Summary
Human pluripotent stem cells (hPSCs) offer a renewable source of cells that can be expanded indefinitely and differentiated into virtually any type of cell in the human body, including neurons. This opens up unprecedented possibilities to study neuronal cell and developmental biology and cellular pathology of the nervous system, provides a platform for the screening of chemical libraries that affect these processes, and offers a potential source of transplantable cells for regenerative approaches to neurological disease. However, defining protocols that permit a large number and high yield of neurons has proved difficult. We present differentiation protocols for the generation of distinct subtypes of neurons in a highly reproducible manner, with minimal experiment-to-experiment variation. These neurons form synapses with neighboring cells, exhibit spontaneous electrical activity, and respond appropriately to depolarization. hPSC-derived neurons exhibit a high degree of maturation and survive in culture for up to 4–5 months, even without astrocyte feeder layers.
Highlights
-
•
Efficient, large-scale differentiation of hPSC to distinct neuronal subtypes
-
•
Differentiated neurons can be maintained in spinner flasks for extended time periods
-
•
Derived neurons are functionally mature and form an extensive synaptic network
-
•
Attractive platform for in vitro modeling of different forms of neurological disease
Ruben and colleagues present new protocols for large-scale production of cortical and motor neurons from human pluripotent stem cells. The obtained neurons are functional, survive an extended period of time in culture without the use of feeder layers, and constitute a promising platform for in vitro modeling of neurological diseases.
Introduction
With the seminal discovery of human pluripotent stem cells (hPSCs) (Thomson et al., 1998, Takahashi et al., 2007), human cells that would be difficult or impossible to obtain can be produced using in vitro cell-culture techniques. This in turn has raised hopes that hPSCs can be used to study and treat different forms of disease, including neurological and neuropsychiatric disorders (Dolmetsch and Geschwind, 2011, Fox et al., 2014, Han et al., 2011, Imaizumi and Okano, 2014, Kanning et al., 2010, Liu and Zhang, 2010, Mariani et al., 2015). However, a key step in the utilization of hPSCs for these purposes is the ability to obtain cell types of interest. This has often proved to be challenging for several reasons including neural diversity, culture-to-culture and line-to-line variability, and limitations on large-scale cell production.
Several methods have been described to obtain neurons of specific subtypes through differentiation of hPSCs, either via formation of three-dimensional (3D) embryoid bodies (EBs) or using monolayers as starting material (Amoroso et al., 2013, Boissart et al., 2013, Boulting et al., 2011, Eiraku and Sasai, 2012, Eiraku et al., 2008, Espuny-Camacho et al., 2013, Hu and Zhang, 2009, Kim et al., 2014, Li et al., 2009, Qu et al., 2014, Shi et al., 2012, Zeng et al., 2010). An alternative approach is transcriptional programming, whereby the forced overexpression of a cocktail of transcription factors instructs PSCs, fibroblasts, or other cell populations to adopt a specific neuronal fate (Hester et al., 2011, Vierbuchen et al., 2010). These methods have provided important insights into human neurogenesis and the pathogenesis of neurodevelopmental disorders, but they have limitations. For instance, EB-based protocols generally have comparatively low efficiencies (10%–40%) and require a relatively long time in culture to generate functional motor neurons. In addition, the neurons generated often require cellular feeder layers to survive for longer times in culture (Hu and Zhang, 2009, Boulting et al., 2011, Amoroso et al., 2013). Moreover, EB methods typically result in the formation of spheres of cells varying in size and shape, leading to differences in the kinetics and efficiency of differentiation within individual plates and from experiment to experiment. Monolayer-based protocols for the generation of both cortical and motor neurons have also been published, with recent work describing improved efficiencies (Qu et al., 2014). However, a disadvantage of this adherent monolayer-based protocol is that the neurons need to be passaged, and successful long-term culture after replating has not been described. Another common theme in the field has been the problem of obtaining mature cells from hPSCs. It has been shown that maintaining differentiated cells in culture can be challenging, thereby precluding experiments studying aspects of cellular functions that take longer times to manifest (Bellin et al., 2012, Grskovic et al., 2011).
Recently, a 3D culture system that yields brain tissue from hPSCs in the form of neural organoids has been described (Bershteyn and Kriegstein, 2013, Lancaster et al., 2013, Sasai, 2013). These organoids produce neurons organized in a manner reminiscent to what is seen in distinct anatomical structures within the mammalian CNS. At least some of the neurons in the organoids are functional, and this method has thereby offered a promising approach to study neurodevelopmental mechanisms and disorders. However, at this point, formation of neural organoids is not a process that is fully controlled. Another promising recent report based on a scaffold-free plate-based 3D method used to generate spheroids showed the possibility of yielding functional neurons with properties of deep and superficial cortical neurons (Pasca et al., 2015). However, this method may be difficult to implement for large-scale production of neurons and also generates cellular structures that are large enough to be potentially subject to necrosis in the core regions of the spheroids.
Here, we describe a method for large-scale production of neurons from multiple lines of human embryonic stem cells (hESCs) and human induced PSCs. We show that our method, based on the differentiation of 3D hPSC spheres maintained in suspension in spinner flasks (hereafter referred to as spin cultures), gives a higher purity and larger absolute number of cells, and has the potential to make functional neurons that can be maintained in culture for extended periods of time. Importantly, we have applied this method to the production of both cortical neurons of multiple types and spinal cord motor neurons. For both neuronal subtypes, we were able to document not only expression of appropriate marker genes but also several characteristics of mature neurons. The obtained neurons respond robustly to depolarization, form synapses as determined by punctate staining with antibodies against established synaptic proteins, and exhibit spontaneous neural activity assessed by electrophysiology and fluctuations in intracellular Ca2+ levels. Neurons generated from spheres also exhibited increased survival after dissociation compared with EB-derived neurons, and could be kept in culture without glial feeder layers for up to 5 months without compromising their functional properties. Taken together, these data illustrate that this protocol robustly generates large numbers of functional neurons that can reach a high degree of maturation, and should offer an improved cellular source of human neurons for disease modeling and drug screening.
Results
Spin-Culture hPSCs Form Spheres that Adopt a Cortical Identity after Dual SMAD Inhibition
It has previously been shown that hPSCs can be cultured in 3D spheres, consisting of hundreds of individual hPSCs, in a suspension-based system that can be readily scaled to permit the production of hundreds of millions of cells per culture (Ismadi et al., 2014, Otsuji et al., 2014, Pagliuca et al., 2014). Moreover, recent work has shown that hPSCs grown under such conditions can be differentiated into pancreatic β cells that were more mature than those produced in standard two-dimensional cultures (Pagliuca et al., 2014).
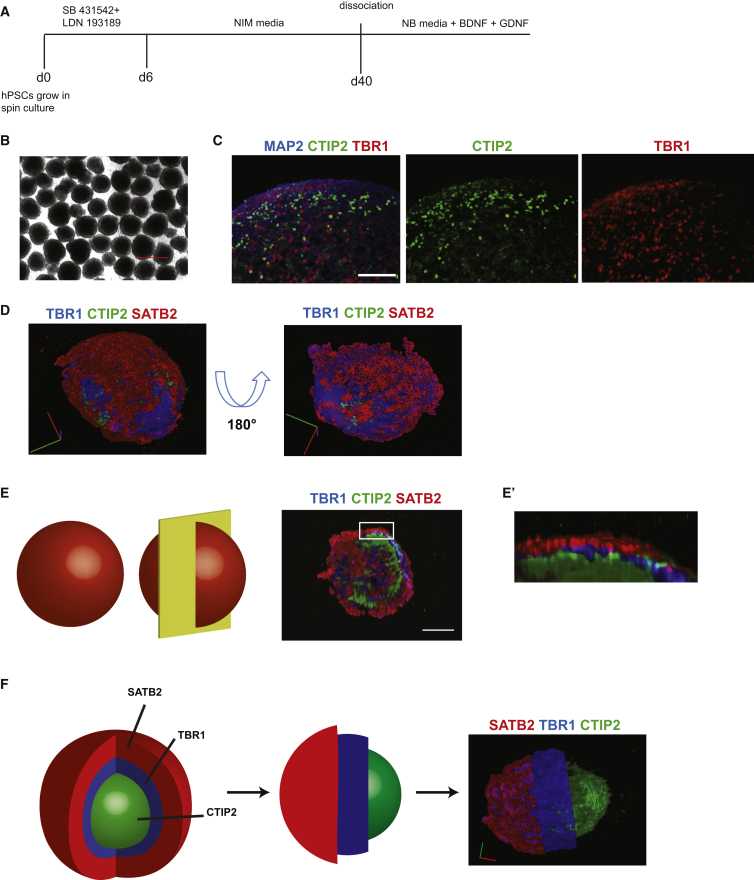
To explore the potential for generating cortical neurons using suspension-based cultures, we initially employed a differentiation protocol based on inhibition of the activin/transforming growth factor β (TGF-β) and bone morphogenetic protein (BMP) signaling pathways (Chambers et al., 2009) (illustrated schematically in Figure 1A). Prior to differentiation, the size and general appearance of hPSC spheres was quite homogeneous, and virtually all cells expressed the pluripotency markers OCT4 and NANOG (Figures S1A and S1B). During differentiation, spheres remained homogeneous in size (Figure 1B), and qRT-PCR analysis of RNA extracted from cells collected at different time points showed a rapid loss of expression of the pluripotency genes OCT4 and NANOG, and gradual expression of cortical progenitor, and then neuronal genes (Figure S2). The neural progenitor genes OTX1 and OTX2, and FOXG1, an established marker of cortical progenitor cells, reached maximum levels of expression around day 11 of differentiation, while the temporal expression profile of genes specific for mature cortical layers seemed to resemble the sequence seen during embryonic development, with TBR1 (a specific marker of neurons in cortical layer VI) appearing first, CTIP2 (layer V) thereafter, and SATB2 and CUX1 (layers II, III, and IV) at later time points (Figure S2).
Figure 1.
3D Neuronal Spheres from hPSC Lines Adopt a Predominantly Cortical Identity by Default
(A) Schematic illustration of the differentiation strategy used to obtain cortical neurons from 3D spheres. d, day; BDNF, brain-derived neurotrophic factor; GDNF, glial cell-derived neurotrophic factor.
(B) Representative bright-field image of HUES8-derived cortical spheres at day 15 of differentiation.
(C) Representative images of immunohistochemistry on sections of BJSiPS-derived cortical spheres at day 50 of differentiation, stained with antibodies specific for MAP2 (blue), TBR1 (red), and CTIP2 (green). Scale bar represents 200 μm.
(D–F) Representative results from a BJSiPS-derived cortical sphere cleared by SeeDB at day 50 of differentiation, stained with antibodies specific for TBR1 (blue), CTIP2 (green), and SATB2 (red), and imaged by LSFM. Photos in (D) show 3D images of the same sphere from two different angles. Scale bars in red and green represent 200 μm. Schematic images in (E) illustrate the virtual section of the sphere. The low-magnification 3D image in (E) shows the distribution of neurons inside the sphere expressing markers specific for distinct cortical subtypes. Scale bar represents 200 μm. (E′) is a higher-magnification image of the boxed area in (E). Schematic illustrations in (F) indicate how the sphere was virtually processed to generate a partially sectioned sphere.
Scale bars in green and red represent 100 μm. See also Figures S1 and S2; Movies S1A–S1C.
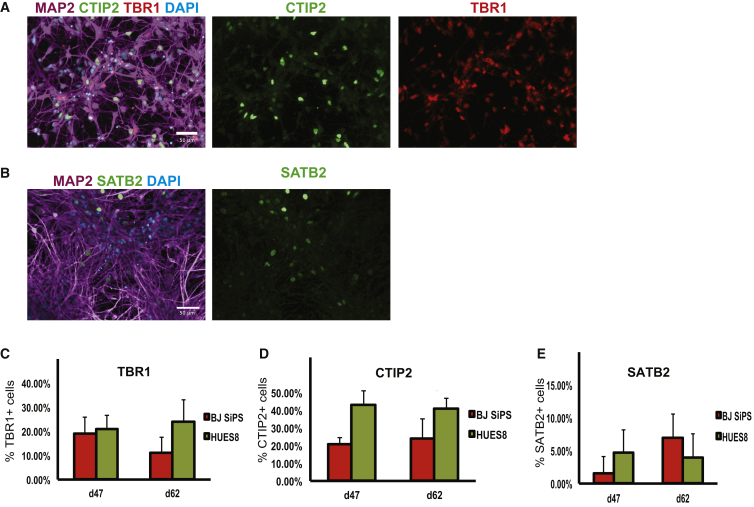
At day 50 of differentiation, spheres were collected and processed for immunocytochemistry using antibodies for cortical and synaptic markers. The results showed that spheres express the pan-neuronal marker MAP2, cortical neuron markers TBR1, CTIP2, and SATB2, and the synaptic marker SYNAPSIN1 (Figures 1C, S4A, and S4B). Immunocytochemistry performed on sections of spheres indicated that neurons positive for markers of the different cortical layers were not arranged randomly, but rather appeared in groups expressing markers of particular cortical layers. To further substantiate this notion, we performed 3D light-sheet fluorescence microscopy (LSFM) after clearing the spheres using the SeeDB method (Ke et al., 2013), allowing us to obtain a 3D image of stained spheres. The data confirmed that neurons expressing markers for similar cortical layer identities were organized in clusters rather than being mixed in a random, stochastic manner (Figures 1D–1F and S1C; Movies S1A–S1C). To obtain results with better cellular resolution to more readily quantify the efficiency and kinetics of neuronal differentiation, we dissociated, plated, and cultured spheres differentiated for 40 days for an additional 7 or 22 days. Immunocytochemistry revealed the presence of neurons expressing the cortical markers TBR1, CTIP2, and SATB2 (Figures 2A and 2B). Unbiased quantification revealed that cultures with up to 70% of all cells expressing one of these three markers could be obtained (Figures 2C–2E; Table S1). The remaining cells were positive for the neural progenitor markers OTX1 and OTX2 and the proliferation marker Ki67, consistent with the notion that these cells are residual cortical progenitors that have not yet differentiated to post-mitotic cortical neurons. Furthermore, we found that the number of Ki67-positive cells declined with increasing time in culture, suggesting that the progenitors would ultimately be depleted from the culture (Figure S3). We did not detect any cells stained using an antibody raised against the glial marker GFAP, arguing against the presence of any astroglia in these cultures. Importantly, the spin-culture protocol facilitates the generation of large numbers of neurons. From one 300-ml spinner flask we routinely obtained 3 × 109 neurons. Importantly, presumably because the continuous agitation provided optimal oxygenation and nutrient penetration, this spin-based method allows cells in spheres to thrive for months without any signs of having necrotic cores.
Figure 2.
Neurons from Dissociated Cortical Spheres Maintain the Differentiated Phenotype in Culture
(A and B) Representative images of immunocytochemistry of neurons dissociated at day 40 from BJSiPS-derived cortical spheres, plated and cultured for 7 (A) and 22 days (B) after dissociation. Cells were stained using antibodies specific for MAP2 (purple), CTIP2 (green), and TBR1 (red) in (A) and antibodies specific for MAP2 (purple) and SATB2 (green) in (B). Nuclei were counterstained with DAPI. Scale bars represent 50 μm.
(C–E) Quantification of the relative proportion of cortical subtypes obtained from experiments in different hPSC lines. Cells were dissociated and plated at day 40, and processed for immunocytochemistry with the indicated antibodies 7 and 22 days after dissociation. Error bars indicate SD from three independent experiments. See also Figure S3; Table S1.
These results show that hPSC spheres in spinner flasks differentiate efficiently to neuronal subtypes with neurons acquiring a predominantly cortical identity in the absence of patterning factors. Cortical cells expressing markers of different layers were readily identified and clustered together within the 3D spheres, with CTIP2-expressing neurons predominantly residing centrally, SATB2-expressing neurons on the outside of the spheres, and TBR1-positive neurons sandwiched in between the other two populations. When dissociated and cultured as a monolayer, the neurons maintained expression of cortical markers.
Dissociated Cortical Neurons Form Spontaneously Active Neuronal Networks and Exhibit Stimulation-Induced Activity
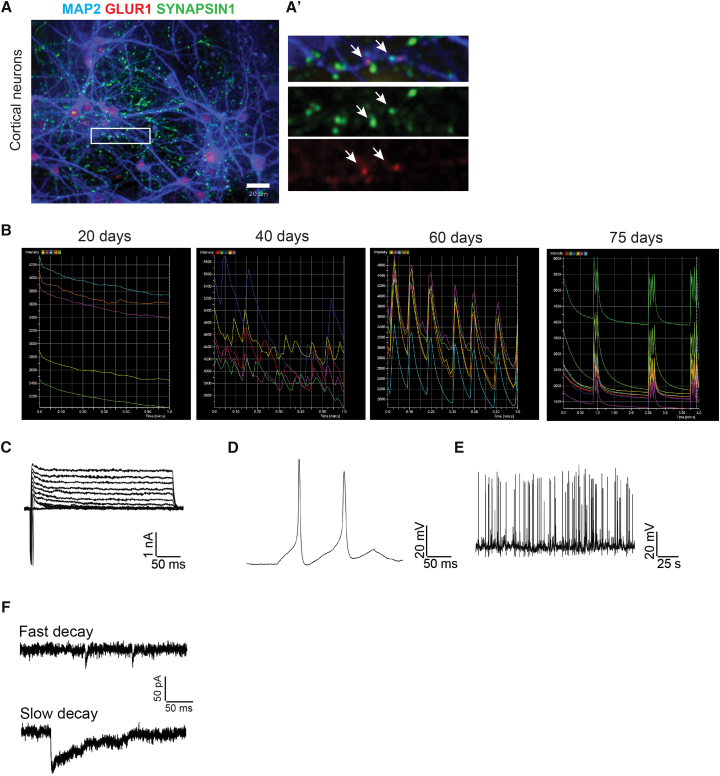
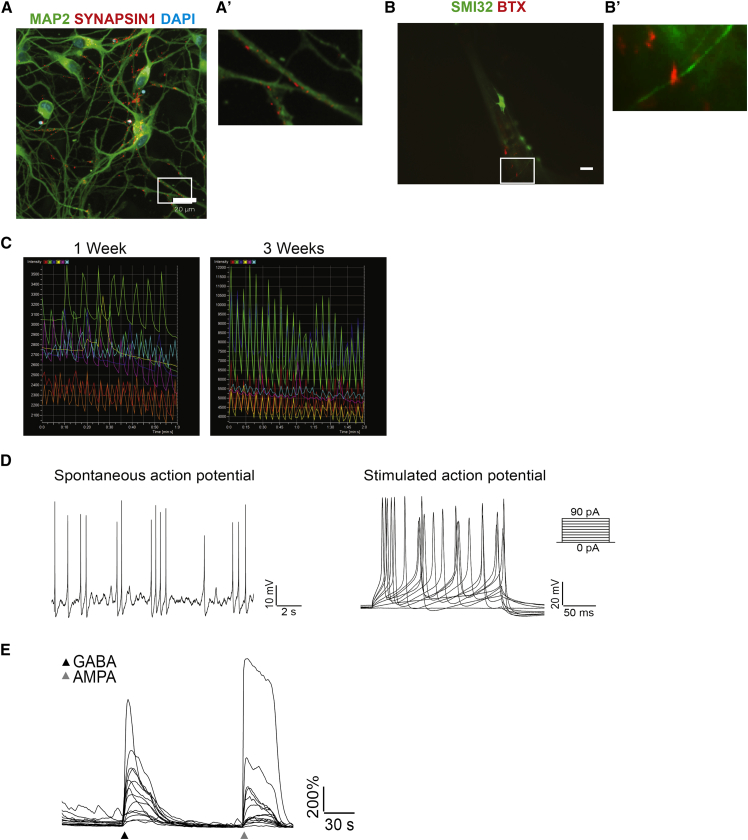
We thereafter sought to further characterize dissociated post-mitotic neurons. We analyzed these neurons for synaptic markers using antibodies specific for MAP2, the pre-synaptic marker SYNAPSIN1, and the post-synaptic marker GLUR1, specific for glutamatergic synapses. The results showed the presence of distinct SYNAPSIN1- and GLUR1-immunoreactive puncta distributed along the MAP2-positive dendrites. As we and others have observed in monolayer cultures of hPSC-derived neurons, SYNAPSIN1 and GLUR1 immunoreactivity were not always completely overlapping, but immunoreactive puncta were frequently found in close apposition, suggesting the formation of synaptic connections (Figure 3A).
Figure 3.
Dissociated Cortical Neurons Are Spontaneously Active
(A) Representative immunocytochemistry of dissociated neurons from HUES8-derived cortical spheres stained with antibodies specific for MAP2 (blue), SYNAPSIN1 (green), and GLUR1 (red). Images in (A′) constitute a higher magnification of the boxed area in (A). Arrows in A′ indicate putative synapses.
(B) Representative results from experiments in which BJSiPS-derived cortical spheres were dissociated at day 40. Three days after dissociation, the plated neurons were transduced with AAV Syn:GCaMP6s, and time-lapse microscopy was employed to record fluctuations in fluorescence over time (20, 40, 60, and 75 days after dissociation).
(C–F) Electrophysiological recordings of HUES8-derived cortical neurons 6 weeks after dissociation from spheres. (C) Representative traces showing voltage-gated sodium and potassium currents. (D) Cortical neurons fired action potentials upon current injection (30 pA). (E) Representative traces showing spontaneous firing of action potentials. (F) Sample traces showing spontaneous excitatory post-synaptic currents (fast decay) and inhibitory post-synaptic currents (slow decay) with a holding potential of −70 mV.
To seek evidence for functional synapses more directly, we asked whether hPSC-derived cortical neurons exhibit spontaneous electrical activity. We first monitored intracellular Ca2+ dynamics as a surrogate for electrical activity by performing time-lapse microscopy on cultures of dissociated neurons transduced with an adeno-associated virus expressing the genetically encoded Ca2+ sensor GCaMP6s under the control of the human synapsin promoter (AAV Syn:GCaMP6s) (Chen et al., 2013). Cells were transduced 3 days after dissociation and imaged every 10 days. Early after dissociation, neurons exhibited low levels of activity which increased over time, and all cultures of cortical neurons exhibited synchronized Ca2+ waves 60 days after dissociation (Figure 3B; Movies S2 and S3). To further examine the electrophysiological characteristics of neurons from dissociated cortical spheres, we conducted patch-clamp recordings. After 6 weeks in culture, cortical neurons showed typical voltage-gated Na+ and K+ currents (Figure 3C) and were able to fire action potentials upon current injection (Figure 3D). Spontaneous firing of action potentials and post-synaptic currents (Figures 3E and 3F) was also observed in most cells recorded (4 of 5), showing that cortical neurons obtained from dissociated spheres are able to mature into functional, electrophysiologically active cells.
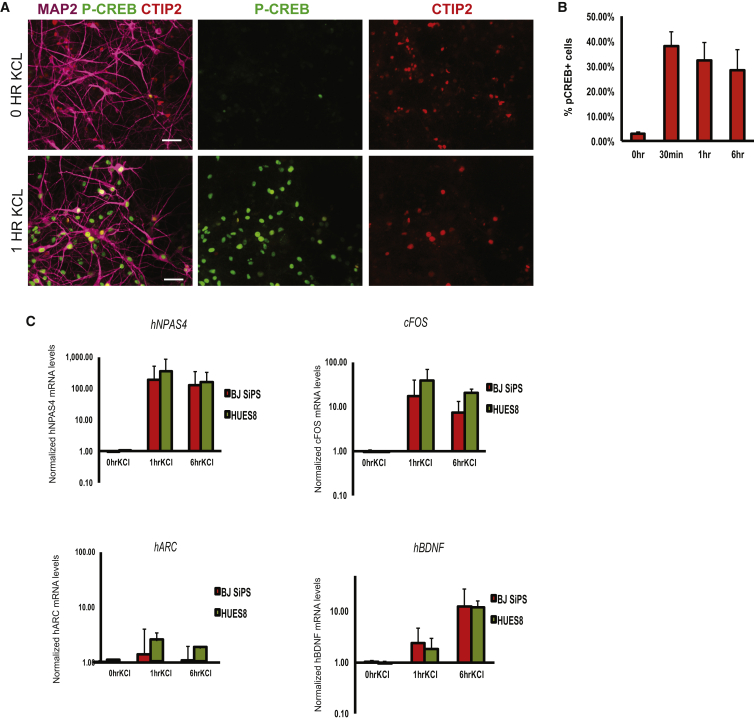
It is well known that synaptic activation through depolarization induces a transcriptional response in neurons (Ebert and Greenberg, 2013, Flavell et al., 2008, Hong et al., 2008, Lin et al., 2008). A key event in many types of neurons, including cortical neurons, is the phosphorylation of the transcription factor CREB. To examine whether neurons from dissociated cortical spheres respond to depolarization in this way, we treated them with high levels of extracellular KCl. Immunostaining with a phospho-CREB-specific antibody revealed robust phosphorylation of CREB in these cultures (Figures 4A and 4B; Table S2). We then performed transcriptional analyses of KCl-stimulated cortical neurons and found substantially increased transcription of known activity-regulated genes, including the early-induced genes NPAS4, cFOS, and ARC by 1 hr after stimulation and the late-induced gene BDNF after 6 hr (Figure 4C). These results are similar to those observed using freshly isolated rodent cortical neurons (Flavell et al., 2008, Hong et al., 2008, Lin et al., 2008). Similar results were also obtained from KCl stimulation of whole intact spheres after 20 days of differentiation (Figure S4).
Figure 4.
Dissociated Cortical Neurons Respond to Depolarization
(A) Representative images of immunocytochemistry of HUES8-derived cortical neurons 20 days after dissociation from spheres, before and after stimulation by KCl. Cells were stained using antibodies specific for MAP2 (purple), CTIP2 (red), and phosphorylated forms of CREB (green). Scale bars represent 50 μm.
(B) Quantification of the relative proportion of neurons staining positive in immunocytochemistry experiments using an antibody specific for phospho-CREB. Error bars indicate SD from three independent experiments.
(C) Quantification of expression of NPAS4, cFOS, ARC, and BDNF by qRT-PCR, following stimulation by KCl. Error bars indicate SD from three independent experiments.
See also Table S2.
Taken together, these data demonstrate that cortical neurons obtained from hPSCs cultured and differentiated as spheres in spinner flasks are electrophysiologically active and form neural networks in culture. Moreover, the neurons display voltage-gated Na+ and K+ currents, intracellular Ca2+ dynamics, and activity-induced gene expression characteristic of cortical neurons, further underlining their maturation into functional cortical neurons.
Motor Neuron Production from Spin-Culture hPSCs
In the next set of experiments, we explored whether the spheres would respond to exogenously supplied factors by differentiating to other neuronal subpopulations. To accomplish this, we set out to modify standard motor neuron differentiation protocols (schematic in Figure 5A). By supplying retinoic acid and sonic hedgehog (Wichterle et al., 2002), spheres rapidly differentiating into motor neurons were obtained. qRT-PCR analyses revealed a transient peak in expression of the neuronal progenitor gene SOX1, followed by expression of the motor neuron-specific genes ISL1 and HB9 (Figure S5). At later time points, increasing expression of the biosynthetic enzyme choline acetyltransferase (ChAT), necessary for the synthesis of acetylcholine, and the mature pan-neuronal marker MAP2 was detected (Figure S5). When analyzed by immunocytochemistry at day 20 of differentiation, cells in spheres expressed ISL1, MAP2, ChAT, and SYNAPSIN1 (Figures 5B, S6A, and S6B).
Figure 5.
3D Neuronal Spheres from Human PSC Lines Differentiate into Motor Neurons in the Presence of Patterning Factors
(A) Schematic illustration of differentiation strategy used to obtain motor neurons from 3D spheres. CNTF, ciliary neurotrophic factor.
(B) Representative images of immunohistochemistry on sections of 1016A-derived spheres differentiated towards the motor neuron fate and stained with antibodies specific for MAP2 (red) and ISL1 (green) at day 20 of differentiation. Nuclei were counterstained with DAPI (blue). Scale bar represents 50 μm.
(C) Representative images of immunocytochemistry of 1016A-derived motor neurons from dissociated spheres plated and cultured for 7 days after dissociation. Cells were stained using antibodies specific for MAP2 (purple) and ISL1 (green). Nuclei were counterstained with DAPI (blue). Scale bar represents 50 μm.
(D) Quantification of the relative proportion of the different motor neurons subtypes obtained from experiments in different hPSC lines. Cells were dissociated and plated at day 15, cultured for 1 week, and processed for immunocytochemistry with an antibody specific for ISL1. Error bars indicate SD from three independent experiments.
(E) Representative immunocytochemistry of 1016A-derived neurons from spheres stained with antibodies specific for MAP2 (purple), ISL1 (green), and ChAT (red). Nuclei were counterstained with DAPI. Scale bar represents 50 μm.
(F) Quantification of the relative proportion of ChAT-positive cells obtained from experiments in two different hPSC lines. Cells were dissociated and plated at day 15, cultured for 1 week, and processed for immunocytochemistry with an antibody specific for ChAT. Error bars indicate SD from three independent experiments.
After 15 days of differentiation, motor neuron spheres were dissociated, plated, and cultured under feeder-free conditions for an additional week. Immunocytochemistry revealed that up to 85% of the dissociated cells expressed ISL1, with consistent results obtained in different cell lines and low experiment-to-experiment variability (Figures 5C and 5D; Table S3). We also found that the vast majority of cells in the cultures were immunopositive for ChAT (Figures 5E and 5F; Table S3). Cells from dissociated motor neuron spheres also exhibited punctate SYNAPSIN1 immunoreactivity along MAP2-positive neurites (Figure 6A). We also performed experiments in which spin-culture-derived motor neurons were cultured with human myotubes. Staining with fluorescently labeled α-bungarotoxin (BTX), a toxin that specifically binds to post-synaptic acetylcholine receptors, labeled regions of contact between motor neurons and muscle fibers (Figure 6B), indicating formation of neuromuscular junctions.
Figure 6.
Dissociated Motor Neurons Are Spontaneously Active
(A) Representative immunocytochemistry of 1016A-derived neurons from dissociated motor neuron spheres dissociated and plated at day 15, cultured for 1 week, and stained with antibodies specific for MAP2 (green) and SYNAPSIN1 (red). Nuclei were counterstained with DAPI (blue). (A′) is an image captured at higher magnification of the boxed area in (A). Scale bar represents 20 μm.
(B) Representative immunocytochemistry of HUES8-derived motor neurons spheres seeded at day 15 on top of cultures of human myotubes. After 1 week the co-culture was stained with antibodies specific for SMI32 (green) and fluorescently labeled α-bungarotoxin (BTX) (red). Scale bar represents 20 μm. (B′) is an image captured at higher magnification of the boxed area in (B).
(C) Representative results from experiments in which HUES8-derived motor neurons were dissociated from spheres at day 15 and transduced with AAV Syn:GCaMP6s 3 days later. Time-lapse microscopy was employed to record fluctuations in fluorescence over time 1 and 3 weeks after transduction.
(D) Electrophysiological recordings of HUES8-derived neurons from motor spheres analyzed 12 weeks after dissociation. Left: representative traces showing spontaneous action potential firing. Right: representative traces showing evoked action potential firing upon current injection from 0 to 90 pA in 10 pA increments.
(E) Representative traces showing normalized fluorescence intensity of GCaMP6s after GABA and AMPA (both at 100 μM) application in HUES9-derived motor neurons cultured for 3 weeks after sphere dissociation.
See also Figure S6.
Dissociated Motor Neurons Exhibit Stimulation-Induced Activity and Form Spontaneously Active Neural Networks
To further address the functionality of the motor neurons that we produced, we characterized their physiological properties. First, we confirmed that the cells were capable of responding to high KCl depolarization by phosphorylating CREB (Figures S6C and S6D). Next, we asked whether these motor neurons exhibited spontaneous electrical activity in culture. As before, we monitored intracellular Ca2+ dynamics in cultures of dissociated motor neurons transduced with AAVSyn:GCaMP6s. Cells were transduced 3 days after dissociation and imaged every week. As early as 1 week after dissociation, motor neurons already exhibited significant levels of activity, which increased over time. Neurons again displayed more synchronicity in their firing patterns at later times, suggesting the formation of neural networks, and with groups of motor neurons exhibiting synchronized Ca2+ dynamics 3 weeks after dissociation (Figure 6C; Movie S4).
Patch-clamp recordings were performed to investigate the electrophysiological characteristics of motor neurons from dissociated spheres. Motor neurons were found to fire action potentials spontaneously and upon current injection even after up to 3 months in culture (Figure 6D, see also Figure S7). Furthermore, GCaMP6s-mediated monitoring of intracellular Ca2+ dynamics showed that motor neurons responded to both γ-aminobutyric acid (GABA) and α-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA), as might have been expected (Figure 6E).
Thus, the sphere differentiation protocol was successfully modified to allow the generation of billions of motor neurons from one single differentiation experiment. The yield of motor neurons was very efficient, up to 85%, with little variation between individual experiments, and sphere-derived motor neurons could be cultured without glial feeder layers. Moreover, the motor neurons expressed ChAT and were physiologically active.
Increased Survival and Function of Motor Neurons Obtained through the Spin-Culture Protocol Compared with EB-Derived Motor Neurons
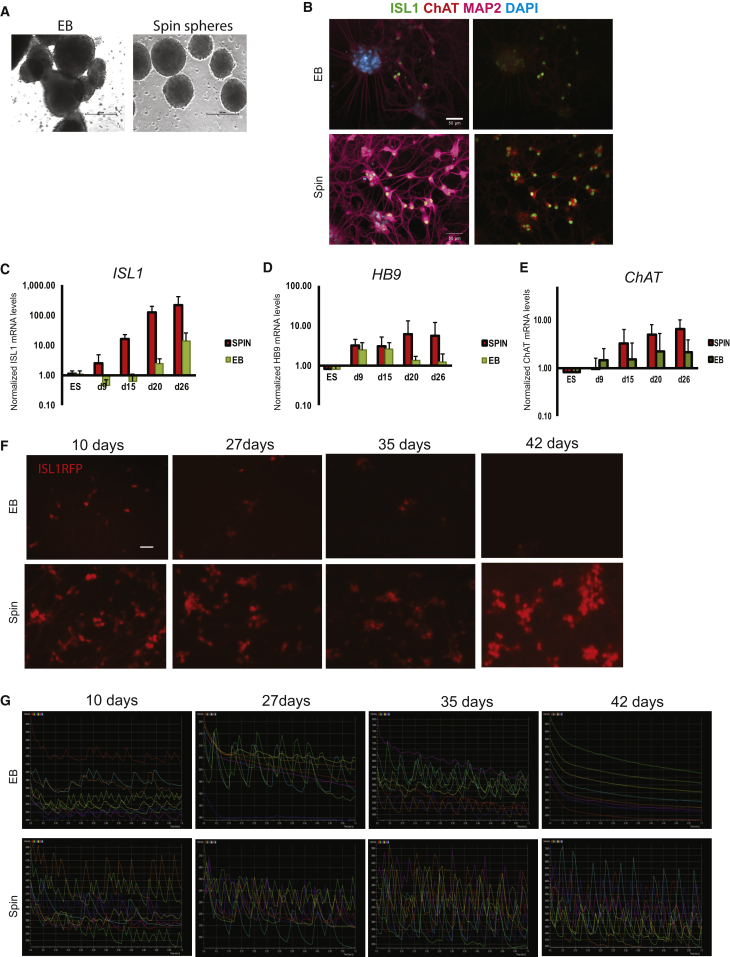
Throughout the course of the experiments presented, we noted a marked increase in survival of spin-culture-derived neurons compared with EB-derived neurons in all assays requiring manipulations such as dissociation, fluorescence-activated cell sorting, and viral transduction. In an attempt to address this in a quantifiable manner, we generated motor neurons in parallel using two methods (but with exactly the same medium composition), namely spin-culture differentiation and EB formation in low-attachment tissue-culture plates using protocols standard in our laboratory (Ng et al., 2015, Yang et al., 2013). Motor neuron spheres in spin culture were uniform and homogeneous, whereas motor neuron differentiation using EBs led to formation of cell clumps exhibiting marked heterogeneity with respect to size, shape, and general appearance (Figures 7A, S7I, S7J, and S7K). Expression analyses by qRT-PCR of cells undergoing differentiation at different time points revealed that motor neuron spheres exhibited higher levels of expression of ISL1, HB9, and ChAT compared with conventional EB-derived motor neurons (Figures 7C–7E). When spheres and EBs were dissociated and neurons plated, immunocytochemistry showed that spin-culture-derived motor neurons co-expressed ISL1, MAP2, and ChAT to a higher degree than EB-derived motor neurons (Figure 7B).
Figure 7.
Sphere-Derived Human Motor Neurons Survive Longer in Culture and Maintain Functionality throughout Extended Time in Culture
(A) Representative bright-field images of HUES9 EBs (left) and HUES9 spheres generated by spin culture (right) at day 6 of motor neuron differentiation.
(B) Representative images of immunocytochemistry of dissociated HUES9-derived motor neurons obtained through EB formation (top panels) or spin-culture differentiation (bottom panels), stained after 7 days of culture with antibodies against MAP2 (purple), ISL1 (green), and ChAT (red). Nuclei were counterstained with DAPI (blue). Scale bar represents 50 μm.
(C–E) Analysis of expression levels of ISL1, HB9, and ChAT by qRT-PCR in cells undergoing differentiation through EB formation (green bars) or spin culture (red bars) at different time points. Error bars indicate SD from three independent experiments.
(F) Representative images of dissociated H9ISL1RFP-reporter line motor neurons (Bu et al., 2009) from EBs (top panel) and spin spheres (bottom panel), plated under feeder-free conditions and imaged after 10, 27, 35, and 42 days of culture. Scale bar represents 50 μm.
(G) Representative results from experiments where H9ISL1RFP-reporter line motor neurons generated using either EB (top panel) or spin sphere (bottom panel) methodology were dissociated and transduced with AAV Syn:GCaMP6s 3 days after dissociation. Time-lapse microscopy was employed to record fluctuations in fluorescence over time (10, 27, 35, and 42 days after transduction).
See also Figure S7.
Next, we performed a set of experiments in which we monitored the appearance and functionality of cultures of motor neurons derived from spin-culture spheres or using EB methodology. Irrespective of plating conditions, motor neurons derived from spin-culture spheres survived for a longer period of time in culture. The difference was particularly striking on coated tissue-culture plates (Figure 7F), where motor neurons from dissociated spin-culture spheres could be maintained in culture, without glial feeder layers, for several months, which in our hands proved to be impossible using motor neurons obtained through EB formation. In experiments where motor neurons were plated and transduced with genetically encoded Ca2+ sensors, motor neurons derived from spin-culture spheres exhibited higher amplitude and frequency in levels of fluorescence compared with EB-derived motor neurons (Figure 7G). With extended time in culture, the amplitude and frequency of fluctuations in Ca2+ increased in the spin-culture-derived motor neurons, whereas EB-derived motor neurons exhibited a loss in Ca2+ fluctuations (Figure 7G). These results are consistent with the notion that motor neurons derived from spin-culture spheres survive better and maintain their functionality during extended periods of culture.
To formally address the maturation of the different motor neuron populations, we performed comparisons using whole-cell patch recordings (Figures S7A–S7F). Our recordings of action potentials and voltage-gated Na/K currents showed that spin-culture-derived motor neurons were able to fire faster action potentials and larger Na/K currents than EB-derived motor neurons (Figures S7A–S7F). In addition, calcium imaging experiments suggest that spin-culture-derived motor neurons express functional GABA receptors and begin to undergo the transition from GABA excitation to inhibition earlier than EB-derived motor neurons (Figures S7G–S7H), which indicate a faster maturation time course (Ben-Ari, 2002). These data suggest that spin-culture-derived motor neurons may mature faster than EB-derived motor neurons with regard to their electrophysiological characteristics, although additional experiments in more hPSC lines are required to address whether this is a general phenomenon.
Discussion
In the last 3 years, several groups have shown that neural organoids with features of the mammalian CNS can be generated (Bershteyn and Kriegstein, 2013, Lancaster et al., 2013, Sasai, 2013). This innovative approach has opened up the possibility of studying basic mechanisms of the developmental biology of the human nervous system (Camp et al., 2015, Otani et al., 2016) and neurodevelopmental disorders (Mariani et al., 2015). However, it is important to note that there are limitations to this model system. Neural organoids are produced from hPSCs through formation of neural progenitors that are aggregated individually in Matrigel and then transferred to spinner flasks for further development. The resulting organoids are highly complex structures containing many different types of neurons organized into areas exhibiting features of distinct regions of the mammalian CNS, including the cerebral cortex. However, neural differentiation in organoids is quite variable with respect to both the exact type of neuronal subtype and the structures produced, as well as the efficiency of the differentiation process.
In this paper, we present a method based on 3D growth of cells from hPSC to differentiated neurons. Importantly, this protocol does not rely on isolation and manipulation of an intermediate cell population in Matrigel. In contrast, we culture cells as spheres starting from the pluripotent state and only subject the spheres to differentiation cues by changing the composition of the culture medium. For both cortical neurons and motor neurons produced with this method, we were able to document not only expression of appropriate marker genes but also several hallmarks of mature, functional neurons, including synapse formation, expression of functional receptors for neurotransmitters, spontaneous electrical activity, and appropriate expression of activity-induced genes.
This system has several advantages. First, it is simple to obtain a high absolute number of cells—in the range of billions even in a single flask. This is of importance for downstream applications such as high-throughput screening, where cell numbers are a limiting factor. For example, our laboratory routinely performs chemical screens on hPSC-derived neurons in 384-well plates, where 15,000 neurons are plated in each well (Yang et al., 2013). One single differentiation of hPSC-derived spheres in spinner flasks as described herein would be sufficient for approximately 350 × 384-well plates, thereby greatly reducing the time and effort required to produce the cells necessary to conduct such experiments. Second, spheres cultured in the spinner flasks appear uniform in size and shape during the differentiation, reducing experiment-to-experiment variation. Third, spin-culture differentiation results in increased relative numbers of cortical neurons and motor neurons. Fourth, after dissociation of the differentiated spheres, we were able to culture both types of neurons without the use of feeder layers for extended periods of time, as long as 4 months or more. This is in stark contrast to neurons obtained from EBs, where cells start to detach from the plate at relatively early times.
In this report we have focused on generating cortical and motor neurons, but we believe it is likely that future adaptations of the sphere protocol to generate other distinct neuronal subtypes will prove to be advantageous compared with other methods. In addition, establishing imaging and physiological assays to cells maintained in three dimensions should be possible in the near future so that functional connections among populations of neurons can be maintained and analyzed (rather than studied after dissociation). Future work will also determine whether neurons maintained in suspension for extended periods can achieve a greater degree of maturation, of major importance in the study of late-onset neurodegenerative disease. Finally, it will be important to understand whether suspension culture conditions can be adapted to allow for faster generation of neurons and glial cells. If this is the case, one of the main problems in using hPSC-derived neurons, that of prolonged times required for cell production, can be circumvented.
Experimental Procedures
Human Pluripotent Stem Cell Lines
All experiments with the human ESC lines were reviewed and approved by the Harvard Embryonic Stem Cell Oversight Committee. The use of the wild-type iPS cell lines by the Rubin lab was reviewed by the Harvard Committee on the Use of Human Subjects (the Harvard IRB) and determined to not constitute human subjects research.
Media
mTeSR (STEMCELL Technologies); KSR medium: 15% KSR (Invitrogen), KO DMEM (Invitrogen), L-glutamine (Gibco), 100× non-essential amino acids (NEAA) (Gibco), 100× penicillin-streptomycin (Gibco), and 1000× β-mercaptoethanol (Gibco); neural induction medium (NIM): DMEM/F12 (Invitrogen), 100× N2 supplement (Gibco), 50× B27 supplement (Life Technologies) (for cortical differentiation we substituted regular B27 for B27 supplement without vitamin A [Life Technologies]), 100× Glutamax (Gibco), 100× NEAA (Gibco), 100× penicillin-streptomycin (Gibco), 125× glucose (J.T. Baker), and (only for motor neurons) 0.2 mM ascorbic acid (Sigma); neurobasal (NB) medium: Neurobasal (Invitrogen), 100× N2 supplement (Gibco), 50× B27 supplement (Life Technologies) (for cortical differentiation we substituted regular B27 for B27 supplement without vitamin A [Life Technologies]), 100× Glutamax (Gibco), 100× NEAA (Gibco), 100× penicillin-streptomycin (Gibco) (for motor neurons, the medium was supplemented with 125× glucose [J.T. Baker] and 0.2 mM ascorbic acid [Sigma]).
hPSC Adaption and Maintenance in Spinner Flasks
hPSCs were cultured in 500-ml disposable spinner flasks (Corning, VWR) on a nine-position stir plate (Dura-Mag) at a speed of 70 rpm, in a 37°C incubator with 5% CO2. Prior to adaption to spinner-flask culture, hPSCs were expanded in 10-cm dishes (Corning) and dissociated to single cells with Accutase (STEMCELL Technologies) for 5 min incubation at 37°C, when they were approaching confluency. 180 million individual pluripotent stem cells were seeded into a spinner flask in 180–200 ml of mTeSR medium (STEMCELL) supplemented with 5 μM ROCK inhibitor Y-27632 (STEMCELL). Spheres formed spontaneously, and after 2 days the culture medium was changed. The cells were maintained as undifferentiated pluripotent spheres in spin culture, with medium changes every day. Every third day, when the spheres were approximately 50–100 μm in diameter, cells were passaged by incubating at 37°C for 6 min in a 1:1 mixture of Accutase and PBS, and by mechanically dissociating the cells. Cells were filtered and reseeded into the flask at the same initial density as described above. Medium was changed by taking the flasks off the stir plate, allowing the cells to settle to the bottom of the flask and the medium to be changed. At day 1 of differentiation, medium was changed to mTeSR with the activin/TGF-β inhibitor SB431542 (R&D Systems) (10 μM) and the BMP inhibitor LDN193189 (Stemgent) (1 μM) (hereafter referred to as “dual SMAD inhibition”). Subsequent steps of the differentiation protocol (as described below) use the same method for changing media.
Differentiation Conditions
Cortical Differentiation
For cortical differentiation from days 2 to 10, spheres were gradually adapted to NIM through a dilution series of KSR and NIM, with dual SMAD inhibition maintained until day 7. Medium was changed as follows: days 2–4: 100% KSR medium with the Wnt signaling inhibitor XAV939 (2 μM) (Stemgent); day 5: 75% KSR medium, 25% NIM; day 7: 50% KSR medium, 50% NIM; day 9: 25% KSR medium, 75% NIM. From days 11 to 21, cultures were maintained in 100% NIM (Maroof et al., 2013). From day 21 onward, cultures were maintained in NB medium with 10 ng/ml brain-derived neurotrophic factor (BDNF) and 10 ng/ml glial cell-derived neurotrophic factor (GDNF) (both from R&D).
Motor Neuron Differentiation
For motor neuron differentiation from days 2 to 10, spheres were gradually adapted to NIM through a dilution series of KSR and NIM, with dual SMAD inhibition maintained until day 6. Medium was changed as follows: day 2: 100% KSR; day 3: 100% KSR + 1 μM retinoic acid (RA) (Sigma); day 5: 75% KSR, 25% NIM + RA, and 10 ng/ml BDNF (R&D); day 6: 50% KSR, 50% NIM + RA, 1 μM Smoothened agonist (SAG) (Curis), and BDNF; day 8: 25% KSR, 75% NIM + RA, SAG, and BDNF. From days 10 to 15, cultures were maintained in 100% NIM + RA, SAG, BDNF, and 2.5 μM DAPT (R&D). From day 15 onward, cultures were kept in NB + BDNF, GDNF, and 10 ng/ml ciliary neurotrophic factor (R&D).
During differentiation, spheres were collected at different time points and RNA was extracted for qRT-PCR analysis. For examination of cortical differentiation, spheres were dissociated with Accutase and plated onto Matrigel-coated plates at day 13 and analyzed the following day for expression of cortical progenitor markers by immunofluorescence. Spheres were also dissociated at day 40 with 0.05% trypsin (Gibco) and plated onto poly-D-lysine/laminin-coated (EMD Millipore/Life Technologies) plates. After 28 and 43 days of additional culture, cells were analyzed as described below. For motor neuron differentiation, spheres were dissociated with 20 units of papain per milliliter in Earle’s balanced salt solution supplemented with 1 mM L-cysteine, 0.5 mM EDTA (Worthington), and DNAse. 60,000 cells/well were plated onto 96-well poly-D-lysine/laminin-coated plates at day 15 (see Supplemental Experimental Procedures for a detailed protocol for dissociation). Before dissociation, both cortical and motor neuron spheres were approximately 200 μm in diameter. Neurons were analyzed after one additional week in culture after dissociation.
Statistics
All experiments were performed in three independent biological replicates unless stated otherwise. A two-tailed Student's t test was used to calculate statistical significance.
Author Contributions
A.R. conceived and designed the study, carried out experiments, collected and analyzed data, and wrote the manuscript; G.R. and D.C.R. carried out experiments, and collected and analyzed data; C.S. carried out experiments, collected and analyzed data, and wrote the manuscript; F.P. contributed reagents, carried out experiments, and analyzed data; F.R., K.W., and C.B. carried out experiments; L.D. analyzed the data, Q.P.P. contributed reagents and helped with establishing spinner cultures; E.M.H. analyzed data and wrote the manuscript; L.L.R. conceived and designed the study and wrote the manuscript.
Acknowledgments
We would like to thank Douglas Melton for advice on the spinner method and for other helpful suggestions; Douglas Richardson from the Harvard Center for Biological Imaging (CBI) for assistance with LSFM; Gabriella Boulting and Bulent Ataman for helpful discussions; Laurence Daheron of the Harvard Stem Cell Institute iPS cell core and members of the Harvard Stem Cell Institute Histology Core for invaluable assistance; Jane Lalonde for editorial assistance; and Geraldine Jowett for technical help. We would also like to thank all the members of the Rubin laboratory for helpful discussions. We are grateful to Douglas Melton for providing the human ESCs HUES9, HUES8, and the 1016A human iPSCs; Kevin Eggan for the HUES3 hESCs; and Kenneth Chien for the H9ISL1RFP hESCs. BJ SiPSCs were generated by the HSCI iPSC Core. This work was supported by grants from the SMA Foundation, NINDS (P01NS066888), Ellison Medical Foundation, Hoffman La-Roche, Inc., the Stanley Center for Psychiatric Research at the Broad Institute, and the Harvard Stem Cell Institute. F.D.P. is a Banting Fellow and E.M.H. is a fellow of the Wenner-Gren Foundation.
Published: June 14, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, three tables, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.05.010.
Supplemental Information
References
- Amoroso M.W., Croft G.F., Williams D.J., O'Keeffe S., Carrasco M.A., Davis A.R., Roybon L., Oakley D.H., Maniatis T., Henderson C.E. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 2013;33:574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M., Marchetto M.C., Gage F.H., Mummery C.L. Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y. Excitatory actions of GABA during development: the nature of the nurture. Nat. Rev. Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- Bershteyn M., Kriegstein A.R. Cerebral organoids in a dish: progress and prospects. Cell. 2013;155:19–20. doi: 10.1016/j.cell.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissart C., Poulet A., Georges P., Darville H., Julita E., Delorme R., Bourgeron T., Peschanski M., Benchoua A. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl. Psychiatry. 2013;3:e294. doi: 10.1038/tp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting G.L., Kiskinis E., Croft G.F., Amoroso M.W., Oakley D.H., Wainger B.J. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Brauninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R., Geschwind D.H. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D.H., Greenberg M.E. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Sasai Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc. 2012;7:69–79. doi: 10.1038/nprot.2011.429. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Espuny-Camacho I., Michelsen K.A., Gall D., Linaro D., Hasche A., Bonnefont J., Bali C., Orduz D., Bilheu A., Herpoel A. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Flavell S.W., Kim T.K., Gray J.M., Harmin D.A., Hemberg M., Hong E.J., Markenscoff-Papadimitriou E., Bear D.M., Greenberg M.E. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox I.J., Daley G.Q., Goldman S.A., Huard J., Kamp T.J., Trucco M. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy for treating disease. Science. 2014;345:1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M., Javaherian A., Strulovici B., Daley G.Q. Induced pluripotent stem cells-opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- Han S.S., Williams L.A., Eggan K.C. Constructing and deconstructing stem cell models of neurological disease. Neuron. 2011;70:626–644. doi: 10.1016/j.neuron.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Hester M.E., Murtha M.J., Song S., Rao M., Miranda C.J., Meyer K., Tian J., Boulting G., Schaffer D.V., Zhu M.X. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol. Ther. 2011;19:1905–1912. doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E.J., McCord A.E., Greenberg M.E. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Zhang S.C. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat. Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi Y., Okano H. Modeling human neurological disorders with induced pluripotent stem cells. J. Neurochem. 2014;129:388–399. doi: 10.1111/jnc.12625. [DOI] [PubMed] [Google Scholar]
- Ismadi M.Z., Gupta P., Fouras A., Verma P., Jadhav S., Bellare J., Hourigan K. Flow characterization of a spinner flask for induced pluripotent stem cell culture application. PLoS One. 2014;9:e106493. doi: 10.1371/journal.pone.0106493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanning K.C., Kaplan A., Henderson C.E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- Ke M.T., Fujimoto S., Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Kim D.S., Ross P.J., Zaslavsky K., Ellis J. Optimizing neuronal differentiation from induced pluripotent stem cells to model ASD. Front. Cell Neurosci. 2014;8:109. doi: 10.3389/fncel.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.J., Zhang X., Johnson M.A., Wang Z.B., Lavaute T., Zhang S.C. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Bloodgood B.L., Hauser J.L., Lapan A.D., Koon A.C., Kim T.K., Hu L.S., Malik A.N., Greenberg M.E. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S.C. Human stem cells as a model of motoneuron development and diseases. Ann. N. Y Acad. Sci. 2010;1198:192–200. doi: 10.1111/j.1749-6632.2010.05537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Coppola G., Zhang P., Abyzov A., Provini L., Tomasini L., Amenduni M., Szekely A., Palejev D., Wilson M. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof A.M., Keros S., Tyson J.A., Ying S.W., Ganat Y.M., Merkle F.T., Liu B., Goulburn A., Stanley E.G., Elefanty A.G. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.Y., Soh B.S., Rodriguez-Muela N., Hendrickson D.G., Price F., Rinn J.L., Rubin L.L. Genome-wide RNA-Seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell. 2015;17:569–584. doi: 10.1016/j.stem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T., Marchetto M.C., Gage F.H., Simons B.D., Livesey F.J. 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell. 2016;18:467–480. doi: 10.1016/j.stem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuji T.G., Bin J., Yoshimura A., Tomura M., Tateyama D., Minami I., Yoshikawa Y., Aiba K., Heuser J.E., Nishino T. A 3D sphere culture system containing functional polymers for large-scale human pluripotent stem cell production. Stem Cell Rep. 2014;2:734–745. doi: 10.1016/j.stemcr.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gurtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q., Li D., Louis K.R., Li X., Yang H., Sun Q., Crandall S.R., Tsang S., Zhou J., Cox C.L. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat. Commun. 2014;5:3449. doi: 10.1038/ncomms4449. [DOI] [PubMed] [Google Scholar]
- Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Yang Y.M., Gupta S.K., Kim K.J., Powers B.E., Cerqueira A., Wainger B.J., Ngo H.D., Rosowski K.A., Schein P.A., Ackeifi C.A. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013;12:713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Guo M., Martins-Taylor K., Wang X., Zhang Z., Park J.W., Zhan S., Kronenberg M.S., Lichtler A., Liu H.X. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS One. 2010;5:e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.