Abstract
Carbapenems, “last resort” β-lactam antibiotics, are inactivated by zinc-dependent metallo-β-lactamases (MBLs). The host innate immune response withholds nutrient metal ions from microbial pathogens by releasing metal-chelating proteins such as calprotectin. We show that metal sequestration is detrimental for the accumulation of MBLs in the bacterial periplasm, since these enzymes are readily degraded in their non-metallated form. However, the New Delhi Metallo-β-lactamase (NDM-1) is able to persist under conditions of metal depletion. NDM-1 is a lipidated protein anchored to the outer membrane of Gram-negative bacteria. Membrane-anchoring contributes to the unusual stability of NDM-1 and favors secretion of this enzyme in outer membrane vesicles (OMVs). OMVs containing NDM-1 can protect nearby populations of bacteria from otherwise lethal antibiotic levels, and OMVs from clinical pathogens expressing NDM-1 can carry this MBL and the blaNDM gene. We show that protein export into OMVs can be targeted, providing possibilities of new antibacterial therapeutic strategies.
Antibiotic resistance is one of the most serious public health threats plaguing our world1,2. This unwelcome clinical development is rapidly accelerating as the result of the global spread of antibiotic resistance genes. Carbapenems are “last resort” β-lactam antibiotics employed to combat multi-drug resistant bacteria. However, their therapeutic efficacy is challenged by the dissemination of genes encoding enzymes able to inactivate carbapenems (i.e., carbapenemases) among Gram-negative Enterobacteriaceae3. There are two types of carbapenemases, serine-β-lactamases (SBLs) and metallo-β-lactamases (MBLs). SBLs employ an active site serine residue for substrate hydrolysis, whereas MBLs require the binding of Zn(II) cofactors for activity4. MBLs are the most difficult to overcome; in contrast to SBLs, clinical inhibitors have not yet been developed for these Zn(II)-dependent carbapenemases.
MBLs are periplasmic enzymes, and worldwide spread of these resistance factors is accelerated by horizontal gene transfer4,5. Among MBLs, the New Delhi Metallo-β-lactamase (NDM-1) has manifested the fastest and largest geographical spread, involving now more than 70 countries6. The blaNDM gene was detected not only in patient samples, but also in 30% of surface waters and seepage in New Delhi7, in >60% of environmental waters in Bangladesh, and in hospital sewage8 and wastewater treatment plants in China9. This scenario reveals an unprecedented environmental dissemination of resistance determinants in regions covering half of the human population. Surprisingly, this dissemination is not related to the spread of specific clones, specific plasmids, or a single genetic structure10.
Clinically relevant MBLs employ two Zn(II) ions as cofactors, which are essential for binding and hydrolyzing β-lactam antibiotics4,11. In Gram-negative bacteria, MBLs are exported through the SecA/SecYEG system to the periplasm, where they fold and bind the Zn(II) ions12. Periplasmic Zn(II) levels are not regulated in most bacteria, thus depending on the Zn(II) availability in the extracellular environment13. This situation becomes critical during infection, since an innate immune response is elicited which involves sequestration of metal ions to starve bacteria of these essential nutrients14. Calprotectin (CP) is an abundant metal-chelating protein released at sites of infection by neutrophils. CP binds Zn(II) with high affinity and thereby prevents microbial acquisition of this essential nutrient15,16. Therefore, to overcome this metal-withholding response, MBLs must withstand conditions of limited Zn(II) availability in the host. Recent studies have shown that the active sites of MBLs are exquisitely tuned to optimize Zn(II) binding in the periplasm11,17.
Here we demonstrate that metal deprivation not only hampers MBL activity, but also affects protein stability. Apo MBLs (devoid of metal ions) formed under zinc-limiting conditions are readily degraded in the periplasm, implying that Zn(II) is essential for protein folding in the periplasm. In contrast, NDM-1 resists degradation under Zn(II)-limiting conditions. We show that the unusual stability of NDM-1 is a result of the protein being lipidated and anchored to the outer membrane of Gram-negative bacteria. Membrane-anchoring also enables secretion of NDM-1 in outer membrane vesicles (OMVs). These membrane-enclosed “bacterial packs”, shed by all Gram-negative bacteria, have been associated with cellular communication and bacterial pathogenesis18. We show that OMVs containing NDM-1 display carbapenemase activity and can protect nearby populations of carbapenem-susceptible bacteria. We report that OMVs from clinical pathogens expressing NDM-1 can carry the protein and the blaNDM gene. Lastly, we also reveal that targeting protein lipidation can prevent protein export to vesicles, providing a basis for possible therapeutic approaches.
Results
Zn(II) starvation reduces periplasmic levels of MBLs
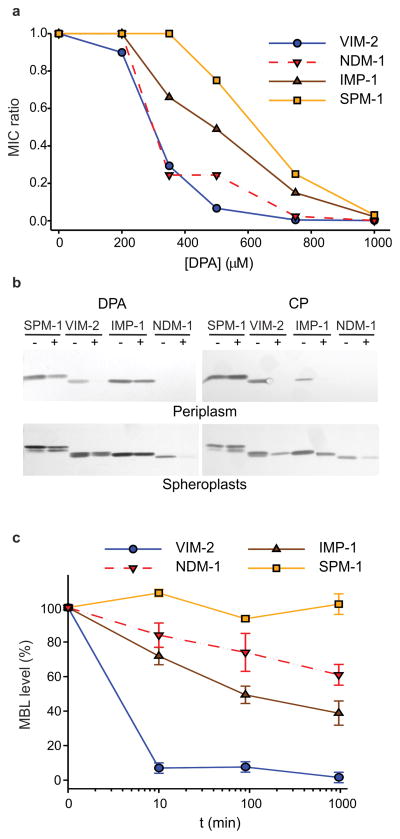
Zn(II) limitation in the extracellular medium impairs MBL-mediated resistance by reducing the levels of this essential cofactor in the bacterial periplasm19. This phenomenon can be elicited by adding metal-chelating agents to the growth medium. In order to compare the relative susceptibilities of different MBLs to zinc-limiting conditions, we targeted four clinically relevant MBLs (NDM-1, VIM-2, IMP-1 and SPM-1) to the periplasmic space of Escherichia coli with their native peptide leaders. The proteins were expressed fused to a C-terminal Strep-tag to allow for immunodetection and quantitation. Strep-tagging did not affect the resistance profile of any of the four MBLs employed (Supplementary Results, Supplementary Table 1). We then evaluated the resistance conferred by these MBLs in E. coli to two clinically relevant β-lactam antibiotics (cefotaxime and imipenem) upon addition of the metal-chelating agent dipicolinic acid (DPA). The minimum inhibitory concentration (MIC) values of these antibiotics were highly dependent on Zn(II) availability (Fig. 1a). Among the three MBLs prevalent in Enterobacteriaceae (IMP-1, VIM-2, NDM-1), IMP-1 was more refractory to the addition of DPA compared to NDM-1 or VIM-2, being able to confer antibiotic resistance at relatively high concentrations of the chelating agent. Resistance by SPM-1 (a MBL intrinsic to Pseudomonas aeruginosa) was reduced only at higher concentrations of DPA (Fig. 1a, Supplementary Fig. 1a and Supplementary Table 2). In principle, these different susceptibilities could be attributed to distinct metal-binding affinities of each MBL; however, Western blot analysis showed that periplasmic accumulation of MBLs (i.e., SPM-1, VIM-2, IMP-1) decreased under Zn(II) limitation and that these changes varied among the different proteins (Fig. 1b).
Figure 1. Zn(II) deprivation reduces bacterial antibiotic resistance and causes degradation of MBLs in the Escherichia coli periplasm.
(a) Relative MIC values of cefotaxime for E. coli cells expressing VIM-2, NDM-1, IMP-1 or SPM-1 in growth medium supplemented with different concentrations of the metal chelator DPA. MIC ratio values were calculated as described in the Methods section. Data correspond to three independent experiments, with standard errors ≤16% of each data point. (b) Western blot analysis of MBL levels in cells grown under conditions equivalent to those used for MIC assays, with (+) and without (–) addition of 350 μM DPA or 250 μg/mL CP to the growth medium. Original Western Blots are displayed in Supplementary Figure 11 (c) Relative MBL levels in the bacterial periplasm as a function of time after addition of 1000 μM DPA. Protein levels were quantified from Western blots (Supplementary Fig. 1b), normalized to control samples not treated with DPA, and plotted as the % of initial protein content remaining after treatment with DPA. Data correspond to three independent experiments and are shown as mean ± s.e.m.
VIM-2 could not be detected in periplasmic extracts of cells treated with DPA, correlating with the loss of resistance conferred by this enzyme under zinc-deprivation conditions (Fig. 1a, b). On the other hand, SPM-1 and IMP-1 levels in the periplasm were affected to a lesser extent, in agreement with the reduced effect of intermediate concentrations of DPA on the resistance conferred by these MBLs. Similar results were obtained when Zn(II) sequestration was induced by addition of CP to the medium (Fig. 1b), confirming that the phenomenon arises from restricting the external pool of available Zn(II) regardless of the chelating agent. NDM-1 was not detected in the periplasmic fraction (Fig. 1b) since this enzyme localizes to the bacterial outer membrane20. NDM-1 levels in spheroplasts were affected by DPA, but the impact on protein levels was not as drastic as observed for VIM-2 in the periplasm (Fig. 1b) even though DPA affects the antibiotic resistance profiles of these enzymes to comparable degrees (Fig. 1a).
The reduced MBL levels induced by Zn(II) depletion can be attributed to impaired expression levels or to protein degradation. To assess the impact of Zn(II) deprivation on protein levels during bacterial growth, we treated liquid cultures of E. coli cells expressing MBLs with DPA and quantified MBL levels in periplasm, spheroplasts and membrane fractions at different time points. Under these conditions, VIM-2 levels rapidly decreased in the periplasm. SPM-1 levels, in contrast, were not reduced even after 16 h of incubation following DPA addition, and IMP-1 showed an intermediate behavior (Fig. 1c and Supplementary Fig. 1b). This phenomenon of decreased protein levels was exclusive to MBLs and, hence, not a result of generalized degradation of periplasmic proteins (Supplementary Fig. 5). Moreover, cytoplasmic levels (evaluated from spheroplasts) of MBL precursors were unaltered (Supplementary Fig. 1b and 6). We conclude that the impaired antibiotic resistance observed upon Zn(II) starvation is a result of MBL degradation in the periplasm.
We also determined the amount of NDM-1 sequestered in membrane fractions. In contrast with the pattern observed in the antibiotic resistance profile upon addition of DPA, protein levels of NDM-1 were moderately reduced (Fig. 1c and Supplementary Fig. 1b). Indeed, NDM-1 levels in the membrane fractions after 16 h of growth were higher than those of IMP-1 expressed in the periplasm. As a result of this finding, we next investigated whether the different cellular localization of NDM-1 and IMP-1 MBLs accounts for this observation.
Membrane-anchoring increases the in vivo stability of MBLs
In E. coli, NDM-1 is an outer-membrane lipoprotein20. Given that NDM-1 is widely found in both Enterobacteriaceae and non-fermenting bacteria (e.g., P. aeruginosa, Acinetobacter baumannii), we investigated its localization by cellular fractionation experiments both in E. coli and P. aeruginosa cells expressing NDM-1 (Supplementary Figs. 3 and 4). In both cases, the membrane fractions displayed carbapenemase activity that was inhibited by the metal chelator ethylenediaminetetraacetic acid (EDTA). Neither β-lactamase activity nor detectable NDM-1 was present in the soluble fraction. Treatment of membranes with potassium chloride (KCl) or sodium carbonate (Na2CO3) did not release NDM-1, whereas exposure to Triton X-100 produced a soluble fraction of detergent micelles that contained NDM-1. These detergent micelles possessed carbapenemase activity (Supplementary Fig. 3). We conclude that NDM-1 is membrane associated in both E. coli and P. aeruginosa and is anchored by hydrophobic interactions. A sucrose gradient experiment confirmed that NDM-1 is localized to the outer membrane in both organisms (Supplementary Figs. 4a–b). To further examine the cellular localization of NDM-1, we first subjected whole cells to limited proteolysis and observed negligible change in NDM-1 levels as a result of proteinase K treatment. In contrast, NDM-1 was susceptible to degradation by proteinase K in permeable spheroplasts (Supplementary Fig. 4c), showing that NDM-1 is localized to the inner leaflet of the outer membrane in both E. coli and P. aeruginosa.
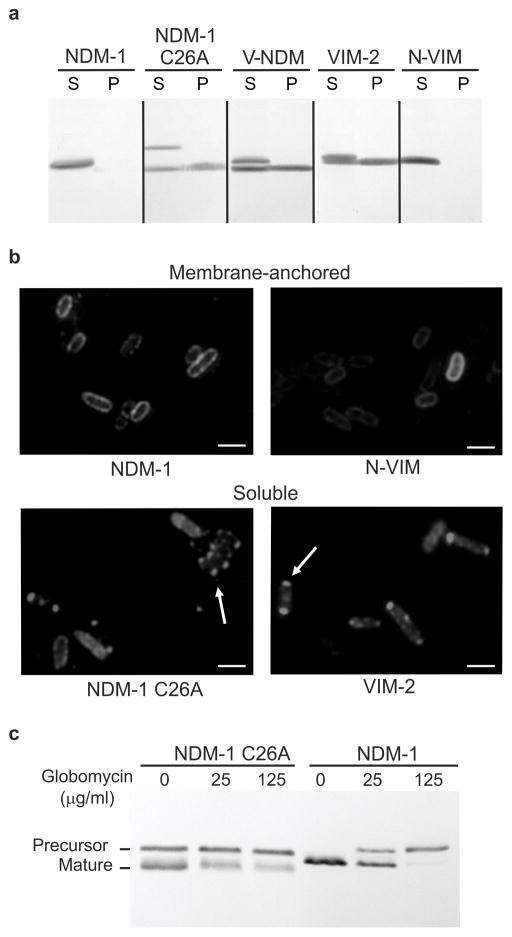
This cellular localization has been attributed to the presence of a canonical lipidation amino acid sequence LSGC (lipobox) proximal to the NDM-1 signal peptide20. In this sequence, the reactive thiol moiety of the Cys residue becomes lipidated (Supplementary Fig. 4d). Lipoboxes are recognized and processed by the Lgt-LspA-Lnt-Lol system in bacteria, which also targets lipidated proteins to the membrane21. We investigated how membrane anchoring affects β-lactamases through the construction of mutants of NDM-1 and VIM-2 with altered cellular localizations (Supplementary Fig. 2a). Expression of a NDM-1 variant in which the lipidation site was removed by mutation of the anchoring Cys residue (C26A) resulted in accumulation of soluble NDM-1 in the periplasm (Fig. 2a). The same was observed for V-NDM, a chimera of NDM-1 and the N-terminus of the soluble β-lactamase VIM-2 (including its signal peptide) (Fig. 2a). In contrast, expression of N-VIM, a chimera of VIM-2 and the N-terminal signal peptide of NDM-1, afforded a membrane-anchored variant of VIM-2 (Fig. 2a). We verified that N-VIM is associated to the outer membrane by cellular fractionation (Supplementary Fig. 4), further demonstrating the role of the lipobox in membrane anchoring and targeting. Using immunofluorescence microscopy we show different localization patterns for membrane-anchored and soluble MBLs. Membrane anchored MBLs appear evenly distributed along the periphery of the cell, whereas the soluble MBLs accumulate at the cell poles, possibly reflecting the formation of inclusion bodies22 (Fig. 2b). Moreover, we utilized globomycin, an inhibitor of LspA that selectively blocks prolipoprotein processing23, to evaluate the different processing pathways adopted by NDM-1 and NDM-1 C26A (Fig. 2c). Treatment of liquid cultures of E. coli expressing NDM-1 with globomycin led to accumulation of NDM-1 as prolipoprotein, whereas no effect was observed in cells expressing the soluble NDM-1 C26A variant. These results further establish the lipoprotein character of NDM-1 as well as the lipidation pathway.
Figure 2. The cellular localization of MBLs is determined by their N-terminal region.
(a) Western-blot analysis of NDM-1, NDM-1 C26A, V-NDM, VIM-2 and N-VIM variants in spheroplasts (S) and periplasmic fractions (P) obtained from Escherichia coli. (b) Immunofluorescence microscopy of soluble and membrane-anchored variants of NDM-1 and VIM-2. E. coli cells expressing the MBLs (fused to a C-terminal Strep tag sequence) were fixed, probed with anti-Strep tag and Cy3-conjugated anti-IgG antibodies, and visualized by fluorescence microscopy. Arrows indicate putative inclusion bodies at cell poles. The negative control with E. coli cells not expressing any MBL gave no detectable fluorescence signal. Scale bars correspond to 3 μm. (c) NDM-1 and NDM-1 C26A levels by Western blot in whole cells expressing MBLs after exposure to 0, 25 or 125 μg/mL globomycin. Original Western Blots for panels (a) and (c) are displayed in Supplementary Figure 11.
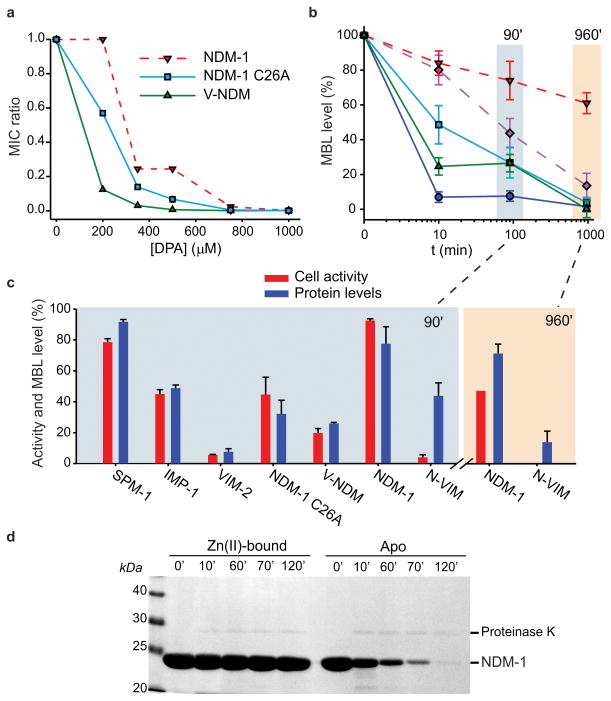
The variants of NDM-1 and VIM-2 were active and capable of conferring resistance to E. coli cells against five β-lactam antibiotics (Supplementary Table 3). The soluble variants of NDM-1, albeit conferring similar or higher levels of resistance in lysogeny broth (Supplementary Table 3), were more susceptible to diminished Zn(II) levels than native NDM-1 (the opposite was observed for VIM-2, Supplementary Fig. 2b–c). A similar impact of Zn(II) deprivation on the MIC values of these variants was observed using either DPA or CP as chelators (Fig. 3a, Supplementary Fig. 2b and Supplementary Table 4). We studied the effect of Zn(II) starvation on the protein levels of these variants (Fig. 3b). After a 16-h exposure to Zn(II) starvation, the soluble periplasmic variants of NDM-1, C26A and V-NDM, were almost fully degraded, whereas membrane-bound NDM-1 persisted (Fig. 3b). Soluble VIM-2 was almost completely degraded at 10 min, whereas its membrane-bound variant N-VIM was only reduced by 50% at 90 min. These data clearly show that membrane binding stabilizes MBLs by preventing protein degradation under conditions of Zn(II) starvation.
Figure 3. Membrane-anchoring protects NDM-1 from degradation upon Zn(II) deprivation.
(a) Relative MICs of cefotaxime for E. coli cells expressing wild type or mutant variants of NDM-1 in growth medium supplemented with different concentrations of the metal chelator DPA. Data correspond to three independent experiments, with standard errors ≤16% of each data point. (b) MBL levels in periplasmic (solid lines) or membrane (dashed lines) fractions as a function of time after addition of 1000 μM DPA, relative to levels in untreated control samples grown in the same conditions. Data correspond to three independent experiments and are shown as mean ± s.e.m. (c) Comparison between whole cell imipenem hydrolysis rates and periplasmic MBL levels after 90-minute incubation with 1000 μM DPA. Values are relative to untreated control samples grown in the same conditions, and protein levels correspond to those of Fig. 3b. The same data was determined for membrane bound variants after 960 minutes. Data correspond to three independent experiments are shown as mean ± s.e.m. Equal amounts of total protein were loaded on gels for each sample. (d) Limited proteolysis of purified recombinant NDM-1 (a soluble variant lacking the lipobox sequence), in its Zn(II)-bound or apo form. Aliquots were taken at various time intervals after addition of 2.5 μg proteinase K to solutions of 200 μg protein, and analyzed by SDS-PAGE. Original gel is displayed in Supplementary Figure 11.
Zn(II) protects MBLs from degradation in the periplasm
To assess the impact of Zn(II) starvation on the specific activity of the soluble (periplasmic) and membrane-anchored MBL variants, we measured the carbapenemase activity of whole living cells treated with DPA for 90 min or 16 h. We compared the carbapenemase activities of these DPA-treated cultures to the activities of cultures that were not treated with DPA (Fig. 3b and c). At 90 min, the decrease of carbapenemase activity paralleled the reduction of MBL expression levels in most cases (Fig. 3c). This result suggests that the detected MBLs in the DPA-treated samples correspond mostly to the active, Zn(II)-bound forms. Consequently, inactive apo-MBLs do not accumulate upon metal depletion. In contrast, N-VIM showed a low specific activity, suggesting accumulation of the inactive apo form. This observation became more evident after 16 h, when the membrane-bound variants (native NDM-1 and N-VIM) reveal accumulation of the apo forms. These results indicate that membrane anchoring stabilizes MBLs, allowing for accumulation of these proteins as apoenzymes within the periplasmic space by protecting them from degradation (Fig. 3c).
Subsequently, we explored the relative stabilities of the metallated and apo variants by exposing purified recombinant soluble NDM-1 (lacking the membrane-anchoring Cys) to limited proteolysis by proteinase K. Zn(II)-bound NDM-1 resisted proteolysis, whereas the apo form was readily degraded (Fig. 3d). This in vitro experiment demonstrates that soluble apo MBLs are more susceptible to proteolysis, accounting for the decrease in protein levels in the periplasm.
We conclude that the Zn(II) cofactor is not only essential for the hydrolytic activity of MBLs, but that is also essential for preventing protein degradation of soluble MBLs in the periplasm. For membrane-anchored variants, which can be accumulated as apo MBLs, this protective role of Zn(II) is not as relevant.
Lipidated NDM-1 is secreted in outer membrane vesicles
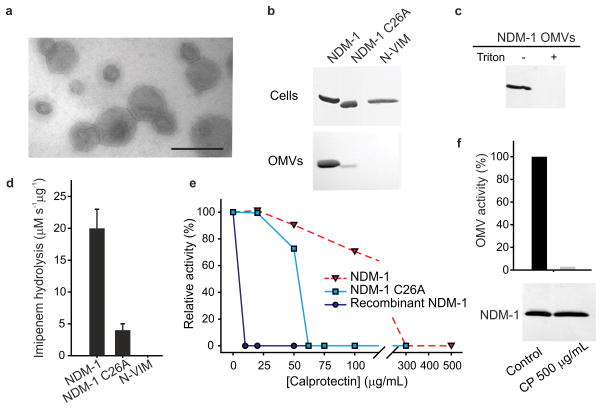
The SBLs BRO-1 from the human pathogen Moraxella catarrhalis and PenA from Burkholderia pseudomallei are the only lipidated membrane-anchored β-lactamases reported apart from NDM-1 in Gram-negative bacteria24,25. M. catarrhalis, a common cause of respiratory infections, releases outer membrane vesicles (OMVs) carrying membrane-bound BRO-126. OMVs also harbor periplasmic β-lactamases27–29, and possibly plasmids encoding the genes for these enzymes, as reported for OXA-2430, the latter of which may allow for horizontal gene transfer. Based on these observations, we purified OMVs from E. coli strains expressing NDM-1. Imaging by transmission electron microscopy (TEM) confirmed that the vesicle preparations were homogeneous, and the vesicles exhibited diameters ranging from 30 to 90 nm (Fig. 4a). Moreover, the OMVs presented protein profiles characteristic of outer membrane components, devoid of detectable cytoplasmic contaminants (Supplementary Fig. 7).
Figure 4. Membrane-anchoring favors secretion of NDM-1 into outer membrane vesicles (OMVs).
(a) Negative stain TEM of OMVs from E. coli cells expressing NDM- 1. (b) MBL levels in whole cells expressing NDM-1, NDM-1 C26A or N-VIM and OMVs purified from their cell culture supernatants. Gel lanes were loaded with equal amounts of total protein. (c) Western blot of intact (–) and lysed OMVs (+, treated with 0.1 % Triton X-100) upon treatment with 100 μg/mL proteinase K. (d) Imipenem hydrolysis by OMVs purified from E. coli cells expressing NDM-1, NDM-1 C26A and N-VIM. Data correspond to three independent experiments and are shown as mean ± s.e.m. (e) Effect of calprotectin on the imipenemase activity of OMVs (0.8 μg) containing NDM-1 or NDM-1 C26A, compared to recombinant NDM-1 (6 nM). Data correspond to mean values from two independent experiments. (f) Effect of calprotectin on NDM-1 levels in OMVs. NDM-1 OMVs were incubated for 20 minutes at 37ºC in 10 mM HEPES, 200 mM NaCl at pH 7.4 with or without addition of calprotectin 500 μg/mL. Top, relative imipenem hydrolysis rates of OMVs; bottom, Western blot showing NDM-1 levels in OMVs. Data correspond to mean values from two independent experiments. Original Western Blots for panels (b), (c) and (f) are displayed in Supplementary Figure 11.
NDM-1 was present in the OMVs, as shown by immunoblot analysis (Fig. 4b). The soluble periplasmic C26A variant was also present in the OMVs, albeit at markedly reduced levels. This observation contrasts the similar cellular expression levels of the wild-type protein and C26A variant (Fig. 4b). Immunoblots of OMVs isolated from cells expressing membrane-anchored N-VIM did not reveal the presence of this variant, indicating that N-VIM was not localized into these vesicles (Fig. 4b). Taken together, these observations support the notion that membrane binding enriches the β-lactamase content in the vesicles and that secretion of NDM-1 into OMVs is selectively favored. NDM-1 was able to resist treatment with proteinase K in intact OMVs, whereas the enzyme was readily degraded in detergent-lysed vesicles (Fig. 4c). We conclude that NDM-1 is internally located in OMVs, thus being protected from extracellular proteases.
The β-lactamase activity of the OMVs was evaluated using imipenem as a substrate. The carbapenemase activity was five-fold larger for OMVs harboring NDM-1 compared to C26A-containing OMVs (Fig. 4d). β-Lactamase activity was not detected in OMVs produced by E. coli cells expressing the N-VIM variant. Next, we examined the β-lactamase activity of these OMVs under conditions of Zn(II) deprivation elicited by addition of CP. The carbapenemase activity of OMVs carrying NDM-1 was resistant up to 100 μg/mL of CP (Fig. 4e), whereas OMVs containing the soluble C26A variant were inactivated at ~5-fold lower levels of CP. Moreover, samples of purified soluble NDM-1 were highly susceptible to CP-mediated inactivation. These experiments show that the OMV matrix exerts a protective role on membrane-anchored NDM-1 upon Zn(II) starvation. Full inactivation of OMV-packaged NDM-1 at high concentrations of CP did not alter the protein levels in the vesicles (Fig. 4f), revealing that NDM-1 is stable as an apo protein within OMVs. We conclude that secretion inside OMVs ensures NDM-1 integrity in the extracellular space.
OMVs carrying NDM-1 protect susceptible bacteria
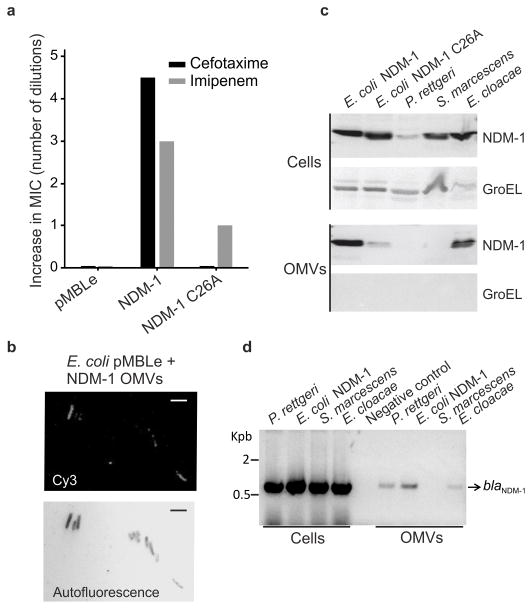
We next tried to evaluate whether the OMVs containing NDM-1 can alter the antibiotic susceptibility of bacteria not producing β-lactamases. E. coli cells susceptible to β-lactam antibiotics were incubated with OMVs containing NDM-1. Treated cells showed an 8- to 32-fold increase in antibiotic resistance against imipenem and cefotaxime, respectively (Fig. 5a), compared to untreated cells. The susceptible phenotype was restored after plating the cells in fresh LB agar. The presence of the blaNDM-1 gene was not found in these cells by polymerase chain reaction (PCR), indicating that the increase in resistance was not due to a gene transfer event. In contrast, immunofluorescence microscopy revealed the presence of NDM-1 associated with OMV-treated cells (Fig. 5b) which endows them with lactamase activity (Supplementary Fig. 8). This result suggests that the transient increase in resistance results from the presence of NDM-1 protein derived from the vesicles.
Figure 5. NDM-1-carrying OMVs provide carbapenem resistance to susceptible bacteria.
(a) Relative MIC values (shown as number of dilutions over control strain) of β-lactam-sensitive cells after treatment with OMVs purified from cells expressing native NDM-1, NDM-1 C26A or control (pMBLe). Data correspond to mean values from two independent experiments. (b) Immunofluorescence microscopy analysis of β-lactam-sensitive E. coli cells (not expressing MBLs) after 1 h incubation with OMVs purified from cells expressing NDM-1. Cells were washed from OMVs, fixed and probed with anti-Strep tag and Cy3-conjugated anti-IgG antibodies for fluorescence microscopy. Above, Cy3 visualization; below, cells detected by autofluorescence. The negative control with E. coli cells not expressing any MBL gave no detectable fluorescence signal. Scale bars correspond to 3 μm (c) Immunodetection of NDM-1 in whole cells and OMVs from clinical strains P. rettgeri, S. marcescens and E. cloacae compared to lab E. coli expressing NDM-1 and NDM-1 C26A. The presence of cytoplasmic contaminants in vesicle preps was assessed by Western blot of GroEL. (d) PCR detection of NDM-1 gene in whole cells and OMVs. Original Western Blots and gels for panels (c), and (d) are displayed in Supplementary Figure 11.
In order to explore the clinical impact of these observations, we purified OMVs from blaNDM-harboring clinical isolates of Providencia rettgeri, Serratia marcescens and Enterobacter cloacae. We identified the presence of NDM-1 by Western blot in OMVs isolated from E. cloacae, but not in OMVs from P. rettgeri or S. marcescens (Fig. 5c). Moreover, we purified DNA from the lumen of these vesicles (that were pre-treated with DNAse I to degrade possible exogenous DNA) and analyzed the samples by PCR. The blaNDM gene was amplified from P. rettgeri and E. cloacae samples, as well as from the E. coli lab strain, but not from S. marcescens (Fig. 5d). Thus, these data suggest that the dissemination of NDM-1 can be fostered by OMVs released by clinical pathogens at both the gene and protein levels.
Discussion
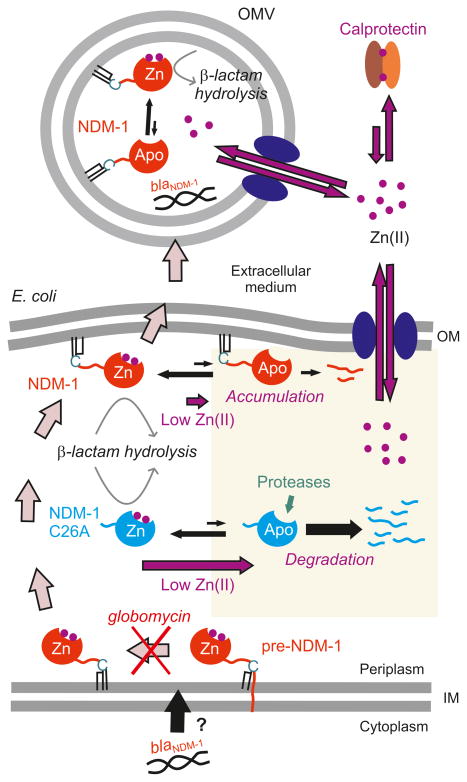
The MBL NDM-1 has become one of the most prevalent carbapenemases worldwide only a few years after its discovery. Here we show that membrane anchoring confers NDM-1 several selective advantages that may have played a crucial role in its success as a resistance determinant. Figure 6 depicts how outer membrane-anchoring favors secretion of NDM-1 in OMVs and prolongs its efficacy by enhancing protein stability under Zn(II) deprivation by the host.
Figure 6. Membrane-anchoring assures long-term survival of NDM-1.
Membrane- anchoring increases protein stability under Zn(II) deprivation, as elicited by host calprotectin during infection, and favors protein (together with blaNDM-1) secretion into OMVs, providing transient resistance to nearby populations of bacteria.
MBLs bind the Zn(II) ions in the periplasm12. Indeed, compartmentalization of metal binding is crucial to elicit metal binding selectivity31,32. Our Zn(II) starvation experiments revealed that periplasmic levels of MBLs are regulated by external Zn(II) availability. Periplasmic amounts of SPM-1 and NDM-1 remained stable upon Zn(II) limitation, whereas VIM-2 levels decreased below the detection limit. These results are in agreement with previous studies demonstrating that SPM-1, a MBL intrinsic to P. aeruginosa, is particularly optimized for conferring antibiotic resistance at low Zn(II) concentrations19. Immunodetection and cell activity measurements suggest that soluble MBLs can accumulate in the bacterial periplasm only as metal-bound species. Thus, we attribute the decrease in protein levels to rapid degradation of the apo proteins formed during Zn(II) starvation. Protease accessibility experiments indeed show that a soluble NDM-1 variant is more susceptible to degradation in its apo form. Therefore, Zn(II) binding is not only essential for MBL activation, but also for in vivo stability. This conclusion is unexpected, since apo MBLs have been characterized as relatively stable variants, being amenable to structure determination33–35. Moreover, the stabilizing effect of the Zn(II) ions in purified proteins has been largely overlooked, despite thermodynamic data supporting this notion19,36–38. These results challenge the hypothesis suggesting that MBLs could be prevalent in the apo form in the absence of substrate39 and highlight the essentiality of Zn(II) binding to MBLs in the periplasm. To the best of our knowledge, this work is the first report of a catalytic zinc site essential for in vivo stabilization of a metalloprotein.
Optimization of the Zn(II)-binding affinity has been identified as an important trait gained along the evolutionary pathway of MBLs11,17. Membrane anchoring represents an alternative mechanism providing protein stability under Zn(II) starvation. This phenomenon was observed also for a chimeric membrane-anchored variant of the lactamase VIM-2 that is more stable than the native, soluble enzyme. NDM-1 accumulates as an inactive apo variant, suggesting that membrane-anchoring limits degradation of apo MBLs, possibly by restricting the accessibility to proteases and/or protein aggregation.
During infection, pathogens are exposed to large amounts of CP (with extracellular levels >500 μg/mL40), and other zinc-chelating proteins such as S100A12 (calgranulin C, 5% of neutrophil cytosolic protein)41 and S100A7 (psoriasin, abundant in the skin)42. Therefore, MBLs must endure Zn(II) starvation to efficiently protect bacteria from an antibiotic treatment. In this way, lipidation constitutes an important trait allowing NDM-1 to resist the action of the host innate immune system. The BRO serine-β-lactamase, despite containing a lipobox, is present both as a membrane-bound and a soluble form, with ratios depending on the bacterial host25. NDM-1 is only found in the outer membrane of E. coli and P. aeruginosa, suggesting that this localization provides a stronger evolutionary advantage to MBLs compared to SBLs.
VIM-2 is one of the most clinically successful metallo-β-lactamases, despite being sensitive to metal deprivation. We posit two hypotheses to account for this apparent paradox: (i) Bacterial strains depending on VIM-2 to survive in the presence of β-lactam antibiotics may only persist and cause infection at locales where the Zn(II) deprivation response is limited, or (ii) pathogens harboring VIM-2 may overcome metal-withholding by the host. Indeed, P. aeruginosa PA01 and PA14 strains are able to resist Zn(II) starvation conditions. Moreover, some prokaryotes use ribosomal proteins as a Zn(II) store43. Redistribution of Zn(II) within different stores may allow for MBL metallation in the periplasm when extracellular Zn(II) is restricted by the host.
We also show that NDM-1 is selectively secreted into OMVs as a consequence of outer membrane-anchoring. This conclusion stems from the observation that neither soluble NDM-1 C26A nor membrane-anchored N-VIM were efficiently secreted into OMVs. The lower secretion of NDM-1 C26A into OMVs was not entirely unexpected. In E. coli, outer membrane proteins represent a larger proportion of the protein composition of OMVs than those from the periplasm44 (Supplementary Fig. 7). In contrast, the exclusion from OMVs of the lipid anchored OM protein N-VIM was surprising, possibly due to a cargo selection mechanism45,46.
Secretion of NDM-1 into OMVs illuminates another relevant evolutionary advantage: NDM-1-containing OMVs can protect microbes located in the immediate environment that would be otherwise susceptible to β-lactam antibiotics. This protection takes place by protein transport mediated by the vesicles. We note that these assays were performed with relatively high OMVs concentrations. However, OMV levels vary widely among species, reaching levels as high as 162 mg/L47, and pathogenic bacteria produce large amounts of vesicles during infection48,49. Therefore, the quantity of OMVs used here may mimic conditions met during the infection process.
OMVs protect packaged DNA and proteins from the extracellular environment; thus, OMVs have been proposed to act as vehicles for DNA and protein delivery18. Indeed, here we show that these vesicles shield NDM-1 from the action of extracellular proteases and from the Zn(II) starvation elicited by the immune system.
The worldwide dissemination of the blaNDM gene is not related to the spread of specific clones, plasmids, or a single genetic structure10. We propose that the transient resistance conferred by NDM-1-carrying OMVs may enhance the chances for horizontal gene transfer. This process can be particularly favored in environments like the lung, where the viscous nature of the airway secretions does not readily permit plasmid transfer. The finding of both NDM-1 protein and the blaNDM gene in OMVs secreted by clinical pathogens supports this hypothesis.
The conservation of the lipobox sequence in all NDM alleles (Supplementary Fig. 9) suggests that this evolutionary advantage is common to all known NDMs. Moreover, lipoboxes are present in MBLs from all three subclasses (Supplementary Fig. 10 and Supplementary Table 5), mostly endogenous MBLs from environmental bacteria. Thus, strains harboring lipidated MBLs may represent a reservoir of carbapenem resistance genes with enhanced potential for dissemination, which couples protein transfer via OMVs to horizontal gene transfer. Globomycin blocks NDM-1 processing (Fig. 2c), trapping the enzyme as an inner membrane-associated precursor23 and preventing NDM-1 secretion into vesicles, revealing a possible strategy to prevent dissemination of NDM variants.
These findings create an unprecedented opportunity to design novel antibacterial strategies including MBL inhibition approaches enhancing site-specific metal chelation elicited by the immune system together with interference of bacterial lipidation. A combination of these approaches may offer promise for prolonging the lifespan of the current antibiotics.
Online Methods
Bacterial strains and reagents
Escherichia coli DH5α was used for construction/expression of plasmid pMBLe, microbiological and biochemical studies. Pseudomonas aeruginosa PAO1 was used for expression of the pMBLe-blaNDM-1 construct in cellular fractionation studies. P. aeruginosa 48-1997A50, Acinetobacter junii M797851, P. aeruginosa COL-152 and plasmid pET26-blaNDM-1, kindly provided by Dr. James Spencer (University of Bristol, UK), were used as the sources of blaSPM-1, blaIMP-1, blaVIM-2 and blaNDM-1 genes respectively. Clinical isolates producing NDM-1 used in OMV studies (Providencia rettgeri 15758, Serratia marcescens 17468 and Enterobacter cloacae 17464) were kindly provided by Alejandra Corso (ANLIS “Dr. C. G. Malbran”, Argentina). Unless otherwise noted, all strains were grown aerobically at 37 °C in lysogeny broth (LB) medium supplemented with gentamicin 20 μg/mL when necessary. Chemical reagents were purchased from Sigma-Aldrich, molecular biology enzymes from Promega, and primers from Invitrogen.
Plasmid constructs for expression of MBLs in E. coli and P. aeruginosa
Plasmid pMBLe was constructed with the aim of expressing the different bla genes either in E. coli or P. aeruginosa, retaining the native peptide leader of each lactamase. A C-terminal Strep-tag sequence was added to standardize protein quantitation by Western blot. MBL expression was induced with low concentrations of IPTG (10–50 μM) (Sigma-Aldrich, ≥99% (TLC)) in order to avoid over-expression artifacts, intending to mimic lactamase levels in clinical strains (Figure 5c).
The region of plasmid pTGR1153 containing lacIQ, Ptac and transcription terminators was amplified with primers SacIFw (5’ GTCTAGTGAGCTCCTCTCTCAAGGGCATCG 3’) and NcoIRv (5’ GGAATTAAAGGTACCATGGTTAATTAATATATACTAG 3’) and subcloned into the SacI and NcoI sites of broad-spectrum plasmid pBBR1-MCS5 GmR 54, obtaining the pMBLe plasmid. Full-length bla genes (including their native peptide leaders) were amplified with or without addition of a C-terminal Strep-tag sequence (for comparative protein detection and quantification), and subcloned into NdeI and HindIII (EcoRI in the case of blaIMP-1) sites of pMBLe. All PCRs were carried out using Platinum ® Pfx DNA Polymerase (Invitrogen) with the following thermal cycle: 3 min at 95°C, 30 cycles of 15 s at 95°C, 30 s at 55°C and 1 min at 68ºC, and 10 min at 68ºC. Primers used for this procedure were IMP1NdeIFw (5’ GACATCATATGAGCAAGTTATCTGTATTC 3’), IMP1EcoRIRv (5’ GACGTGAATTCTTAGTTGCTTGGTTTTGATGG 3’) and IMP1StEcoRIRv (5’ GACGTGAATTCCTACTTTTCGAATTGTGGGTGAGACCAGTT GCTTGGTTTTGATGGTTTTTTAC 3’) for blaIMP-1; NDM1NdeIFw (5’ TATACATATGGAATTGCCCAATATTATGCACC 3’), NDM1HindIIIRv (5’ GACGTAAGCTTCTAGCGCAGCTTGTCGGC 3’) and NDM1StHindIIIRv (5’ GACGTAAGCTTCTACTTTTCGAATTGTGGGTGAGACCAGCGCAGCTTGTCGGC 3’) for blaNDM-1; VIM2NdeIFw (5’ GACATCATATGTTCAAACTTTTGAGTAAGTTATTGGTC 3’), VIM2HindIIIRv (5’ GACGTAAGCTTCTACTCAACGACTGAGCGATTTGTG 3’) and VIM2StHindIIIRv (5’ GACGTAAGCTTCTACTTTTCGAATTGTGGGTGAGACCACTCAACGACTGAGCGATTTGTG 3’) for blaVIM-2; SPM1NdeIFw (5’ GTACGTCATATGAATTCACCTAAATCGAGAGC 3’), SPM1HindIIIRv (5’ GTACGTAAGCTTCTACAGTCTCATTTCGCCAAC 3’) and SPM1StHindIIIRv (5’ GTACGTAAGCTTCTACTTTTCGAATTGTGGGTGAGACCACAGTCTCATTTCGCCAAC 3’) for blaSPM-1. The resulting plasmids were introduced into E. coli DH5α or P. aeruginosa PAO1 as previously described55, and expression of bla was induced by addition of 10 μM IPTG. Addition of the Strep-tag at the C-terminus did not affect MBL ability to confer resistance. The minimal inhibitory concentration (MIC) values of cefotaxime and imipenem were identical for strains expressing the wild-type or tagged variants of bla genes (Supplementary Table 1). All experiments were therefore performed in the Strep-tag containing variants.
Construction of MBL mutants
NDM-1 C26A variant gene was generated from pMBLe-blaNDM-1 by site-directed mutagenesis as previously described17 using primers NDM-1-C26AFw (5’ CATTGATGCTGAGCGGGGCGATGCCCGGTGAAATC 3’) and NDM-1-C26ARv (5’ GATTTCACCGGGCATCGCCCCGCTCAGCATCAATG 3’). V-NDM (NDM-1 in which the first 47 amino acids are replaced by the first 42 residues of VIM-2) and N-VIM (VIM-2 in which the first 42 amino acids are replaced by the first 47 residues of NDM-1) were constructed by overlap extension PCR using overlapping primers VIM2-B (5’ ATTCGGTGCGAGCTGGCGGAAAACCAGATCCCCGACCGGAATTTCGC 3’), NDM1-C (5’ GATCTGGTTTTCCGCCCAGCTCGCACCG 3’), NDM1-D (5’ ACCATCGGCAATCTGGTAAAGCCGGACCTCGCCAAACCGTTGGTCGCC 3’), VIM2-E (5’ GAGGTCCGGCTTTACCAGATTGCCG 3’) with external primers VIM2NdeIFw, NDM1NdeIFw, VIM2StHindIIIRv and NDM1StHindIIIRv. The PCR-amplified genes were subcloned into plasmid pMBLe as described above. All constructs were verified by DNA sequencing (University of Maine, USA).
MBL detection and β-lactamase activity measurements
MBL protein levels were determined by SDS-PAGE followed by Western blot with Strep-Tag® II monoclonal antibodies (at 1:1000 dilution from 200 μg/ml solution) 29 (Novagen) and immunoglobulin G-alkaline phosphatase conjugates (at 1:3000 dilution). Protein band intensities were quantified from PVDF membranes with ImageJ software56 and converted to relative protein amounts through a calibration curve constructed under the same experimental conditions. β-Lactamase activity was measured in a JASCO V-670 spectrophotometer at 30ºC in 20 mM Tris pH 8.0 (for living cells) or 10 mM HEPES, 200 mM NaCl pH 7.4 (for membranes and OMVs), in 0.1 cm cuvettes, using 400 μM imipenem as a substrate. Imipenem hydrolysis was monitored at 300 nm (Δε300nm = −9,000 M−1cm−1). In all cases, periplasmic, membrane and OMVs samples were normalized according to total protein concentrations, as quantified with the Pierce® BCA Protein Assay Kit (Thermo Scientific). Western blots with antibodies detecting periplasmic maltose binding protein (Rockland Immunochemicals) and cytoplasmic RNA polymerase (BioLegend) were performed as loading controls for periplasmic extracts and spheroplasts, respectively.
Cellular fractionation and analysis of NDM-1 localization
Periplasm and spheroplasts preparations
Extraction of periplasmic proteins was performed as previously described12. Briefly, 2–3 mL of E. coli (or P. aeruginosa) pMBLe-bla cultures were pelleted and cells were washed once with 20 mM Tris, 150 mM NaCl, pH 8.0. The washed cells were resuspended in 20 mM Tris, 0.1 mM EDTA, 20% w/v sucrose, 1 mg/mL lysozyme (from chicken egg white, Sigma-Aldrich, protein ≥90%), 0.5 mM PMSF, pH 8 (resuspension volume was normalized according to the formula V = 100 μL x OD600 x Vc, where Vc is the starting volume of culture sample), incubated with gentle agitation at 4ºC for 30 min, and finally pelleted, with the periplasmic extract in the supernatant. The pellet consisting of spheroplasts was washed in 20 mM Tris, 0.1 mM EDTA, 20% w/v sucrose, pH 8 and resuspended in the same volume of this buffer.
Total membranes
A 20-mL aliquot of E. coli (or P. aeruginosa) pMBLe-bla culture (OD600 1) induced for 2 h with 50 μM IPTG at 37ºC, was pelleted, resuspended in 10 mM HEPES, 200 mM NaCl, 1 mM PMSF, pH 7.4, and disrupted by sonication. Cell debris were removed by centrifugation at 14,000xg and 4ºC for 20 min, and membranes pelleted by ultracentrifugation at 150,000xg and 4ºC for 1 h. Membrane preparations were washed, ultracentrifuged, resuspended in the same buffer and quantified by determination of the total protein content using the Pierce® BCA Protein Assay Kit.
Solubilization of membrane protein
Membrane proteins were sequentially extracted as previously described57. In brief, total membranes were pelleted by ultracentrifugation at 150,000xg and 4ºC for 1 h, resuspended in 1 M KCl, incubated for 30 min on ice and ultracentrifuged as before, obtaining loosely associated peripheral proteins in supernatant. The remaining membranes were then treated with 0.1 M Na2CO3 for 30 min on ice to release peripheral proteins associated by strong electrostatic interactions, pelleted and finally treated with Triton X-100 (0.0016 and 0.23% w/v) (Sigma-Aldrich, laboratory grade), to extract integral or hydrophobically-associated proteins in detergent micelles.
Separation of inner and outer membranes
Spheroplasts prepared from 250-mL cultures (OD600 = 1) of E. coli (or P. aeruginosa) harboring either pMBLe-blaNDM-1 or pMBLe-blaN-VIM were resuspended in 20 mM Tris, 1 mM PMSF, pH 8 and disrupted by sonication. Cell debris was removed by centrifugation at 14,000xg and 4ºC for 20 min, membrane vesicles pelleted by ultracentrifugation at 150,000xg and 4ºC for 1 h, and resuspended in 20% w/v sucrose, 20 mM Tris, pH 8. Membrane vesicles were loaded on top of a continuous sucrose gradient (30 to 55% w/v) in 20 mM Tris, pH 8 and ultracentrifuged for 16 h and 4ºC at 160,000xg in a Beckman SW 40 Ti rotor. Fractions along the gradient were analyzed by SDS-PAGE, Western blot and β-lactamase activity to assess the localization of NDM-1 or N-VIM. Inner and outer membrane vesicles were identified by NADH oxidase activity as previously described57, and mass spectrometry characterization of protein from excised gel bands (trypsin digestion followed by MS/MS, performed at Mass Spectrometry Laboratory for Protein Sequencing, The Cleveland Clinic Foundation, USA).
Analysis of NDM-1 orientation in outer membranes
Spheroplasts and whole cell suspensions were treated with Proteinase K (Fungal, Invitrogen, >20 U/mg) as previously described58, and analyzed by SDS-PAGE and immunoblotting.
Inhibition of lipoprotein processing
Liquid cultures of E. coli pMBLe-blaNDM-1 or pMBLe-blaNDM-1 C26A were grown at 37ºC until OD600 0.8. Globomycin (Sigma-Aldrich, ≥98% (HPLC)) was added at a final concentration of 0, 25 or 125 μg/ml and growth was continued for 10 min at 28ºC. Finally, lactamase expression was induced for 1 h at 28ºC with 100 μM IPTG and aliquots taken for Western blot analysis.
Minimum inhibitory concentration (MIC) determinations
Cefotaxime and imipenem MIC determinations were performed in LB medium using the agar macrodilution method according to CLSI guidelines59. In order to measure the effect of Zn(II) availability on antibiotic resistance, the growth medium was supplemented with varying concentrations of the metal chelator dipicolinic acid (DPA) (Merck, >98%). Imipenem MIC determinations in the presence of human calprotectin were performed in LB medium supplemented with 0, 100 or 300 μg/mL CP, using the broth microdilution method59. CP used in these studies was overexpressed and purified as previously described15. The antibacterial activity of the purified protein was confirmed against Escherichia coli ATCC 2592215. In all cases, bla expression was induced with 10 μM IPTG.
Relative MICs as plotted in Figures 1A, 4A and Supplementary Figure 1 were calculated as (MICMBL – MICcontrol)/(MICMBL + 0 μM DPA – MICcontrol + 0 μM DPA), where MICMBL and MICcontrol refer to values measured for E. coli DH5α pMBLe-bla or pMBLe, respectively, under each set of conditions, and MICMBL + 0μM DPA and MICcontrol + 0μM DPA, the corresponding values in media without addition of DPA.
Effect of external Zn(II) depletion in MBL protein levels and living cells β-lactamase activity
E. coli pMBLe-bla cells were grown at 37ºC up to an OD600 of 0.4. MBL expression was induced by the addition of 50 μM IPTG, and growth was continued at 37ºC for 1 h, and then at 20ºC for 2 h (to slow down cellular metabolism). At this time, cultures were divided in two equal parts. One portion was treated with 1000 μM DPA and the other kept as an untreated control, and both cultures were grown at 20ºC. Aliquots of DPA-treated and untreated cultures were taken at different time intervals after DPA addition: 0, 10 (only for DPA-treated), 90 and 960 min, and periplasmic extraction performed as described to yield the corresponding periplasm and spheroplast fractions. Imipenem hydrolysis was measured in living cells from the aliquots taken at 90 and 960 min from DPA-treated and control samples. In brief, cells pelleted from 1 mL culture were washed and resuspended in 300 x OD600 μL of Chelex 100-treated (manufacturer protocol) 20 mM Tris, pH 8 buffer. β-Lactamase activity was measured from 5 μL of cell suspension in 300 μL reaction media. Data were corrected by subtraction of basal hydrolysis rates observed for non-MBL-producing control strain. Protein and activity values in DPA-treated samples were reported relative to the corresponding values in untreated samples.
Detection of cellular MBLs by immunofluorescence microscopy
E. coli cells (either lactamase-expressing or OMV treated) were washed twice with PBS, fixed with 0.5 ml of 3% paraformaldehyde solution in PBS for 10 min, and permeabilized with 0.1% Triton X-100 solution in PBS for 5 min. Subsequently, cells were incubated with Strep-Tag II monoclonal antibodies (1:100) and detected by incubation with anti-mouse Cy3-conjugated secondary antibodies (1:1000) (Invitrogen). Cells were mounted with SlowFadeAntifade reagent (Molecular Probes) and finally analyzed in a Nikon Eclipse E800 fluorescence microscope (Nikon, Japan).
Expression and purification of soluble NDM-1
The NDM-1 gene coding for residues 39 to 270 was cloned into a modified version of pET28 plasmid, kindly provided by Dr. Rodolfo Rasia (IBR, CONICET-UNR) in which the thrombin cleavage site is replaced by a TEV protease cleavage site. The protein was overexpressed in E. coli BL21(DE3). The bacterial culture was grown at 37 ºC in M9 minimal medium until it reached OD600 0.6. Then, protein expression was induced by addition of 0.5 mM IPTG and 0.5 mM ZnSO4 to the medium. Cells were incubated overnight at 18 ºC, harvested by centrifugation and resuspended in lysis buffer (50 mM Tris, 200 mM NaCl, pH 8.0). The cells were lysed by sonication and the insoluble material was removed by centrifugation. The protein was purified using Ni-Sepharose affinity chromatography, the His-tag was cleaved by treatment with His6-tagged TEV protease (Sigma-Aldrich, manufacturer protocol), and the tag was removed by a second chromatographic step with the Ni-Sepharose resin in which the protease was also retained. The purified protein was concentrated using a 10-kDa MW cut-off Centricon device (Millipore, Bedford, MA, USA). Protein concentrations were determined from the absorbance at 280 nm using a molar absorption coefficient (ε280) of 28,000 M−1 cm−1 (calculated from aromatic residues using Expasy ProtParam, available at web.expasy.org/protparam/). Apo NDM-1 was obtained by two rounds of dialysis of the purified holoprotein (ca. 200 μM) against 100 volumes of 10 mM HEPES, 200 mM NaCl, 20 mM EDTA, pH 7.5 over a 12-h period under stirring. EDTA was removed from the resulting apoenzyme solution by three dialysis steps against 100 volumes of 10 mM HEPES, 1 M NaCl, pH 7.5, Chelex 100, and three dialysis steps against 100 volumes of 10 mM HEPES, 200 mM NaCl, pH 7.5, Chelex 100. All buffer solutions used to prepare the apoenzymes were treated by extensive stirring with Chelex 100 (Bio-Rad)11.
Limited proteolysis of holo and apo recombinant NDM-1
Aliquots (200 μg) of pure recombinant holo or apo NDM-1 were treated with 2.5 μg proteinase K in 10 mM Tris, 5 mM CaCl2, pH 8 at 16ºC. Aliquots were taken at different times, the reaction was quenched with 5 mM PMSF, and samples were subjected to SDS-PAGE and Coomassie blue staining.
Purification of Outer Membrane Vesicles (OMVs)
OMVs were purified from early stationary phase cultures of E. coli and clinical strains. Briefly, 250 mL of LB medium was inoculated with 3 mL of saturated E. coli pMBLe-bla culture, grown at 37ºC up to OD600 of 0.4, induced with 20 μM IPTG, and growth continued overnight with agitation. In the case of clinical strains, 250 mL of LB broth was inoculated with 3 mL of saturated culture and grown for 5 h at 37ºC. The cells were harvested and the supernatant filtered through a 0.45-μm membrane (Millipore). Ammonium sulfate was added to the filtrate at a concentration of 55% w/v, followed by incubation with stirring at 4ºC for 3 h (E. coli) or overnight (clinical strains). Precipitated material was separated by centrifugation at 12,800xg for 10 min, resuspended in 10 mM HEPES, 200 mM NaCl, pH 7.4 and dialyzed overnight against >100 volumes of the same buffer. Next, the samples were filtered through a 0.45-μm membrane, layered over an equal volume of 50 % w/v sucrose solution and ultracentrifuged at 150,000xg for 1 h and 4ºC. Pellets, containing OMVs, were washed once with 10 mM HEPES, 200 mM NaCl, pH 7.4 and further purified by sucrose isopycnic density gradient ultracentrifugation in equal buffer. Fractions containing OMVs (those possessing an SDS-PAGE pattern corresponding to the main outer membrane proteins) were pooled, washed with 10 mM HEPES, 200 mM NaCl, pH 7.4 and stored at –80ºC until use. 10 microliters of a 300 μg/ml OMV preparation was plated on LB agar and determined to be sterile. OMV yields were of ~200 μg protein/250 mL culture. Quality of vesicle preps was assessed by negative staining transmission electron microscopy, as described elsewhere30. Briefly, vesicles were fixed in 4% glutaraldehyde in 10 mM HEPES, 200 mM NaCl, pH 7.4 for 30 min at room temperature, stained with 2% uranyl acetate and visualized in a Zeiss EM 109T Transmission Electron Microscope. Finally, the absence of cellular contaminants due to cell lysis during culture growth or cell manipulation was verified by immunodetection of cytoplasmic GroEL in OMV preps. Additionally, OMVs and samples from different fractions of E. coli pMBLe-blaNDM-1 (cytoplasmic, periplasmic, inner-membrane and outer-membrane58,60) were analyzed by SDS-PAGE and their protein patterns compared. GroEL antibodies were kindly provided by Dr. Alejandro Viale (IBR).
Localization of NDM-1 in OMVs
A 16-μg aliquot of intact or previously lysed OMVs were incubated with 100 μg proteinase K in 10 mM Tris, 5 mM CaCl2, pH 8 at 25ºC. Aliquots were taken at 10 and 60 min, protease was inactivated with 5 mM PMSF, and samples analyzed by SDS-PAGE and immunoblotting. OMV lysis was performed by treatment with 0.125% v/v Triton X-100 for 30 min at 37°C, as previously described30.
Evaluation of β-lactam resistance transfer by OMVs
β-lactam sensitive E. coli cells, carrying empty pMBLe plasmid, were grown at 37ºC in 8 mL LB medium to an OD600 of 0.5, centrifuged at 6,000xg for 10 min and resuspended in 240 μL of 10 mM HEPES, 200 mM NaCl, pH 7.4. 1 μL of this suspension was mixed with 8 μg of NDM-1, NDM-1 C26A or non-MBL-producing OMVs, and incubated statically for 1 h at 37ºC. The mixture was diluted 1/10 in 10 mM HEPES, 200 mM NaCl, pH 7.4 and 3 μL drops used for MIC determinations in LB-agar plates.
Effect of calprotectin on OMV lactamase activity
OMVs harboring NDM-1 or NDM-1 C26A (0.8 μg) were incubated for 2 min at 30ºC in 10 mM HEPES, 200 mM NaCl, pH 7.5 buffer with different amounts of calprotectin (0–300 μg/mL). After incubation, 500 μM imipenem was added and the remnant β-lactamase activity was determined spectrophotometrically. The same experiment was also performed with purified soluble recombinant NDM-1, adjusting the protein concentration to match the activity of the NDM-1 OMV sample in absence of calprotectin.
Purification of DNA from vesicles
OMV samples (10 μg) were treated with 50 μg/mL DNase I (Sigma) for 1 h at 37ºC to hydrolyze surface-associated DNA and free DNA in suspension. Reactions were stopped by incubation for 10 min at 80ºC. DNase-treated vesicles were then lysed with 0.125% Triton X-100 solution for 30 min at 37°C, and DNA was purified by the use of Wizard® SV Gel and PCR Clean-Up System (Promega). The DNA was quantified by using a Nanodrop 2000 (Thermo Scientific). Average yields were as follows: E. coli pMBLe-blaNDM-1 13 ± 4 ng/μg OMV, P. rettgeri 59 ± 30 ng/μg OMV, S. marcescens 28 ± 9 ng/μg OMV, E. cloacae 51 ± 14 ng/μg OMV.
PCR amplification of blaNDM-1 was performed using 50 ng DNA as template, with primers NDM1NdeIFw and NDM1HindIIIRv, as already described. Amplifications were carried out from two independent preparations of vesicle DNA. Negative controls indicated that DNA contamination did not occur.
Bioinformatic search of lipobox-containing lactamases
Homologues to subclass B1, B2 and B3 MBLs were extracted from the NCBI non-redundant protein database, and depurated for further analysis. In the case of B1 and B2 MBLs, sequences not containing the typical motifs HXHXD and GGC, for B1, or NXHXD and GNC, for B2, were discarded. In a similar way, B3 homologues not possessing the HXHXDH motif and sequence identity >25% to GOB-1 were excluded. Afterwards, proteins containing the lipobox consensus sequence [LVI][ASTVI][GAS][C] within their first 50 residues were selected and analyzed with DOLOP (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/) and LipoP (http://www.cbs.dtu.dk/services/LipoP/) lipoprotein predictors. Positive sequences were aligned with the structure-assisted multiple sequence alignment tool Expresso (http://www.tcoffee.org/) and phylogenetic trees were constructed (LG substitution model and 100 bootstraps) using the maximum likelihood algorithm PhyML, available at http://www.phylogeny.fr. Unrooted trees were drawn with DrawTree from the Phylip package (available at http://www.evolution.genetics.washington.edu/phylip.html).
Supplementary Material
Acknowledgments
We thank Dr. James Spencer (University of Bristol, UK), Hugo Menzella (University of Rosario, Argentina) and Rodolfo Rasia (IBR, CONICET-UNR, Argentina) for providing plasmids pET26-blaNDM-1, pTGR11 and pET28-TEV respectively, Alejandra Corso (ANLIS, Argentina) for providing clinical strains, and Alejandro Viale (IBR) for GroEL antibodies. This research was supported by grants from the National Institutes of Health (R01AI100560) to AJV and RAB and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) to AJV. RAB acknowledges support from NIH under award numbers R01AI072219, R01AI063517, and R01AI100560 and by funds and/or facilities provided by the Louis Stokes Cleveland Department of Veterans Affairs Medical Center and the VISN 10 Geriatric Research, Education and Clinical Care Center (VISN 10) of the Department of Veterans Affairs. EMN thanks support from the Kinship Foundation (Searle Scholars Program) and MIT Department of Chemistry.
Footnotes
Author contributions
LJG and GB performed the microbiological, molecular biology and biochemical experiments. TGN purified calprotectin. LJG, GB, TGN, EMN, RAB and AJV analyzed and discussed data. LJG, GB, RAB, and AJV wrote the paper, and all authors discussed the results and commented on the manuscript.
Competing financial interests statement
The authors declare that there are no conflicts of interest.
References
- 1.Hede K. Antibiotic resistance: An infectious arms race. Nature. 2014;509:S2–3. doi: 10.1038/509S2a. [DOI] [PubMed] [Google Scholar]
- 2.CDC. Antibiotic Resistance Threats in the United States 2013. Centers for Disease Control and Prevention; Atlanta, GA: 2013. [Google Scholar]
- 3.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowder MW, Spencer J, Vila AJ. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 5.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dortet L, Poirel L, Nordmann P. Worldwide Dissemination of the NDM-Type Carbapenemases in Gram-Negative Bacteria. Biomed Res Int. 2014;2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, et al. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One. 2014;8:e64857. doi: 10.1371/journal.pone.0064857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y, et al. Proliferation of Multidrug-Resistant New Delhi Metallo-β-lactamase Genes in Municipal Wastewater Treatment Plants in Northern China. Environ Sci Technol Lett. 2014;1:26–30. [Google Scholar]
- 10.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother. 2011;55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez JM, et al. Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat Chem Biol. 2012;8:698–700. doi: 10.1038/nchembio.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran-Barrio J, Limansky AS, Viale AM. Secretion of GOB metallo-β-lactamase in Escherichia coli depends strictly on the cooperation between the cytoplasmic DnaK chaperone system and the Sec machinery: completion of folding and Zn(II) ion acquisition occur in the bacterial periplasm. Antimicrob Agents Chemother. 2009;53:2908–2917. doi: 10.1128/AAC.01637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerasi M, Ammendola S, Battistoni A. Competition for zinc binding in the host-pathogen interaction. Front Cell Infect Microbiol. 2013;3:108. doi: 10.3389/fcimb.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brophy MB, Hayden JA, Nolan EM. Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J Am Chem Soc. 2012;134:18089–18100. doi: 10.1021/ja307974e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbin BD, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 17.Meini MR, Tomatis PE, Weinreich DM, Vila AJ. Quantitative Description of a Protein Fitness Landscape Based on Molecular Features. Mol Biol Evol. 2015;32:1774–1787. doi: 10.1093/molbev/msv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez LJ, Moreno DM, Bonomo RA, Vila AJ. Host-specific enzyme-substrate interactions in SPM-1 metallo-β-lactamase are modulated by second sphere residues. PLoS Pathog. 2014;10:e1003817. doi: 10.1371/journal.ppat.1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King D, Strynadka N. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011;20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun. 2011;79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rokney A, et al. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J Mol Biol. 2009;392:589–601. doi: 10.1016/j.jmb.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Hussain M, Ichihara S, Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980;255:3707–3712. [PubMed] [Google Scholar]
- 24.Randall LB, Dobos K, Papp-Wallace KM, Bonomo RA, Schweizer HP. Membrane Bound PenA β-lactamase of Burkholderia pseudomallei. Antimicrob Agents Chemother. 2015;60:1509–14. doi: 10.1128/AAC.02444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bootsma HJ, van Dijk H, Verhoef J, Fleer A, Mooi FR. Molecular characterization of the BRO β-lactamase of Moraxella (Branhamella) catarrhalis. Antimicrob Agents Chemother. 1996;40:966–972. doi: 10.1128/aac.40.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaar V, Nordstrom T, Morgelin M, Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother. 2011;55:3845–3853. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bomberger JM, et al. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devos S, et al. The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front Microbiol. 2015;6:298. doi: 10.3389/fmicb.2015.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaar V, Uddback I, Nordstrom T, Riesbeck K. Group A streptococci are protected from amoxicillin-mediated killing by vesicles containing β-lactamase derived from Haemophilus influenzae. J Antimicrob Chemother. 2014;69:117–120. doi: 10.1093/jac/dkt307. [DOI] [PubMed] [Google Scholar]
- 30.Rumbo C, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z, Gunasekera TS, Spadafora L, Bennett B, Crowder MW. Metal content of metallo-β-lactamase L1 is determined by the bioavailability of metal ions. Biochemistry. 2008;47:7947–7953. doi: 10.1021/bi8004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y, et al. Structure of apo- and monometalated forms of NDM-1--a highly potent carbapenem-hydrolyzing metallo-β-lactamase. PLoS One. 2011;6:e24621. doi: 10.1371/journal.pone.0024621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez JM, Buschiazzo A, Vila AJ. Evidence of adaptability in metal coordination geometry and active-site loop conformation among B1 metallo-β-lactamases. Biochemistry. 2010;49:7930–7938. doi: 10.1021/bi100894r. [DOI] [PubMed] [Google Scholar]
- 35.Borra PS, et al. Crystal structures of Pseudomonas aeruginosa GIM-1: active-site plasticity in metallo-β-lactamases. Antimicrob Agents Chemother. 2013;57:848–854. doi: 10.1128/AAC.02227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez Valladares M, et al. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry. 1997;36:11534–11541. doi: 10.1021/bi971056h. [DOI] [PubMed] [Google Scholar]
- 37.Selevsek N, et al. Zinc ion-induced domain organization in metallo-β-lactamases: a flexible “zinc arm” for rapid metal ion transfer? J Biol Chem. 2009;284:16419–16431. doi: 10.1074/jbc.M109.001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Periyannan G, Shaw PJ, Sigdel T, Crowder MW. In vivo folding of recombinant metallo-β-lactamase L1 requires the presence of Zn(II) Protein Sci. 2004;13:2236–2243. doi: 10.1110/ps.04742704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wommer S, et al. Substrate-activated zinc binding of metallo-β-lactamases: physiological importance of mononuclear enzymes. J Biol Chem. 2002;277:24142–24147. doi: 10.1074/jbc.M202467200. [DOI] [PubMed] [Google Scholar]
- 40.Johne B, et al. Functional and clinical aspects of the myelomonocyte protein calprotectin. Mol Pathol. 1997;50:113–123. doi: 10.1136/mp.50.3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haley KP, et al. The Human Antimicrobial Protein Calgranulin C Participates in Control of Helicobacter pylori Growth and Regulation of Virulence. Infect Immun. 2015;83:2944–2956. doi: 10.1128/IAI.00544-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glaser R, et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 43.Pederick VG, et al. ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci Rep. 2015;5:13139. doi: 10.1038/srep13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kesty NC, Kuehn MJ. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J Biol Chem. 2004;279:2069–2076. doi: 10.1074/jbc.M307628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haurat MF, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286:1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veith PD, et al. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res. 2014;13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- 47.Santos S, et al. Outer membrane vesicles (OMV) production of Neisseria meningitidis serogroup B in batch process. Vaccine. 2012;30:6064–6069. doi: 10.1016/j.vaccine.2012.07.052. [DOI] [PubMed] [Google Scholar]
- 48.Irazoqui JE, et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitagawa R, et al. Biogenesis of Salmonella enterica serovar typhimurium membrane vesicles provoked by induction of PagC. J Bacteriol. 2010;192:5645–5656. doi: 10.1128/JB.00590-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy TA, Simm AM, Toleman MA, Jones RN, Walsh TR. Biochemical characterization of the acquired metallo-β-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2003;47:582–587. doi: 10.1128/AAC.47.2.582-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fiorilli G, et al. Emergence of metallo-β-lactamases in Acinetobacter spp clinical isolates from Argentina. Rev Esp Quimioter. 2010;23:100–102. [PubMed] [Google Scholar]
- 52.Marchiaro P, et al. Biochemical characterization of metallo-β-lactamase VIM-11 from a Pseudomonas aeruginosa clinical strain. Antimicrob Agents Chemother. 2008;52:2250–2252. doi: 10.1128/AAC.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ravasi P, Peiru S, Gramajo H, Menzella HG. Design and testing of a synthetic biology framework for genetic engineering of Corynebacterium glutamicum. Microb Cell Fact. 2012;11:147. doi: 10.1186/1475-2859-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 55.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X, Brown T, Tai PC. Identification and characterization of protease-resistant SecA fragments: secA has two membrane-integral forms. J Bacteriol. 1998;180:527–537. doi: 10.1128/jb.180.3.527-537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190:7523–7531. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard - Ninth Edition. CLSI Document M07-A9. Clinical Laboratory Standards Institute; Wayne, PA: 2012. [Google Scholar]
- 60.Park M, et al. Isolation and characterization of the outer membrane of Escherichia coli with autodisplayed Z-domains. Biochim Biophys Acta. 2015;1848:842–847. doi: 10.1016/j.bbamem.2014.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.