Abstract
Many psychiatric drugs act on multiple targets and therefore require screening assays that encompass a wide target space. With sufficiently rich phenotyping, and a large sampling of compounds, it should be possible to identify compounds with desired mechanisms of action based on their behavioral profiles alone. Although zebrafish (Danio rerio) behaviors have been used to rapidly identify neuroactive compounds, it remains unclear exactly what kind of behavioral assays might be necessary to identify multi-target compounds such as antipsychotics. Here, we developed a battery of behavioral assays in larval zebrafish to determine if behavioral profiles could provide sufficient phenotypic resolution to identify and classify psychiatric drugs. Using the antipsychotic drug haloperidol as a test case, we found that behavioral profiles of haloperidol-treated animals could be used to identify previously uncharacterized compounds with desired antipsychotic-like activities and multi-target mechanisms of action.
Introduction
Polygenic psychiatric disorders, such as schizophrenia, will likely require systems-modulating therapeutics, which are difficult to identify without complex in vivo readouts. The most effective antipsychotic drugs bind to many receptors in the nervous system and unlike “magic bullet” drugs (including many antibiotics and some chemotherapeutics that act on single molecular targets), antipsychotics are thought to act via poly-pharmacology on many targets simultaneously1. The prototypes of most antipsychotic drugs including chlorpromazine, haloperidol, and clozapine were originally discovered via their behavioral phenotypes in vivo2. It has been difficult, but not impossible3, to identify antipsychotics and other multi-target drugs using traditional target-based assays on isolated receptors in vitro4. Given that there are no known biomarkers for most psychiatric disorders, behavior phenotyping is an attractive endpoint for central nervous systems (CNS) drug screens. However the time, space, and financial resources required for high-throughput (HT) behavioral drug screening using traditional animal models are prohibitive.
Antipsychotic drugs, including haloperidol and clozapine bind to a wide range of targets including dopamine, serotonin, histamine and adrenergic receptors that collectively contribute to their efficacy and their side effects1,5,6. Haloperidol, a typical antipsychotic drug and potent D2 antagonist is known to bind at least 20 molecular targets in the human brain1. Used to treat patients since the 1960s, haloperidol is one of the most efficacious therapeutics to treat schizophrenia and has been designated a core drug on the WHO Model List of Essential Medicines7,8. Clozapine, an atypical antipsychotic, binds more tightly to serotonin receptors9 and causes fewer extrapyramidal side effects10, but can also cause rare and fatal agranulocytosis and myocarditis as well as seizures11. Attempts to develop better antipsychotics by enhancing potency and selectivity at specific receptors have been largely unsuccessful1,12, underscoring the unmet need for compounds with multi-target mechanisms. Identifying compounds with antipsychotic-like phenotypes may elucidate new therapeutic mechanisms and accelerate the development of therapeutics with improved safety and side effect profiles.
With sufficiently rich behavioral phenotyping, and a large sampling of compounds, it should be possible to identify neuroactive compounds that possess desired multi-target mechanisms of action. Despite their differences, the receptors, cell types and neuronal architectures that underlie human and zebrafish CNS functions are highly conserved13,14. Antipsychotics, antidepressants and anxiolytics affect swimming patterns in adult and larval zebrafish via conserved molecular targets15–18. Although zebrafish behaviors have been used to rapidly identify neuroactive compounds and predict their mechanisms of action, such predictions have so far been limited to compounds with relatively simple mechanisms, like enzyme inhibitors13,14,19–21. It remains unclear whether larval assays can provide sufficient phenotypic resolution to identify and classify compounds with more complex multi-target mechanisms such as antipsychotics.
Results
A battery of scalable behavioral assays
To generate rich behavioral profiles that can resolve complex and subtle differences between compounds, we devised a battery of 10 behavioral assays in larval zebrafish. The battery included two acoustic stimulus response (ASR) assays, five visual stimulus response (VSR) assays and three assays that combined acoustic and visual stimulus responses (AVSR). Together, these assays used five different stimuli including red light (600 nm), blue light (420 nm), violet light (405 nm), low magnitude sound (60 dB) and high magnitude sound (70 dB) (Supplementary Results, Supplementary Fig. 1). These stimuli were presented in various contexts and combinations that elicited robust and reproducible patterns of activity as measured by a motion index (MI) that quantifies the amount of larval zebrafish motor activity in each well of a 96-well plate (Fig. 1a). For example, in the ASR1 assay, low-magnitude acoustic stimuli elicited weak motor responses. In the ASR2 assay, high-magnitude acoustic stimuli elicited strong motor responses that habituated over time. The VSR assays contained various combinations of violet, blue and red stimuli. We found that violet light increased motor activity, whereas red or blue light reduced motor activity (Fig. 1a). In the AVSR assays, acoustic and light stimuli were combined to diversify stimulus contexts. These zebrafish assays were not intended to simulate any specific behavioral phenotype or disorder in humans; the molecular mechanisms and neuronal circuitry that control these responses are still incompletely understood. However, we hypothesized that the wide variety of stimuli and contexts would provide a means to reproducibly elicit behavioral signatures that are uniquely affected by specific classes of neuroactive compounds such as antipsychotic drugs.
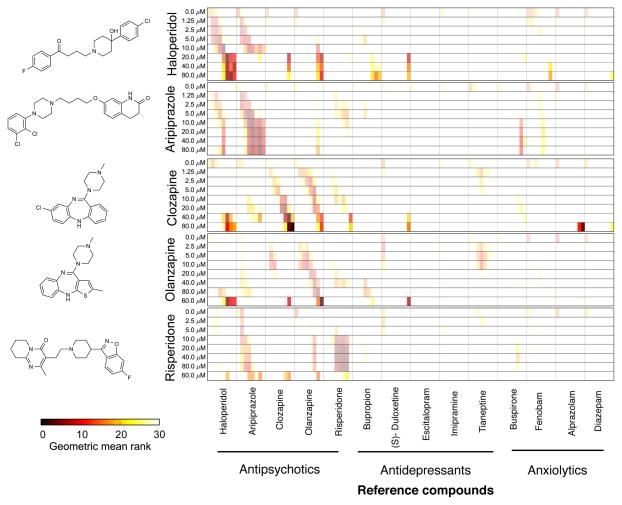
Figure 1. Antipsychotics and other psychiatric drugs affect zebrafish behavior.
(a) Plot showing the average motion index (MI) of control animals as a function of time. Assay names and their order in the battery are indicated as x-axis labels. The MI time series (black) is the average of 48 time-series, each obtained from a well containing 8 larvae and the shaded area (gray) covers ± 3*sem. All wells were located on the same 96-well plate. (b) Line plots showing phenotypic distance (y-axis) of the indicated compounds from DMSO control phenotypes at the indicated concentration (x-axis). The phenotypic distance of the DMSO phenotype from itself (0μM) is arbitrarily set to 0.1. The phenotype for each condition (compound & concentration) is the average of 36 time-series obtained from 36 wells screened over 3 daily experiments of 12 wells each. Each well contained 8 larvae. The phenotypic distances shown are distances between average phenotypes, so effectively n=1. Marker size represents, for each phenotype, the MI averaged over time, i.e. the area under the curve for the MI time series. Colors represent different classes of psychiatric drugs (Blue, antipsychotics; yellow, antidepressants; purple, anxiolytics). (c) Multi-dimensional scaling representation of the pairwise distances between MIs of animals treated with the indicated compounds. Larger marker sizes indicate greater concentrations. Gray circles represent equal volumes of DMSO. Each data point is the average of the same 36 time series used for panel b.
Psychiatric drugs alter zebrafish behaviors
To determine how different classes of psychiatric drugs affected the various zebrafish behaviors assessed by this battery of assays, we generated a reference set of behavioral profiles from animals treated with 14 psychiatric drugs in three therapeutic classes including antipsychotics, antidepressants and anxiolytics (Supplementary Table 1). These 14 drugs were selected to test the behavioral battery against a range of compounds with diverse structures and mechanisms that span a range of therapeutic activities. To quantify how these treatments affected behavior, we compared both the shape and magnitude of their behavior profiles to control profiles. Most compounds changed the shape of the behavioral profile, increasing the phenotypic distance from control animals in a dose-dependent manner (Fig. 1b, y-axis). At high concentrations, many compounds also caused a dose-dependent decrease in the average MI magnitude (Fig. 1b, marker size). Only one drug out of 14 (tianeptine, an antidepressant) failed to show an effect possibly due to poor bioavailability in zebrafish. Interestingly, duloxetine, another antidepressant, reduced the average MI magnitude at low concentrations, but then brought it back toward normal levels at the highest concentrations tested. Perhaps this pattern reflects duloxetine’s engagement of different targets at different concentrations, a well-established phenomenon of small molecule drugs22. These data indicate that different psychiatric medicines had measurable affects on larval zebrafish in the high-throughput battery of behavioral assays.
To determine if different classes of psychiatric drugs caused behavioral phenotypes that are characteristic of that class, we calculated all pairwise distances between the behavioral profiles of animals treated with compounds in each therapeutic class, and represented the data set in two dimensions using multi-dimensional scaling (Fig. 1c). We found that behavioral profiles of antipsychotics (at various concentrations) tended to cluster more closely with other antipsychotics (at various concentrations) than with antidepressants or anxiolytics. In addition, antidepressants and anxiolytics also appeared to cluster into distinct behavioral groups, although the phenotypic differences between them were less pronounced (Fig. 1c). Because the behavioral profiles of antipsychotic-treated animals clustered together, this suggests that the battery of behavioral assays captured class-specific effects of antipsychotic drugs.
Antipsychotic drugs cause specific behavioral profiles
To determine if these behavioral profiles could be used to discriminate between different types of psychiatric drugs, we calculated phenotypic distances between each behavioral profile in the reference set (Supplementary Table 1). We then used the average behavioral profile of each compound at each concentration to rank all other profiles by their similarity to the query profile. We refer to this kind of analysis as a “phenoBlast” because the process of ranking compounds based on their behavioral similarity is reminiscent of using the popular BLAST algorithm23 to identify related nucleotide sequences (although the underlying algorithms are unrelated). We found that the behavioral profiles of haloperidol-treated animals were most similar to profiles from other animals independently treated with haloperidol on different days (Fig. 2). Haloperidol profiles also resembled the profiles caused by aripiprazole (at low concentrations) as well as clozapine and olanzapine (at high concentrations) (Fig. 2). Similarly, the behavioral profiles of aripiprazole-treated animals matched most closely the behavioral profiles of animals treated with aripiprazole and haloperidol (Fig. 2). Profiles from animals treated with the atypical antipsychotic clozapine were most similar to profiles from other wells treated with clozapine or with olanzapine (Fig. 2). Olanzapine-treated wells were most similar to other wells treated with olanzapine (Fig. 2). Finally, risperidone profiles were similar to both risperidone and aripiprazole (Fig. 2). These data indicated that the behavioral profiles of antipsychotic drugs were most similar to the profiles of other antipsychotic drugs, rather than the profiles of animals treated with other psychiatric medicines (Fig. 2). Like antipsychotic profiles, the behavioral profiles caused by antidepressants and anxiolytics also preferentially matched profiles from within the same mechanistic class (Supplemenatry Fig. 2). These data suggest that the behavioral profiles generated by the phenotyping battery reflect specific drug mechanisms and may be useful for identifying uncharacterized compounds with similar mechanisms and phenotypes.
Figure 2. Antipsychotic drugs cause specific behavioral profiles in the test battery.
Chemical structures (left) of each reference compound queried and heat maps (right) showing a matrix comparing queries (rows) and reference compounds (columns). Note that each cell of the matrix is divided into eight segments representing increasing concentrations of the indicated reference compound (left to right). Heat map color represents the phenotypic similarity ranking as indicated in the color bar (see Methods). Color saturation represents the magnitude of the behavioral distance from DMSO controls, such that darker colors represent larger magnitudes. Each condition (compound & concentration) is the average of 36 time-series obtained from 36 wells screened over 3 daily experiments of 12 wells each. Each well contained 8 larvae.
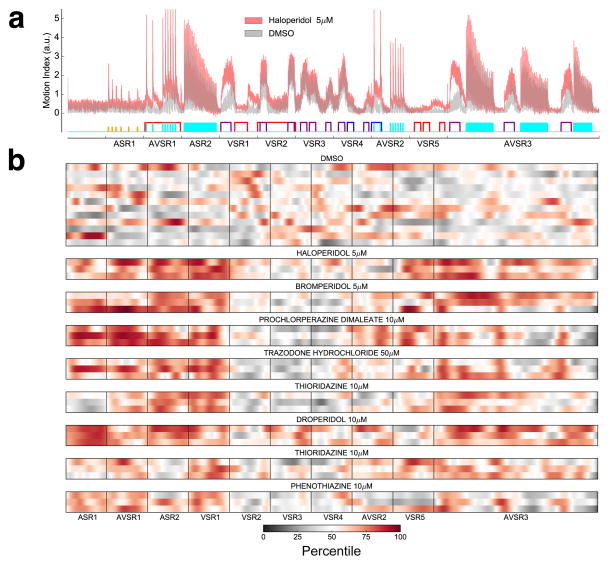
Among the compounds in the test set, behavioral profiles from haloperidol treated animals were among the most effective at recalling other antipsychotic drugs (Fig. 2). To further understand how haloperidol affected zebrafish behavior, we analyzed zebrafish treated with different concentrations (1.25–80 μM). We found that haloperidol reduced motor activity at very high concentrations (20– 80 μM) as shown previously in both zebrafish and mice18,24 (Fig. 1b). Unexpectedly, at lower concentrations, haloperidol (2.5 –10 μM) changed the shape of the behavioral profile without reducing its average magnitude (Fig. 1b). To analyze this low concentration phenotype more closely, we treated half of a 96-well plate (48 wells) with haloperidol (5 μM) and the other half with DMSO (equal volume) to generate high-resolution average behavioral profiles of each treatment condition (Fig. 3a). Given that haloperidol is known to reduce locomotor activity in larval zebrafish18, we were surprised to find that haloperidol (5 μM) increased MI magnitude at many points in the time series. Hierarchical clustering of behavioral profiles based on pair-wise phenotypic distances revealed that haloperidol and DMSO treated wells clustered into different groups (Supplementary Fig. 3), indicating that this behavioral phenotype is robust and reproducible. To determine if other antipsychotic drugs can also stimulate zebrafish motor behavior, we analyzed five additional typical antipsychotic drugs including two butyrophenones (bromperidol and droperidol) and three phenothiazines (prochlorperazine, thioridazine and phenothiazine). We found that all five of these antipsychotics also cause haloperidol-like phenotypes (Fig. 3b). These data suggest that many antipsychotic drugs cause similar patterns of behavior and that behavioral profiling in larval zebrafish may be useful for identifying compounds with antipsychotic-like activity.
Figure 3. Haloperidol causes complex behavioral phenotypes in the zebrafish.
(a) Average MI of animals treated with haloperidol (5μM) or DMSO (n= 48 wells per condition). (b) Heat maps showing the effect of indicated antipsychotic compounds on zebrafish motor activity. For every drug treatment, each of the three rows represents a single well (8 larvae). The x-axis indicates time and specific assays administered as shown in panel a. Color indicates percentile ranking of the motion index relative to DMSO controls (8 wells, each containing 8 larvae).
PhenoBlast for haloperidol-like compounds
Based on its robust and reproducible behavioral phenotypes, we sought to determine if haloperidol’s behavioral profile could be used to identify additional compounds with antipsychotic-like phenotypes and mechanisms. We screened a 24,760 compound-library (including 4,300 known bioactive compounds and 20,000 uncharacterized compounds) and more than 5,000 DMSO controls, and compiled the behavioral profiles into a database. Using the average profile of three haloperidol (5 μM)-treated wells to query the database, we identified the top 100 hit compounds that caused haloperidol-like phenotypes (Supplementary Fig. 4a, Supplementary Table 2). Among these 100 hit compounds were 23 annotated bioactive drugs including 9 antipsychotics and antipsychotic-like compounds, 4 antihistamines, 10 compounds with other annotations and 5 false positive DMSO-treated control wells (Supplementary Fig. 4b). The top-ranked hit compound, bromperidol, is a close structural and functional analog of haloperidol25. Among the 9 known antipsychotic drugs identified in the screen, 6 were butyrophenone derivatives and 3 were tricyclics. The fourth ranked hit compound, DO 897/99, has been under investigation for treatment of schizophrenia26 and the fourteenth ranked compound, lidoflazine, is structurally related to the diphenylbutylpiperidine class of antipsychotics (e.g. amperozide, clopimozide, fluspirilene and pimozide). Haloperidol has antihistamine activity27, suggesting a reason why antihistamines were identified in the screen. It is unclear why other known bioactive compounds (of various annotated mechanisms) were identified among the top hits (Supplementary Table 2). These compounds may reflect poorly understood haloperidol activities or be false positives. Because the phenoBlast identified many known antipsychotic and antipsychotic-like compounds among the top hits, we hypothesized that some of the 72 uncharacterized compounds might also have antipsychotic-like activities.
Antipsychotic-related target predictions
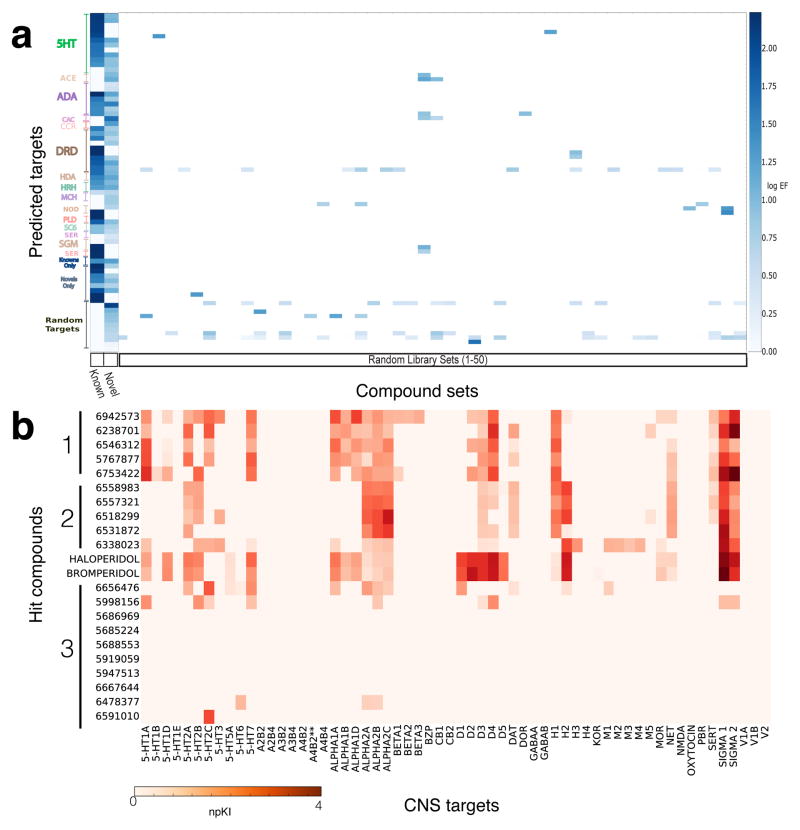
To determine if the hit compounds acted via haloperidol-like mechanisms, we predicted receptors for both the hit compounds and the known antipsychotics identified in the screen using the similarity ensemble approach (SEA)28. For both compound sets, we found that the top 20 SEA predictions were predominantly schizophrenia-related targets including dopamine and serotonin receptors (Supplementary Table 3). Strikingly, out of the 1,873 possible targets surveyed by SEA, the dopamine D2 and serotonin 2a receptors were the top two predicted targets in both sets. These data suggest that many of the uncharacterized hit compounds act through classic antipsychotic-like mechanisms. We next asked to what extent the known antipsychotics and the uncharacterized hit compounds shared a distinct predicted-target profile, as compared to the underlying compound library. Using a guilt-by-association “enrichment factor” (EF) metric29, we compared EFs of targets predicted for the known and uncharacterized compounds against target EFs calculated for 1,000 size-matched control groups randomly selected from the compound library (Fig. 4a, Supplementary Fig. 5). We observed a strong overlapping target signature between the known and uncharacterized compounds, versus no consistent target pattern for the controls. As before, the known and uncharacterized groups both achieved exceptional enrichment for serotonin and dopamine receptors, with an emphasis on serotonin type 2 and dopamine D2 receptors (Fig. 4a, Supplementary Table 4). The library-wide EF metric also uncovered shared enrichments for adrenergic receptors (ADA, ADR), voltage-dependent T-type calcium channels (CAC), and melanin-concentrating hormone receptors (MCH). Interestingly, both sets of compounds were also predicted to target sigma receptors (SGM), which were recently identified as a key target in zebrafish freezing behavior (see companion paper by Rennekamp et al.) and are implicated in a variety of physiological processes and in schizophrenia30. Although most predicted targets overlapped between known and uncharacterized groups, certain targets such as cytochrome P450 2J2 (CYP2J2) linked solely to the known antipsychotics, while telomerase reverse transcriptase linked solely to the uncharacterized compounds. These target predictions may represent off-target activities or possible new modes of action. Together, these data suggest that many of the uncharacterized hit compounds may act through similar targets as known antipsychotic drugs, including the sigma-1 receptor.
Figure 4. Hit compounds show haloperidol-like target profiles.
(a) Library-wide target signatures for known antipsychotics and uncharacterized compound hits as calculated by SEA. Heat map of shared target enrichments for known and uncharacterized groups, as compared to 1,000 randomly-selected size-matched groups from the underlying screening library. Out of 1,873 possible targets, fewer than 58 were significantly enriched for the known or uncharacterized groups. The y-axis shows the 58 enriched targets for the known and uncharacterized groups. 50 random groups are shown for context. Significantly enriched target classes for known and uncharacterized groups were: 5-HT: serotonin receptors, ADA and ADR: alpha and beta adrenergic receptors, CAC: voltage-dependent T-type calcium channels, DRD: dopamine receptors, HDA: histone deacetylases, HRH: histamine receptors, SGM: sigma receptors; for others see Supplementary Table 4 for full list. Enrichment factors (EF) of ≥ 2.0 and q-value ≤ 1e-10 are shown in blue, on a log scale. (b) Binding affinity profiles (as computed npKi values) of 22 uncharacterized hit compounds (rows) at 60 human and rodent CNS receptors in vitro (columns) in addition to haloperidol and bromperidol controls.
Hit compounds show antipsychotic-like binding profiles
To test SEA target predictions, we measured the binding affinity of a subset of 22 uncharacterized hit compounds at 60 human and rodent CNS targets in vitro31 (Fig. 4b). These compounds were selected based on phenotypic strength and structural diversity. For comparison, we also determined the binding profiles of haloperidol and bromperidol, two close structural analogs with very similar binding affinity profiles. We found that many hit compounds share a similar binding profile to haloperidol (Fig. 4b, Groups 1 and 2). Interestingly, other hit compounds showed little to no binding at any of the receptors tested, suggesting that these compounds may represent false positive hits in the screen or antipsychotic-like compounds with unknown mechanisms of action (Fig. 4b, Group 3). Together, these data indicate that many compounds with haloperidol-like phenotypes in larval zebrafish also have haloperidol-like receptor binding profiles in vitro.
Identification of finazines
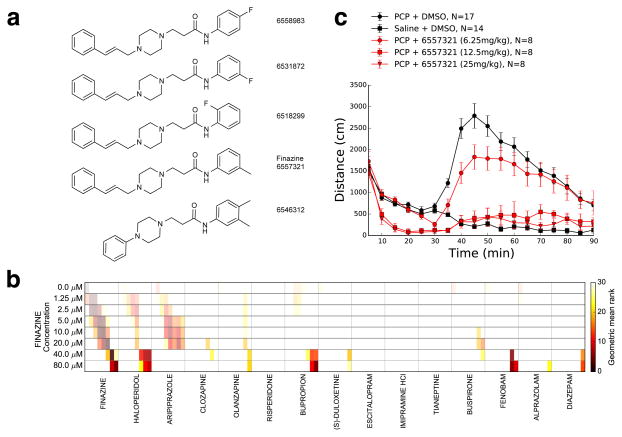
To understand the structural relationships between haloperidol and the hit compounds, we analyzed their Tanimoto similarities. We found that the uncharacterized hit compounds, including those with very similar target binding profiles, were structurally distinct from haloperidol (< 30% Tanimoto similarity) (Supplementary Fig. 6). These compounds clustered into several structural classes. We focused our attention on a cluster of five very closely related hit compounds (61%–89% Tanimoto similarity), that share a piperazine-containing maximum common substructure and that we named “finazines” (Fig. 5a). Only 38 compounds in the library of more than 20,000 contained this substructure. As expected, each of the individual finazines shared a very similar receptor-binding profile as haloperidol in vitro (Fig. 4b, Group 2), suggesting that these compounds share similar mechanisms of action. Binding patterns between haloperidol and the finazines showed both similarities and differences. Like haloperidol, the finazines bound to serotonin-2, adrenergic alpha-2, dopamine, histamine and sigma receptors. However, unlike haloperidol, the finazines showed stronger binding at the dopamine and serotonin transporters (DAT and SERT) and weaker binding serotonin-1, alpha-1, and dopamine receptors. Because these compounds had similar structures, phenotypes and binding profiles, we chose to focus further experiments on a single member of the group (6657321) that we refer to as “finazine” to differentiate it from other members of the structural class.
Figure 5. Finazine phenocopies haloperidol in zebrafish and in mice.
(a) The five compound structures in the finazine cluster (arrow). (b) Heat map showing the phenotypic similarity rank of 14 psychiatric drugs (columns) relative to the finazine query at each indicated concentration (rows). Each cell in the matrix is divided into 8 segments to represent different concentrations, and similarity rank is indicated in the color bar. (c) Plot of mouse locomotor activity as measured by distance traveled (y-axis) during the psychostimulant (PCP)-induced locomotor assay. Different treatments are as indicated; DMSO indicates the vehicle control and 6557321 indicates finazine. PCP was administered at 30 min (arrow). Values are mean ± sem.
To confirm that finazine causes haloperidol-like behaviors, we retested this compound and used its average behavioral profile at each concentration to query the reference database of psychiatric drugs. We found that finazine-treated animals behaved most similarly to other animals also treated (independently) with finazine, or to animals treated with two typical antipsychotic drugs: haloperidol and aripiprazole (Fig. 5b, Supplementary Fig. 7). These data indicate that finazine caused robust and reproducible antipsychotic-like phenotypes in the zebrafish. By contrast, the finazine profiles were less similarity to atypical antipsychotics (clozapine, olanzapine or risperidone), antidepressants or anxiolytics. Because finazine caused haloperidol-like behaviors in zebrafish, we hypothesized that finazine may also cause haloperidol-like phenotypes in mice.
Finazine suppresses PCP-induced hyperactivity in mice
The zebrafish behavioral profiles described above do not resemble psychosis. Given the low face validity of the zebrafish model, we sought to test its predictive validity for identifying compound with translatable effects in a psychostimulant-induced schizophrenia model in mice. In humans, acute administration of psychostimulants such as phencyclidine (PCP), a NMDA receptor antagonist, induces psychosis-like symptoms resembling schizophrenia32. In mice, PCP induces a hyperlocomotion phenotype that can be reversed by haloperidol and other typical and atypical antipsychotic drugs33,34. Reversal of PCP-induced hyper locomotion is a standard screening assay for both typical and atypical antipsychotics35,36. To determine if finazine might have antipsychotic-like activity in mammals we injected three groups of mice with graded doses (25, 12.5 and 6.25 mg/kg). Thirty minutes after compound treatment, mice were injected with PCP (5 mg/kg). We found that, like haloperidol, finazine reduced PCP-induced hyperactivity in mice (Fig. 5c). Baseline locomotor activity was unaffected in mice treated with the lowest dose of finazine (6.25 mg/kg). By contrast, at the two higher doses (12.5 and 25 mg/kg) finazine-treated mice exhibited a decrease in baseline locomotor activity, similar to what was observed in zebrafish and mice treated with high doses of haloperidol33. At high concentrations, many antipsychotic drugs, including haloperidol, cause extrapyramidal side effects in humans and catalepsy in rodents37. To further understand how finazine affects mouse behavior, we performed a modified Irwin observational battery38 and catalepsy test. At its lowest effective doses finazine (6.5 and 12.5 mg/kg), caused less catalepsy than haloperidol (at 1 mg/kg) (Supplementary Fig. 8). In addition, mice treated with finazine showed no evidence of lethality, convulsions, excitation, abnormal gait, jumps, writhes, stereotypy, or head twitches, although they did exhibit tremor and Straub tail responses (Supplementary Table 5). As expected for any hit compound, this observation suggests that substantial medicinal chemistry would be necessary to reduce side effects and develop finazine for any potential therapeutic use.
Discussion
Genomic databases and their search algorithms are fundamental tools in bioinformatics research. However, quantitative search tools for multi-dimensional phenotyping remain a key challenge in behavioral phenomics39. The basic local alignment search tool (BLAST) allows researchers to query nucleotide and protein databases for evolutionarily related sequences23,40. A conceptually similar bioinformatics tool, the Connectivity map, allows researchers to search for functional connections between compounds, genes and disease states using gene expression signatures41. Although researchers have developed BLAST-like approaches for searching cell division phenotypes in C. elegans42 and morphological phenotypes in yeast43, the phenoBlast concept has been difficult to apply to behavioral datasets which are typically smaller, more variable and less systematic than gene-expression databases. Here, we have used similarity metrics between motor activity profiles to describe a large collection of small molecules and identify antipsychotic-like compounds. This battery of high-throughput behavioral assays has enabled us to systematically quantify effects of thousands of compounds on vertebrate motor activity. The quantity and quality of the dataset have permitted us to conduct behavior-based connectivity mapping, using metrics conceptually related to those previously used in other fields of bioinformatics 41. We have designed phenoBlast to be modular. Data from additional assays can be easily added and would likely improve the resolution of each phenotypic profile. These need not be limited to behavioral or motion analyses44. For example, automated morphological phenotyping, and full brain calcium imaging are two rapidly developing areas of zebrafish phenotyping with great potential for improving resolution of behavioral profiles45,46. Together, all of these assays have great potential for addressing current limitations in multi-dimensional phenotypic analyses. Here, we have applied the phenoBlast approach to haloperidol. In the future, it will be interesting to determine if the approach can be applied to additional classes of neuroactive compounds such as atypical antipsychotics, antidepressants or anxiolytics.
Although phenotypic approaches to neuroactive drug discovery in zebrafish may address some limitations of in vitro screening assays there are also many caveats including insufficient phenotypic resolution and biological differences between humans and zebrafish. Although it is estimated that approximately 80% of human disease genes have a zebrafish ortholog47, molecular differences between these receptors may have important pharmacological effects. For example, the delta opioid receptor shows different pharmacology in mammals and zebrafish due to a single inactivating amino acid substitution in the ligand-binding site33. Because we still do not fully understand haloperidol’s mechanisms of action or the mechanisms underlying zebrafish behaviors, it is possible that some haloperidol-induced phenotypes in the fish may relate to off-target effects with little therapeutic value in humans. Such limitations are part of all model systems48. For the haloperidol-like compounds, such as the finazines, future studies are required to fully assess their potential as therapeutic candidates. Antipsychotic agents, like haloperidol, often cause extrapyramidal and other side-effects during chronic exposure49. Our data in mice suggest that finazine does not cause catalepsy at the same concentration that reduces background motor activity in mice. However, finazine does cause other side effects including tremor and the Straub tail response, suggesting that further optimization and safety testing would be necessary before contemplating any further exploration of potential therapeutic utility.
Historically, psychiatric drug candidates have shown high failure rates in clinical trials as compared to candidates in other therapeutic fields50. Several reasons account for this high failure rate including the requirement that CNS drugs pass the blood-brain barrier, the polygenetic nature of CNS disorders (which likely require multi-receptor rather than “magic bullet” drugs), and limited understanding of the therapeutic mechanisms of clinically efficacious drugs12. Using a phenotype-based behavioral readout during the initial phase of CNS drug discovery may increase the likelihood that hit compounds will meet these criteria. In summary, we have developed a phenoBlast approach for the rapid querying of in vivo phenotypic similarities among thousands of structurally diverse small molecules. The approach is unbiased, structure- and target-blind, and based solely on compounds’ behavioral effects. Not surprisingly, many compounds that share phenotypic signatures also share structural similarities and target-binding profiles. Thus, the phenoBlast approach provides a way to both validate structure and target-based hypotheses and also to discover structurally and mechanistically neuroactive compounds with multi-target mechanisms.
Online Methods
Aquaculture and chemical treatments
Fertilized eggs (up to 20,000 embryos per day) were collected from group matings of Ekkwill zebrafish (Danio rerio). Embryos were raised in hatching jars at 28 °C on a 14/10-hour light/dark cycle until 3 days post fertilization (d.p.f.), then transferred to an incubator under the same conditions until 7 d.p.f. Groups of approximately 8–10 larvae (7 d.p.f.) were distributed into the wells of clear flat-bottom 96-square-well plates filled with E3 medium (300 μl). Larvae were then incubated at 25 °C on the bench top for 1 h prior to chemical treatment and subsequent experiments. Larval zebrafish are of indeterminate sex. All zebrafish protocols were approved by the UCSF Institutional Animal Care and Use Committee (Authorization Protocol number: AN107525-02B).
Chemical libraries and treatments
The Actiprobe library (TimTec Corporation) contains 10,000 compounds dissolved in DMSO at a stock concentration of 1mg/ml (~3 mM). The Chembridge library (Chembridge Corporation) contains 10,000 compounds dissolved in DMSO at a stock concentration of 1 mM. The Spectrum Collection (MicroSource Discovery) contains 2,320 compounds dissolved in DMSO at a concentration of 10mM. The Prestwick library (Prestwick Chemical) contains 1,280 approved drugs dissolved in DMSO at a stock concentration of 10 mM. The Neurotransmitter library (Biomol International; cat. No. 2810) contains 700 compounds dissolved in DMSO at a stock concentration of 10 mM. All compounds were diluted in E3 buffer and screened at 10 μM final concentration and < 1% DMSO. Negative controls were treated with an equal volume of DMSO. Stock solutions were added directly to zebrafish in the wells of a 96-well plate, mixed and allowed to incubate for 1 h at room temperature before behavioral evaluation in the Behavioral Battery of assays. Reordered hit compounds were dissolved in DMSO and added to wells as described above. Each drug in the reference set was tested at seven concentrations, twelve wells per concentration, and in triplicate (on different days). Concentration ranges were identified by range finding experiments.
Chemoinformatics
We used the similarity ensemble approach (SEA) algorithms to predict candidate molecular targets for each compound as previously described51. Instant JChem was used for structure database management and substructure searching, Instant JChem 14.7.14.0, 2014, ChemAxon (http://www.chemaxon.com). Chemical similarities were computed as Tanimoto similarities using rdKit52. SEA enrichment factor (EF) calculations were calculated as described53. We calculated EF’s for targets predicted for each compound within two preset groups (8 known antipsychotics, 72 uncharacterized antipsychotic-like compounds from the screen) against baseline groups of random compounds. As the baseline we enumerated 1,000 groups, each comprising 72 compounds randomly selected from the entire screening library. To be included in the enrichment analysis, target predictions had to achieve SEA p-values ≤ 1e-10, and we required at least 2 target-group pairs for the known antipsychotics and at least 5 such pairs for the larger uncharacterized compounds and for the random compound sets. We retained target-group predictions with EF > 2.0 and q-values ≤ 1e-10, which left 58 enriched targets across the known and uncharacterized antipsychotic sets. Only 182 targets total (including the targets for the random groups) passed these enrichment filters, out of the total of 1,873 SEA target predictions possible. Of the 1,000 random baseline groups, 63% did not have any target passing these enrichment filters whatsoever.
CNS receptor profiling
For known psychoactive compounds, binding profiles were downloaded from the PDSP Ki database54. Ki values greater than 10,000 nM or missing were set to 10,000 nM. Normalized Ki (npKi) values were computed as described55. For uncharacterized hit compounds, in vitro binding assay and Ki data were generated by the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP), contract no. HHSN-271-2008-00025-C (NIMH PDSP) as described. Complete assay details are found online at (http://pdsp.med.unc.edu/PDSP%20Protocols%20II%202013-03-28.pdf).
Automated behavioral phenotyping in zebrafish
We captured digital videos of zebrafish in a battery of 10 behavioral assays containing combinations of acoustic and light-based stimuli. Digital videos of animals in 96-well plates were recorded at 25 or 34 frames per second using an AVT Pike 210 camera mounted on a 0.08x telecentric lens (Optoengineering) such that the entire 96-well plate was captured in each frame. Stimuli were applied to all wells of the entire 96-well plate simultaneously. Light stimuli were generated using high-intensity LEDs (LEDNGIN) in red (650 nm, 11 μW/mm2), blue (560 nm, 18 μW/mm2) and violet (400 nm, 11 μW/mm2) wavelengths. Low magnitude (60 dB) and high magnitude (70 dB) acoustic stimuli were generated using push-style solenoids to tap the stage. Stimulus control and data management were accomplished using computer scripts (Matlab). The 96 well plate was illuminated from below with infrared (850 nm) LEDs. The behavioral battery, including video capturing and raw data processing, took approximately 10 minutes per plate. Approximately 10–20 plates could be analyzed per camera-day. To quantify behavioral activity elicited by the stimuli, a motion index (MI) was calculated by frame differencing within the region of interest around each well of the plate. The MI was calculated by: MI= sum(abs(framen-framen−1)). This MI correlates with the overall amount of motor activity observed in the well. Complete stimulus protocols for each assay are included in Supplementary Table 6 and 7.
Phenotype quantification, phenoBlast ranking and statistics
A motion index (MI) is calculated for each well by frame differencing digital video of the behavioral battery. This MI time series is referred to as a “behavioral profile”, and consists of MI values at a total of 10,500 time points. To quantify the phenotypic distance between two time series v and w we used the angular distance d = arcos(v.w/(||v|| ||w||))/pi, where ‘.’ is the canonical inner product and ‘|| ||’ is its induced norm. The reference data set of psychoactive drugs was replicated three times over different days (see below) and, in order to mitigate batch effects, query phenotypes from a given replicate were only allowed to find matching phenotypes in the other two replicates. When querying with the time series of compound x at concentration c1 from replicate A, we ranked all phenotypes in the data set by increasing distance to this query time series and we indicated as r(y,c2,B|x,c1,A) the ranking of compound y at concentration c2 from replicate B. We then computed the heatmap phenoBlast ranking as geometricMean [r(y,c2,B|x,c1,A), r(y,c2,C|x,c1,A), r(y,c2,A|x,c1,B), r(y,c2,C|x,c1,B), r(y,c2,A|x,c1,C), r(y,c2,B|x,c1,C)]. When considering the overall amount of motion for a given time series, we first normalized the baseline by subtracting the 5th percentile MI, and then averaged the resulting MIs over time. Hierarchical clustering was performed using the complete-linkage algorithm. The haloperidol query for the high-throughput screen was calculated by averaging the time series from three replicate haloperidol-treated wells. The 29,760 compounds in the screen were ranked based on their phenoBlast score, defined as the L1 distance between each of the 29,760 time series (s) from the haloperidol query (q): d = sumi(|si −qi|).
The size of a phenotypic effect is compound-dependent and dose-dependent. Compounds in the chemical library were screened in a single well and concentration, with the understanding that only compounds with strong phenotypic would emerge as hits. At the end of the screen it became apparent that phenotypic effects similar to the effects produced by haloperidol at 5uM were enough to be selected as preliminary hits. The characterization of the hit list in and the estimate of false positive rates helped guide power analysis in follow-up studies. For the reference compounds we dedicated an entire 96-well plate per compound to avoid phenotypic similarity being driven by two or more compounds being tested on the same plate. Most compounds go from inactive to sedative/lethal within a 50-fold concentration range. Based on this observation we designed dose-response plates to test 7 concentrations per compound increasing the concentrations by multiplicative factors of 2, covering concentrations from x to 64 x, where x is a compound-dependent concentration determined in preliminary experiments. The 8th row on a plate was dedicated to DMSO-treated control wells. We then used all the 12 wells on a row to generate time series to mitigate the effect of random bursts of larval activity that add noise to the time series for single wells. Three replicates of the dose-response plates described above were screen on three different days, both to estimate and to mitigate the effects of day-to-day variability.
No animal samples were excluded after performing the experiments. Larvae were randomly transferred from their hatching basins to the 96-well plates. Compounds were assigned random identifiers (mixing pharmacological classes), sorted by identifier, and screened according to that order. The investigators were blinded during the screen and the analysis of the chemical library compounds and not blinded during the screen and the analysis of the reference compounds.
Code Availability
Code availability by request
Mouse phenotyping
Male C57BL/6J mice (9–10 weeks at testing) were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were group-housed 4 per cage in Techniplast ventilated cages and were maintained on a 12/12-hr light/dark cycle (lights on 0700 EST). The room temperature was maintained at 20–23°C with relative humidity at approximately 50%. Food and water were available ad libitum for the duration of the study, except during testing and all testing was conducted during the light phase of the light dark cycle. The behavioral tests were conducted according to established protocols approved by the Harvard Medical Area (HMA) Standing Committee on Animals IACUC in AALAC-accredited facilities, and in accordance with the Guide to Care and Use of Laboratory Animals (National Institutes of Health 1996). Locomotor activity was measured in Plexiglas square chambers (27.3 x 27.3 x 20.3 cm; Med Associates Inc.) surrounded by infrared photobeam sources and detectors, as the total distance traveled (cm) assessed by infrared beam breaks. Mice were tested under ambient light and data were collected by Med Associates software. Mice were injected with 10% DMSO vehicle or finazine (6.25, 12.5, or 25 mg/kg in 10% DMSO) and locomotor activity was monitored for 30 minutes (baseline total distance). Mice were then administered saline vehicle or PCP (5 mg/kg) and activity was measured for an additional 60 minutes. Antagonism of PCP-induced hyperactivity was used as the measure of antipsychotic efficacy. All compounds were administered by intraperitoneal (IP) injection in a volume of 10 ml/kg. Locomotor activity was measured as total distance traveled (cm), assessed via infrared beam breaks. Locomotion prior to PCP administration (baseline, 0–30 m) and locomotion post PCP administration (PCP, 30–60 m) were analyzed by one-way analysis of variance (ANOVA) with finazine (0, 6.25, 12.5, 25) as the independent variable. All significant effects were followed up with the Fisher’s PLSD post hoc test. An effect was considered significant if p<0.05 (Statview for Windows, Version 5.0).
Modified Irwin observational battery and catalepsy testing
To test for catalepsy, C57BL/6J males (Jackson Laboratory) of approximately 9 weeks of age were injected IP with 10 ml/kg haloperidol (Sigma Aldrich) or finazine1 prepared in 10% DMSO in saline. As a control, 10% DMSO was used as a vehicle injection. Recording of time in a cataleptic position was carried out at 30, 60, 90, 120, and 180 min after injection. Catalepsy was defined as the amount of time spent on a horizontal steel rod (~0.5 cm in diameter, positioned ~4.5 cm above the surface of the testing surface). The time during which each mouse maintained this position was recorded up to a maximum of 2 min. Following each cataleptic measurement, mice were observed for clinical signs. The following signs were recorded as present or absent for each mouse at each time point: Lethality, Convulsions, Tremor, Straub Tail, Excitation, Abnormal Gait, Jumps, Writhes, Stereotypy, Head Twitches. Data were analyzed by ANOVA followed by Fisher’s LSD post hoc test.
Source and purity of compounds
Psychiatric drug reference compounds were purchased from the following sources at the indicated purity. Haloperidol (Sigma-Aldrich, NA), Aripiprazole (Sigma-Aldrich, >98%), Clozapine (Tocris, >99%), Olanzapine (Sigma-Aldrich, >98%), Risperidone (Sigma-Aldrich, >98%), Bupropion (Sigma-Aldrich, >98%), (S)-Duloxetine (Sigma-Aldrich, >98%), Excitalopram (Sigma-Aldrich, >98%), Imipramine (Sigma-Aldrich, >98%), Tianeptine (Sigma-Aldrich, >98%), Buspirone (Sigma-Aldrich, NA), Fenobam (Sigma-Aldrich, >98%), Alprazolam (Sigma-Aldrich, NA), Diazepam (Sigma-Aldrich, NA).
Supplementary Material
Acknowledgments
We thank members of our research groups for helpful advice. This work was supported by NIH grants K01MH091449, U01MH105027 and R01AA022583 (DK), R44GM093456 (MJK), the Glenn Foundation Award for Research in Biological Mechanisms of Aging (DK and MJK), T32EB009383 and T32GM008284 (LG), R01MH086867 and R21MH085205 (RTP), and the Charles and Ann Sanders MGH Research Scholar Award (RTP).
Footnotes
AUTHOR CONTRIBUTIONS
GB performed the behavior-based chemical screen and preliminary analysis of the data. AR analyzed the finazine compounds. AV designed statistical analyses to profile the reference set and analyze all the zebrafish data. LG performed target prediction and enrichment calculations and interpreted data with MJK. MM, EF, JT, PL, DL, TE and GC performed the zebrafish behavioral profiling and interpreted the data. PJL performed the rodent work and analyzed the data with BJC. XPH performed the target binding assays and analyzed the data with BLR. SK and EP designed the psychiatric drug reference set. RTP and DK designed the experiments and wrote the paper. All authors analyzed the data and edited the manuscript.
Competing financial interests
AJR, DK and RTP declare competing financial interests in the form of a pending patent application, No. PCT/US2015/037755, covering the finazine compounds described in this manuscript. AV, TE, GC and DL are full time employees of Teleos Therapeutics. DK and RTP consult for Teleos Therapeutics. SK and EP are full time employees of F. Hoffmann – La Roche Ltd.
References
- 1.Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 2.Kokel D, Peterson RT. Chemobehavioural phenomics and behaviour-based psychiatric drug discovery in the zebrafish. Brief Funct Genomic Proteomic. 2008;7:483–490. doi: 10.1093/bfgp/eln040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnard J, et al. Automated design of ligands to polypharmacological profiles. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul SM, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 5.López-Muñoz F, Alamo C. The consolidation of neuroleptic therapy: Janssen, the discovery of haloperidol and its introduction into clinical practice. Brain research bulletin. 2009;79:130–141. doi: 10.1016/j.brainresbull.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Hippius H. The history of clozapine. Psychopharmacology (Berl ) 1989;99(Suppl):S3–5. doi: 10.1007/BF00442551. [DOI] [PubMed] [Google Scholar]
- 7.Conn PJ, Roth BL. Opportunities and challenges of psychiatric drug discovery: roles for scientists in academic, industry, and government settings. Neuropsychopharmacology. 2008;33:2048–2060. doi: 10.1038/sj.npp.1301638. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. WHO model list of essential medicines: 18th list, April 2013. 2013 [Google Scholar]
- 9.Meltzer HY. An overview of the mechanism of action of clozapine. J Clin Psychiatry. 1994;55(Suppl B):47–52. [PubMed] [Google Scholar]
- 10.Lieberman JA, et al. The effects of clozapine on tardive dyskinesia. Br J Psychiatry. 1991;158:503–510. doi: 10.1192/bjp.158.4.503. [DOI] [PubMed] [Google Scholar]
- 11.Lieberman JA, et al. Clozapine-induced agranulocytosis: non-cross-reactivity with other psychotropic drugs. J Clin Psychiatry. 1988;49:271–277. [PubMed] [Google Scholar]
- 12.Hyman SE. Revolution stalled. Science Translational Medicine. 2012;4:155cm11–155cm11. doi: 10.1126/scitranslmed.3003142. [DOI] [PubMed] [Google Scholar]
- 13.Wolman MA, Jain RA, Liss L, Granato M. Chemical modulation of memory formation in larval zebrafish. Proc Natl Acad Sci USA. 2011;108:15468–15473. doi: 10.1073/pnas.1107156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baraban SC, Dinday MT, Hortopan GA. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat Commun. 2013;4:2410. doi: 10.1038/ncomms3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bencan Z, Sledge D, Levin ED. Buspirone, chlordiazepoxide and diazepam effects in a zebrafish model of anxiety. Pharmacology Biochemistry and Behavior. 2009;94:75–80. doi: 10.1016/j.pbb.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-Patiño MA, Yu L, Cabral H, Zhdanova IV. Anxiogenic effects of cocaine withdrawal in zebrafish. Physiology & Behavior. 2008;93:160–171. doi: 10.1016/j.physbeh.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Egan RJ, et al. Understanding behavioral and physiological phenotypes of stress and anxiety in zebrafish. Behavioural Brain Research. 2009;205:38–44. doi: 10.1016/j.bbr.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giacomini NJ, Rose B, Kobayashi K, Guo S. Antipsychotics produce locomotor impairment in larval zebrafish. Neurotoxicol Teratol. 2006;28:245–250. doi: 10.1016/j.ntt.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Rihel J, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokel D, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6:231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokel D, et al. Photochemical activation of TRPA1 channels in neurons and animals. Nat Chem Biol. 2013;9:257–263. doi: 10.1038/nchembio.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bymaster FP, et al. Comparative affinity of duloxetine and venlafaxine for serotonin and norepinephrine transporters in vitro and in vivo, human serotonin receptor subtypes, and other neuronal receptors. Neuropsychopharmacology. 2001;25:871–880. doi: 10.1016/S0893-133X(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Boulay D, et al. Haloperidol-induced catalepsy is absent in dopamine D(2), but maintained in dopamine D(3) receptor knock-out mice. Eur J Pharmacol. 2000;391:63–73. doi: 10.1016/s0014-2999(99)00916-4. [DOI] [PubMed] [Google Scholar]
- 25.Dubinsky B, et al. Bromperidol, a new butyrophenone neuroleptic: a review. Psychopharmacology (Berl ) 1982;78:1–7. doi: 10.1007/BF00470578. [DOI] [PubMed] [Google Scholar]
- 26.Lecrubier Y. A partial D3 receptor agonist in schizophrenia. European Neuropsychopharmacology. 2003;13:S167–S168. [Google Scholar]
- 27.Peroutka SJ, Synder SH. Relationship of neuroleptic drug effects at brain dopamine, serotonin, alpha-adrenergic, and histamine receptors to clinical potency. Am J Psychiatry. 1980;137:1518–1522. doi: 10.1176/ajp.137.12.1518. [DOI] [PubMed] [Google Scholar]
- 28.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 29.Lounkine E, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi T, Su T. The sigma receptor: evolution of the concept in neuropsychopharmacology. Curr Neuropharmacol. 2005;3:267–280. doi: 10.2174/157015905774322516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth BL, Lopez E, Patel S, Kroeze WK. The Multiplicity of Serotonin Receptors: Uselessly Diverse Molecules or an Embarrassment of Riches? Neuroscientist. 2000;6:252–262. [Google Scholar]
- 32.Jentsch JD, Roth RH. The Neuropsychopharmacology of Phencyclidine: From NMDA Receptor Hypofunction to the Dopamine Hypothesis of Schizophrenia. Neuropsychopharmacology. 1999;20:201–225. doi: 10.1016/S0893-133X(98)00060-8. [DOI] [PubMed] [Google Scholar]
- 33.Gleason SD, Shannon HE. Blockade of phencyclidine-induced hyperlocomotion by olanzapine, clozapine and serotonin receptor subtype selective antagonists in mice. Psychopharmacology (Berl ) 1997;129:79–84. doi: 10.1007/s002130050165. [DOI] [PubMed] [Google Scholar]
- 34.Freed WJ, Bing LA, Wyatt RJ. Effects of neuroleptics on phencyclidine (PCP)-induced locomotor stimulation in mice. Neuropharmacology. 1984;23:175–181. doi: 10.1016/s0028-3908(84)80011-8. [DOI] [PubMed] [Google Scholar]
- 35.Porsolt RD, Moser PC, Castagné V. Behavioral indices in antipsychotic drug discovery. J Pharmacol Exp Ther. 2010;333:632–638. doi: 10.1124/jpet.110.166710. [DOI] [PubMed] [Google Scholar]
- 36.Castagné V, Moser PC, Porsolt RD. Preclinical behavioral models for predicting antipsychotic activity. Adv Pharmacol. 2009;57:381–418. doi: 10.1016/S1054-3589(08)57010-4. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman DC, Donovan H. Catalepsy as a rodent model for detecting antipsychotic drugs with extrapyramidal side effect liability. Psychopharmacology (Berl ) 1995;120:128–133. doi: 10.1007/BF02246184. [DOI] [PubMed] [Google Scholar]
- 38.Irwin S. Comprehensive observational assessment: Ia. A systematic, quantitative procedure for assessing the behavioral and physiologic state of the mouse. Psychopharmacologia. 1968;13:222–257. doi: 10.1007/BF00401402. [DOI] [PubMed] [Google Scholar]
- 39.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 40.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 42.Gunsalus KC, Yueh WC, MacMenamin P, Piano F. RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 2004;32:D406–10. doi: 10.1093/nar/gkh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin K, et al. PhenoM: a database of morphological phenotypes caused by mutation of essential genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:D687–94. doi: 10.1093/nar/gkr827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kokel D, Rennekamp AJ, Shah AH, Liebel U, Peterson RT. Behavioral barcoding in the cloud: embracing data-intensive digital phenotyping in neuropharmacology. Trends Biotechnol. 2012;30:421–425. doi: 10.1016/j.tibtech.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahrens MB, et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardo-Martin C, et al. High-throughput in vivo vertebrate screening. Nature Methods. 2010;7:634–636. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGonigle P. Animal models of CNS disorders. Biochem Pharmacol. 2014;87:140–149. doi: 10.1016/j.bcp.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 49.Arana GW. An overview of side effects caused by typical antipsychotics. Journal of clinical psychiatry. 2000 [PubMed] [Google Scholar]
- 50.Pangalos MN, Schechter LE, Hurko O. Drug development for CNS disorders: strategies for balancing risk and reducing attrition. Nat Rev Drug Discov. 2007;6:521–532. doi: 10.1038/nrd2094. [DOI] [PubMed] [Google Scholar]
- 51.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 52.Landrum G. RDKit: Open-source cheminformatics. < http://www.rdkit.org>.
- 53.Lounkine E, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth BL, Lopez E, Patel S, Kroeze WK. The Multiplicity of Serotonin Receptors: Uselessly Diverse Molecules or an Embarrassment of Riches? Neuroscientist. 2000;6:252–262. [Google Scholar]
- 55.Besnard J, et al. Automated design of ligands to polypharmacological profiles. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.