Summary
A widely shared view reads that mesenchymal stem/stromal cells (“MSCs”) are ubiquitous in human connective tissues, can be defined by a common in vitro phenotype, share a skeletogenic potential as assessed by in vitro differentiation assays, and coincide with ubiquitous pericytes. Using stringent in vivo differentiation assays and transcriptome analysis, we show that human cell populations from different anatomical sources, regarded as “MSCs” based on these criteria and assumptions, actually differ widely in their transcriptomic signature and in vivo differentiation potential. In contrast, they share the capacity to guide the assembly of functional microvessels in vivo, regardless of their anatomical source, or in situ identity as perivascular or circulating cells. This analysis reveals that muscle pericytes, which are not spontaneously osteochondrogenic as previously claimed, may indeed coincide with an ectopic perivascular subset of committed myogenic cells similar to satellite cells. Cord blood-derived stromal cells, on the other hand, display the unique capacity to form cartilage in vivo spontaneously, in addition to an assayable osteogenic capacity. These data suggest the need to revise current misconceptions on the origin and function of so-called “MSCs,” with important applicative implications. The data also support the view that rather than a uniform class of “MSCs,” different mesoderm derivatives include distinct classes of tissue-specific committed progenitors, possibly of different developmental origin.
Key words: bone marrow stromal cell, mesenchymal stem cell, skeletal progenitors, myogenic progenitors, transplantation, differentiation, in vivo assays, hematopoietic microenvironment
Highlights
-
•
CD146+ “MSCs” from different tissues exhibit different transcriptional profiles
-
•
CD146+ “MSCs” from different tissues have different differentiation capacities
-
•
CD146+ “MSCs” from different tissues organize blood vessels and become pericytes
Bianco, Riminucci, Robey, and colleagues have provided evidence that “mesenchymal stem/stromal cells” (according to current “jargon”), derived from different sources (bone marrow, muscle, cord blood) vary widely in their transcriptional profile, and their differentiation capacity as assessed by in vitro assays and in vivo transplantation assays. Bone marrow “MSCs” form bone and support hematopoiesis, but are not myogenic; muscle “MSCs” are spontaneously myogenic, but do not form bone; cord blood “MSCs” are inherently chondrogenic, and do form bone, but do not support hematopoiesis. However, despite their significant differences, “MSCs” from different sources are capable of forming pericytes when co-transplanted with endothelial cells in vivo, resulting in the development of functional blood vessels. These findings are important not only in understanding the biology of specific tissues, but also from an applicative clinical angle.
Introduction
The anatomical identity of mesenchymal stem/stromal cells (“MSCs,” the current “jargon”), their phenotype, distribution in different tissues, lineage, physiological functions, and biological properties represent one of the most controversial and confusing areas in stem cell biology. At this time, two quite distinct descriptions of “MSCs” are found in the literature. One, which emanates from ∼50 years of widely reproduced experimental work in vivo, sees “MSCs” as the same biological object previously known as cultured bone marrow stromal cells (BMSCs); these cells are unique to bone marrow (BM), and include a subset of physically identifiable clonogenic, multipotent, self-renewing progenitors of skeletal tissues, and skeletal tissues only (Bianco et al., 2013). This progenitor is endowed with the unique capacity to organize the hematopoietic microenvironment and the hematopoietic stem cell niche (Bianco, 2011, Friedenstein et al., 1982). The other view sees “MSCs” as progenitors of multiple tissues beyond the range of skeletal tissues, such as skeletal muscle (Caplan, 1991, Caplan, 2008, Crisan et al., 2008). The demonstration that “MSCs” are perivascular cells in BM (Sacchetti et al., 2007) was later extrapolated to claim that in virtually all tissues, pericytes (identified as CD34−/CD45−/CD146+ cells) would represent “MSCs” (Caplan, 2008, Crisan et al., 2008). Hence, these broadly multipotent progenitors, essentially defined by in vitro assays (Dominici et al., 2006, Pittenger et al., 1999) that are neither specific nor stringent, would be found in multiple tissues well beyond BM (e.g., skeletal muscle, fat, placenta, umbilical cord) (Caplan, 2008, da Silva Meirelles et al., 2006).
Definition of the origin, anatomy, biological properties, and function of so-called “MSCs” has obvious implications, both for understanding their biology and for their use in potential therapies. Notably, assuming that “MSCs” with identical differentiation properties can be isolated from virtually every tissue would imply that multiple tissues are equally suitable cell sources for the regeneration of multiple tissues. On the other hand, the assumption that “MSCs” are the ex vivo counterpart of pericytes would lend support to the view that a number of non-progenitor functions (Bianco et al., 2013) of “MSCs” (anti-inflammatory, immunomodulatory, trophic), claimed to be of major import for therapy of a number of unrelated disorders (Caplan and Correa, 2011), are traceable to an identifiable and ubiquitous in vivo cell type. Nonetheless, pericytes are only defined by anatomy, and currently no experimental data support the notion that they represent a distinct lineage (Armulik et al., 2011, Diaz-Flores et al., 2009). In addition, their role in tissue injury and repair is pleiotropic and spans multiple distinct processes including inflammation; furthermore, their participation in the repair of tissues (e.g., through the formation of scar tissue) does not necessarily coincide with a regenerative function.
We previously identified a minimal surface phenotype suited not only to enrich the archetypal human “MSCs” in uncultured BM cell suspensions, but also to correlate their ex vivo-assayed clonogenic capacity with their in situ identity and in vivo fate following transplantation (Sacchetti et al., 2007). As applied to the study of BMSCs, this led to identification of “MSCs” as subendothelial, perivascular CD146+ cells on BM sinusoids, and also provided evidence for their self-renewal in vivo, which had long been the missing evidence to support the claim that BMSCs indeed include a subset of bona fide stem cells, rather than multipotent progenitors (Bianco et al., 2013, Sacchetti et al., 2007). Using an identical approach to prospectively isolate “MSCs” from a variety of non-BM tissues, Crisan and co-workers later reported that a ubiquitous population of highly myogenic and skeletogenic CD146+ cells, coinciding with “MSCs,” is found in association with microvessels of skeletal muscle and other tissues, lending support to the view of pericytes as a uniform, widely distributed population of cells that can be explanted and cultured as “MSCs” (Caplan, 2008, Caplan and Correa, 2011, Crisan et al., 2008). However, striated muscle and skeletal lineages such as bone, cartilage, and marrow fat diverge early in development, and no common progenitor of bone and muscle is found in prenatal life past the time of sclero-myotome specification in somites (Applebaum and Kalcheim, 2015). The notion of a common postnatal progenitor of bone and muscle, therefore, would be at odds with established tenets in developmental biology (Bianco and Robey, 2015).
We show here that MCAM/CD146-expressing stromal cells from different human tissues diverge radically from their BM counterparts in differentiation potency and transcriptional profile, reflective of their different developmental origin. While BM-derived “MSCs”/pericytes are natively skeletogenic but not myogenic, muscle-derived “MSCs”/pericytes are inherently myogenic but not natively skeletogenic, and appear to represent a subset of cells with functional features of satellite cells, but not their characteristic anatomical location. We further show that prenatal, cord blood-borne “MSCs” in turn exhibit a distinct transcriptional and potency profile, and an inherent cartilage commitment, which diverge markedly from that of postnatal BM-derived “MSCs.” Finally we show that, irrespective of the postnatal tissue source of these perivascular cells or from fetal blood, these committed progenitors of mesoderm derivatives can associate with nascent blood vessels (BVs) in vivo and be recruited to a mural cell fate. However, a system of committed and self-renewing progenitors with distinct native potency, and not a uniform, equipotent class of “MSCs” is associated with microvascular walls in postnatal mesoderm-derived tissues as reported previously for bone/marrow (Sacchetti et al., 2007), and as shown herein for muscle. Pericyte recruitment from preexisting local progenitors is a simple developmental process that explains the very existence of such progenitors in postnatal life and their tissue-specific properties.
Results
The Phenotype of “MSCs” In Vitro Does Not Reflect Cell Identity and Function
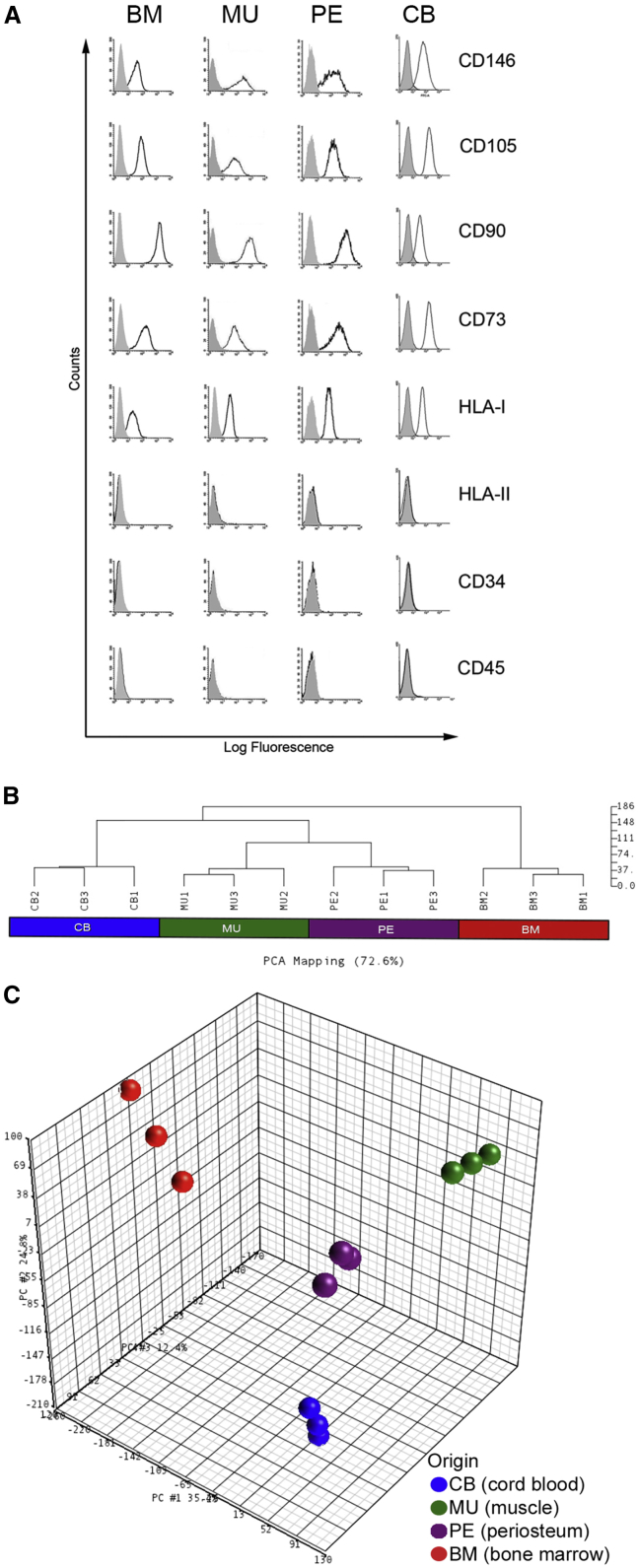
Stromal cell strains were established from four different tissue sources: BM, skeletal muscle (MU), periosteum (PE), and perinatal cord blood (CB). For all postnatal tissue sources, clonogenic cells were prospectively isolated based on a minimal surface phenotype as previously described for human BMSCs (CD34−/CD45−/CD146+); colonies of CB stromal cells were established as described previously (Kluth et al., 2010, Kogler et al., 2004). Of note, CD146 identified a clonogenic subset in MU (presented below) and PE (data not shown), as it does in BM. Multiclonal strains derived from growth of the originally explanted cells were then expanded under identical basal culture conditions that do not support the growth of endothelial cells or induce differentiation. All resulting cell strains exhibited the canonical in vitro cell-surface markers regarded as characteristic of “MSCs” (Figure 1A).
Figure 1.
Cell Surface and Transcriptomic Comparison of “MSCs” from Different Tissues
(A) Fluorescence-activated cell sorting (FACS) analysis of multiclonal BM (bone marrow), MU (muscle), PE (periosteum) and CB (cord blood) cells (representative of one from least three independent experiments). All strains express the canonical in vitro phenotype of “MSCs” and CD146 (isotype controls indicated in gray).
(B and C) Unsupervised clustering of gene-expression profiling data of CD146+ cells purified from BM, CB, MU, and PE (three independent samples for each cell type). Unsupervised analysis was performed to investigate whether there was evidence for native groupings of samples based on correlations between gene-expression profiles. Results of the hierarchical clustering (B) and the principal component analysis (C) revealed that gene-expression profiles of CD146+ cells are clearly separated by the origin factor. See Supplemental Experimental Procedures for further details of statistical analyses.
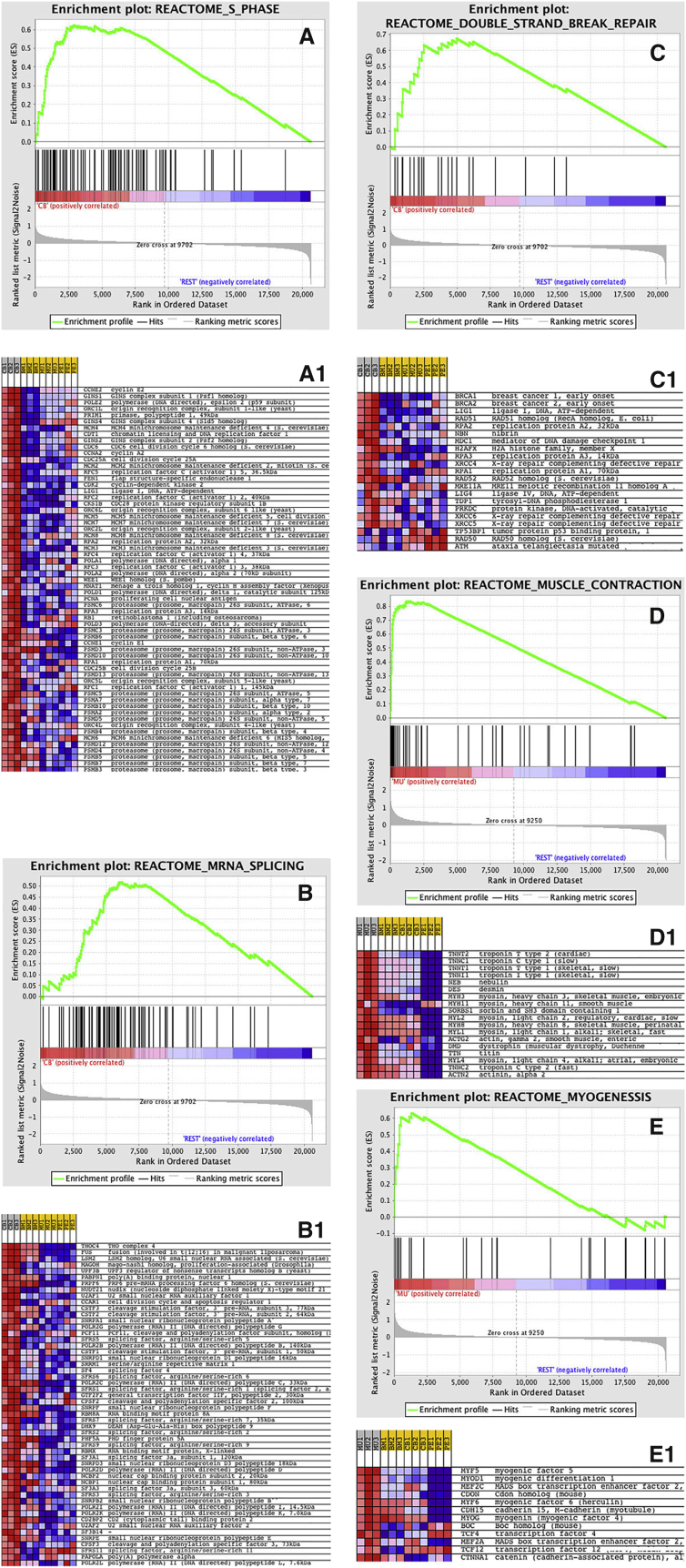
To determine the specificity and functional significance of the cell-surface phenotype of “MSCs,” widely regarded as a defining feature of “MSCs” across tissues, we performed gene-expression profiling using Affimetrix technology. Both unsupervised hierarchical clustering (Figure 1B) and principal component analysis (Figure 1C) revealed that gene-expression profiles of “MSCs” are clearly separated by an “origin” factor, indicating the lack of specificity and sensitivity of the widely used “minimum” surface phenotype. ANOVA-based supervised analysis selected 1,614 class-specific, differentially expressed genes (Table S1) showing a fold difference >3 and a false discovery rate q value of <0.05. While perinatal CB cells are characterized by over-representation of genes related to proliferation and cell-cycle regulation (Figures 2A–2C), the postnatal MU cells are characterized by over-representation of tissue-specific genes related to their tissue origin (Figures 2D and 2E), including the myogenic transcription factor, PAX7 (Figure S1A). Tables S2 and S3 show the first 100 enriched gene sets for CB and MU classes, respectively, while Figures 2A1–2E1 show enrichment plots and heatmaps for selected gene sets. The over-represented gene sets coming from gene set enrichment analysis (GSEA) (Subramanian et al., 2005) support the notion that prospectively purified CB “MSCs” are highly proliferative, since the majority of gene sets enriched in this phenotype are related to proliferation, S phase, RNA and DNA synthesis, or DNA repair. On the other hand, prospectively purified MU “MSCs” are clearly characterized by the over-representation of gene sets specifically related to either muscle development or muscle differentiated function (muscle contraction, muscle development, and energy metabolism). PE and BM expression profiling was analyzed in the same way, but no gene sets were statistically significantly enriched in PE versus CB, BM, and MU, or in BM versus PE, CB, and MU. However, a number of genes enriched in BM and PE cells was identified (Table S4). Furthermore, genes associated with hematopoietic support, a defining feature of BM cells, were over-represented in BM cells compared with CB, MU, and PE cells (Figure S2A).
Figure 2.
Enrichment Plots and Heat Maps of Selected Gene Sets for Cord Blood- and Muscle-Derived CD146+ Cells
(A–C1) Prenatal CB (cord blood) “MSCs” are characterized by the over-representation of many genes related to proliferation and cell-cycle regulation.
(D and E) Postnatal MU (muscle) “MSCs” are characterized by the over-representation of tissue-specific genes related to their tissue origin, specifically by genes regulating muscle contraction, muscle development, and energy metabolism. The over-represented gene sets from GSEA showed that prenatal CB CD146+ cells are enriched in gene sets related to proliferation S phase, RNA and DNA synthesis, or DNA repair.
For each enriched gene set, the gene expression is also represented as a blue-pink o’gram in (A1), (B1), and (C1) (CB “MSCs”), and (D1) and (E1) (MU “MSCs”). See Supplemental Experimental Procedures for further details of statistical analyses.
“MSCs” from Different Sources Have Radically Different Differentiation Properties
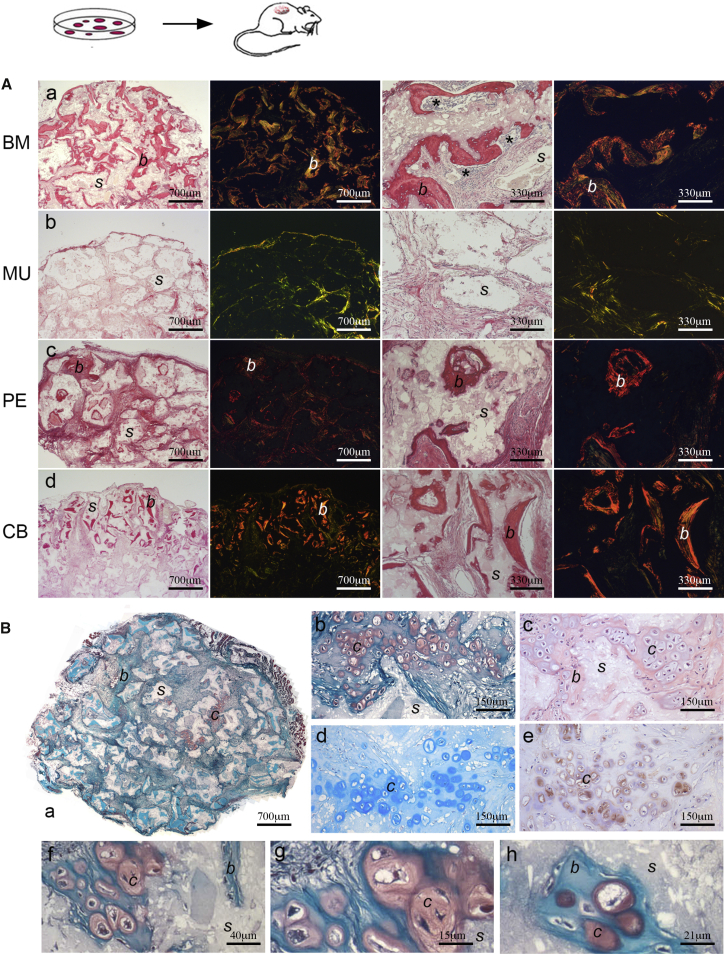
BM “MSCs,” prospectively sorted as CD34−/CD45−/CD146+ and grown under basal conditions that do not induce differentiation, regularly form bone and establish the hematopoietic microenvironment when transplanted heterotopically using an osteoconductive carrier (Sacchetti et al., 2007) (Figure 3Aa). Cells sorted based on the same phenotype from BM and other tissues, including MU, were later reported to be highly myogenic both in vitro and in vivo, in addition to sharing the ability to differentiate in culture toward skeletal lineages (Crisan et al., 2008), based on widely used artificial differentiation assays. In vitro, Alizarin red S and von Kossa staining cannot distinguish between dystrophic calcification induced by dead and dying cells versus matrix mineralization, or calcium phosphate precipitates generated by cleavage of β-glycerophosphate (a component of osteogenic medium) by alkaline phosphatase (ALP), which is expressed by many types of stromal cells. In vivo transplantation of MU “MSCs” of identical surface phenotype as BM “MSCs” revealed no spontaneous in vivo osteogenic potential (Figure 3Ab). Likewise, cells established in culture from skin, adipose tissue, and amniotic fluid, all sharing the in vitro phenotype of “MSCs,” regularly failed to form any histology-proven bone (Figure S2), whereas PE “MSCs” did form bone in vivo, as previously reported (Sacchetti et al., 2007; Figure 3Ac). Using the same in vivo assay and carrier, CB “MSCs” formed histology-proven bone of donor origin (Figure 3Ad, human Lamin A/C-positive osteocytes, not shown). Surprisingly, they also generated Safranin O+, Alcian blue+, COL2+ hyaline cartilage intermingled with bone in the same assay (three of three strains from different donors, and one of three single colony-derived strain), but never established a hematopoietic microenvironment (Figures 3Ba–3Bh). This result was unique, as previously we have never seen BM “MSCs” make cartilage in this ceramic-based assay. Failure to generate cartilage under these conditions has been interpreted as a need for a hypoxic environment for chondrogenesis in vivo, which is not provided in an open transplantation system permissive for vascularization. While hypoxia undoubtedly contributes to chondrogenesis (as in fracture callus formation; e.g., Hirao et al., 2006), our results suggest that there are also cell-intrinsic factors at play in CB-derived chondrogenesis based on their more primordial, fetal origin (Bianco and Robey, 2015).
Figure 3.
In Vivo Transplantation of CD146+ “MSCs” Derived from Different Tissues
(A) In vivo osteogenic differentiation of BM (bone marrow, a), MU (muscle, b), PE (periosteum, c), and CB (cord blood, d) cells (representative results of one from at least three transplants). Sirius red (columns 1 and 3) stains bone (labeled b) intensely due its high collagen content. Polarized light (columns 2 and 4) shows the distribution of Sirius red-stained collagen fibers in bone. When CD146+ “MSC” strains were transplanted, using the same in vivo assay with hydroxyapatite/tricalcium phosphate (HA/TCP) as a scaffold (labeled s), osteogenic potential was restricted to BM-, PE-, and CB-derived cells. MU cells regularly failed to form any histologically identifiable bone. PE and CB “MSCs” formed bone, but never established a hematopoietic microenvironment. BM “MSCs” formed bone and established the hematopoietic microenvironment (asterisks in a, third panel), which is assessed by determining the presence of extravascular blood cells such as megakaryocytes and granulocytes, as seen under high-power microscopy (not shown). Scale bars represent 700 μm.
(B) Clonal strains of CB CD146+ “MSCs” transplanted using HA/TCP as a scaffold (representative results from one of at least three transplants) generated abundant bone and hyaline cartilage (labeled c). a, b, f–h: Safranin O (stains chondrocytes and cartilage orange due to their proteoglycan content) and Fast green (stains bone matrix deep green-blue); c: H&E, whereby unlike bone which stains pink with eosin, cartilage stains light blue; d: Alcian blue (stains cartilage matrix); e: anti-type II collagen (an essential cartilage component) immunohistochemistry. Scale bars represent 700 μm (a), 150 μm (b–e), 40 μm (f), 15 μm (g), and 21 μm (h).
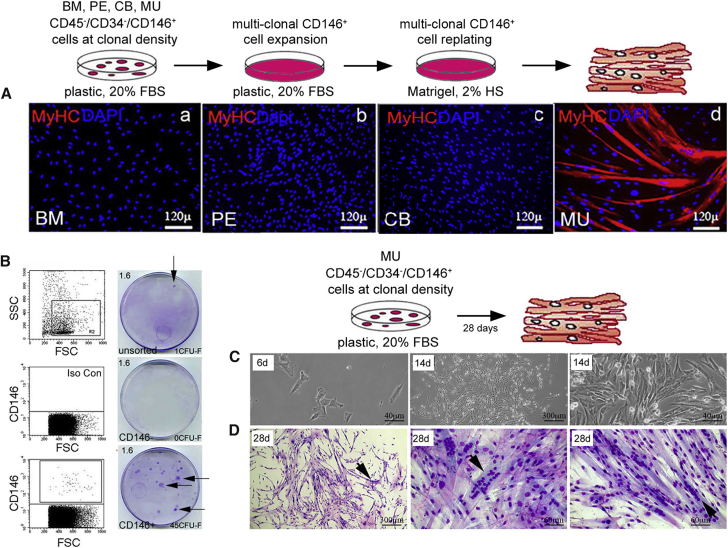
The myogenic capacity of the same cell strains was tested under stringent conditions (i.e., in the absence of exogenous myoblasts in vitro or endogenous myoblasts in vivo [Sherwood et al., 2004], of demethylating agents such as 5-azacytidine [Wakitani et al., 1995]). MU “MSCs” cultured without myoblasts in canonical myogenic conditions (2% horse serum on Matrigel) (Dellavalle et al., 2007) readily and efficiently generated myotubes in culture, whereas no myogenesis was observed under identical conditions with non-MU-derived stromal cells (Figure 4A). With these non-MU “MSCs,” myotubes could only be obtained in co-cultures with myoblasts (mouse C2C12 cells). Notably, the vast majority of nuclei incorporated in the newly formed myotubes were murine, indicating that human non-MU “MSCs” contribute only marginally to myogenesis in vitro even in permissive co-culture differentiation assays (data not shown). In cultures of MU CD146+ cells (Figure 4C) highly efficient myogenesis was observed, even in colonies established by single CD146+ cells (Figure 4B), and occurred spontaneously as the colonies became dense (13/32; 41%) as observed with bona fide myoblasts (Figure 4D).
Figure 4.
In Vitro Myogenic Assay of CD146+ “MSCs” Derived from Different Tissues
(A) Conventional in vitro differentiation assays were conducted using multiclonal “MSCs” from BM (bone marrow, a), PE (periosteum, b), CB (cord blood, c), and MU (muscle, d) cells, which were first expanded on plastic with 20% FBS, and then replated on Matrigel with 2% horse serum (representative results from one of at least three independent experiments). With MU CD146+ “MSCs,” extensive formation of myotubes expressing specific myogenic markers were observed. No myogenic differentiation was observed with any non-MU-derived cell strain. MyHC, myosin heavy chain. Scale bar represents 120 μm.
(B) Freshly isolated MU CD146+ cells were plated at clonal density (1.6 cells/cm2, indicated in upper left corner). At clonal density, MU CD146+ but not MU CD146− cells formed discrete fibroblastic colonies (arrows point to colonies, number of colonies formed indicated in lower right corners, Giemsa stain) (representative results from one of least three independent experiments).
(C) Cell morphology in single colonies generated by CD45−/CD34−/CD146+ muscle-derived clonogenic cells at 6 days (scale bar, 40 μm) and 14 days (scale bars, 300 μm and 40 μm) in culture, in 20% serum, and on plastic (results from one experiment).
(D) Three such colonies of MU cells, one at low power (scale bar, 300 μm) and two at high power (scale bar, 60 μm), at 28 days without passage. With time and without passage, colonies become dense and 13 of 32 (41%) underwent spontaneous differentiation. Multinucleated myotubes are obvious after Giemsa staining (arrows).
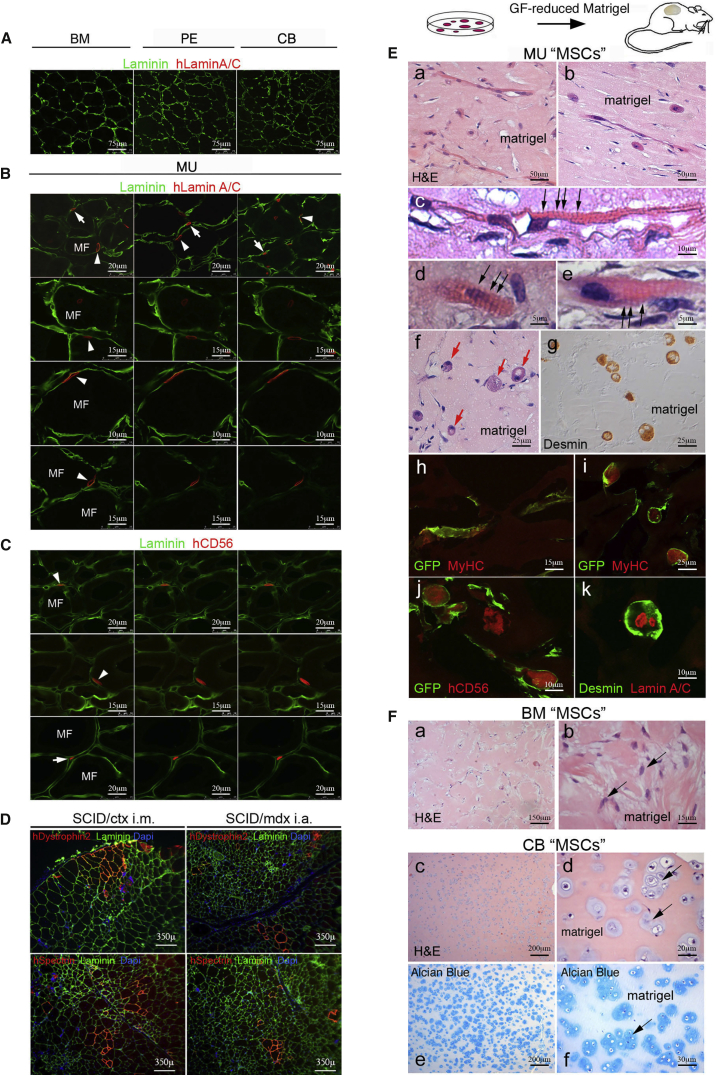
Local transplantation of CD146+ BM, PE, or CB cells into cardiotoxin (CTX)-injured tibialis anterior muscle of SCID/beige (Figure 5A) or SCID/mdx mice (data not shown) (Dellavalle et al., 2007) failed to contribute to regenerating muscle. In contrast, CD146+ MU cells revealed an efficient incorporation of donor nuclei (Figures 5B; Tables S5A and S5B), and expression of human muscle proteins in regenerating myofibers (Figure 5C [CD56], Figure 5D [Dystrophin 2, Spectrin], and Table S5C). Subsequently, enzyme-released cells of the harvested injected TA were used to perform secondary MU colony-forming efficiency (CFE) assays (Supplemental Experimental Procedures; Figure S3). Human cells were isolated based on hCD44, hCD90, and hCD146 expression and, after brief expansion (2 weeks), reanalyzed by fluorescence-activated cell sorting (FACS) for hCD44, hCD90, and hCD146 expression. The human cells were then isolated by MiniMacs (Miltenyi) and were replated in culture at clonal density. All colonies harvested at 2 weeks were uniformly positive for hCD146 and negative for hCD56 and hCD34, demonstrating that hCD146+ pericytes isolated from the injected muscle were the source of the secondary MU colony-forming unit-fibroblasts (CFU-Fs), indicative of self-renewal.
Figure 5.
Intramuscular and Ectopic In Vivo Transplantation of CD146+ “MSCs” Derived from Different Tissues
(A) Human BM (bone marrow), PE (periosteum), and CB (cord blood) derived cells in orthotopic transplantation into cardiotoxin (CTX)-injured muscle in SCID/beige mice after 4 weeks failed to contribute to generation of muscle cells or regenerating myofibers as would be detected by anti-human Lamin A/C (red) (laminin, green). Scale bar represents 75 μm.
(B) Human CD34–/CD56–/CD146+ muscle-derived cells were injected intramuscularly into the left tibialis anterior of 2-month-old female SCID/beige mice, injured 1 day earlier by an intramuscular injection of CTX. After 4 weeks transplanted human MU cells, identified by expression of human Lamin A/C (red), were distributed to the interstitium (arrows), but also below the basement membrane (green) on the surface of myofibers (MF) in a satellite cell-like position (arrowheads). Scale bars represent 10 μm, 15 μm, or 20 μm as indicated.
(C) Cells in a characteristic satellite cell-like position (arrowheads and arrow) express human CD56 (red). Scale bars represent 15 μm or 20 μm as indicated.
(D) Clusters of myofibers expressing human Dystrophin 2 and Spectrin were generated by MU cells by in vivo transplantation into SCID/beige/CTX-treated and SCID/mdx mice (DAPI: nuclear stain). Scale bar represents 350 μm.
(E) In a heterotopic transplantation assay, human CD34–/CD56–/CD146+ MU cells, transduced to express a cell-surface GFP label, were suspended in growth factor-reduced Matrigel (labeled matrigel in panels) without HUVECs and injected into the epifascial space of the back of SCID/beige mice, and harvested 3 weeks later. Extensive formation not only of myotubes (a, b, and h) expressing human-specific myogenic markers, Desmin (g and k), myosin heavy chain (MyHC, h and i), and CD56 (j), but even of striated myofibers (c–e, black arrows) and non-conventional muscle structures (f, red arrows, i–k) were observed (H&E, a–f). Scale bars represent 5 μm, 10 μm, 15 μm, 25 μm, or 50 μm as indicated.
(F) No myogenic differentiation was observed with BM (stained blue with H&E in a, b, arrows) or any non-MU cell strain (lack of myotubes and myofibers). In contrast, CB CD146+ “MSCs” spontaneously generated hyaline cartilage (stained light blue with H&E in c, d [arrows] and Alcian blue in e, f [arrows]) when transplanted in Matrigel as a carrier. No ingrowth of BVs into the Matrigel plug was associated with the extensive cartilage differentiation observed.
All data shown are representative results from one of at least three independent experiments. Scale bars represent 15 μm, 20 μm, 30 μm, 150 μm, or 200 μm as indicated.
The canonical in vivo muscle regeneration assay relies in large part on fusion between donor cells and host myoblasts, which implies recruitment of the former to a newly forming myotube where they will be reprogrammed by host myogenic factors. Even though CB, PE, and BM cells failed to differentiate under these conditions, a more stringent in vivo myogenesis assay requires the exclusion of such recruiting myoblasts. To this end, we suspended equal numbers of MU (transduced to express a cell-surface GFP label), BM, or CB cells with identical surface phenotype, grown in culture under identical basal conditions that did not induce differentiation, in growth factor-reduced Matrigel, and injected the resulting suspension into the epifascial space of the back of SCID/beige mice. Plugs harvested at 3 weeks demonstrated that MU cells had generated myotubes, myofibers, and a unique spheroidal syncytia expressing a differentiated muscle phenotype (Figure 5E, desmin, MyHC, CD56). The myosac phenotype may result from spontaneous contraction of the myotube poorly adhering to the substrate and obviously not constrained by tendons. BM cells, while remaining viable, did not generate bone, cartilage, or muscle. CB cells, which formed bone and cartilage upon transplantation with an osteoconductive carrier, generated hyaline cartilage but not bone in Matrigel (Figure 5F).
MU CD146+ Cells Are Perivascular, Committed Myogenic Progenitors
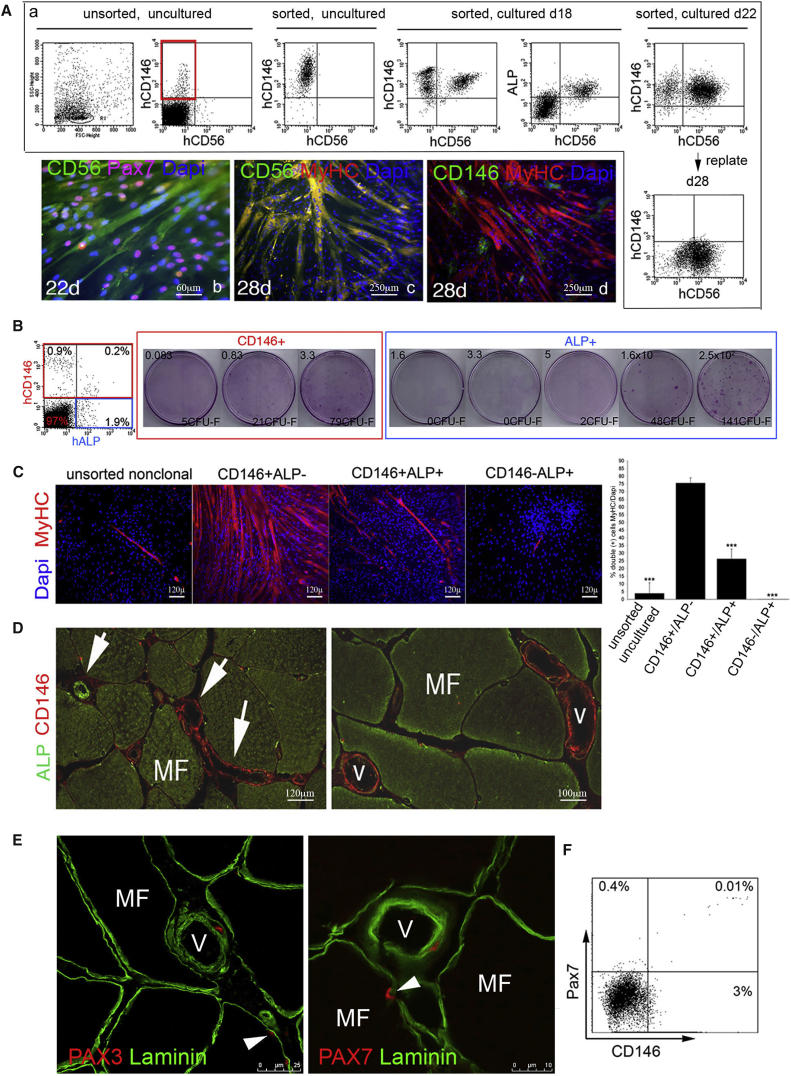
These data revealed a strong, spontaneous myogenic capacity specific for MU CD146+ cells, fully consistent with the high expression of myogenic regulatory factors and genes reflecting the differentiated function of muscle (Figure S1B). Cell sorting, immunolocalization, and CFE assays identified MU CD146+ cells as distinct from CD56+/CD146− satellite cells and CD34+/CD146+ endothelial cells (Figure S4 and Tables S6A–S6C). In cultures established from highly purified CD56−/CD146+ cells, CD146 expression was turned off over time while expression of CD56, PAX7, ALP (previously reported to be a pericyte marker in muscle; Dellavalle et al., 2007], MyoD, Myogenin were turned on (Figures 6A and S1C). Only a small subset of clonogenic, myogenic CD146+ cells co-expressed ALP (Figure 6B and Table S6D), and myogenic cells were highly enriched in the CD146+/ALP− fraction (Figure 6C). In situ, ALP and CD146 were expressed in a mutually exclusive fashion in precapillary arterioles and postcapillary venules, respectively, suggesting that the venous compartment represents the largest repository of perivascular CD146+ cells that can be assayed as myogenic progenitors (Figure 6D). Immunolocalization studies further revealed the presence of a relatively rare population of adventitial cells expressing muscle regulatory factor proteins (PAX3, PAX7) in postcapillary venules (Figure 6E). Freshly isolated MU CD146+ cells also co-expressed PAX7 by FACS (Figure 6F) and by RT-PCR (Figure S1B).
Figure 6.
Induction of CD56 Expression, Colony-Forming Efficiency and Myogenic Differentiation of FACS Sorted Muscle-Derived “MSCs,” and In Vivo Localization of ALP, CD146, and PAX 7
(A) Induction of the satellite cell marker, CD56, in cultures of CD34−/CD56−/CD146+ cells by FACS and fluorescent immunocytochemistry. Red box indicates the population of CD146+/CD56− cells used to establish the cultures. By FACS, co-expression of CD56 in a subset of CD146- and ALP-expressing cells is detectable after 18 days in culture, and the percentage of CD146+/CD56+ cells increases over time. Upon replating cultured CD146+/CD56+ cells at high density with 20% FBS, CD146 expression is turned off, whereas CD56+ expressing cells remain detectable. By fluorescent immunocytochemistry, subconfluent cultures express CD56 (green) and PAX7 (purple when colocalized with the blue nuclear stain, DAPI) (22 days in culture), and when replated at high density with 20% FBS, myotubes with numerous nuclei (DAPI staining) express myosin heavy chain (MyHC, red), co-localizing with CD56 (green) resulting in yellow. At a terminal stage of differentiation (26–28 days in culture), CD146 expression is barely detectable. Scale bars represent 60 μm or 250 μm as indicated.
(B) FACS of collagenase-released cell suspensions of muscle. Co-expression of CD146 and ALP, in human MU cell suspensions before culture. CFE assay of FACS-sorted CD146−/ALP+ cells and CD146+/ALP+ cell subsets. CD146+/ALP+ and CD146−/ALP+ cells were plated at different cell densities. Numbers (upper left of each panel) indicate the number of cells plated/cm2.
(C) When unsorted cell strains, sorted CD146+/ALP−, CD146+/ALP+, and CD146−/ALP+ total collagenase-released cells (bar graph is representative of one of at least three independent experiments, ∗∗∗p < 0.001 compared with CD146+/ALP− cells) were plated at clonal density and induced to myogenic differentiation (2% horse serum on Matrigel), high numbers of myotubes expressing skeletal muscle-specific myosin heavy chain (MyHC, red) are found in freshly sorted cultures enriched in MU CD146+/ALP− cells. Only rare myotubes are observed in cultures of CD146+/ALP+ cells. Scale bar represents 120 μm.
(D) Immunolocalization analysis of the distribution of CD146 and ALP expression in intact muscle revealed that while ALP cells (green, arrow, upper left) were predominantly expressed in precapillary arterioles, CD146 cells (red, arrows, center and lower right) were predominantly expressed in large venules (v). Scale bars represent 100 μm or 120 μm as indicated.
(E) A subset of vascular-wall-associated cells was found to express PAX3 (red, left panel) and PAX7 (red, right panel) in intact adult skeletal MU. Immunoreactivity was distributed to the surface of myofibers (MF) in a satellite cell-like position (arrowhead) underneath the basement member (laminin, green), and to BV walls (V). Scale bars represent 10 μm or 25 μm as indicated.
(F) Freshly isolated, uncultured CD146+ cells were found to co-express the satellite cell marker, PAX7.
All data shown are representative results from one of at least three independent experiments.
Generation of Perivascular Cells from Different Types of Stromal Cells In Vivo
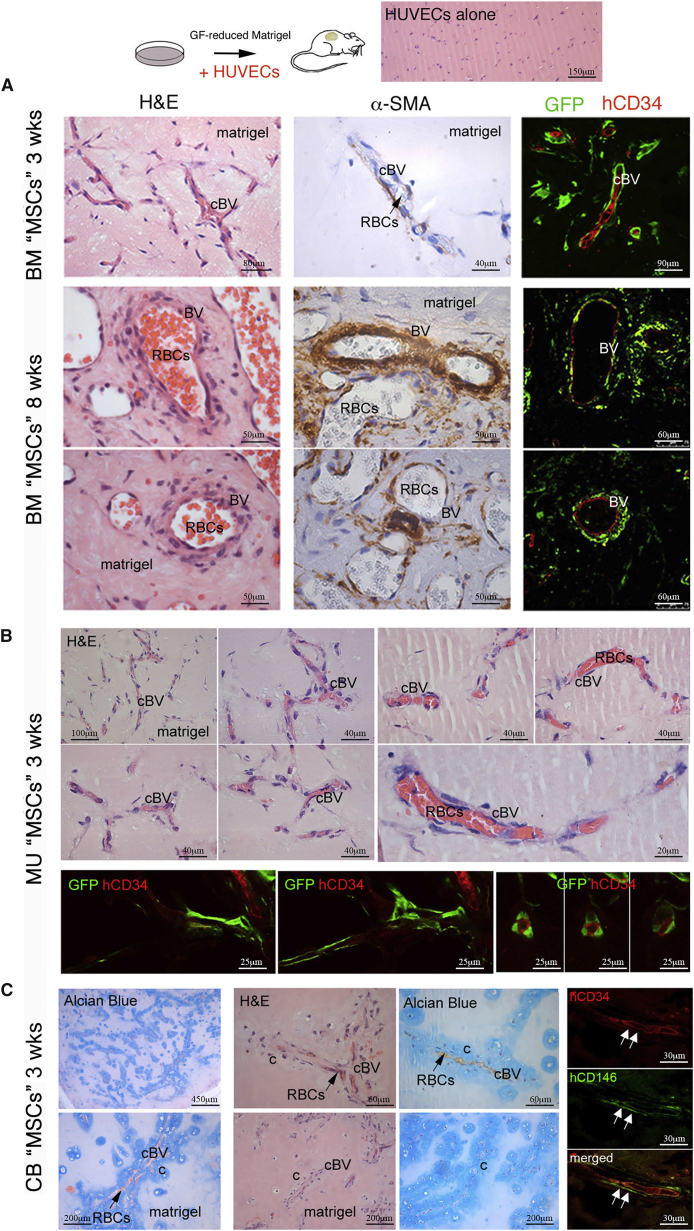
In heterotopic transplants made with osteoconductive carriers, BM-derived skeletal stem cells can guide and organize the formation of BVs, with which a subset of them ultimately associates (Sacchetti et al., 2007). To test the ability of BM- and non-BM-derived progenitors to guide and organize nascent BVs under simplified conditions that bar the formation of bone, we co-transplanted BM- and non-BM-derived progenitors in Matrigel, along with human umbilical vein endothelial cells (HUVECs). With cell-surface GFP-labeled BM-derived progenitors, this resulted in the formation of an extensive network of capillary-like BVs at 3 weeks. Their wall was composed of two cell layers: an inner hCD34+ endothelial layer (red), and an outer, discontinuous layer of mural cells (green) made by transplanted BM progenitors, expressing CD146 and α-smooth muscle actin (α-SMA) (Figure 7A). In transplants harvested at 8 weeks, extensive maturation of the BV wall was observed, as indicated by the formation of thick-walled artery- and vein-like BVs, with a multilayered GFP+, α-SMA+ smooth muscle coat. The BVs were functional and perfused by host blood (Figure 7A). Experiments performed with cell-surface GFP-labeled MU CD146+ cells also resulted in the formation of a capillary lattice, which was less extensive than with BM CD146+ cells, but similarly well organized (Figure 7B, quantitated in Figure S5). No obvious differentiation of muscle structures were observed in these transplants, at variance with transplants of MU CD146+ cells without HUVECs, suggesting that the contribution to BV formation and myogenesis could be alternative fates dictated by endothelial cells. With CB CD146+ cells, again the formation of functional BVs was observed. Strikingly, these BVs, lined by a continuous hCD34+ endothelial layer, were coated by a thin layer of subendothelial CD146+ mural cells, and an outer coat of differentiated chondrocytes (Figure 7C), making these BVs a unique kind of cartilage-armored BV, not known to occur in natural tissues. Knockdown of CD146 in cell-surface GFP-labeled BM progenitors (as previously described by Sacchetti et al., 2007) (Supplemental Experimental Procedures) partially interfered with BV organization, resulting in BVs of irregular size and shape, often devoid of a lumen, and devoid of a mural cell coat (Figure S6). Due to the ability of BM, MU, and CB cells to form perivascular cells, we examined the transcriptome data (including PE cells, although not used for this assay) for additional markers that are associated with pericytes (e.g., Armulik et al., 2011). A variety of pericyte markers were expressed by all four cell populations, but the pattern of expression varied from one cell type to another, consistent with their diverse developmental origins (Figure S7).
Figure 7.
In Vivo Transplantation of CD146+ “MSCs” Derived from Different Tissues in Conjunction with HUVECs
(A) Co-transplant of cell-surface GFP-labeled BM-derived progenitors in Matrigel (labeled as matrigel) along with HUVECs resulted in the formation of an extensive network of capillary-like blood vessels (cBV) at 3 weeks as demonstrated by H&E staining. By fluorescence microscopy and immunostaining, their wall was made by two cell layers: an inner endothelial layer as marked by the endothelial marker, hCD34+ (red), and an outer, discontinuous layer of mural cells made by transplanted, GFP-tagged BM progenitors. Transplants harvested at 8 weeks exhibited extensive maturation of the blood vessel (BV) wall as indicated by the formation of thick-walled artery- and vein-like BVs (H&E staining). These vessels were functional as indicated by perfusion with host blood, as shown by the presence of red blood cells (RBCs, black arrow). The vessels were coated with a multilayer of α-SMA+ cells demonstrated by immunohistochemistry (middle column illuminated by differential interference contrast microscopy to see the disc-shaped host RBCs in the vessels). By fluorescence microscopy and immunostaining, a multilayer of GFP+ cells surrounding hCD34+ endothelial cells (red) were observed. Scale bars represent 40 μm, 50 μm, 60 μm, 80 μm, or 90 μm as indicated.
(B) Similar experiments with cell-surface GFP-labeled MU CD146+ progenitors transplanted with HUVECs also resulted in the formation of a capillary lattice (cBV) at 3 weeks, which was less extensive than with BM cells, but similarly well organized and functional, as demonstrated by perfusion with host RBCs. As with BM cells, GFP-labeled MU cells were seen surrounding hCD34+ endothelial cells (red). Scale bars represent 20 μm, 25 μm, 40 μm, or 100 μm as indicated.
(C) Heterotopic co-transplantation of CB cells in Matrigel along with HUVECs resulted in the formation of an extensive network of capillary-like BVs (cBV) at 3 weeks, which were functional and perfused by host RBCs (black arrows). By fluorescent immunohistochemistry, their wall was made by an inner continuous hCD34+ endothelial layer (red, white arrows), coated by a subluminal layer of thin, hCD146+ mural cells (green, white arrows) and an outer coat of chondrocytes and cartilage matrix (labeled c as identified by H&E [light blue staining] and the cartilage stain, Alcian blue), making these BVs a unique kind of cartilage-armored BV. Scale bars represent 30 μm, 60 μm, 200 μm, or 450 μm as indicated.
Discussion
A widely accepted view in the literature holds that “MSCs” can be defined by rather loose and non-specific in vitro properties, and exhibit identical functional and differentiation properties regardless of their tissue source. This view, complemented by the notion that “MSCs” would coincide with ubiquitous pericytes, was recently reinforced by the claim that MU-derived pericytes would be both myogenic and skeletogenic, and exhibit an in situ surface phenotype similar to the one characteristic of BMSCs (i.e., of a skeletal stem cell origin) (Bianco, 2014). We have shown here that CD34−/CD45−/CD146+ “MSCs” isolated from different tissues have inherently distinct transcriptomic signatures and differentiation capacities. By gauging their native differentiation potential with a variety of stringent differentiation assays, we demonstrated that human BM CD146+ cells are inherently geared to generate bone and BM stroma that support hematopoiesis and include adipocytes, but are not myogenic, and are not spontaneously chondrogenic in vivo; MU “MSCs” with an identical cell-surface phenotype are not inherently skeletogenic, and are inherently myogenic; and CB “MSCs” are not myogenic, but are chondro-osteoprogenitors most likely due to their fetal origin (Bianco and Robey, 2015) and are the only kind of human “MSCs” ever shown to actually form cartilage consistently in open heterotopic transplants in vivo, independent of any ex vivo induction to cartilage differentiation as applied in the widely used pellet culture assay. CB cells are unlike other cells that are found in the umbilical cord, based on their cell-surface markers, their ability to differentiate into adipocytes and cartilage, and expression of associated markers, and they have a distinct expression pattern of HOX genes compared with other umbilical cord derivatives (Bosch et al., 2012, Liedtke et al., 2010, Liedtke et al., 2016). BM- and MU-derived clonogenic progenitors of mesoderm derivatives are associated with BV walls in situ and include cells in a position characteristic of mural cells/pericytes, but also include vascular-wall cells that do not necessarily exhibit anatomical features of pericytes proper, for example, adventitial cells in muscular veins. More strikingly, fetal chondro-osteoprogenitors that circulate in CB, by definition, are neither associated with BV walls nor represent pericytes. The origin of these cells remains to be elucidated. It is conceivable, however, that skeletal progenitors located at sites of active skeletal growth can accidentally spill over into the bloodstream. Nonetheless, independent of their origin, native differentiation potencies, gene-expression profiles, and in situ anatomical positioning, perinatal and postnatal mesoderm progenitors of different origins can dynamically associate with, and organize, nascent BVs as shown here using a simple in vivo assay.
Our data reveal that in lieu of a uniform, broadly multipotent, and equipotent class of ubiquitous “MSCs,” perinatal and postnatal assayable progenitors found in CB, skeletal tissues, and skeletal muscle comprise a varied system of tissue-specific progenitors. Each member of the system is committed to a specific differentiation range. CB mesenchymal progenitors are committed to cartilage and bone formation, reminiscent of the potential of prenatal skeletal progenitors to generate skeletal rudiments made of cartilage and bone. Postnatal BM skeletal progenitors are instead committed to bone and BM stroma formation, but not to cartilage, mirroring an adult structure of skeletal segments. Myogenic and skeletogenic potential are mutually exclusive, as dictated by a specific set of master transcriptional regulators.
Our data are consistent with recent studies showing that in vivo, transplants of BM cells were able to form bone and support marrow formation, while white adipose-derived stromal cells (WAT), cells from umbilical cord (UC), and skin fibroblasts (SF) were not (Kaltz et al., 2008, Reinisch et al., 2015). Importantly, the “MSCs” derived from cord blood (CB) used in our study must be clearly distinguished from fibroblastic cells derived from UC “MSCs” in the Reinisch study; UC cells fail to differentiate in vitro and in vivo (Kaltz et al., 2008, Liedtke et al., 2016, Reinisch et al., 2015), differ in their typical HOX expression pattern (Liedtke et al., 2010, Liedtke et al., 2016), and have a different molecular transcriptomic signature lacking relevant integrin-binding sialoprotein (IBSP) expression (Bosch et al., 2012). Differences in our study include culture in 20% fetal bovine serum (FBS) and use of a ceramic carrier that favors direct osteogenesis (no evidence of endochondral bone formation). In Reinisch et al. (2015), BM cells were grown in 10% human platelet lysate and transplanted with a Matrigel equivalent (In Vitro Vasculogenesis Assay Kit; Millipore), and bone formed through an endochondral process. Another study investigated expression of μRNAs by BM cells, WAT cells, and CB cells at a multiclonal level (Ragni et al., 2013). While the μRNA patterns between the cells types were similar, a small number of highly differentially expressed μRNAs were identified, which could affect the expression of many genes. Of note, in vitro osteogenic, adipogenic, and chondrogenic differentiation assays were performed in both of these studies, and significant differences were noted between the cell types. Nonetheless, the authors of both papers concluded that an in vitro mesengenic process was at play in all cell types. However, in vitro analyses of osteogenic differentiation are not predictive of in vivo transplantation (e.g., Phillips et al., 2014). For chondrogenesis, 3D cultures are needed to clearly see bona fide chondrocytes lying in lacuna, surrounded by a matrix that stains purple with toluidine blue (metachromasia) as shown by Reinisch et al. (2015). Close examination of this study suggests that there was little or no chondrogenic differentiation of WAT, UC, or SFs compared with BM cells. In Ragni et al. (2013), chondrogenesis was difficult to assess based on Alcian blue-stained 2D cultures and expression of a limited number of markers, which is insufficient to confirm chondrogenic differentiation.
Even if updated to equate “MSCs” with pericytes (Caplan, 2008, Crisan et al., 2008), the widespread concept of broadly and uniformly multipotent “MSCs” that are invariant in anatomical space and developmental time leaves the developmental origin of such cells unaddressed, and in fact collides with certain fundamental tenets of developmental biology, such as the early segregation of inherent osteogenic and myogenic potential into different lineages (Bianco and Robey, 2015). Our data now reveal a simple mechanism whereby assayable skeletogenic or myogenic progenitors can be established in different tissues. As previously postulated (Bianco et al., 2008, Bianco et al., 2013), we have now directly shown that committed progenitors of different origin and differentiation potential can physically associate with nascent BV walls in an experimental assay, and be recruited to a mural cell fate. In an earlier study, the most robust contribution of grafted mesoderm progenitor cells in quail chick chimeras was, in fact, to the adventitia of BVs of any caliber (Minasi et al., 2002). In development, somite-derived cells associate with, and are incorporated into, the wall of the dorsal aorta (Esner et al., 2006). Pericytes originate from the recruitment of non-endothelial, mesenchymal cells to vascular walls (Armulik et al., 2011, Hellstrom et al., 1999, Hirschi and D'Amore, 1996, Jain, 2003, Lindahl et al., 1997). Obviously, the same mechanism may operate in different tissues and at different developmental times. General mechanisms regulating mural cell recruitment have largely been elucidated, and provide the background against which to seek additional determinants operating in the retention of quiescent mesoderm progenitors in a vascular niche. CD146 itself may represent a player in this scenario, consistent with its nature as a cell-adhesion molecule (Shih, 1999), and with evidence provided here that its knockdown perturbs the establishment of properly structured BVs in vivo.
The direct interaction of committed progenitors with endothelial walls of nascent BV can be related to their arrested proliferation and differentiation, leading to retention, within specific tissues, of a compartment of progenitors with predefined commitment, and a potential for further growth defined by their replicative history prior to incorporation into BV walls. The long-known heterogeneity of clonogenic mesodermal progenitors (CFU-Fs) in proliferative potential ex vivo might find in this view a simple explanation.
Our data have additional implications of applied nature. Prospective isolation of putative “MSCs” from different tissues, based on the use of the minimal effective surface phenotype CD34−/CD45−/CD146+ (Sacchetti et al., 2007), leads to isolation of committed tissue-specific progenitors, not of broadly multipotent, equivalent cells. Hence, design of cell-therapy strategies for specific tissues must remain cognizant of the specificity of the isolatable progenitors. In fact, reliance on this specificity leads to highly effective isolation of tissue-specific progenitors. This is best illustrated by the case of skeletal muscle, where we have shown that CD146+ cells are far more enriched in spontaneously myogenic cells than previously identified populations of putative non-satellite cell classes (Dellavalle et al., 2007). Importantly, we have shown that these cells, which do express the satellite lineage marker, PAX7, and behave like satellite cells in in vitro and in vivo assays (Montarras et al., 2005), may in fact represent subsets of the same original population, recruited to distinct anatomical niches based on distinct, local cell-cell interactions, randomly directing individual progenitors to either the surface of developing fibers (satellite cells) or the surface of nascent BVs (myogenic pericytes, or else ectopic satellite cells). Consistent with this hypothesis, proximity of one anatomical locale to the other can amount, in fact, to less than one cell diameter. However, it is also possible that the pericytic MU CD146+ cells are more primitive than satellite cells based on the fact that they give rise to CD56+ satellite cells (Figure 5C). Also of note, MU CD146+ cells do not coincide with PICs (interstitial populations of PW1+/PAX7− cells; Mitchell et al., 2010), based on the fact that MU CD146+ cells are PAX7+, or FAPs (fibro/adipogenic progenitors) that cannot generate myofibers (Joe et al., 2010), while MU CD146+ clearly do.
In summary, we have provided a rigorous analysis of mesodermal progenitors isolated from perinatal and postnatal tissues that reveals their inherent differentiation potential. These data underscore the fact that a ubiquitous “MSC” with identical differentiation capacities does not exist, and thus the “MSC” terminology should be abandoned for the sake of clarity. Furthermore, we have demonstrated that irrespective of their inherent differences, tissue-specific mesodermal progenitors are capable of being recruited to a mural cell fate, providing a plausible mechanism by which pericytes are formed, and how they serve as a source of local stem/progenitor cells.
Experimental Procedures
Cell Isolation and Culture
Samples were obtained with informed consent according to institutionally approved protocols. BM-, PE-, and CB-derived cells, dermal fibroblasts, adipose-derived stromal cells, amniotic fluid cells, and HUVECs were grown as described in Supplemental Experimental Procedures.
For MU cells, normal muscle samples (1–30 × 102 mg) from 17 human adult patients (25–65 years old) were enzymatically dissociated as described in Supplemental Experimental Procedures. The resulting single-cell suspensions were used to obtain cultures of unfractionated cells, or for sorting of CD146+ cells, CD146+/CD56+ cells, CD146+/CD34+ cells, or CD146+/ALP+ cells. Unsorted and sorted cells were plated in basal medium (α minimum essential medium [αMEM; Invitrogen] with 20% FBS [Invitrogen], 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin). For non-clonal cultures, cells were seeded at 1.6 × 103–1.6 × 105 cells/cm2 in 75-cm2 flasks or in 100-mm dishes (Becton Dickinson). For multiclonal cultures (combining multiple colonies), cells were seeded into 100-mm dishes at 1.6 cells/cm2, and formation of discrete colonies was scored after 14 days. Non-clonal cultures were passaged when subconfluent; multiclonal cultures were passaged on day 14.
Cell Sorting and Flow Cytometry
Surface markers were assessed using a FACSCalibur flow cytometer with CellQuest software (BD Biosciences) as reported previously (Sacchetti et al., 2007) with mouse anti-human monoclonal antibodies (Table S7A) and isotype controls. CD146+, CD146+/CD56+, CD146+/CD34+ cell subsets, and CD146+/ALP+ subsets were separated using a FACS DIVAntageSE flow cytometer (BD Labware). CD146+, CD146+/CD56+, CD146+/CD34+, and CD146+/ALP+ fractions were separated using a MiniMACS magnetic column separation unit (Miltenyi Biotec) according to the manufacturer's instructions.
Gene-Expression Profiling and Data Analysis
Total RNA was isolated from multiclonal cultures of CD146+ cell populations from three independent cultures after 2 weeks of culture in basal medium (see above), and processed as described in Supplemental Experimental Procedures. Raw data of gene-expression profiling were submitted to the GEO repository (GEO: GSE69991).
Gene Set Enrichment Analysis
GSEA was performed using GSEA software (http://www.broadinstitute.org/gsea/index.jsp) (Subramanian et al., 2005) as described in Supplemental Experimental Procedures.
Colony-Forming Efficiency Assays
CFE assays were performed with different cell fractions obtained by cell sorting. CD146+ cells were seeded in basal medium (see above) at 1.6–3.3 cells/cm2, CD146+/CD34+ and CD146+/CD56− cells were seeded at 1.6–8.3 cells/cm2, and CD146+/ALP+ were seeded at 0.083–3.3 cells/cm2. CD146− (CD56+ or CD34+ or ALP+) unsorted cells were seeded at an equivalent or higher density (1.6–1.6 × 105 cells/cm2) and grown under identical conditions. After 14 days, CFE was determined as the mean number ± SD of colonies (>50 cells)/102–105 cells initially plated. ALP cytochemistry was done using naphthol-AS-phosphate as substrate and Fast Blue BB as coupler.
In Vitro Differentiation Assays
Spontaneous myogenic differentiation was assessed by plating cells onto Matrigel-coated dishes, with DMEM/2% horse serum, or on plastic with αMEM/20% FBS at clonal density. After 7 days, cultures were fixed and labeled for immunofluorescence with a monoclonal antibody against striated MyHC. Myogenic efficiency was estimated as the percentage of DAPI+ nuclei found within myosin-positive myotubes. Fluorescence images were obtained using an Eclipse TE2000 Inverted Microscope (Nikon). Data were compared by ANOVA.
In Vivo Transplantation Assays
All animal procedures were approved by the relevant institutional committees.
Heterotopic Bone Formation
Constructs of test cells and osteoconductive material (hydroxyapatite/tricalcium phosphate [HA/TCP; Zimmer]) were transplanted into the subcutaneous tissue of SCID/beige mice (CB17.Cg-PrkdcscidLystbg-J/Crl; Charles River), using an established assay (Krebsbach et al., 1997, Sacchetti et al., 2007).
Orthotopic Myogenesis
CTX model: MU- BM-, and CB-derived cell populations (1 × 106) were injected intramuscularly into the left tibialis anterior of 2-month-old female SCID/beige mice injured 1 day earlier by an intramuscular injection of cardiotoxin (CTX; Latoxan) (Dellavalle et al., 2007). Muscles were examined 4 weeks following transplantation. SCID/mdx model: MU- and BM-derived cell populations were injected via the femoral artery of 2-month-old female SCID/mdx dystrophic mice (C57BL/10ScSn-mdx/J; Jackson Laboratory) as described by Dellavalle et al. (2007). Two injections of 5 × 105 cells at a 15-day interval were performed, and animals were euthanized 15 days after the last injection (30 days in total). The injected tibialis anterior muscles were analyzed for myogenic markers by immunofluorescence, with uninjected contralateral muscles serving as controls.
Heterotopic Differentiation, Matrigel
1 × 106 cells from multiclonal cultures of MU, BM, and CB cells were suspended in 1 ml of Matrigel Growth Factor-Reduced (BD Biosciences Labware), either alone or mixed with an equal number of HUVECs (Cambrex). Aliquots (∼0.7 ml) were injected in the subcutaneous tissue of the back of SCID/beige mice, and transplants were harvested after 20 days. In some experiments, cells from multiclonal cultures of MU and BM cells were transduced with GFP-lentiviral vectors.
Lentiviral Vectors
Lentiviral vectors for GFP expression and CD146 silencing were produced and used as described previously (Piersanti et al., 2010, Sacchetti et al., 2007). Sorted CD146+ cells from BM and MU were transduced with GFP-lentiviral vectors at an MOI of 5 and cultured for 2 weeks in basal medium (see above) before use. CD146 silencing is described in Supplemental Experimental Procedures.
Immunohistochemistry Studies
Heterotopic and orthotopic transplants were processed as described in Supplemental Experimental Procedures. All experiments using mice were performed under institutionally approved protocols. All primary antibodies used for immunolocalization studies are listed in Table S7B.
RT-PCR
Conditions used in this study are described in Supplemental Experimental Procedures, and primers are listed in Table S7C.
Acknowledgments
Sadly, Prof. Paolo Bianco passed away while this manuscript was in revision. We dedicate this study to him, for all of his outstanding contributions throughout the years, and his complete dedication to bringing clarity to this area of research. This work was supported by Telethon (Grant GGP09227), MIUR, Fondazione Cenci Bolognetti, Ministry of Health of Italy, EU (PluriMes consortium, FP7-HEALTH-2013-INNOVATION-1—G.A. 602423), Sapienza University of Rome to P.B.; Fondazione Roma to P.B. and M.R.; Bilateral grant DFG-KO2119/8-1 to P.B. and G.K.; by Telethon (Grant TCP07004) to M.S.; and by DIR, NIDCR, of the IRP, NIH, DHHS to P.G.R.
Published: June 14, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.05.011.
Contributor Information
Pamela G. Robey, Email: probey@dir.nidcr.nih.gov.
Mara Riminucci, Email: mara.riminucci@uniroma1.it.
Supplemental Information
References
- Applebaum M., Kalcheim C. Mechanisms of myogenic specification and patterning. Results Probl. Cell Differ. 2015;56:77–98. doi: 10.1007/978-3-662-44608-9_4. [DOI] [PubMed] [Google Scholar]
- Armulik A., Genove G., Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Bianco P. Bone and the hematopoietic niche: a tale of two stem cells. Blood. 2011;117:5281–5288. doi: 10.1182/blood-2011-01-315069. [DOI] [PubMed] [Google Scholar]
- Bianco P. “Mesenchymal” stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:677–704. doi: 10.1146/annurev-cellbio-100913-013132. [DOI] [PubMed] [Google Scholar]
- Bianco P., Robey P.G. Skeletal stem cells. Development. 2015;142:1023–1027. doi: 10.1242/dev.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Robey P.G., Simmons P.J. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Cao X., Frenette P.S., Mao J.J., Robey P.G., Simmons P.J., Wang C.Y. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J., Houben A.P., Radke T.F., Stapelkamp D., Bunemann E., Balan P., Buchheiser A., Liedtke S., Kogler G. Distinct differentiation potential of “MSC” derived from cord blood and umbilical cord: are cord-derived cells true mesenchymal stromal cells? Stem Cells Dev. 2012;21:1977–1988. doi: 10.1089/scd.2011.0414. [DOI] [PubMed] [Google Scholar]
- Caplan A.I. Mesenchymal stem cells. J. Orthop. Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- Caplan A.I. All MSCs are pericytes? Cell Stem Cell. 2008;3:229–230. doi: 10.1016/j.stem.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Caplan A.I., Correa D. The MSC: an injury drugstore. Cell Stem Cell. 2011;9:11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan M., Yap S., Casteilla L., Chen C.W., Corselli M., Park T.S., Andriolo G., Sun B., Zheng B., Zhang L. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- da Silva Meirelles L., Chagastelles P.C., Nardi N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- Dellavalle A., Sampaolesi M., Tonlorenzi R., Tagliafico E., Sacchetti B., Perani L., Innocenzi A., Galvez B.G., Messina G., Morosetti R. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat. Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- Diaz-Flores L., Gutierrez R., Madrid J.F., Varela H., Valladares F., Acosta E., Martin-Vasallo P., Diaz-Flores L., Jr. Pericytes. Morphofunction, interactions and pathology in a quiescent and activated mesenchymal cell niche. Histol. Histopathol. 2009;24:909–969. doi: 10.14670/HH-24.909. [DOI] [PubMed] [Google Scholar]
- Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Esner M., Meilhac S.M., Relaix F., Nicolas J.F., Cossu G., Buckingham M.E. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133:737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- Friedenstein A.J., Latzinik N.W., Grosheva A.G., Gorskaya U.F. Marrow microenvironment transfer by heterotopic transplantation of freshly isolated and cultured cells in porous sponges. Exp. Hematol. 1982;10:217–227. [PubMed] [Google Scholar]
- Hellstrom M., Kalen M., Lindahl P., Abramsson A., Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- Hirao M., Tamai N., Tsumaki N., Yoshikawa H., Myoui A. Oxygen tension regulates chondrocyte differentiation and function during endochondral ossification. J. Biol. Chem. 2006;281:31079–31092. doi: 10.1074/jbc.M602296200. [DOI] [PubMed] [Google Scholar]
- Hirschi K.K., D'Amore P.A. Pericytes in the microvasculature. Cardiovasc. Res. 1996;32:687–698. [PubMed] [Google Scholar]
- Jain R.K. Molecular regulation of vessel maturation. Nat. Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- Joe A.W., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltz N., Funari A., Hippauf S., Delorme B., Noel D., Riminucci M., Jacobs V.R., Haupl T., Jorgensen C., Charbord P. In vivo osteoprogenitor potency of human stromal cells from different tissues does not correlate with expression of POU5F1 or its pseudogenes. Stem Cells. 2008;26:2419–2424. doi: 10.1634/stemcells.2008-0304. [DOI] [PubMed] [Google Scholar]
- Kluth S.M., Buchheiser A., Houben A.P., Geyh S., Krenz T., Radke T.F., Wiek C., Hanenberg H., Reinecke P., Wernet P. DLK-1 as a marker to distinguish unrestricted somatic stem cells and mesenchymal stromal cells in cord blood. Stem Cells Dev. 2010;19:1471–1483. doi: 10.1089/scd.2010.0070. [DOI] [PubMed] [Google Scholar]
- Kogler G., Sensken S., Airey J.A., Trapp T., Muschen M., Feldhahn N., Liedtke S., Sorg R.V., Fischer J., Rosenbaum C. A new human somatic stem cell from placental cord blood with intrinsic pluripotent differentiation potential. J. Exp. Med. 2004;200:123–135. doi: 10.1084/jem.20040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach P., Kuznetsov S.A., Satomura K., Emmons R.V., Rowe D.W., Robey P.G. Bone formation in vivo: comparison of osteogenesis by transplanted mouse and human marrow stromal fibroblasts. Transplantation. 1997;65:1059–1069. doi: 10.1097/00007890-199704270-00003. [DOI] [PubMed] [Google Scholar]
- Liedtke S., Buchheiser A., Bosch J., Bosse F., Kruse F., Zhao X., Santourlidis S., Kogler G. The HOX Code as a “biological fingerprint” to distinguish functionally distinct stem cell populations derived from cord blood. Stem Cell Res. 2010;5:40–50. doi: 10.1016/j.scr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Liedtke S., Sacchetti B., Laitinen A., Donsante S., Klockers R., Laitinen S., Riminucci M., Kogler G. Low oxygen tension reveals distinct HOX codes in human cord blood-derived stromal cells associated with specific endochondral ossification capacities in vitro and in vivo. J. Tissue Eng. Regen. Med. 2016 doi: 10.1002/term.2167. [DOI] [PubMed] [Google Scholar]
- Lindahl P., Johansson B.R., Leveen P., Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- Minasi M.G., Riminucci M., De Angelis L., Borello U., Berarducci B., Innocenzi A., Caprioli A., Sirabella D., Baiocchi M., De Maria R. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development. 2002;129:2773–2783. doi: 10.1242/dev.129.11.2773. [DOI] [PubMed] [Google Scholar]
- Mitchell K.J., Pannerec A., Cadot B., Parlakian A., Besson V., Gomes E.R., Marazzi G., Sassoon D.A. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat. Cell Biol. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Phillips M.D., Kuznetsov S.A., Cherman N., Park K., Chen K.G., McClendon B.N., Hamilton R.S., McKay R.D., Chenoweth J.G., Mallon B.S. Directed differentiation of human induced pluripotent stem cells toward bone and cartilage: in vitro versus in vivo assays. Stem Cells Transl. Med. 2014;3:867–878. doi: 10.5966/sctm.2013-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersanti S., Remoli C., Saggio I., Funari A., Michienzi S., Sacchetti B., Robey P.G., Riminucci M., Bianco P. Transfer, analysis, and reversion of the fibrous dysplasia cellular phenotype in human skeletal progenitors. J. Bone Miner. Res. 2010;25:1103–1116. doi: 10.1359/jbmr.091036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Ragni E., Montemurro T., Montelatici E., Lavazza C., Vigano M., Rebulla P., Giordano R., Lazzari L. Differential microRNA signature of human mesenchymal stem cells from different sources reveals an “environmental-niche memory” for bone marrow stem cells. Exp. Cell Res. 2013;319:1562–1574. doi: 10.1016/j.yexcr.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Reinisch A., Etchart N., Thomas D., Hofmann N.A., Fruehwirth M., Sinha S., Chan C.K., Senarath-Yapa K., Seo E.Y., Wearda T. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood. 2015;125:249–260. doi: 10.1182/blood-2014-04-572255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Sherwood R.I., Christensen J.L., Conboy I.M., Conboy M.J., Rando T.A., Weissman I.L., Wagers A.J. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–554. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Shih I.M. The role of CD146 (Mel-CAM) in biology and pathology. J. Pathol. 1999;189:4–11. doi: 10.1002/(SICI)1096-9896(199909)189:1<4::AID-PATH332>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakitani S., Saito T., Caplan A.I. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve. 1995;18:1417–1426. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.