealed that the increase in mtDNA copy number in PAECs exposed to hypoxia was completely attenuated in cells overexpressing hOgg1.
Fig. 5.
Impact of Ogg1 overexpression on hypoxia-induced oxidative damage to mitochondrial genome in rat pulmonary artery endothelial cells (PAECs). Quantitative Southern blot analysis of oxidative damage in coding and D-loop regions of mtDNA in rat PAECs transfected either with empty vector (PAEC+V) or with Ogg1 construct (PAEC+OGG) in normoxic (N) conditions and after hypoxia exposure for 1, 3, 6 and 24 hours (H1 – H24). TOP: Representative Southern blot analysis of Fpg-treated (+) or untreated (−) samples to reveal oxidatively modified purines. BOTTOM: Calculated data expressed as ratio between Fpg treated and not treated samples showing the fraction of mtDNA without oxidative damage in above mentioned experimental groups. Mean + SE, N = 4 – 5, *P < 0.05, significantly different from normoxic controls.
Fig. 6.
Impact of Ogg1 overexpression on hypoxia-induced mtDNA replication in rat pulmonary artery endothelial cells (PAECs). LEFT: Representative slot-blot analysis of mtDNA from rat PAECs transfected either with empty vector (PAEC+V) or with Ogg1 construct (PAEC+OGG) in normoxic (N) conditions and after hypoxia exposure for 1, 3, 6 and 24 hours (H1 – H24). RIGHT: Reduced results of hybridization band intensities normalized to nuclear DNA. Mean ± SE, N = 5 – 6, *P < 0.05, significantly different from normoxic controls.
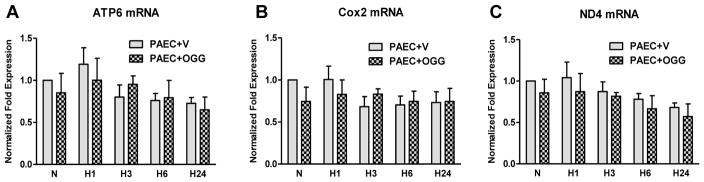
Since mechanisms of replication and transcription of the mtDNA are tightly connected, we analyzed the abundance of three mitochondria-encoded transcripts, ATP6, Cox2, and ND4, using real time RT-PCR. The results of these experiments are shown in Figure 7. Neither hypoxia, nor hOgg1 overexpression, had significant effects on the selected mitochondrial mRNAs, although there was some tendency toward decrease in abundance of some transcripts at 24 h hypoxic exposure.
Fig. 7.
Transcription of mitochondrial genes in rat pulmonary artery endothelial cells (PAECs) transfected either with empty vector (PAEC+V) or with Ogg1 construct (PAEC+OGG) in normoxic (N) conditions and after hypoxia exposure for 1, 3, 6 and 24 hours (H1 – H24). Results of quantitative real time RT-PCR analysis revealing the accumulation of mRNA transcripts of mitochondrial genes: ATP6 (A), Cox2 (B) and ND4 (C). Abundance of mitochondrial mRNA was normalized to 28S RNA expression. Mean ± SE, N = 5–13.
Overexpression of hOgg1 attenuates TFAM binding to both D-loop and coding regions of mtDNA in rat PAECs
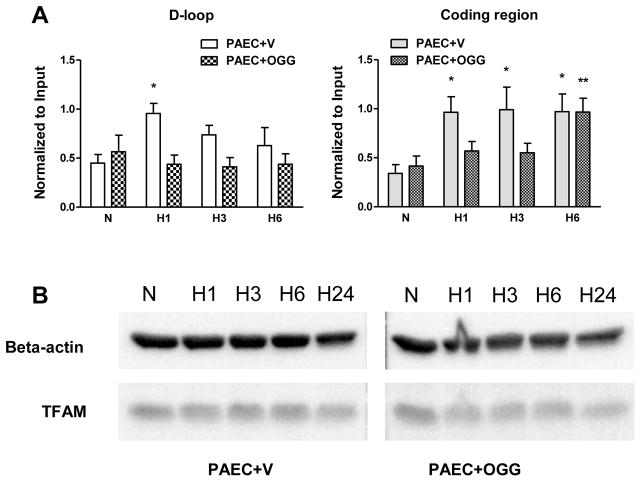
To explore a possible mechanism whereby mtDNA oxidation during hypoxia regulates mtDNA replication, we determined if mtDNA oxidative modifications impacted TFAM binding to the mtDNA. Since TFAM preferentially binds to oxidized DNA, we expected that enhanced mtDNA repair in cells overexpressing hOgg1 should affect its binding. A modification of the ChIP assay - mtDNA/protein cross-linking and immunoprecipitation analysis - was performed on vector- and hOgg1-transfected rat PAECs after cell exposure to hypoxia for 1, 3 and 6 hours using an antibody to TFAM. The results of the PCR analysis, using primers for coding and D-loop regions and precipitated DNA as a template, are depicted in the Figure 8A. Surprisingly, in vector-transfected PAECs, TFAM binding to the D-loop region of mtDNA increased only transiently after 1 hour of hypoxia, whereas its binding to the coding region of mtDNA was elevated at all times in hypoxic PAECs when compared to normoxic cells. Mitochondria-targeted hOgg1 overexpression attenuated TFAM binding to both the D-loop and coding regions of mtDNA. The results of Western blot analyses showed that hypoxia did not change TFAM abundance in total cell lysates (Figure 8B).
Fig. 8.
TFAM abundance and binding to mtDNA in rat pulmonary artery endothelial cells (PAECs) transfected either with empty vector (PAEC+V) or with Ogg1 construct (PAEC+OGG) in normoxic (N) conditions and after hypoxia exposure for 1, 3, 6 and 24 hours (H1 – H24). (A) Impact of Ogg1 overexpression on TFAM binding to the D-loop and coding regions of mtDNA in rat PAECs. Reduced data represent results of mtDNA/protein cross-linking and immunoprecipitation analysis performed as described in MATERIALS AND METHODS. The assay was performed using antibody to TFAM, and precipitated DNA was subjected to PCR with primers for D-loop and coding (ATP6 site) regions of mtDNA. Intensity of PCR bands was normalized to input. Mean ± SE, N = 4 – 8, *P < 0.05, significantly different from normoxic PAEC+V, **P < 0.05, significantly different from normoxic PAEC+OGG. (B) Representative Western blot analysis of TFAM abundance in total cell lysates of rat PAECs.
DISCUSSION
Hypoxia, as a fundamental stimulus, which complicates many cardiopulmonary, infectious and neoplastic disorders, displays complex and sometimes controversial action on cells and mitochondria. Indeed, the cell’s reaction to hypoxia depends on many factors: cell origin, nutrient conditions, duration and severity of hypoxia [50]. In some cases hypoxia appears as a damaging factor, inducing autophagy, inflammation, cell injury, and apoptosis [50–52]. But in many tissues, especially in the vasculature, hypoxia, by up-regulating VEGF expression, acts as a powerful stimulus for cell proliferation, vasculogenesis and angiogenesis [53–56]. The effect of hypoxia on mtDNA replication and mitochondrial biogenesis is also complex and may depend on many factors, including sex specificity [57]. Although some cell types respond to hypoxia by reducing their mitochondrial biogenesis and mtDNA content [58–60], mtDNA copy number is reported to increase under hypoxic conditions in many other tissues, including brain, liver, heart, placenta, sperm and blood cells [23–25, 61–67]. Hypoxia, like many other factors that affect mitochondrial biogenesis, utilizes mitochondria-generated ROS as second messengers [68–71]. In the context of ROS-mediated nuclear gene expression, a surprising target of free radicals is specific DNA sequences in gene promoters, and emerging evidence suggests that a process of “controlled oxidative DNA damage and repair” may be necessary for normal transcriptional regulation [6, 7, 10, 11, 15, 72–76]. In this model, ROS generated during signaling lead to oxidative DNA “damage” and activation of base excision repair in key promoter sequences. Complete execution of the repair entails the formation and subsequent re-ligation of DNA strand breaks; such breaks have profound effects on DNA topology, flexibility, and conformation, which may contribute to regulation of promoter function and gene expression.
Could a similar model for ROS-dependent transcriptional activation be operative in the mitochondrial genome, triggering mtDNA replication and transcription via formation and repair of oxidative base lesions in the mtDNA transcriptional regulatory region the – D-loop? Such a mechanism could be involved in either mtDNA replication or transcription, since initiation of both processes is mediated by the same transcription machinery [30]. Results of the present study support this intriguing idea. Whereas in our earlier research we focused mostly on the “common” deletion region of the mtDNA [9], in the present work we searched for oxidative damage in two discrete, functionally relevant mtDNA regions: the D-loop region with flanking sequences, and the coding region of the mitochondrial genome.
We were particularly interested in the D-loop region of the mtDNA because of its exquisite sensitivity to oxidative damage [34, 35]. In addition, it has been reported that base oxidation in the model oligonucleotides enhance DNA binding affinity for TFAM, the main factor for regulation of the transcription and replication of the mtDNA [36]. Our experiments on PAECs showed that hypoxia-induced transient oxidative damage to mtDNA was localized in the D-loop region and was absent in the coding region of the molecule. This mtDNA oxidation temporarily coincided with a transient increase in TFAM binding and an elevation in mtDNA copy number, suggesting the potential role of oxidative modifications in the D-loop region for the TFAM binding and mtDNA replication.
Being part of the High Mobility Group domain family proteins, TFAM is capable of introducing specific structural alterations in DNA; in the mtDNA D-loop region, binding of TFAM is believed to unwind [77, 78] and bend the promoter sequences [79]. The enhanced bending of the promoter DNA imparted by the C-terminal tail is considered as a critical component of the ability of TFAM to initiate transcription and subsequent mtDNA replication machinery [29, 79].
The requirement for a TFAM-induced conformational change in the D-loop is central to our model. For TFAM binding to initiate conformational changes in the D-loop, it must overcome the intrinsic stiffness of the sequence. In mammalian cells, the circular mtDNA is covalently closed and adopts a supercoiled configuration [80]. Replication of mtDNA, like any other closed-circular DNA, requires cycles of nicking and closing of strands in parental mtDNA to relieve tensional stress caused by supercoiling [81]. Recently it was shown that TFAM binding to mtDNA increases its flexibility which can be explained by local denaturation of the DNA [82]. We have observed hypoxia-induced formation of oxidative mtDNA base modifications in the D-loop region in a very narrow time interval (1–3 hrs of hypoxia) that might be sufficient for the transient increased binding of TFAM to this region. We suggest that this increased TFAM binding temporarily decreases stiffness in the regulatory sequence, increases its flexibility, and changes its conformation. This, in turn, may reduce the steric obstruction and facilitate assembly of other components of the replication machinery, which results in increased mtDNA replication. We can also suggest involvement of DNA repair enzymes in the regulation of mtDNA replication, since D-loop oxidative lesions under hypoxic exposure were transient in character and were eventually repaired. Such a possibility is supported by some existing data on potential involvement of DNA repair enzymes in mitochondrial and nuclear transcription [7, 12, 83].
To explore the importance of sequence-specific oxidation of the mtDNA for TFAM binding and initiation of mtDNA replication, we overexpressed the mitochondria-targeted human DNA repair glycosylase Ogg1 as described previously. Increased Ogg1 expression should enhance DNA repair capacity, eliminate oxidized guanines in the DNA, and thereby decrease TFAM binding to the sequence. As predicted, hOgg1 over-expression eliminated hypoxia-induced oxidative damage in D-loop region of the mitochondrial genome in rat PAECs and attenuated TFAM binding to both D-loop and coding regions of mtDNA, which resulted in prevention of hypoxia-induced mtDNA replication. Interestingly, Ogg1 overexpression suppressed TFAM binding not only to the oxidized D-loop sequence, but also to the mtDNA coding region, which, unlike the D-loop, was not oxidatively modified by hypoxia. These results may indicate that transient oxidative modifications in the regulatory sequence not only increase TFAM binding to the D-loop region, but also facilitate further TFAM binding to the rest of the mtDNA molecule. Elimination of such transient site-specific mtDNA oxidation decreases the amount of TFAM bound to the entire mtDNA molecule and prevents its replication. Interestingly, TFAM has the unique ability to bind to the promoter region in a sequence-specific manner and to non-promoter DNA in a non sequence-specific manner [84, 85]. The mechanisms of TFAM binding to promoter and non-promoter regions are not understood, but according to our results, preferential oxidation of D-loop region in response to stimuli could be the key factor in orchestrating TFAM interaction with other regions of the mtDNA that leads to its replication.
It is important to emphasize that changes in TFAM binding and mtDNA replication occur without increases in TFAM expression, suggesting that binding of TFAM to the promoter region of the mtDNA, but not its abundance, is a key event in initiation of mtDNA replication. Although it has been reported that mtDNA copy number is directly proportional to total TFAM levels [86], several lines of evidence show that the link between TFAM abundance and mtDNA replication might be more complicated. For example, Noack et al. showed that mild oxidative stress led to proliferation of the mtDNA, but did not change TFAM protein levels [87]. Another study with transient overexpression of TFAM demonstrated that increase in TFAM abundance in the cell is sufficient to stimulate mtDNA transcription, but not sufficient to increase mtDNA copy number [88].
Interestingly, despite the fact that hypoxia increased mtDNA copy number, changes in mRNA transcripts of the examined mitochondrial proteins did not occur. It has been reported that mtDNA copy number is regulated by cellular proliferation [89] and, though not studied here, previous reports show that prolonged hypoxia induces proliferation of pulmonary endothelial cells [53–55]. We consider the observed increase in mtDNA copy number only as an early stage of mitochondrial biogenesis on the threshold of cell proliferation. Initiation of mitochondrial DNA replication uses the same pathways as employed for transcription initiation; therefore the processes of replication and transcription must be highly coordinated. Recently it has been proposed that replication and transcription are mutually exclusive processes in human mitochondria that allow the corresponding machineries to avoid the detrimental consequences of a head-on collision [32]. It was suggested that human transcription elongation factor serves as a molecular switch that allows the organelles either to replicate the mtDNA and regulate its copy number or to elevate its transcription rates. The modest effect of hypoxia on mtDNA transcription observed in our experiments may indicate that, prior to hypoxia-induced cell proliferation, PAECs undergo mtDNA replication without altering the transcription rate.
In summary, our observations suggest that hypoxia causes oxidative modifications that are prominent in the D-loop region of the mitochondrial genome of the PAECs and that such DNA modifications may be important for TFAM binding and subsequent mtDNA replication. These data support our hypothesis that controlled oxidative DNA damage and repair in the D-loop region of the mitochondrial genome are required for mitochondrial DNA replication in hypoxia. The concept that oxidative mtDNA damage might be important for mitochondrial proliferation is supported indirectly by other recent findings. For example, the robust mitochondrial biogenesis that occurs during the postnatal cardiac development in rodents is accompanied by oxidative mtDNA damage and repair [83]. Here too, it was suggested that up-regulation of DNA damage response genes may be involved in control of mitochondrial biogenesis.
The present results are significant because they extend the emerging concept that controlled DNA damage and repair also contribute to regulation of mtDNA replication. In addition, our findings point to a molecular link between a normal, ROS-dependent process and the mtDNA instability characteristic of a variety of diseases including cancer, cardiovascular disease, diabetes and neurodegenerative diseases.
HIGHLIGHTS.
A new mechanism, by which hypoxia triggers mtDNA replication, is proposed.
Hypoxia-induced mtDNA oxidative modifications are prominent in the D-loop region.
Such modifications are important for TFAM binding and subsequent mtDNA replication.
Overexpression of Ogg1 prevents hypoxia-induced oxidation of the D-loop region.
Ogg1 overexpression attenuates TFAM binding and mtDNA replication under hypoxia.
Acknowledgments
We acknowledge Dr. Mikhail Alexeyev and Mrs. Viktoriya Pastukh from Gene Delivery Core for their work on generation and production of lentiviral vectors. We also appreciate the assistance of Mrs. Gina Capley Bardwell and Ms. Mita Patel. This study was funded by National Heart, Lung, and Blood Institute Grants R21 HL102789 (PI M. Ruchko), R01 HL113614 (PI M. Gillespie), and R01 HL058234 (PI M. Gillespie).
Abbreviations
- ANOVA
analysis of variance
- ATP6
ATP synthase subunit 6
- ChIP
chromatin immunoprecipitation
- Cox2
cytochrome c oxidase subunit 2
- DIG
digoxigenin
- Fpg
formamidopyrimidine DNA glycosylase
- mtDNA
mitochondrial DNA
- ND4
NADH dehydrogenase subunit 4
- Ogg1
8-oxoguanine glycosylase
- PAECs
pulmonary artery endothelial cells
- PBS
phosphate-buffered saline
- ROS
reactive oxygen species
- SE
standard error
- TE
Tris-EDTA
- TFAM
mitochondrial transcription factor A
- VEGF
vascular endothelial growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Viktor M. Pastukh, Email: vpastukh@southalabama.edu.
Olena M. Gorodnya, Email: gorodnya@southalabama.edu.
Mark N. Gillespie, Email: mgillesp@southalabama.edu.
Mykhaylo V. Ruchko, Email: mruchko@southalabama.edu.
BIBLIOGRAPHY
- 1.Freeman BA, Crapo JD. Biology of disease: free radicals and tissue injury. Lab Invest. 1982;47:412–426. [PubMed] [Google Scholar]
- 2.Dimauro S, Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005;37:222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hailer-Morrison MK, Kotler JM, Martin BD, Sugden KD. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003;42:9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 6.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 7.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 8.Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishko V, Solomon M, Breit JF, Killilea DW, Ledoux SP, Wilson GL, Gillespie MN. Hypoxia promotes oxidative base modifications in the pulmonary artery endothelial cell VEGF gene. FASEB J. 2001;15:1267–1269. doi: 10.1096/fj.00-0755fje. [DOI] [PubMed] [Google Scholar]
- 10.Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: impact on DNA integrity and gene expression. FASEB J. 2005;19:387–394. doi: 10.1096/fj.04-2805com. [DOI] [PubMed] [Google Scholar]
- 11.Pastukh V, Ruchko M, Gorodnya O, Wilson GL, Gillespie MN. Sequence-specific oxidative base modifications in hypoxia-inducible genes. Free Radic Biol Med. 2007;43:1616–1626. doi: 10.1016/j.freeradbiomed.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 12.Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, Gillespie MN. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1367–1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruchko MV, Gorodnya OM, Pastukh VM, Swiger BM, Middleton NS, Wilson GL, Gillespie MN. Hypoxia-induced oxidative base modifications in the VEGF hypoxia-response element are associated with transcriptionally active nucleosomes. Free Radic Biol Med. 2009;46:352–359. doi: 10.1016/j.freeradbiomed.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breit JF, Ault-Ziel K, Al-Mehdi AB, Gillespie MN. Nuclear protein-induced bending and flexing of the hypoxic response element of the rat vascular endothelial growth factor promoter. FASEB J. 2008;22:19–29. doi: 10.1096/fj.07-8102com. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie MN, Wilson GL. Bending and breaking the code: dynamic changes in promoter integrity may underlie a new mechanism regulating gene expression. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1–3. doi: 10.1152/ajplung.00275.2006. [DOI] [PubMed] [Google Scholar]
- 16.Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136:507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 17.Lee HC, Lu CY, Fahn HJ, Wei YH. Aging- and smoking-associated alteration in the relative content of mitochondrial DNA in human lung. FEBS Lett. 1998;441:292–296. doi: 10.1016/s0014-5793(98)01564-6. [DOI] [PubMed] [Google Scholar]
- 18.Pesce V, Cormio A, Fracasso F, Vecchiet J, Felzani G, Lezza AM, Cantatore P, Gadaleta MN. Age-related mitochondrial genotypic and phenotypic alterations in human skeletal muscle. Free Radic Biol Med. 2001;30:1223–1233. doi: 10.1016/s0891-5849(01)00517-2. [DOI] [PubMed] [Google Scholar]
- 19.Masuyama M, Iida R, Takatsuka H, Yasuda T, Matsuki T. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim Biophys Acta. 2005;1723:302–308. doi: 10.1016/j.bbagen.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci. 2011;89:65–71. doi: 10.1016/j.lfs.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Suliman HB, Carraway MS, Welty-Wolf KE, Whorton AR, Piantadosi CA. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J Biol Chem. 2003;278:41510–41518. doi: 10.1074/jbc.M304719200. [DOI] [PubMed] [Google Scholar]
- 22.Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348(Pt 2):425–432. [PMC free article] [PubMed] [Google Scholar]
- 23.Costa LE, Boveris A, Koch OR, Taquini AC. Liver and heart mitochondria in rats submitted to chronic hypobaric hypoxia. Am J Physiol. 1988;255:C123–129. doi: 10.1152/ajpcell.1988.255.1.C123. [DOI] [PubMed] [Google Scholar]
- 24.Gutsaeva DR, Carraway MS, Suliman HB, Demchenko IT, Shitara H, Yonekawa H, Piantadosi CA. Transient hypoxia stimulates mitochondrial biogenesis in brain subcortex by a neuronal nitric oxide synthase-dependent mechanism. J Neurosci. 2008;28:2015–2024. doi: 10.1523/JNEUROSCI.5654-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HM, Greeley GH, Jr, Englander EW. Sustained hypoxia modulates mitochondrial DNA content in the neonatal rat brain. Free Radic Biol Med. 2008;44:807–814. doi: 10.1016/j.freeradbiomed.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carraway MS, Suliman HB, Kliment C, Welty-Wolf KE, Oury TD, Piantadosi CA. Mitochondrial biogenesis in the pulmonary vasculature during inhalational lung injury and fibrosis. Antioxid Redox Signal. 2008;10:269–275. doi: 10.1089/ars.2007.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samper E, Morgado L, Estrada JC, Bernad A, Hubbard A, Cadenas S, Melov S. Increase in mitochondrial biogenesis, oxidative stress, and glycolysis in murine lymphomas. Free Radic Biol Med. 2009;46:387–396. doi: 10.1016/j.freeradbiomed.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Kasiviswanathan R, Collins TR, Copeland WC. The interface of transcription and DNA replication in the mitochondria. Biochim Biophys Acta. 2012;1819:970–978. doi: 10.1016/j.bbagrm.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 32.Agaronyan K, Morozov YI, Anikin M, Temiakov D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science. 2015;347:548–551. doi: 10.1126/science.aaa0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mambo E, Gao X, Cohen Y, Guo Z, Talalay P, Sidransky D. Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc Natl Acad Sci U S A. 2003;100:1838–1843. doi: 10.1073/pnas.0437910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothfuss O, Gasser T, Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2009;38:e24. doi: 10.1093/nar/gkp1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, Kasai H, Kohno K. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem Biophys Res Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 37.Clark DW, Phang T, Edwards MG, Geraci MW, Gillespie MN. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic Biol Med. 2012;53:51–59. doi: 10.1016/j.freeradbiomed.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L530–535. doi: 10.1152/ajplung.00255.2004. [DOI] [PubMed] [Google Scholar]
- 39.Santos RX, Correia SC, Zhu X, Smith MA, Moreira PI, Castellani RJ, Nunomura A, Perry G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer’s disease. Antioxid Redox Signal. 2013;18:2444–2457. doi: 10.1089/ars.2012.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim Biophys Acta. 2012;1819:979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muftuoglu M, Mori MP, de Souza-Pinto NC. Formation and repair of oxidative damage in the mitochondrial DNA. Mitochondrion. 2014;17:164–181. doi: 10.1016/j.mito.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Ruchko MV, Gorodnya OM, Zuleta A, Pastukh VM, Gillespie MN. The DNA glycosylase Ogg1 defends against oxidant-induced mtDNA damage and apoptosis in pulmonary artery endothelial cells. Free Radic Biol Med. 2011;50:1107–1113. doi: 10.1016/j.freeradbiomed.2010.10.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobson AW, Grishko V, LeDoux SP, Kelley MR, Wilson GL, Gillespie MN. Enhanced mtDNA repair capacity protects pulmonary artery endothelial cells from oxidant-mediated death. Am J Physiol Lung Cell Mol Physiol. 2002;283:L205–210. doi: 10.1152/ajplung.00443.2001. [DOI] [PubMed] [Google Scholar]
- 44.Rachek LI, Grishko VI, Musiyenko SI, Kelley MR, LeDoux SP, Wilson GL. Conditional targeting of the DNA repair enzyme hOGG1 into mitochondria. J Biol Chem. 2002;277:44932–44937. doi: 10.1074/jbc.M208770200. [DOI] [PubMed] [Google Scholar]
- 45.Druzhyna NM, Hollensworth SB, Kelley MR, Wilson GL, Ledoux SP. Targeting human 8-oxoguanine glycosylase to mitochondria of oligodendrocytes protects against menadione-induced oxidative stress. Glia. 2003;42:370–378. doi: 10.1002/glia.10230. [DOI] [PubMed] [Google Scholar]
- 46.Chouteau JM, Obiako B, Gorodnya OM, Pastukh VM, Ruchko MV, Wright AJ, Wilson GL, Gillespie MN. Mitochondrial DNA integrity may be a determinant of endothelial barrier properties in oxidant-challenged rat lungs. Am J Physiol Lung Cell Mol Physiol. 2011;301:L892–898. doi: 10.1152/ajplung.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pastukh VM, Zhang L, Ruchko MV, Gorodnya O, Bardwell GC, Tuder RM, Gillespie MN. Oxidative DNA damage in lung tissue from patients with COPD is clustered in functionally significant sequences. Int J Chron Obstruct Pulmon Dis. 2011;6:209–217. doi: 10.2147/COPD.S15922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 49.Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ. Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci U S A. 2005;102:13915–13920. doi: 10.1073/pnas.0502878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 51.Rouschop KM, Ramaekers CH, Schaaf MB, Keulers TG, Savelkouls KG, Lambin P, Koritzinsky M, Wouters BG. Autophagy is required during cycling hypoxia to lower production of reactive oxygen species. Radiother Oncol. 2009;92:411–416. doi: 10.1016/j.radonc.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 52.Gurevich RM, Regula KM, Kirshenbaum LA. Serpin protein CrmA suppresses hypoxia-mediated apoptosis of ventricular myocytes. Circulation. 2001;103:1984–1991. doi: 10.1161/01.cir.103.15.1984. [DOI] [PubMed] [Google Scholar]
- 53.Tucci M, Hammerman SI, Furfaro S, Saukonnen JJ, Conca TJ, Farber HW. Distinct effect of hypoxia on endothelial cell proliferation and cycling. Am J Physiol. 1997;272:C1700–1708. doi: 10.1152/ajpcell.1997.272.5.C1700. [DOI] [PubMed] [Google Scholar]
- 54.Toby IT, Chicoine LG, Cui H, Chen B, Nelin LD. Hypoxia-induced proliferation of human pulmonary microvascular endothelial cells depends on epidermal growth factor receptor tyrosine kinase activation. Am J Physiol Lung Cell Mol Physiol. 2010;298:L600–606. doi: 10.1152/ajplung.00122.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porter KM, Kang BY, Adesina SE, Murphy TC, Hart CM, Sutliff RL. Chronic hypoxia promotes pulmonary artery endothelial cell proliferation through H2O2-induced 5-lipoxygenase. PLoS One. 2014;9:e98532. doi: 10.1371/journal.pone.0098532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conway EM, Collen D, Carmeliet P. Molecular mechanisms of blood vessel growth. Cardiovasc Res. 2001;49:507–521. doi: 10.1016/s0008-6363(00)00281-9. [DOI] [PubMed] [Google Scholar]
- 57.Sharma J, Johnston MV, Hossain MA. Sex differences in mitochondrial biogenesis determine neuronal death and survival in response to oxygen glucose deprivation and reoxygenation. BMC Neurosci. 2014;15:9. doi: 10.1186/1471-2202-15-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliveira PH, Boura JS, Abecasis MM, Gimble JM, da Silva CL, Cabral JM. Impact of hypoxia and long-term cultivation on the genomic stability and mitochondrial performance of ex vivo expanded human stem/stromal cells. Stem Cell Res. 2012;9:225–236. doi: 10.1016/j.scr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Hoppeler H, Vogt M, Weibel ER, Fluck M. Response of skeletal muscle mitochondria to hypoxia. Exp Physiol. 2003;88:109–119. doi: 10.1113/eph8802513. [DOI] [PubMed] [Google Scholar]
- 60.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 61.Carabelli J, Burgueno AL, Rosselli MS, Gianotti TF, Lago NR, Pirola CJ, Sookoian S. High fat diet-induced liver steatosis promotes an increase in liver mitochondrial biogenesis in response to hypoxia. J Cell Mol Med. 2011;15:1329–1338. doi: 10.1111/j.1582-4934.2010.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin W, Signore AP, Iwai M, Cao G, Gao Y, Chen J. Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke. 2008;39:3057–3063. doi: 10.1161/STROKEAHA.108.520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo Y, Liao W, Chen Y, Cui J, Liu F, Jiang C, Gao W, Gao Y. Altitude can alter the mtDNA copy number and nDNA integrity in sperm. J Assist Reprod Genet. 2011;28:951–956. doi: 10.1007/s10815-011-9620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo Y, Lu G, Chen Y, Liu F, Xu G, Yin J, Gao Y. Long-term cycles of hypoxia and normoxia increase the contents of liver mitochondrial DNA in rats. Eur J Appl Physiol. 2013;113:223–232. doi: 10.1007/s00421-012-2414-9. [DOI] [PubMed] [Google Scholar]
- 65.Zungu M, Alcolea MP, Garcia-Palmer FJ, Young ME, Essop MF. Genomic modulation of mitochondrial respiratory genes in the hypertrophied heart reflects adaptive changes in mitochondrial and contractile function. Am J Physiol Heart Circ Physiol. 2007;293:H2819–2825. doi: 10.1152/ajpheart.00806.2006. [DOI] [PubMed] [Google Scholar]
- 66.Lattuada D, Colleoni F, Martinelli A, Garretto A, Magni R, Radaelli T, Cetin I. Higher mitochondrial DNA content in human IUGR placenta. Placenta. 2008;29:1029–1033. doi: 10.1016/j.placenta.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 67.Lacedonia D, Carpagnano GE, Crisetti E, Cotugno G, Palladino GP, Patricelli G, Sabato R, Foschino Barbaro MP. Mitochondrial DNA alteration in obstructive sleep apnea. Respir Res. 2015;16:47. doi: 10.1186/s12931-015-0205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishida I, Kubo H, Suzuki S, Suzuki T, Akashi S, Inoue K, Maeda S, Kikuchi H, Sasaki H, Kondo T. Hypoxia diminishes toll-like receptor 4 expression through reactive oxygen species generated by mitochondria in endothelial cells. J Immunol. 2002;169:2069–2075. doi: 10.4049/jimmunol.169.4.2069. [DOI] [PubMed] [Google Scholar]
- 69.Paddenberg R, Ishaq B, Goldenberg A, Faulhammer P, Rose F, Weissmann N, Braun-Dullaeus RC, Kummer W. Essential role of complex II of the respiratory chain in hypoxia-induced ROS generation in the pulmonary vasculature. Am J Physiol Lung Cell Mol Physiol. 2003;284:L710–719. doi: 10.1152/ajplung.00149.2002. [DOI] [PubMed] [Google Scholar]
- 70.Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 71.Yoboue ED, Devin A. Reactive oxygen species-mediated control of mitochondrial biogenesis. Int J Cell Biol. 2012;2012:403870. doi: 10.1155/2012/403870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haince JF, Rouleau M, Poirier GG. Transcription. Gene expression needs a break to unwind before carrying on. Science. 2006;312:1752–1753. doi: 10.1126/science.1129808. [DOI] [PubMed] [Google Scholar]
- 73.Gillespie MN, Pastukh V, Ruchko MV. Oxidative DNA modifications in hypoxic signaling. Ann N Y Acad Sci. 2009;1177:140–150. doi: 10.1111/j.1749-6632.2009.05036.x. [DOI] [PubMed] [Google Scholar]
- 74.Gillespie MN, Pastukh VM, Ruchko MV. Controlled DNA “damage” and repair in hypoxic signaling. Respir Physiol Neurobiol. 2010 doi: 10.1016/j.resp.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 76.Amente S, Lania L, Avvedimento EV, Majello B. DNA oxidation drives Myc mediated transcription. Cell Cycle. 2010;9:3002–3004. doi: 10.4161/cc.9.15.12499. [DOI] [PubMed] [Google Scholar]
- 77.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 78.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 79.Malarkey CS, Bestwick M, Kuhlwilm JE, Shadel GS, Churchill ME. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J, Kadlubar FF, Chen JZ. DNA supercoiling suppresses real-time PCR: a new approach to the quantification of mitochondrial DNA damage and repair. Nucleic Acids Res. 2007;35:1377–1388. doi: 10.1093/nar/gkm010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robberson DL, Clayton DA. Replication of mitochondrial DNA in mouse L cells and their thymidine kinase - derivatives: displacement replication on a covalently-closed circular template. Proc Natl Acad Sci U S A. 1972;69:3810–3814. doi: 10.1073/pnas.69.12.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farge G, Laurens N, Broekmans OD, van den Wildenberg SM, Dekker LC, Gaspari M, Gustafsson CM, Peterman EJ, Falkenberg M, Wuite GJ. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor A. Nat Commun. 2012;3:1013. doi: 10.1038/ncomms2001. [DOI] [PubMed] [Google Scholar]
- 83.Pohjoismaki JL, Boettger T, Liu Z, Goffart S, Szibor M, Braun T. Oxidative stress during mitochondrial biogenesis compromises mtDNA integrity in growing hearts and induces a global DNA repair response. Nucleic Acids Res. 2012;40:6595–6607. doi: 10.1093/nar/gks301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 87.Noack H, Bednarek T, Heidler J, Ladig R, Holtz J, Szibor M. TFAM-dependent and independent dynamics of mtDNA levels in C2C12 myoblasts caused by redox stress. Biochim Biophys Acta. 2006;1760:141–150. doi: 10.1016/j.bbagen.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 88.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trinei M, Berniakovich I, Pelicci PG, Giorgio M. Mitochondrial DNA copy number is regulated by cellular proliferation: a role for Ras and p66(Shc) Biochim Biophys Acta. 2006;1757:624–630. doi: 10.1016/j.bbabio.2006.05.029. [DOI] [PubMed] [Google Scholar]