Summary
Odor perception in mammals is mediated by parallel sensory pathways that convey distinct information about the olfactory world. Multiple olfactory subsystems express characteristic seven-transmembrane G-protein coupled receptors (GPCRs) in a one-receptor-per-neuron pattern that facilitates odor discrimination. Sensory neurons of the “necklace” subsystem are nestled within the recesses of the olfactory epithelium and detect diverse odorants; however, they do not express known GPCR odor receptors. Here we report that members of the four-pass transmembrane MS4A protein family are chemosensors expressed within necklace sensory neurons. These receptors localize to sensory endings and confer responses to ethologically-relevant ligands including pheromones and fatty acids in vitro and in vivo. Individual necklace neurons co-express many MS4A proteins and are activated by multiple MS4A ligands; this pooling of information suggests that the necklace is organized more like subsystems for taste than for smell. The MS4As therefore define a distinct mechanism and functional logic for mammalian olfaction.
Introduction
As animals navigate the natural world they encounter an unending variety of small molecules, which are rich sources of information that signify the presence of organisms and salient objects in the environment. The olfactory system detects many of these molecules through odorant receptor proteins expressed by peripheral olfactory sensory neurons (OSNs), which are coupled to higher brain circuits mediating odor perception (Axel, 1995; Ihara et al., 2013). In mammals, the olfactory system is divided into multiple, parallel processing streams made up of anatomically and molecularly distinct sensory neuron populations. The largest subdivision, the main olfactory system, is capable of detecting nearly all volatile odorants and plays key roles in odor discrimination and learning. Smaller subsystems (such as the vomeronasal system) are thought to play a more specialized role in odor perception, discriminating odors of innate significance and releasing specific patterns of reproductive, agonistic, or defensive behavior (Munger et al., 2009).
The main and vomeronasal olfactory systems each express characteristic odorant receptor families that belong to the G-protein coupled receptor (GPCR) superfamily; these receptor families define the specific receptive fields and therefore the function of each subsystem (Buck and Axel, 1991; Dulac and Axel, 1995; Herrada and Dulac, 1997; Liberles and Buck, 2006; Liberles et al., 2009; Matsunami and Buck, 1997; Riviere et al., 2009; Ryba and Tirindelli, 1997). The identification of these receptor genes (including the Odorant Receptors (ORs), Vomeronasal type 1 Receptors, Vomeronasal type 2 Receptors (V2Rs), Formyl Peptide Receptors and the Trace Amine-Associated Receptors) has revealed a key organizational principle: each mature olfactory sensory neuron (with the exception of those within the basal subdivision of the vomeronasal system) expresses just a single receptor gene of the hundreds encoded in the genome (Dalton et al., 2015). This pattern of expression defines specific information channels in the olfactory system, as the axons of those sensory neurons that express the same odorant receptor converge on a small number of insular structures within the olfactory bulb called glomeruli; these glomeruli are differentially recruited as animals sense distinct smells, enabling the brain to discriminate odors detected by the nose (Mori and Sakano, 2011). Individual basal vomeronasal sensory neurons also target specific bulb glomeruli, but express two V2Rs instead of a single receptor (Martini et al., 2001).
While the identification of odorant receptor genes has led to deep insight into the sensory tuning and functional architecture of the main and vomeronasal subsystems, there are additional mammalian subsystems whose modes of odor detection — and therefore function — are less clear. Particularly mysterious is the “necklace” subsystem, which is distinguished by its unusual anatomy: OSNs within this subsystem are concentrated in the recesses of the olfactory epithelium (the “cul-de-sac” regions), and project axons to a ring of 12-40 apparently interconnected glomeruli that encircle the caudal olfactory bulb like beads on a necklace (Juilfs et al., 1997; Shinoda et al., 1989). Necklace sensory neurons and glomeruli respond to a diverse range of chemical stimuli, including gases (such as carbon disulfide and carbon dioxide), pheromones (such as 2,5-dimethylpyrazine (2,5-DMP), 2-heptanone, and E-farnesene), plant-derived odorants, and urinary peptides (Fulle et al., 1995; Hu et al., 2007; Juilfs et al., 1997; Leinders-Zufall et al., 2007; Meyer et al., 2000; Munger et al., 2010; Sun et al., 2009). While many of these ligands have innate significance for the mouse, the specific role of the necklace in mediating odor perception remains unclear.
Intriguingly, necklace OSNs do not express the signaling proteins known to mediate GPCR-based chemotransduction in the rest of the main olfactory epithelium (Juilfs et al., 1997; Meyer et al., 2000). While the ability of the necklace system to detect and behaviorally respond to gases and peptides requires the single pass transmembrane protein Guanylate Cyclase-D (GC-D), which is specifically expressed in all necklace neurons, the remainder of the diverse sensory responses observed in this system are unexplained (Guo et al., 2009; Leinders-Zufall et al., 2007; Sun et al., 2009). These observations suggest that necklace OSNs harbor an as-yet unrecognized receptor type, whose identification could reveal key features of the functional organization and neural logic that governs the necklace system.
Here we show that necklace OSNs express a previously-unidentified class of chemoreceptor encoded by the Ms4a gene family. Each MS4A protein detects a specific subset of ethologically-relevant odorants — including fatty acids and the putative mouse pheromone 2,5-DMP — that stimulate necklace sensory neurons in vivo. Ectopic expression of MS4A proteins is sufficient to confer responses to MS4A ligands upon conventional olfactory neurons. However, unlike all known mammalian olfactory receptors, the Ms4a genes do not belong to the GPCR superfamily and are not expressed in the conventional one-receptor-one-neuron pattern; instead, each Ms4a gene encodes a four-pass transmembrane protein and many Ms4a family members are expressed in every necklace sensory neuron. Taken together, this work defines a new mechanism for mammalian olfaction and identifies a population of atypical olfactory sensory neurons that each express many members of a receptor gene family, suggesting a distinct perceptual role for odor information coursing through the necklace subsystem. Because MS4A proteins are also expressed in chemosensory cells that reside outside the nasal epithelium, these findings suggest a broader role for the MS4A proteins in the detection of chemical cues.
Results
To identify candidate receptor gene families specific to the necklace olfactory system, RNAseq was used to compare transcripts expressed by FACS-isolated GC-D-expressing and conventional OSNs (Figures 1A, 1B, S1A, and S1B). This analysis failed to reveal expression of known odorant receptor families or enrichment of other GPCR subfamilies within necklace sensory neurons. Consistent with this and prior reports, Golf, Adcy3, Cnga2, Cnga4, Trpc2, and Trpm5 — gene products required for odor-related signal transduction in conventional OSNs — also were not expressed in GC-D cells (Munger et al., 2009). To screen for potential non-GPCR odorant receptors, the RNAseq data were filtered to identify transmembrane protein families that exhibit sufficient molecular diversity to potentially interact with a wide range of ligands. As detailed below, this screen identified the Membrane Spanning, 4-pass A (Ms4a) genes, which encode a class of four-transmembrane (4TM) spanning proteins that are structurally distinct from GPCRs (Eon Kuek et al., 2015).
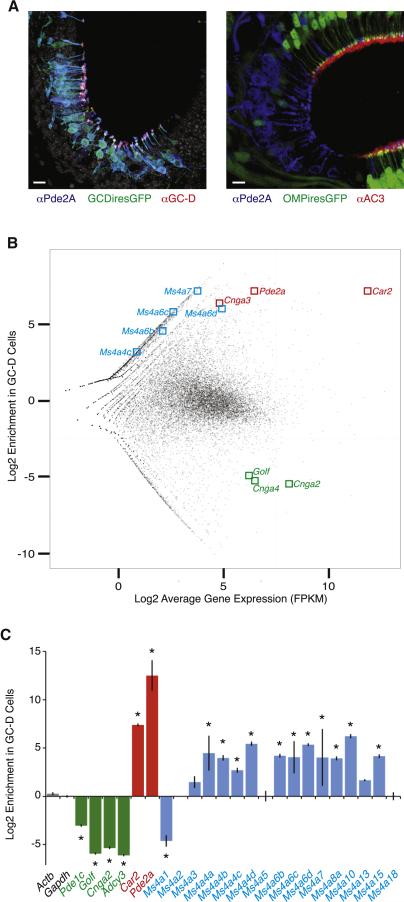
Figure 1.
Expression of Ms4a genes in necklace sensory neurons. A. (Left) The Gucy2d-IRES-TauGFP allele marks necklace sensory neurons expressing PDE2A (blue) and GC-D (red). Grayscale is nuclear counterstain (DAPI). (Right) Pde2a+ necklace sensory neurons reside in epithelial “cul-de-sacs” and do not express the Omp-IRES-GFP allele or the conventional OR signal transduction protein Adenylyl Cyclase3 (red). Scale bars 10 μM. B. Enrichment versus expression plot for every sequenced mRNA in GC-D+ and OMP+ sensory neurons. Each point is a transcript with detectable RNAseq reads, with marker genes associated with GC-D cells (Car2, Pde2a, and Cnga3) and OMP cells (Golf, Cnga2, and Cnga4) labeled in red and green, respectively; reliably detected Ms4a family members are highlighted in blue. C. Nanostring-based mRNA quantification of GC-D cells relative to OMP cells. Marker genes for OMP and GC-D cells are shown in green and red, respectively. Twelve of the 17 annotated Ms4a genes are significantly enriched in GC-D cells relative to OMP cells. * p < 0.05, unpaired t-test. See also Figure S1.
RNAseq revealed transcripts for several Ms4a family members in GC-D-expressing cells (Figure 1B). Because Ms4a transcripts had low RNAseq read counts, the Nanostring single-molecule detection technique was used to unambiguously determine the presence of every member of the Ms4a gene family in GC-D cells (Khan et al., 2011). This analysis revealed the reproducible expression of 12 Ms4a family members (of the 17 members annotated in the mouse genome), demonstrating that GC-D cells express low levels of a specific subset of Ms4a genes (Figure 1C). None of these 12 genes was detected in conventional OSNs above background (data not shown). Notably absent from GC-D cells are the two best-studied Ms4a genes, Ms4a1 and Ms4a2, which have been implicated in calcium signaling downstream of the B-cell receptor and high-affinity Fc-Epsilon receptor, respectively, but whose precise function remains unclear (Bubien et al., 1993; Dombrowicz et al., 1998; Koslowski et al., 2008; Lin et al., 1996; Polyak et al., 2008).
To assess the molecular diversity of the MS4A family, Ms4a genes were identified from representative species of all major mammalian lineages. We then asked how different these genes were within a given species, as amino acid differences are a prerequisite for individual MS4As to interact with distinct odors. Multiple sequence alignments revealed substantial intraspecies diversity amongst the MS4As, particularly within the extracellular domains of the protein, whose length is variable (Figures S2A and S2B). This diversity is comparable to that observed in the third through seventh transmembrane domains in conventional ORs, the regions thought to form ligand-binding pockets that give rise to odorant specificity (Buck and Axel, 1991; Man et al., 2004; Singer, 2000). These findings raise the possibility that each MS4A within a given organism may interact with a distinct set of extracellular cues.
Ms4a genes are found in a single genomic cluster in all queried mammals; this organization, suggestive of tandem duplication, is also found in known chemoreceptor gene families and is thought to facilitate diversification of family members (Figure 2A) (Nei et al., 2008). We therefore also assessed differences between Ms4a genes across species, to identify those regions of the MS4A protein subject to diversifying or purifying selection. MS4A proteins were significantly divergent across evolution (Figure S2C) (Nei et al., 2008); the most rapidly evolving amino acid residues in the MS4A proteins are highly enriched in the predicted extracellular loops, where contact with environmental chemical stimuli could occur (Figure 2B). Bitter taste receptors, which accommodate the specific diet of their host organism, exhibit a similar pattern of diversifying selection (Figure 2B) (Hayakawa et al., 2014; Wooding, 2011). In contrast, members of the Orai family, which encode 4TM proteins and are not thought to detect environmental chemical stimuli, show no signs of diversifying selection (Figure 2B) (Amcheslavsky et al., 2015). The MS4A family therefore exhibits a pattern of expansion and divergence similar to other chemosensors, although the observation that both within- and between-species MS4A sequence variability is enriched within extracellular loops — rather than traditional hydrophobic binding pockets — suggests that if the MS4As detect odors they do so through a distinct domain from conventional GPCR ORs.
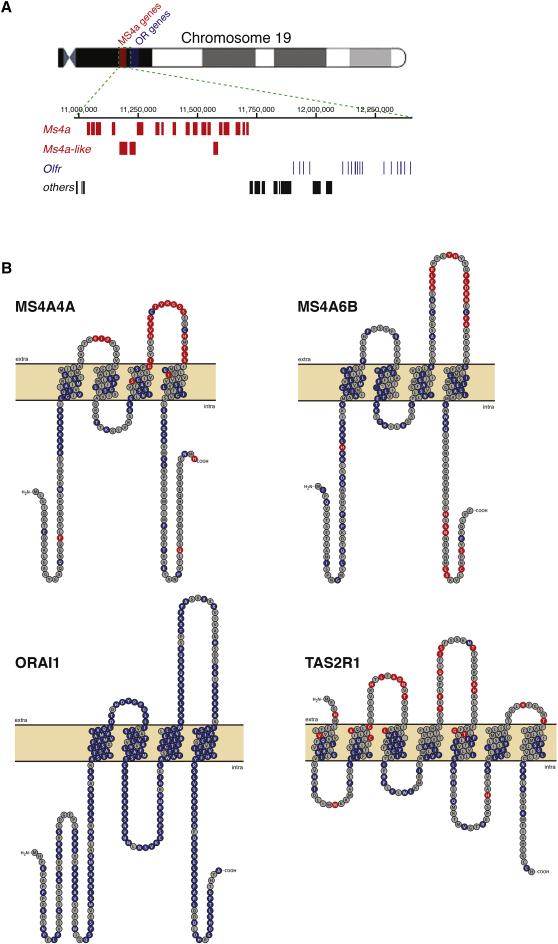
Figure 2.
Genomic clustering and positive selection of the Ms4a gene family. A. Graphical representation of Chromosome 19 of Mus musculus, including the entire Ms4a gene family (red); immediately telomeric to the Ms4a gene cluster resides a large group of conventional mammalian odorant receptor genes (blue). B. Topographical representations of the primary sequences of Mus musculus MS4A4A (top, left), MS4A6B (top, right), ORAI1 (bottom, left), and TAS2R1 (bottom, right). Amino acid residues under strong purifying selection are shown in blue, whereas those under positive selection are shown in red (posterior probability > 0.90, see Experimental Procedures); residues under positive selection are enriched within extracellular loops of MS4A proteins and bitter taste receptors (p = 5.06 X 10−16 and p = 6.76 X 10−7, respectively, hypergeometric test). See also Figure S2.
MS4A Proteins Confer Odor Responses In Vitro
Although no endogenous or natural ligands have been identified for any member of the MS4A family, the specific expression of a molecularly diverse complement of MS4As within olfactory sensory neurons — taken with prior evidence suggesting involvement in calcium signaling — raised the possibility that Ms4a genes encode a novel class of chemoreceptor (Bubien et al., 1993; Dombrowicz et al., 1998; Koslowski et al., 2008; Lin et al., 1996; Polyak et al., 2008). To test whether specific interactions between odors and MS4A proteins induce calcium influx in cells, we heterologously expressed individual MS4A proteins together with the genetically-encoded fluorescent calcium indicator GCaMP6s in HEK293 cells (Figures S3A and S3B); expression of MS4A proteins did not increase the baseline rate of calcium transients (Figures S3C and S3D). Six MS4A proteins (selected for their structural diversity) were exposed to 11 compound mixtures, each of whose constituents shared similar chemical structure. These mixes were designed to cover a broad swath of odor space, and included known ligands for conventional and necklace glomeruli (Gao et al., 2010; Saito et al., 2009). Increases in intracellular calcium were observed during odor exposure for specific MS4A protein/odor mixture pairs, demonstrating that MS4A proteins transduce signals reflecting the presence of extracellular small molecule ligands (Figures 3A and 3B).
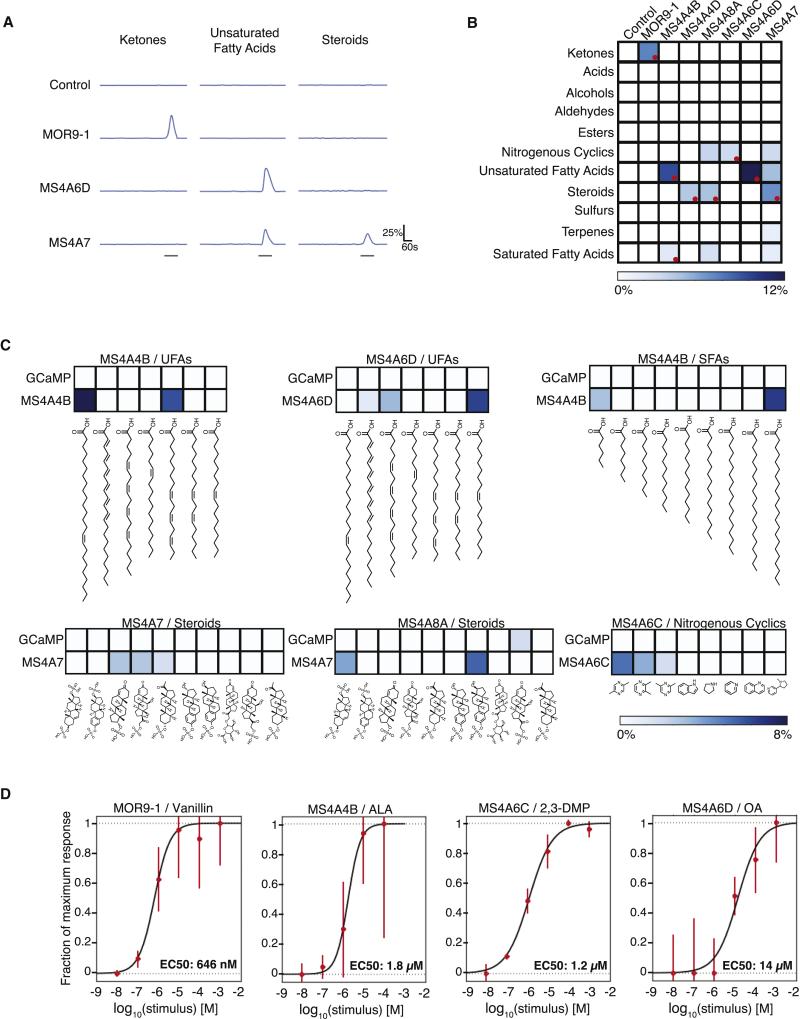
Figure 3.
MS4A proteins confer responses to odorants. A. GCaMP6 fluorescence in response to indicated chemical mixtures in representative HEK293 cells expressing either the indicated MS4A protein or GPCR mOR + G-protein (odor delivery indicated by gray bars after accounting for line and mixing delays, see Experimental Procedures). B. Responses of expressed MS4A protein/odor mixture pairs performed as in A (10 μM per odor, see Supplemental Table 3 for mixture definitions, 97 total compounds). Color code indicates percentage of cells responding (n=3, total cells in experiment > 50,000) after thresholding statistically significant responses (see Experimental Procedures). Deconvolved mixture-MS4A pairs indicated with red circles. C. Deconvolution identifies monomolecular odors that activate each MS4A receptor. Individual odors delivered at 50 μM in liquid phase (n=3, total cells in experiment >68,000) to cells co-expressing GCaMP6s and the indicated MS4A receptor (bottom) or GCaMP6s alone (top). The aggregate percent of cells that responded to each chemical across three independent experiments is color-mapped as in B. SFA: saturated fatty acids, UFA: unsaturated fatty acids. D. Dose response curves reveal low micromolar EC50s for MS4A4B/ALA, MS4A6C/2,3-DMP, and MS4A6D/OA. Each data point represents the mean +/− SEM from at least four independent coverslips. See also Figures S3 and S4.
MS4A sensory responses were specifically tuned to particular odor categories, with responses enriched for long chain fatty acids, steroids, and heterocyclic compounds. To identify individual MS4A ligands, the odorant mixture that evoked the largest response for each MS4A was broken down into its monomolecular constituents (Figure 3C). Individual MS4As conferred responses to a specific subset of odor molecules within a functional class. For example, cells expressing MS4A6C responded to a known ligand for necklace glomeruli, 2,5-dimethylpyrazine (2,5-DMP) as well as to a molecule not previously known to activate the necklace, 2,3-dimethylpyrazine (2,3-DMP) – but only weakly to 2,6-dimethylpyrazine, (2,6-DMP) and not at all to other structurally similar molecules in the parent mixture (Figure 3C). Several additional ligand-receptor relationships were identified, including MS4A4B/alpha-linolenic acid (ALA), MS4A6D/oleic acid (OA), MS4A6D/arachidonic acid (AA), MS4A4D/4-pregnan-11B,21-diol-3,20-dione 21-sulphate, MS4A7/5-pregnan-3A-ol-20-one sulphate, and MS4A8A/4-pregnan-11B,21-diol-3,20-dione,21-sulphate (Figures 3C, S4A, and S4B).
We also asked whether the MS4As and conventional ORs responded to odors with similar kinetics in vitro. No statistically significant differences were observed in time to response onset, time to half-maximal response, or time to peak response either between four queried MS4As or between these MS4As and MOR9-1 (Figure S4C). However, because the bulk calcium imaging assay used to screen MS4A ligands is not optimized for assessing response timing, we verified these results using GCaMP6f (a more rapidly-responding calcium indicator than GCaMP6s), a faster imaging rate, and a stimulus pencil to focally deliver odorants directly above the imaged cells. These experiments revealed that MS4A responses occur seconds after odor presentation, with similar or slightly faster response dynamics to those observed with MOR9-1 (Figures S4D and S4E, see Experimental Procedures). Dose-response curves revealed low micromolar EC50s for three specific MS4A/odor pairs, similar to the EC50 observed for MOR9-1/vanillin (Figure 3D), and well within the range of EC50s typically observed for conventional odorant receptor/ligand pairs in vitro (Saito et al., 2009; Mainland et al., 2015). Depleting extracellular calcium abolished MS4A ligand-dependent calcium transients (Figure S4F). Taken together, these results demonstrate that individual MS4A proteins enable calcium influx in response to specific monomolecular odorants in heterologous cells, with different MS4A proteins conferring responses to different ligands. The simplest explanation for this result is that the MS4A proteins are odorant receptors.
Many Ms4a Genes And Proteins Are Expressed In Each Necklace Sensory Neuron
The functional organization of the mammalian olfactory system depends on each mature OSN expressing just one (or two) of the hundreds of possible olfactory receptor genes (Dalton et al., 2015). We therefore asked whether the MS4As are expressed in a one-receptor-per-neuron pattern within GC-D cells, which would suggest that the necklace system follows a similar functional logic to the main and accessory olfactory systems. Target Ms4a mRNA molecules within dissociated GC-D cells were labeled using a single molecule detection approach (RNAScope), in which messages are detected as diffraction-limited fluorescent puncta whose abundance reflects transcript levels (Wang et al., 2012). RNAScope probes generated multiple puncta in individual GC-D cells for each of the 12 Ms4a genes found by RNA profiling, consistent with the presence of these Ms4a messages in GC-D cells (Figure 4A). Conversely, RNAScope failed to identify Ms4a1, Ms4a2, and Ms4a5 puncta in GC-D cells above the background detection rate, consistent with the absence of these specific Ms4a genes in our earlier RNA profiling experiments (Figures 4B and S5A).
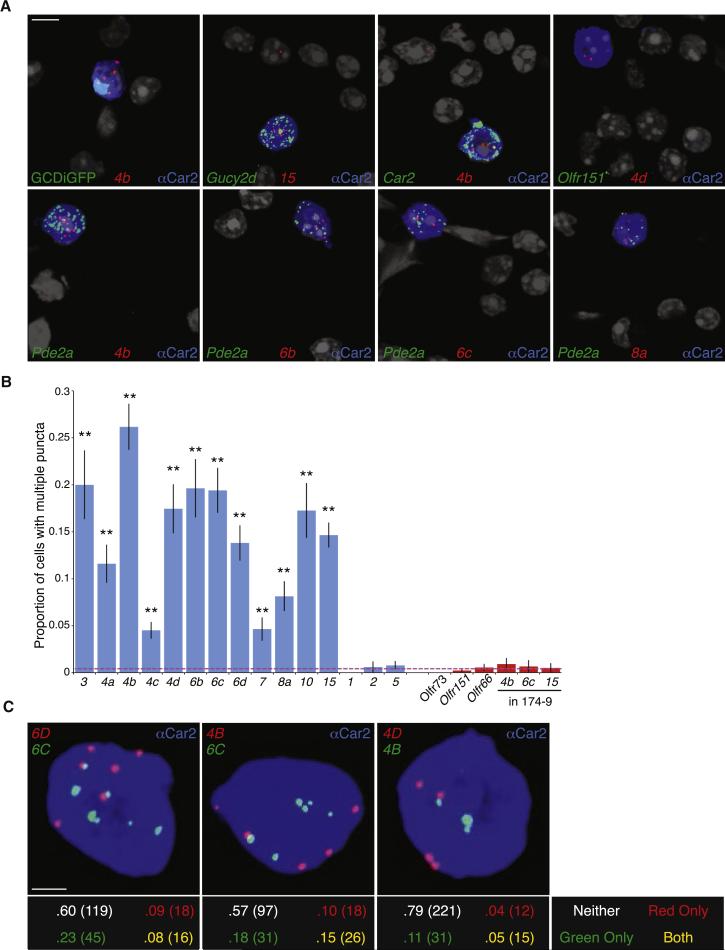
Figure 4.
Multiple Ms4a genes are expressed in each necklace sensory neuron. A. RNAScope single molecule fluorescent in situ hybridization of dissociated olfactory epithelial cells detects Ms4a family members (red). Necklace cells identified via co-labeling with an antibody against Car2 (blue), GFP from the Gucy2d-IRES-TauGFP allele (green, top left) or an RNAScope probe against a necklace marker gene (green, all panels except the top left). Necklace cells are not marked by a probe against the conventional OR gene Olfr151 (top right). Nuclei marked by DAPI (grayscale); cytoplasmic anti-CAR2 signal was saturated to demarcate the entire volume of each GC-D cell. Scale bar 5 μM. B. Proportion of Car2+ necklace OSNs with two or more detected puncta for each Ms4a (blue bars) and Olfr probe (red bars, including Ms4a puncta in OR174-9-IRES-GFP expressing cells; n=3 experiments, between 150-750 cells/probe, error bars are standard error of the proportion). Dashed red line represents the average value of negative controls. All Ms4a probes (other than negative controls Ms4a1, Ms4a2, Ms4a5) give a significantly higher proportion of positive cells than negative controls (p < .01, one-tailed Z test on population proportions). C. Representative images of Car2+ (blue) cells labeled with probes against the indicated Ms4a family members (top panels). The proportion and total number of cells with multiple puncta for neither, one, or both colors was quantified (bottom panels, total cell number in parentheses). Each pair shows significantly more double-positive cells (yellow) than expected (p < .05, Fisher's Exact Test on 2×2 table). Scale bar 2 μM. See also Figure S5.
Using a stringent criterion (in which a cell is counted positive only if it harbors two or more puncta), transcripts for each of the 12 Ms4a genes expressed in GC-D cells were found in 5-30% of GC-D cells (Figure 4B). Moreover, employing a less stringent criterion for Ms4a positivity — in which cells with any Ms4a puncta are considered positive — reveals that individual Ms4a family members may be expressed in >50% of GC-D cells (Figure S5B). Under both of these analyses, the proportions of cells expressing each Ms4a message sum to significantly greater than 100%, raising the surprising possibility that each GC-D cell expresses more than one Ms4a gene. To directly test whether Ms4a genes are co-expressed, we simultaneously labeled cells in two different colors with RNAScope probes recognizing distinct Ms4a genes. Every tested pair revealed a significant rate of cells that were positive for more than one Ms4a gene, demonstrating that unlike conventional ORs, multiple Ms4a genes are co-expressed in individual necklace sensory neurons (Figures 4C, S5C and data not shown). These data are consistent with a model in which each GC-D cell expresses multiple Ms4a genes.
Because different probes could have distinct false negative rates (and because Ms4a transcript levels are low), RNAScope could not be used to definitively determine the number of unique Ms4a genes expressed per necklace OSN (Kim et al., 2015). We therefore asked whether the Ms4a expression pattern was more apparent at the protein level. Peptide antibodies were raised against several MS4As and used to stain the olfactory epithelium. Consistent with every necklace cell expressing all members of the MS4A family (of the subset expressed in GC-D cells), each of five different anti-MS4A antibodies labeled > 95% of GC-D neurons (Figures 5A and 5B). As expected, these antibodies did not label conventional OMP+ OSNs (Figure 5A), and control antibodies against MS4As not detected in necklace neurons by RNA profiling — MS4A1, MS4A2, and MS4A5 — did not label GC-D cells (Figure 5B and data not shown). Purified anti-MS4A antibodies were specific in vivo as assessed by peptide competition, and in vitro as assessed by staining HEK 293T cells overexpressing individual MS4A proteins, although minor cross-reactivity was observed between pairs of highly homologous MS4A proteins (e.g., MS4A6B/MS4A6C and MS4A4B/MS4A4C) (Figures S6A and S6B).
Figure 5.
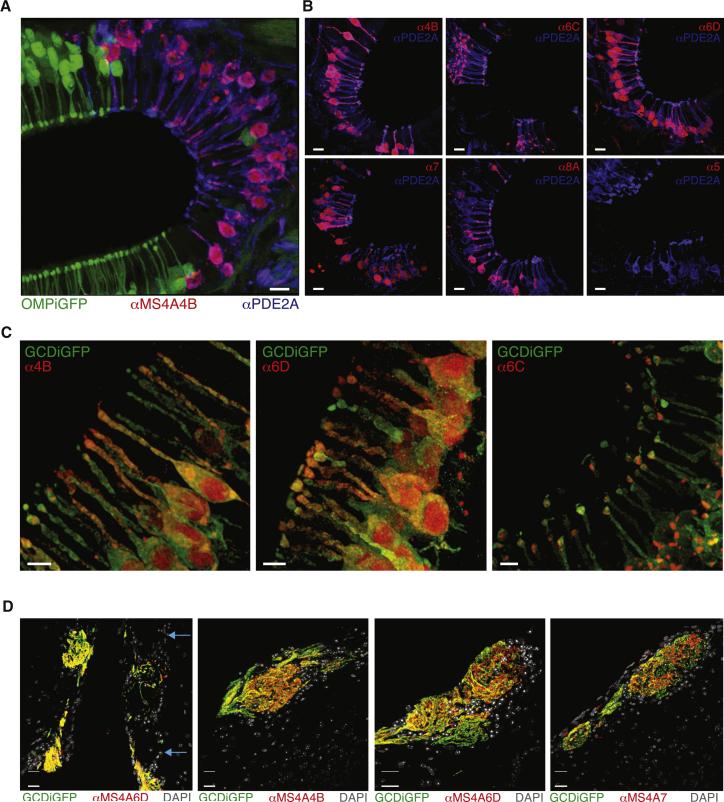
Multiple MS4A proteins are expressed within necklace sensory endings and glomeruli. A. Anti-MS4A4B antibody stains every anti-PDE2A+ cell but no OMP-IRES-GFP+ cells in sections of the olfactory epithelium. Scale bar 10 μM. B. Representative images of immunostaining with antibodies against five different MS4A family members, each of which stains >95% of anti-PDE2A+ necklace cells in epithelial cul-de-sacs; control antibody against MS4A5, which is not detected at the mRNA level in GC-D cells, does not label necklace cells (bottom right). Scale bars 10 μM. C. Anti-MS4A antibodies label dendritic knobs. Many MS4A antibodies also appear to stain perinuclear and nuclear regions; although the origin of this staining, which is eliminated by peptide competition (Figure S6B), is unknown, it may represent MS4A protein trapped within the endoplasmic reticulum or MS4A protein fragments (Cruse et al., 2013). Scale bars 5 μM. D. Anti-MS4A6D staining overlaps with all GCD-IRES-TauGFP+ necklace glomeruli in sections of the olfactory bulb (left). Blue arrows mark non-necklace glomeruli, which are not stained by anti-MS4A6D antibody. Similarly, anti-MS4A4B and anti-MS4A7 antibodies label each necklace glomerulus (right panels). Scale bars 20 μM. See also Figure S6.
If MS4As function as necklace chemoreceptors they must be present at sensory endings where transduction of odorant binding occurs (Barnea et al., 2004). Consistent with this possibility, high resolution imaging demonstrated that each MS4A antibody strongly labeled the dendritic knobs of GC-D cells, with some staining apparent in the cilia as well (Figure 5C). Sensory neurons in the main olfactory epithelium also traffic receptors to their axonal endings, which terminate in glomeruli in the olfactory bulb. To test whether MS4A proteins are similarly localized to axonal endings, olfactory bulb tissue sections were probed with anti-MS4A antibodies, which revealed that each MS4A antibody labeled every necklace glomerulus — but failed to label any conventional glomeruli — within the olfactory bulb (Figure 5D). Taken together, these data demonstrate that MS4A proteins are appropriately positioned within sensory endings to detect chemical cues in the environment, and that every glomerulus in the necklace system receives input from sensory afferents potentially representing information pooled from all the MS4As expressed in the necklace system; this pattern of organization differs sharply from that apparent in the rest of the olfactory system, where individual receptors (or pairs of receptors) define specific glomerular information channels.
Necklace Olfactory Neurons Respond to MS4A Ligands In Vivo
The expression of multiple MS4A family members in GC-D cells predicts that the chemoreceptive fields of individual necklace olfactory neurons in vivo should include the ligands identified for different MS4A proteins in vitro. To address this possibility, the intact olfactory epithelium was explanted and functional responses were assessed by multiphoton microscopy as odorants were delivered in liquid phase. Necklace neurons were labeled with the fluorescent calcium reporter molecule GCaMP3 using the Emx1-Cre driver line; the Emx1 gene was enriched in RNASeq and Nanostring analyses of GC-D neurons, and distinguishes necklace cells from conventional OSNs, which express the related protein EMX2 (Figure S7A and data not shown) (Hirota and Mombaerts 2004). Because Emx1 labels a number of non-GC-D cells in the nasal epithelium (whose identity is unclear), we specifically imaged cul-de-sac regions, where nearly all GCaMP3-positive cells belong to the necklace (Figure S7B), and heuristically identified necklace neurons as those that responded to carbon disulfide, a known necklace ligand whose detection requires the GC-D protein (Munger et al., 2010).
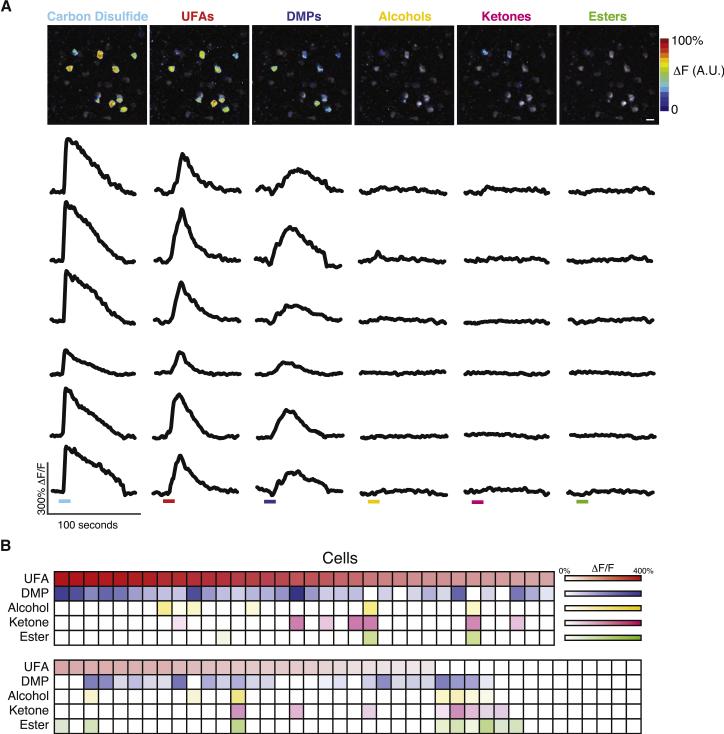
Necklace neurons were activated by a mixture of the unsaturated fatty acids oleic acid and alpha-linolenic acid (UFAs), and by a mixture of 2,3- and 2,5- dimethylpyrazine (DMPs) — two chemical classes that stimulated MS4A proteins in vitro — but rarely by mixtures of ketones, esters, or alcohols, which were not identified as MS4A ligands (Figures 6A, 6B, and 3B). Importantly, UFAs and DMPs do not activate adjacent conventional OSNs, (i.e., those that responded to the control odor mixtures but not to CS2 (data not shown)); necklace cells are therefore specifically tuned to MS4A-activating compounds. These data also demonstrate that single necklace cells respond to multiple compounds that individually activate different MS4A proteins in vitro as oleic acid and alpha-linolenic acid are ligands for MS4A6D and MS4A4B, respectively, and the dimethylpyrazines are ligands for MS4A6C (Figures 6A, 6B, and 3C). Because MS4A proteins exhibit partially overlapping tuning properties to mixtures in vitro, we confirmed that individual necklace cells responded to multiple monomolecular MS4A ligands (data not shown). These results strongly suggest that co-expression of Ms4a genes confers upon each GC-D cell a tuning profile that is broader than that associated with any single MS4A protein.
Figure 6.
Multiple in vitro MS4A ligands activate single necklace cells in vivo. A. Functional responses of individual necklace cells (labeled with Emx1-cre/GCaMP3) to the indicated odorants were heat-mapped (top row) and dF/F traces for six cells (drawn from the heatmapped image) were plotted (bottom rows). Colored bars represent the 10-second odor delivery period (after accounting for line and mixing delays, see Experimental Procedures). All mixtures were at 100 μM total odorant concentration (DMP: 2,3-DMP and 2,5-DMP, UFA: oleic acid and alpha-linolenic acid, ketones, esters, and alcohols as in Supplemental Table 3). Scale bar 10 μM. B. The odor tuning properties of 74 CS2-responsive cells (columns) across eight experiments were quantified, with color coding as in the top panel. DMPs and UFAs each activated significantly more cells (response > 25% dF/F) than each negative control (P < 0.0001, Fisher's Exact Test, corrected for multiple comparisons). Significantly more cells responded to both UFAs and DMPs than expected by chance (P < 0.01, Fisher's Exact Test). See also Figure S7.
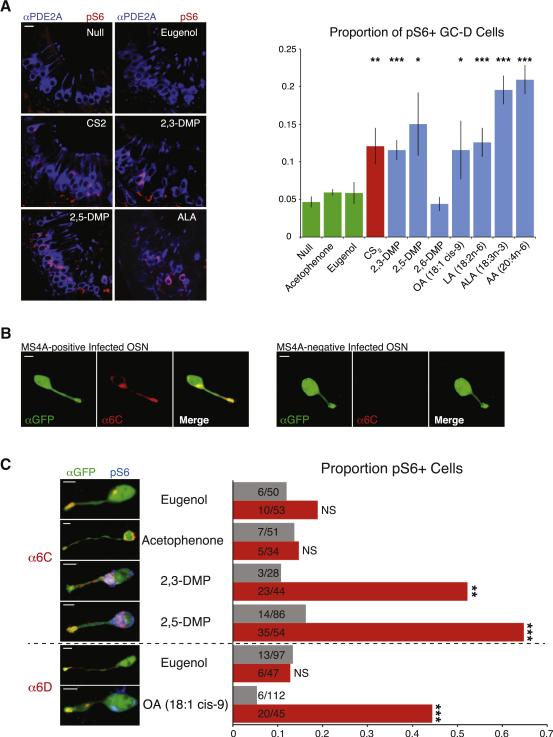
Given that MS4A ligands posess a range of volatilities, we wished to directly demonstrate that these odorants can activate necklace sensory neurons in intact mice. Freely behaving mice were exposed to MS4A ligands in gas phase, and then activation of GC-D cells was measured using an antibody against phosphorylated ribosomal S6 protein, an established marker of prior OSN activity (Jiang et al., 2015). These experiments revealed that the MS4A6C ligands 2,5-DMP and 2,3-DMP, the MS4A4B ligand alpha-linolenic acid, and the MS4A6D ligands oleic acid and arachidonic acid reliably activated GC-D cells in vivo to a similar extent as the positive control carbon disulfide (Figure 7A). Conversely, general odorants (including acetophenone and eugenol) and compounds that only weakly activated MS4A-expressing HEK293 cells (such as 2,6-DMP) did not activate necklace cells above the background rate of plain air (Figure 7A). While prior work had implicated 2,5-DMP as a necklace ligand, 2,3-DMP, arachidonic acid and the fatty acids had not been previously shown to activate this olfactory subsystem. These experiments demonstrate that, in the context of active exploration, necklace sensory neurons respond to ligands for MS4A receptors.
Figure 7.
MS4A ligands activate necklace sensory neurons, and MS4A proteins confer MS4A ligand responses to conventional olfactory sensory neurons in awake, behaving mice. A. Example images of cul-de-sacs from mice exposed to the indicated odorant, immunostained for the necklace cell marker PDE2A (blue) and the neuronal activity marker phospho-S6 (pSerine240/244) (red) (left panels). Quantification of the proportion of pS6+ necklace cells in odor-exposed mice (right panel, mean +/− SEM, n >= 3 independent experiments, * indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.0001, unpaired t-test compared to null exposure). Scale bar 10 μM. B. Olfactory epithelial sections of mice infected with adenovirus carrying an Ms4a6c-IRES-Gfp expression cassette reveal a subset of virally infected cells (green) that also express MS4A6C protein (red). Scale bars 5 μM. C. Representative images (left panels) and quantification (right panel) of phospho-S6 positive, OSNs infected with Ms4a6c-IRES-Gfp or Ms4a6d-IRES-Gfp-expressing adenovirus and exposed to the indicated odorant (DMP = dimethylpyrizine, OA = oleic acid). Gray bars: GFP-positive/ MS4A6C-negative, red bars: GFP-positive/ MS4A6C or MS4A6D-positive cells (n >= 3 animals per odor, ** indicates p < .001, *** p < .0001, Fisher's Exact Test comparing MS4A-positive to MS4A-negative cells for each odorant). Scale bars 5 μM.
It is not clear how soluble ligands such as urinary peptides gain access to necklace sensory neurons within the olfactory epithelium; nevertheless, one highly soluble class of MS4A ligand — the sulfated steroids — also activated the necklace, albeit more weakly than observed for other odorants (control DMSO = 8.7 ± .6 vs. sulfated steroids 1,3,5(10)-estratrien-3,17beta-diol disulphate/1,3,5(10)-estratrien-3,17alpha-diol 3-sulphate 12.9 ± 1.5 percent cells positive, n=4, p<.05 unpaired T test). It is notable that both in explants and in vivo the MS4A ligands and the control ligand carbon disulfide activated a fraction of the necklace sensory neurons. While these partial responses (also previously observed for urinary peptides) (Leinders-Zufall et al., 2007) may reflect specific technical features of these experiments, the consistency of this observation across ligands and preparations raises the possibility that cellular responses in the intact necklace system may be context- or state-dependent.
Altogether these results support a model in which MS4A receptors bind inhaled odorants and induce calcium influx into necklace OSNs. This model predicts that ectopically expressed MS4A protein will confer its in vitro chemosensitivity onto the receptive fields of conventional olfactory neurons. To test this hypothesis directly, mouse nares were irrigated with adenovirus encoding bicistronic Ms4a6c-IRES-GFP transcript, yielding sparse populations of GFP+ OSNs. This approach generated neurons that expressed MS4A6C protein as well as a subset of cells that were infected but did not express MS4A6C, which served as an internal control (Figure 7B); the failure of MS4A6C protein expression in some GFP positive cells may reflect stochastic (and potentially cell type-specific) effects related to ectopic chemosensor expression within mature OSNs (Tsai and Barnea, 2014). Ectopic MS4A6C protein was localized to dendritic endings in infected neurons, suggesting that MS4A proteins associate intrinsically with sensory structures even outside the molecular milieu of the necklace (Figure 7B and data not shown). Quantitative analysis of neural activity (assessed by phosphorylated S6 protein levels, Figure 7C, blue channel) after exposing awake, behaving animals to odors in gas phase revealed that ectopic MS4A6C confers responses to its in vitro ligands 2,3-DMP and 2,5-DMP, but not to the control odorants eugenol and acetophenone (Figure 7C). Similarly, adenovirally-mediated ectopic expression resulted in effective targeting of MS4A6D to sensory endings in conventional olfactory sensory neurons, and conferred specific responses to the MS4A6D ligand oleic acid, but not to the control odorant eugenol (Figure 7C). These results demonstrate that an MS4A protein can directly impart its chemoreceptive properties to generic olfactory neurons in vivo, and furthermore that signaling downstream of MS4A proteins does not require necklace-specific molecular components such as GC-D, PDE2A or CNGA3. The concordance of necklace cell responses with the chemoreceptive fields of MS4A proteins expressed ectopically — both in vitro and in vivo — implies that the necklace subsystem uses the Ms4a family to sense odors.
Discussion
Insects and mammals use similar molecular mechanisms to detect light, heat and several gases, suggesting that solutions to common sensory problems are often conserved (Caterina, 2007; Dhaka et al., 2006; Terakita, 2005). However, peripheral mechanisms for odor detection differ amongst phyla — insects like Drosophila melanogaster deploy several structurally-distinct ionotropic odorant receptor classes to interrogate the chemical world, whereas mammals were thought to detect smells exclusively through metabotropic GPCRs (Silbering and Benton, 2010). Our identification of a non-GPCR family of odorant receptor reveals an unexpected similarity between the mammalian olfactory system and that of insects: both use multiple unrelated receptor types to transduce chemosensory cues into intracellular signals.
Multiple lines of evidence indicate that Ms4a genes encode a novel family of chemoreceptors. Mammalian MS4A proteins are localized to the dendritic endings of olfactory sensory neurons, the site of odorant chemotransduction, and contain hypervariable regions that can potentially interact with diverse extracellular cues. Expression of MS4As in both HEK293 cells in vitro and conventional OSNs in vivo confers specific responses to odorants, indicating that MS4A proteins are sufficient to transduce the binding of extracellular ligands into intracellular signals. Furthermore, individual necklace sensory neurons, which co-express multiple MS4A proteins, each respond in vivo to multiple MS4A ligands identified in vitro. These data demonstrate that the MS4As define a new mechanism and logic for mammalian chemosensation, and are likely responsible for endowing the necklace olfactory subsystem with specific sensory odor tuning properties.
An Alternative Logic Organizes the Necklace Olfactory System
We find that MS4A ligands are enriched for molecules with innate significance for the mouse, including the aversive pheromone 2,5-DMP and several appetitive long chain fatty acids; interestingly, seeds and nuts, which are a major food source for mice in the wild, have high concentrations of those specific fatty acids detected by the MS4As (Sabir et al., 2012). Like the MS4A ligands, many of the molecules previously shown to trigger activity within the necklace olfactory system also have innate meaning for mice; these include carbon dioxide, which mice robustly avoid, and carbon disulfide and the peptides guanylin and uroguanlyin, each of which promotes the social transmission of food preferences (STFP) between mice (Arakawa et al., 2013; Hu et al., 2007; Munger et al., 2010).
The tuning properties of the necklace are determined, in part, by the co-expression of genes encoding multiple MS4A receptors within individual necklace sensory neurons. This pattern of odor receptor gene expression stands in stark contrast to the canonical one-receptor-per-neuron rule that (to a first approximation) organizes the remainder of the mammalian and the entirety of the Drosophila olfactory systems. In those systems, each OR is associated with one or a small number of glomeruli in the brain; in the necklace system, each sensory neuron and glomerulus is, in principle, capable of conveying odor information detected by all of the MS4A receptors expressed in the nose.
The ability of individual necklace neurons to detect multiple innately relevant signals (including gases, peptides, and volatile odors) of widely differing valences suggests two broad models for the perceptual function of the necklace subsystem. First, the necklace could discriminate between odors, albeit through distinct mechanisms from the conventional olfactory system. Odor-evoked differences in activity in individual necklace glomeruli could be generated by subtle differences in MS4A expression within subsets of sensory neurons (but see Figure 5); such differences could also be caused by conventional OSN axons, which may co-mingle with necklace sensory axons in necklace glomeruli (Cockerham et al., 2009; Secundo et al., 2014). Stimulus discrimination could alternatively be achieved through linear or non-linear interactions between intracellular signals downstream of individual MS4A receptors, which could allow the integration or gating of signals arising from distinct odorants and gases detected by single necklace neurons (van Giesen et al., 2016); this process could generate a range of firing rates (depending, for example, on the specific odor components present in a blend) that could then be differentially read-out by the brain.
Second, the main function of the necklace system could be odor detection rather than discrimination. In this model, the necklace system may be organized in a similar manner to the mammalian bitter taste and C. elegans olfactory systems, which co-express receptors with widely-divergent receptive fields in sensory neurons coupled to circuits mediating stereotypical behavioral outputs, such as attraction or aversion (Adler et al., 2000; Troemel et al., 1995). In the case of the necklace system, this singular behavioral output could be aversion, as many of the MS4A ligands have been shown to be contextually aversive, including (despite their nutritional value) fatty acids at high concentrations (Galindo et al., 2012). Alternatively, the necklace could act as an alert system, signaling the presence of salient cues in the environment (whose identity would be disentangled by other sensory systems); such a notification system could be useful for modulating the internal state of the animal in response to particularly relevant external cues, thereby facilitating both innate responses to odors and odor-related learning processes (potentially including STFP).
In either the discrimination or detection model, the physiological function of the necklace could depend upon interactions between intracellular signals downstream of the MS4A receptors and GC-D. Although the MS4As do not obligately require GC-D to promote calcium influx, GC-D enzymatic activity is sensitive to calcium levels in vitro (Duda and Sharma, 2008). This observation raises the possibility that the MS4A and GC-D pathways intersect in some manner, perhaps facilitating coincidence detection between gases and peptides on the one hand, and volatile odors on the other.
Dissecting MS4A Function
The availability of ligands for the MS4As now renders accessible key questions about the biophysics and biochemistry of MS4A function. Previous biochemical studies relied upon clustering antibodies to trigger MS4A1-mediated calcium flux (Janas et al., 2005); this and related work suggested that the MS4As may facilitate increases in intracellular calcium by acting as co-receptors for associated ligand-binding proteins such as the B-cell receptors (Dombrowicz et al., 1998; Eon Kuek et al., 2015). However, interpreting these experiments has been difficult in light of possible gains-of-function imposed by the use of clustering reagents; here we show that small molecule ligands cause an MS4A-dependent influx of extracellular calcium, demonstrating that the MS4A molecules themselves have a receptor function. It is not clear, however, whether the MS4A proteins themselves form a calcium-permeable channel (similar to the Drosophila IR odorant receptors), or whether the MS4As act as ligand-binding co-receptors for an ion channel that is expressed in many cell types (Abuin et al., 2011; Benton et al., 2009).
The underlying molecular mechanisms through which the MS4A proteins interact with ligands are not known. The observation that the molecular diversity within the MS4A family is largely found within extracellular domains — rather than transmembrane domains — suggests that the MS4As interact with odor ligands through biophysical mechanisms that are distinct from those used by conventional ORs. These mechanisms may be similar to those used by mammalian bitter taste receptors, whose diversity is also found largely in extracellular domains (Hayakawa et al., 2014; Woodling, 2011). It is also unclear whether the MS4As, despite being co-expressed in single cells, primarily interact with odorants as homomers (like the bitter taste receptors) or whether they heteromerize in a manner that alters their tuning properties (Howie et al., 2009).
The Four-Pass Transmembrane MS4As: Chemical Detectors Across Cell Types and Species?
Although a number of cellular roles have been suggested for individual MS4A proteins (largely in the context of the immune system), no clear picture has emerged of the core function of the MS4A family across cell types. The finding that multiple MS4As encode olfactory receptors suggests that they act as chemosensors in a range of tissues; this role may be both exteroceptive, serving to detect small molecules from the outside world, and interoceptive, as several of the ligands for MS4A proteins (e.g., oleic acid and arachidonic acid) are used as signaling molecules physiologically. Consistent with this hypothesis, members of the MS4A6/7 subfamily are expressed in microglia, neuroimmune cells responsible for sensing and responding to a variety of endogenous protein and lipid chemosignals in the mammalian brain; human MS4A8B and MS4A12 are expressed in epithelial cells at the lumenal surface of the small and large intestine, tissues that both sense dietary lipids; human MS4A8B is localized specifically in chemosensory cilia in lung cells tasked with probing the sensory environment; and MS4A5 is expressed in mammalian spermatocytes, which chemotax to oocytes (Eon Kuek et al., 2015; Koslowski et al., 2008).
MS4A homologs are present in all mammalian lineages and in many sequenced deuterostomes including osteichthyes (Figure S2C and data not shown), implying that the evolution of the Ms4a genes antedates the advent of the mammalian receptors for taste and for pheromones (Grus and Zhang, 2009). Taken with the finding that the MS4As detect a variety of innately-relevant cues, these observations invite the speculation that the MS4A molecules represent an ancient mechanism for sensing ethologically-salient small molecules in the environment. Testing this hypothesis will require establishing chemosensory roles for Ms4a genes in other species and better definition of those natural ligands that optimally activate the MS4As (given the limits of the synthetic odor panel explored here). It is important to note that all MS4A ligands thus far identified are also detected by other receptor molecules in the smell and taste systems (Isogai et al., 2011; Mamasuew et al., 2011; Oberland et al., 2015). Nevertheless, the maintenance of the MS4A receptor repertoire for more than 400 million years, especially given the evolutionary success of G-protein coupled odor receptors, argues that MS4As play an important — and non-redundant — role in sensory physiology (Zuccolo et al., 2010).
Although the Ms4a genes are conserved across vertebrates, Gucy2d is itself pseudogenized in most primates (Young et al., 2007). It is unclear if the absence of GC-D reflects a disappearance of the necklace system in primates or merely that GC-D became unnecessary for the tasks required of the primate necklace, given the persistence of the Ms4a genes (Young et al., 2007). Similarly, the vomeronasal organ, the sensory epithelium thought to mediate the majority of pheromone responses in rodents, became vestigial approximately 25 million years ago (Zhang and Webb, 2003). Future investigation of the functional role of Ms4a gene families across chordates — and the relevance of interactions between MS4A proteins and ethologically-relevant cues like pheromones and fatty acids — will reveal both common and species-specific roles of the MS4As in processing information from the chemical environment.
Experimental Procedures
Graphical representations of the data are presented as the mean +/− standard error of the mean unless otherwise noted. All mice were obtained from the Jackson Laboratory with the exception of OR174-9-IRES-tauGFP, which was obtained from the Axel lab. For deep sequencing, olfactory epithelia were dissociated using papain and individual cells were FAC sorted; RNA was then isolated using Trizol (Invitrogen) before SmartSeq2-based amplification and Illumina-based sequencing. Nanostring-based RNA quantitation was performed using a modified protocol to optimize for recovery of low-abundance transcripts. MS4A sequences were extracted from the Ensembl and NCBI databases. MS4A proteins were expressed in HEK293 cells using a Tet-regulatable system to control protein expression (Invitrogen); odor delivery was either achieved via bulk exchange or through a focal stimulus pencil placed over specific fields of view, and odor delays were determined using Rhodamine B dye as a surrogate. Odor responses were imaged in both configurations using an Andor Neo sCMOS camera mounted on an Olympus IX83. Single molecule in situ RNA detection was perfomed using RNAScope (Advanced Cell Diagnostics); this protocol was adapted to simultaneously immunostain with either anti-CAR2 or anti-GFP antibodies. Custom antibodies against the MS4A proteins were developed in rabbits or guinea pigs (Covance) and purified using protein A-sepharose and bead-conjugated peptides. Multiphoton-based functional imaging (Prairie Technologies) of explanted olfactory epithelia was performed in superfused carboygenated modified Ringer's solution, with odor delivery in liquid phase controlled by a solenoid valve system (Warner Instruments); image alignment was achieved using custom feature-tracking and homography algorithms. Human adenoviruses were generated by the UNC Viral Core facility. See supplemental information for details regarding experimental methods, reagent sources, and statistical procedures.
Supplementary Material
Acknowledgements
We thank Michael Greenberg, Steve Liberles, Bernardo Sabatini, Rachel Wilson, Elizabeth Hong, Steve Flavell, Eric Griffith, James Schwob and members of the Datta Lab for helpful comments on the manuscript, and Allison Petrosino, Neha Bhagat, and Lauren Coritt for laboratory assistance. We thank Andrew Giessel, Alexander Wiltschko, and Jeffrey Markowitz for coding assistance and Hannah Somhegyi for generation of the graphical abstract. PLG was supported by the Nancy Lurie Marks Foundation, the William Randolph Hearst Fund, the Leonard and Isabelle Goldenson Fund, and the Edward and Anne Lefler Foundation. DMB is supported by a NSF predoctoral fellowship and a Sackler Scholarship in Psychobiology. JML is supported by the European Molecular Biology Organization (ALTF 379-2011), the Human Frontiers Science Program (LT001086/2012), and the Belgian American Educational Foundation. MLB is supported by a NSF predoctoral fellowship. TT is supported by a postdoctoral fellowship from the Alice and Joseph E. Brooks Foundation, the Astellas Foundation for Research on Metabolic Disorders, and the Japan Society for the Promotion of Science postdoctoral fellowship for Research Abroad. SP was supported by a NIDA F31 (DA036922-01) and is a Stuart H.Q. & Victoria Quan Fellow at Harvard Medical School. HH is supported by the Howard Hughes Medical Institute. SRD is supported by fellowships from the Burroughs Wellcome Fund, the Vallee Foundation, the McKnight Foundation, the Khodadad Program, by grants DP2OD007109 and RO11DC011558 from the National Institutes of Health, and the Global Brain Initiative from the Simons Foundation.
Footnotes
Author Contributions
PG, DB, and SRD conceived experiments and wrote the manuscript. PG, DB, MLB, TT, SP, and ACN carried out all wet experiments; evolutionary analysis was performed by JML, FKM, and HH. RK performed RNAseq read quality control and alignment.
References
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A, Wood ML, Yeromin AV, Parker I, Freites JA, Tobias DJ, Cahalan MD. Molecular biophysics of Orai store-operated Ca2+ channels. Biophys J. 2015;108:237–246. doi: 10.1016/j.bpj.2014.11.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H, Kelliher KR, Zufall F, Munger SD. The receptor guanylyl cyclase type D (GC-D) ligand uroguanylin promotes the acquisition of food preferences in mice. Chem Senses. 2013;38:391–397. doi: 10.1093/chemse/bjt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R. The molecular logic of smell. Sci Am. 1995;273:154–159. doi: 10.1038/scientificamerican1095-154. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubien JK, Zhou LJ, Bell PD, Frizzell RA, Tedder TF. Transfection of the CD20 cell surface molecule into ectopic cell types generates a Ca2+ conductance found constitutively in B lymphocytes. J Cell Biol. 1993;121:1121–1132. doi: 10.1083/jcb.121.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Caterina MJ. Transient receptor potential ion channels as participants in thermosensation and thermoregulation. Am J Physiol Regul Integr Comp Physiol. 2007;292:R64–76. doi: 10.1152/ajpregu.00446.2006. [DOI] [PubMed] [Google Scholar]
- Cockerham RE, Puche AC, Munger SD. Heterogeneous sensory innervation and extensive intrabulbar connections of olfactory necklace glomeruli. PLoS One. 2009;4:e4657. doi: 10.1371/journal.pone.0004657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse G, Beaven MA, Ashmole I, Bradding P, Gilfillan AM, Metcalfe DD. A truncated splice-variant of the FcepsilonRIbeta receptor subunit is critical for microtubule formation and degranulation in mast cells. Immunity. 2013;38:906–917. doi: 10.1016/j.immuni.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lomvardas S. Chemosensory receptor specificity and regulation. Annu Rev Neurosci. 2015;38:331–349. doi: 10.1146/annurev-neuro-071714-034145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaka A, Viswanath V, Patapoutian A. Trp ion channels and temperature sensation. Annu Rev Neurosci. 2006;29:135–161. doi: 10.1146/annurev.neuro.29.051605.112958. [DOI] [PubMed] [Google Scholar]
- Dombrowicz D, Lin S, Flamand V, Brini AT, Koller BH, Kinet JP. Allergy-associated FcRbeta is a molecular amplifier of IgE- and IgG- mediated in vivo responses. Immunity. 1998;8:517–529. doi: 10.1016/s1074-7613(00)80556-7. [DOI] [PubMed] [Google Scholar]
- Duda T, Sharma RK. ONE-GC membrane guanylate cyclase, a trimodal odorant signal transducer. Biochem Biophys Res Commun. 2008;367:440–445. doi: 10.1016/j.bbrc.2007.12.153. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Eon Kuek L, Leffler M, Mackay GA, Hulett MD. The MS4A family: counting past 1, 2 and 3. Immunol Cell Biol. 2015 doi: 10.1038/icb.2015.48. [DOI] [PubMed] [Google Scholar]
- Fulle HJ, Vassar R, Foster DC, Yang RB, Axel R, Garbers DL. A receptor guanylyl cyclase expressed specifically in olfactory sensory neurons. Proc Natl Acad Sci U S A. 1995;92:3571–3575. doi: 10.1073/pnas.92.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MM, Voigt N, Stein J, van Lengerich J, Raguse JD, Hofmann T, Meyerhof W, Behrens M. G protein-coupled receptors in human fat taste perception. Chem Senses. 2012;37:123–139. doi: 10.1093/chemse/bjr069. [DOI] [PubMed] [Google Scholar]
- Gao L, Hu J, Zhong C, Luo M. Integration of CO2 and odorant signals in the mouse olfactory bulb. Neuroscience. 2010;170:881–892. doi: 10.1016/j.neuroscience.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol. 2009;26:407–419. doi: 10.1093/molbev/msn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Zhang JJ, Huang XY. Stimulation of guanylyl cyclase-D by bicarbonate. Biochemistry. 2009;48:4417–4422. doi: 10.1021/bi900441v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Suzuki-Hashido N, Matsui A, Go Y. Frequent expansions of the bitter taste receptor gene repertoire during evolution of mammals in the Euarchontoglires clade. Mol Biol Evol. 2014;31:2018–2031. doi: 10.1093/molbev/msu144. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Howie D, Nolan KF, Daley S, Butterfield E, Adams E, Garcia-Rueda H, Thompson C, Saunders NJ, Cobbold SP, Tone Y, et al. MS4A4B is a GITR-associated membrane adapter, expressed by regulatory T cells, which modulates T cell activation. J Immunol. 2009;183:4197–4204. doi: 10.4049/jimmunol.0901070. [DOI] [PubMed] [Google Scholar]
- Hu J, Zhong C, Ding C, Chi Q, Walz A, Mombaerts P, Matsunami H, Luo M. Detection of near-atmospheric concentrations of CO2 by an olfactory subsystem in the mouse. Science. 2007;317:953–957. doi: 10.1126/science.1144233. [DOI] [PubMed] [Google Scholar]
- Ihara S, Yoshikawa K, Touhara K. Chemosensory signals and their receptors in the olfactory neural system. Neuroscience. 2013;254:45–60. doi: 10.1016/j.neuroscience.2013.08.063. [DOI] [PubMed] [Google Scholar]
- Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, Dulac C. Molecular organization of vomeronasal chemoreception. Nature. 2011;478:241–245. doi: 10.1038/nature10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas E, Priest R, Wilde JI, White JH, Malhotra R. Rituxan (anti-CD20 antibody)-induced translocation of CD20 into lipid rafts is crucial for calcium influx and apoptosis. Clin Exp Immunol. 2005;139:439–446. doi: 10.1111/j.1365-2249.2005.02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunami H. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat Neurosci. 2015;18:1446–1454. doi: 10.1038/nn.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juilfs DM, Fulle HJ, Zhao AZ, Houslay MD, Garbers DL, Beavo JA. A subset of olfactory neurons that selectively express cGMP-stimulated phosphodiesterase (PDE2) and guanylyl cyclase-D define a unique olfactory signal transduction pathway. Proc Natl Acad Sci U S A. 1997;94:3388–3395. doi: 10.1073/pnas.94.7.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147:907–921. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- Kim ST, Do IG, Lee J, Sohn I, Kim KM, Kang WK. The NanoString-based multigene assay as a novel platform to screen EGFR, HER2, and MET in patients with advanced gastric cancer. Clin Transl Oncol. 2015;17:462–468. doi: 10.1007/s12094-014-1258-7. [DOI] [PubMed] [Google Scholar]
- Koslowski M, Sahin U, Dhaene K, Huber C, Tureci O. MS4A12 is a colon-selective store-operated calcium channel promoting malignant cell processes. Cancer Res. 2008;68:3458–3466. doi: 10.1158/0008-5472.CAN-07-5768. [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, Cockerham RE, Michalakis S, Biel M, Garbers DL, Reed RR, Zufall F, Munger SD. Contribution of the receptor guanylyl cyclase GC-D to chemosensory function in the olfactory epithelium. Proc Natl Acad Sci U S A. 2007;104:14507–14512. doi: 10.1073/pnas.0704965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, Siltberg-Liberles J, Liberles DA, Buck LB. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. Proc Natl Acad Sci U S A. 2009;106:9842–9847. doi: 10.1073/pnas.0904464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Cicala C, Scharenberg AM, Kinet JP. The Fc(epsilon)RIbeta subunit functions as an amplifier of Fc(epsilon)RIgamma- mediated cell activation signals. Cell. 1996;85:985–995. doi: 10.1016/s0092-8674(00)81300-8. [DOI] [PubMed] [Google Scholar]
- Mainland JD, Li YR, Zhou T, Liu WL, Matsunami H. Human olfactory receptor responses to odorants. Sci Data. 2015;2:150002. doi: 10.1038/sdata.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamasuew K, Hofmann N, Breer H, Fleischer J. Grueneberg ganglion neurons are activated by a defined set of odorants. Chem Senses. 2011;36:271–282. doi: 10.1093/chemse/bjq124. [DOI] [PubMed] [Google Scholar]
- Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004;13:240–254. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini S, Silvotti L, Shirazi A, Ryba NJ, Tirindelli R. Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J Neurosci. 2001;21:843–848. doi: 10.1523/JNEUROSCI.21-03-00843.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Meyer MR, Angele A, Kremmer E, Kaupp UB, Muller F. A cGMP-signaling pathway in a subset of olfactory sensory neurons. Proc Natl Acad Sci U S A. 2000;97:10595–10600. doi: 10.1073/pnas.97.19.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sakano H. How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci. 2011;34:467–499. doi: 10.1146/annurev-neuro-112210-112917. [DOI] [PubMed] [Google Scholar]
- Munger SD, Leinders-Zufall T, McDougall LM, Cockerham RE, Schmid A, Wandernoth P, Wennemuth G, Biel M, Zufall F, Kelliher KR. An olfactory subsystem that detects carbon disulfide and mediates food-related social learning. Curr Biol. 2010;20:1438–1444. doi: 10.1016/j.cub.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Annu Rev Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Oberland S, Ackels T, Gaab S, Pelz T, Spehr J, Spehr M, Neuhaus EM. CD36 is involved in oleic acid detection by the murine olfactory system. Front Cell Neurosci. 2015;9:366. doi: 10.3389/fncel.2015.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak MJ, Li H, Shariat N, Deans JP. CD20 homo-oligomers physically associate with the B cell antigen receptor. Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. J Biol Chem. 2008;283:18545–18552. doi: 10.1074/jbc.M800784200. [DOI] [PubMed] [Google Scholar]
- Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Sabir A, Unver A, Kara Z. The fatty acid and tocopherol constituents of the seed oil extracted from 21 grape varieties (Vitis spp.). J Sci Food Agric. 2012;92:1982–1987. doi: 10.1002/jsfa.5571. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secundo L, Snitz K, Sobel N. The perceptual logic of smell. Curr Opin Neurobiol. 2014;25:107–115. doi: 10.1016/j.conb.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Shinoda K, Shiotani Y, Osawa Y. “Necklace olfactory glomeruli” form unique components of the rat primary olfactory system. J Comp Neurol. 1989;284:362–373. doi: 10.1002/cne.902840304. [DOI] [PubMed] [Google Scholar]
- Silbering AF, Benton R. Ionotropic and metabotropic mechanisms in chemoreception: 'chance or design'? EMBO Rep. 2010;11:173–179. doi: 10.1038/embor.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS. Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem Senses. 2000;25:155–165. doi: 10.1093/chemse/25.2.155. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang H, Hu J, Han J, Matsunami H, Luo M. Guanylyl cyclase-D in the olfactory CO2 neurons is activated by bicarbonate. Proc Natl Acad Sci U S A. 2009;106:2041–2046. doi: 10.1073/pnas.0812220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A. The opsins. Genome Biol. 2005;6:213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Tsai L, Barnea G. A critical period defined by axon-targeting mechanisms in the murine olfactory bulb. Science. 2014;344:197–200. doi: 10.1126/science.1248806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Giesen L, Hernandez-Nunez L, Delasoie-Baranek S, Colombo M, Renaud P, Bruggmann R, Benton R, Samuel AD, Sprecher SG. Multimodal stimulus coding by a gustatory sensory neuron in Drosophila larvae. Nat Commun. 2016;7:10687. doi: 10.1038/ncomms10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Giesen L, Hernandez-Nunez L, Delasoie-Baranek S, Colombo M, Renaud P, Bruggmann R, Benton R, Samuel AD, Sprecher SG. Multimodal stimulus coding by a gustatory sensory neuron in Drosophila larvae. Nat Commun. 2016;7:10687. doi: 10.1038/ncomms10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S. Signatures of natural selection in a primate bitter taste receptor. J Mol Evol. 2011;73:257–265. doi: 10.1007/s00239-011-9481-0. [DOI] [PubMed] [Google Scholar]
- Young JM, Waters H, Dong C, Fulle HJ, Liman ER. Degeneration of the olfactory guanylyl cyclase D gene during primate evolution. PLoS One. 2007;2:e884. doi: 10.1371/journal.pone.0000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci U S A. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. 2010;5:e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.